Abstract
Background
Coronary heart disease is the leading cause of death worldwide. Mitochondrial genetic determinants for the development of this disorder remain less explored.
Methods and Results
We performed a clinical and genetic evaluation and mutational screening of 22 mitochondrial tRNA genes in a cohort of 80 genetically unrelated Han Chinese subjects and 125 members of 4 families with coronary heart disease and 512 Chinese control subjects. This analysis identified 16 nucleotide changes among 9 tRNA genes. Of these, the T5592C mutation creates a highly conservative base pairing (5G‐68C) on the acceptor stem of tRNAGln, whereas the G15927A mutation destabilizes a highly conserved base pairing (28C‐42G) in the anticodon stem of tRNAThr. However, the other tRNA variants were polymorphisms. The pedigrees of BJH24 carrying the T5592C mutation, BJH15, and BJH45 harboring the G15927A mutation exhibited maternal transmission of coronary heart disease. Sequence analysis of their mitochondrial genomes revealed the presence of T5592C or G15927A mutation but the absence of other functionally significant mutations in all matrilineal relatives of these families.
Conclusions
Our previous observations showed that altered structures of tRNAs by these mtDNA mutations caused mitochondrial dysfunction. These may be the first evidence that mtDNA mutations increase the risk of coronary heart disease. Our findings may provide new insights into the pathophysiology of this disorder.
Keywords: coronary heart disease, maternal transmission, mitochondrial tRNA, mutation
Introduction
Coronary heart disease is a leading cause of death worldwide. In particular, coronary heart disease (CHD) annually results in 502 000 deaths in the United States and >700 000 deaths in China.1–2 CHD is a common complex disorder that can be caused by single gene or multifactorial conditions resulting from interactions between environmental and inherited risk factors.3–5 Efforts to identify genetic determinants of CHD have been directed primarily at nuclear genes.6 Genome‐wide association studies in the population of European and Asian ancestries have identified several genetic loci that are associated with risk of CHD.7–9 However, the role of mitochondrial genetic defects in the development of coronary heart disease remains poorly understood.10–11 The human mitochondrial genome encodes 13 peptides for the oxidative phosphorylation system, 2 rRNAs, and 22 tRNAs required for mitochondrial protein synthesis.12 Among these tRNAs, tRNAGlu, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer(UCN), tRNAGln, and tRNAPro reside in the cytosine‐rich light strand; the remaining tRNAPhe, tRNAVal, tRNALeu(UUR), tRNALeu(CUN), tRNAIle, tRNAMet, tRNASer(AGY), tRNATrp, tRNAAsp, tRNALys, tRNAGly, tRNAArg, tRNAHis, and tRNAThr are in the guanine‐rich heavy strand.12–13 Mitochondrial tRNA genes are the hot spots for mutations associated with cardiovascular disorders such as hypertension.14–17 These hypertension‐associated tRNA mutations included the T4291C and A4263G mutations in the tRNAIle gene, the A4435G mutation in the tRNAMet gene, and the A4401G mutation at the junction of the tRNAMet and tRNAGln genes.18–20
It is anticipated that mutations in mitochondrial tRNA genes are associated with coronary heart disease. To investigate the role of mitochondrial genetic defects in the development of coronary heart disease, we carried out a systematic and extended mutational screening of 22 tRNA genes in a cohort of 80 Han Chinese subjects with coronary heart disease. Mutational analysis of these tRNA genes in these subjects identified 16 nucleotide changes in 9 tRNA genes. These tRNA variants were further evaluated by phylogenetic analysis, structure–function relation, and allelic frequency of these variants in the Han Chinese controls from the same region.
Methods
Subjects
A total of 80 genetically unrelated Chinese subjects with coronary heart disease, aged 33 to 79 years old from Beijing, along with some of their family members, were enrolled in this study under an institutional review board–approved protocol of informed consent at the Zhejiang University Institutional Review Board and Ethics Committee of Beijing Anzhen Hospital, China. Members of these families were interviewed and evaluated to identify both personal and medical histories of CHD and other clinical abnormalities. The 512 control DNA samples were obtained from a panel of unaffected Han Chinese individuals from the same area.
Subjects underwent a physical examination and laboratory assessment of cardiovascular disease risk factors. A heart function evaluation and measurement of systolic and diastolic blood pressure of subjects were performed as detailed elsewhere.20 All patients underwent a heart function evaluation by electrocardiography. Signals from the first 10 seconds of the conventional electrocardiography recording were analyzed automatically in software to quantify all major intervals, axes, and voltages as well as ST‐segment levels. The initial candidate criteria used for defining these strictly conventional 12‐lead electrocardiograms were as follows. Eligible patients had ≥10 minutes of ischemic symptoms at rest and presented with 1 of the following: elevated markers of myonecrosis, ≥0.1 mV of ST depression, or diabetes mellitus. Patients were referred for angiography for suspected myocardial ischemia such as myocardial infarction or unstable angina and myocardial infarction as symptoms and increased troponin I, with or without ST elevation on the electrocardiogram. Coronary angiography was performed by the Judkins technique, and images of the coronary tree were obtained in routine, standardized projections. The angiograms were assessed by ≥2 cardiologists. Coronary angiograms were visually evaluated by 3 independent experienced observers according to the clinical review process. Localization and percentage of luminal diameter reduction were documented for any coronary artery with a stenosis. Significant coronary artery stenosis was defined as luminal diameter reduction of 50%. Patients without angiographic lesions were considered the patients without CHD.21
Hypertension, hyperlipidemia, diabetes mellitus, cigarette smoking, and family history for CHD were considered risk factors. Diabetes mellitus was defined as hyperglycemia requiring antidiabetic drugs or fasting blood sugar >126 g/dL. Patients reporting cigarette use during the year prior to examination were considered smokers. Hyperlipidemia was defined as plasma low‐density lipoprotein cholesterol >130 mg/dL or total cholesterol >200 mg/dL or using lipid‐lowering drugs at the time of investigation. Overweight for Chinese subjects was defined as body mass index >24.
Mutational Analysis of Mitochondrial Genome
Genomic DNA was isolated from the whole blood of participants using Puregene DNA Isolation Kits (Gentra Systems, Minneapolis, MN). The fragments spanning all 22 of the tRNA genes of 80 subjects with CHD and 512 control subjects were PCR‐amplified by use of sets of the light‐strand and the heavy‐strand oligonucleotide primers (Table 1). The entire mitochondrial genomes of 3 probands (BJH24‐II‐2, BJH15‐III‐2, and BJH45‐III‐7) were PCR‐amplified in 24 overlapping fragments by use of sets of the light‐strand and the heavy‐strand oligonucleotide primers, as described elsewhere.22 Each fragment was purified and subsequently analyzed by direct sequencing in an ABI 3700 automated DNA sequencer using a Big Dye Terminator Cycle sequencing reaction kit. The resultant sequence data were compared with the updated consensus Cambridge sequence (GenBank accession number: NC_012920).12
Table 1.
Oligonucleotide Primers for Amplification of 22 Human Mitochondrial tRNAs
| Locus | Starting | Ending | Length (bp) | Number of Variants | Primer22 | Primer Sequence 5′‐3′ |
|---|---|---|---|---|---|---|
| tRNAPhe | 577 | 647 | 71 | 5 | 1F/1R | CTCCTCAAAGCAATACACTG/TGCTAAATCCACCTTCGACC |
| tRNAVal | 1602 | 1670 | 69 | 5 | 2F/2R | CGATCAACCTCACCACCTCT/TGGACAACCAGCTATCACCA |
| tRNALeu(UUR) | 3230 | 3304 | 75 | 6 | 4F/4R | AAATCTTACCCCGCCTGTTT/AGGAATGCCATTGCGATTAG |
| tRNAIle | 4263 | 4331 | 69 | 10 | 6F/6R | TGGCTCCTTTAACCTCTCCA/AAGGATTATGGATGCGGTTG |
| tRNAGln | 4329 | 4400 | 72 | 12 | 6F/6R | As above |
| tRNAMet | 4402 | 4469 | 68 | 6 | 6F/6R | As above |
| tRNATrp | 5512 | 5579 | 68 | 8 | 7F/8R | ACTAATTAATCCCCTGGCCC/ACCTAGAAGGTTGCCTGGCT |
| tRNAAla | 5587 | 5655 | 69 | 6 | 8F/8R | CTAACCGGCTTTTTGCCC/ACCTAGAAGGTTGCCTGGCT |
| tRNAAsn | 5657 | 5729 | 73 | 5 | 8F/8R | As above |
| tRNACys | 5761 | 5826 | 66 | 15 | 8F/8R | As above |
| tRNATyr | 5826 | 5891 | 66 | 4 | 8F/8R | As above |
| tRNASer(UCN) | 7446 | 7514 | 69 | 3 | 11F/11R | ACGCCAAAATCCATTTCACT/CGGGAATTGCATCTGTTTTT |
| tRNAAsp | 7518 | 7585 | 68 | 6 | 11F/11R | As above |
| tRNALys | 8295 | 8364 | 70 | 7 | 12F/12R | ACGAGTACACCGACTACGGC/TGGGTGGTTGGTGTAAATGA |
| tRNAGly | 9991 | 10058 | 68 | 10 | 14F/15R | CCCACCAATCACATGCCTAT/AATTAGGCTGTGGGTGGTTG |
| tRNAArg | 10405 | 10469 | 65 | 5 | 15F/15R | TCTCCATCTATTGATGAGGGTCT/AATTAGGCTGTGGGTGGTTG |
| tRNAHis | 12138 | 12206 | 69 | 6 | 18F/18R | TATCACTCTCCTACTTACAG/AGAAGGTTATAATTCCTACG |
| tRNASer(AGY) | 12207 | 12265 | 59 | 11 | 18F/18R | As above |
| tRNALeu(CUN) | 12266 | 12336 | 71 | 5 | 18F/18R | As above |
| tRNAGlu | 14674 | 14742 | 69 | 5 | 21F/21R | GCATAATTAAACTTTACTTC/AGAATATTGAGGCGCCATTG |
| tRNAThr | 15888 | 15953 | 66 | 27 | 22F/23R | TGAAACTTCGGCTCACTCCT/GAGTGGTTAATAGGGTGATAG |
| tRNAPro | 15956 | 16023 | 68 | 6 | 22F/23R | TGAAACTTCGGCTCACTCCT/GAGTGGTTAATAGGGTGATAG |
Structural Analysis
The published secondary structures for the tRNAs were used to define the stem‐and‐loop structure.17,23
Phylogenetic Analysis
A total of 17 vertebrates' mitochondrial DNA sequences were used in the interspecific analysis. These included Bos taurus, Cebus albifrons, Gorilla gorilla, Homo sapiens, Hylobates lar, Lemur catta, Macaca mulatta, Macaca sylvanus, Mus musculus, Nycticebus coucang, Pan paniscus, Pan troglodytes, Papio hamadryas, Pongo abelii, Pongo pygmaeus, Tarsius bancanus, and Xenopus laevis (Genbank; Table 2). The conservation index (CI) was calculated by comparing the human nucleotide variants with 16 other vertebrates. The CI was then defined as the percentage of species from the list of 17 different vertebrates that had the wild‐type nucleotide at that position.
Table 2.
mtDNA Sequence of 17 Vertebrate Species
| Species Name | GenBank Accession Number |
|---|---|
| Homo sapiens | NC_012920 |
| Cebus albifron | NC_002763 |
| Gorilla gorilla | NC_011120 |
| Hylobates lar | NC_002082 |
| Lemur catta | NC_004025 |
| Macaca mulatta | NC_005943 |
| Macaca sylvanus | NC_002764 |
| Nycticebus coucang | NC_002765 |
| Pan paniscus | NC_001644 |
| Pan troglodytes | NC_001643 |
| Papio hamadryas | NC_001992 |
| Pongo pygmaeus | NC_001646 |
| Pongo pygmaeus abelii | NC_002083 |
| Tarsius bancanus | NC_002811 |
| Mus musculus | NC_006914.1 |
| Bos taurus | HM045018.1 |
| Xenopus laevis | NC_001573.1 |
A total of 17 vertebrate mitochondrial DNA sequences were used in the interspecific analysis. These included Bos Taurus, Cebus albifrons, Gorilla gorilla, Homo sapiens, Hylobates lar, Lemur catta, Macaca mulatta, Macaca sylvanus, Mus musculus, Nycticebus coucang, Pan paniscus, Pan troglodytes, Papio hamadryas, Pongo abelii, Pongo pygmaeus, Tarsius bancanus, and Xenopus laevis.
Statistics Analysis
Statistical analyses were performed using the SSPS statistical package, version 16.0, and statistical significance was established at P<0.05. We performed Fisher's exact test to evaluate the difference in mitochondrial tRNA mutations between CHD patients and controls.
Results
Study Samples
The patients in the study samples suffering from coronary heart disease alone or with other clinical phenotypes including hypertension, diabetes mellitus, and cerebrovascular disease, consisted of 57 men and 23 women. Clinical data for these 80 subjects are summarized in Table 3. All participants were Han Chinese from the Beijing area. Of these, 22 subjects only exhibited coronary heart disease. In addition to coronary heart disease, 40 subjects suffered from hypertension, 5 individuals had diabetes mellitus, 9 subjects exhibited hypertension and diabetes mellitus, 2 individuals had diabetes mellitus and cerebrovascular disease, 1 subject exhibited hypertension and cerebrovascular disease, and 1 subject suffered from hypertension, diabetes mellitus, and cerebrovascular disease. The age at onset of coronary heart disease in all participants ranged from 33 to 79 years, with a median age of 61 years. The age at onset of hypertension in 51 subjects varied from 15 to 77 years, with a median age of 48.8 years, whereas the age at onset of diabetes mellitus in 17 subjects ranged from 38 to 71 years, with an average age of 52.8 years. Body mass index of all participants ranged from 18.03 to 31.64, with an average of 25.39. Forty‐two of 80 subjects were smokers. A total of 512 unaffected Han Chinese subjects were obtained from the same area. The age of these participants ranged from 39 to 68 years, with a median age of 57 years.
Table 3.
Summary of Anthropometric, Clinical, and Biochemical Data of 80 Han Chinese Subjects With Coronary Heart Disease
| Subjects | Sex | Age at Onset (year) | Age of Subject (y) | BMI | SBP/DBP (mm Hg) | Smokers | HT | LVH | DM | FBG (mmol/L) | TC (mmol/L) | TG (mmol/L) | HDL‐C (mmol/L) | LDL‐C (mmol/L) | Statins | Angina in Anamnesis | AMI in Anamnesis | Stroke in Anamnesis | Family Anamnesis for AMI | Family Anamnesis for HT | Family Anamnesis for T2DM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BJH1 | M | 76 | 78 | 21.22 | 120/80 | Yes | No | No | No | 5.31 | 6.03 | 1.1 | 1.46 | 4.35 | Yes | Yes | No | Yes | No | Yes | No |
| BJH2 | M | 60 | 60 | 24.57 | 145/70 | Yes | No | No | Yes | 9.19 | 7.08 | 1.44 | 1.06 | 5.73 | No | Yes | No | No | No | No | Yes |
| BJH3 | M | 45 | 46 | 25.16 | 160/100 | Yes | Yes | Yes | No | 4.08 | 6.93 | 1.7 | 1.66 | 4.93 | No | No | No | No | No | Yes | No |
| BJH4 | F | 49 | 52 | 33.75 | 115/65 | No | No | No | No | 4.66 | 5.45 | 1.03 | 1.55 | 3.69 | Yes | Yes | No | No | No | No | No |
| BJH5 | F | 52 | 57 | 25.81 | 135/100 | No | Yes | No | No | 4.44 | 6.65 | 1.21 | 0.99 | 5.42 | No | Yes | No | No | No | No | No |
| BJH6 | M | 66 | 66 | 25.18 | 125/70 | Yes | No | No | Yes | 7.22 | 6.33 | 1.32 | 1.23 | 4.84 | Yes | No | No | No | No | Yes | Yes |
| BJH7 | F | 70 | 73 | 23.92 | 160/100 | No | Yes | Yes | No | 5.06 | 6.41 | 1.08 | 1.45 | 4.74 | Yes | Yes | Yes | No | No | Yes | No |
| BJH8 | M | 60 | 63 | 24.98 | 160/115 | Yes | Yes | Yes | No | 5.32 | 5.21 | 2.11 | 1.02 | 3.77 | Yes | No | No | No | No | Yes | No |
| BJH9 | M | 76 | 77 | 24.57 | 170/100 | Yes | Yes | Yes | Yes | 7.56 | 5.66 | 1.65 | 0.85 | 4.48 | Yes | Yes | No | No | No | No | Yes |
| BJH10 | F | 64 | 64 | 29.38 | 170/100 | No | Yes | Yes | No | 5.11 | 7.66 | 2.65 | 0.97 | 6.16 | Yes | No | No | No | No | Yes | No |
| BJH11 | F | 65 | 68 | 23.42 | 150/90 | No | Yes | No | Yes | 5.52 | 6.33 | 2.66 | 1.66 | 4.14 | Yes | Yes | No | No | No | No | No |
| BJH12 | M | 58 | 58 | 25.25 | 150/110 | Yes | Yes | Yes | No | 4.03 | 6.78 | 1.68 | 1.72 | 4.72 | Yes | Yes | No | No | No | No | No |
| BJH13 | M | 56 | 60 | 23.14 | 150/120 | Yes | Yes | Yes | No | 4.12 | 5.45 | 1.25 | 1.54 | 3.66 | Yes | No | No | No | No | No | No |
| BJH14 | M | 33 | 33 | 26.30 | 150/130 | Yes | Yes | Yes | No | 4.22 | 6.35 | 1.78 | 1.82 | 4.17 | Yes | No | No | No | No | Yes | No |
| BJH15 | M | 50 | 48 | 24.69 | 170/90 | Yes | Yes | Yes | No | 4.35 | 7.12 | 2.31 | 1.02 | 5.64 | Yes | Yes | Yes | No | Yes | No | No |
| BJH16 | M | 60 | 63 | 23.46 | 135/75 | No | No | No | No | 5.21 | 6.56 | 1.54 | 1.03 | 5.22 | Yes | No | No | No | No | No | No |
| BJH17 | M | 58 | 59 | 25.01 | 160/100 | No | Yes | Yes | Yes | 7.54 | 5.68 | 1.68 | 1.87 | 3.47 | Yes | Yes | Yes | No | No | No | No |
| BJH18 | M | 53 | 55 | 29.70 | 160/120 | Yes | Yes | Yes | Yes | 7.2 | 7.98 | 3.12 | 1.66 | 5.70 | No | Yes | No | No | No | Yes | Yes |
| BJH19 | M | 52 | 48 | 26.99 | 130/80 | Yes | No | No | Yes | 15.5 | 6.85 | 1.89 | 1.11 | 5.36 | Yes | No | No | No | No | No | No |
| BJH20 | M | 53 | 54 | 23.05 | 125/75 | Yes | No | No | No | 5.11 | 5.42 | 1.32 | 0.65 | 4.51 | Yes | No | No | No | No | No | No |
| BJH21 | M | 58 | 60 | 21.97 | 145/100 | Yes | Yes | No | Yes | 24.2 | 6.85 | 1.56 | 0.87 | 5.67 | Yes | No | No | No | No | No | No |
| BJH22 | M | 58 | 62 | 27.99 | 130/75 | Yes | No | No | No | 4.12 | 4.65 | 1.08 | 1.12 | 3.31 | Yes | Yes | No | No | No | No | No |
| BJH23 | M | 75 | 75 | 24.21 | 125/75 | No | No | No | No | 4.32 | 5.12 | 1.32 | 1.66 | 3.20 | Yes | Yes | No | No | No | Yes | No |
| BJH24 | F | 49 | 46 | 30.86 | 180/110 | No | Yes | Yes | No | 4.65 | 4.56 | 0.99 | 0.99 | 3.37 | Yes | Yes | Yes | No | Yes | Yes | No |
| BJH25 | M | 60 | 62 | 22.86 | 140/80 | Yes | No | No | No | 5.23 | 5.12 | 1.02 | 1.54 | 3.38 | Yes | No | No | No | No | No | No |
| BJH26 | F | 67 | 70 | 22.04 | 130/75 | No | No | No | No | 5.08 | 6.58 | 1.03 | 1.65 | 4.72 | Yes | Yes | No | No | No | No | No |
| BJH27 | M | 49 | 55 | 26.09 | 180/110 | Yes | Yes | Yes | No | 5.12 | 5.11 | 1.65 | 1.32 | 3.46 | Yes | Yes | No | No | No | Yes | Yes |
| BJH28 | M | Died | Died | 20.52 | 150/90 | Yes | No | No | No | 4.66 | 4.99 | 2.45 | 0.85 | 3.65 | Yes | No | No | Yes | No | No | No |
| BJH29 | M | 70 | 70 | 24.16 | 180/90 | Yes | Yes | Yes | No | 4.87 | 4.58 | 2.12 | 1.66 | 2.50 | Yes | Yes | No | No | No | Yes | No |
| BJH30 | M | 53 | 48 | 27.68 | 170/90 | No | Yes | No | No | 5.03 | 5.68 | 1.06 | 1.24 | 4.23 | Yes | Yes | No | No | No | No | No |
| BJH31 | F | 69 | 70 | 23.28 | 180/90 | No | Yes | Yes | No | 4.66 | 4.21 | 3.12 | 0.99 | 2.60 | Yes | Yes | No | No | No | Yes | Yes |
| BJH32 | F | 79 | 76 | 25.28 | 160/90 | No | Yes | No | No | 5.11 | 5.08 | 2.11 | 1.32 | 3.34 | Yes | Yes | No | No | No | No | No |
| BJH33 | M | 50 | 55 | 22.53 | 170/90 | Yes | Yes | Yes | Yes | 8.9 | 4.26 | 1.04 | 1.34 | 2.71 | Yes | Yes | No | No | No | Yes | No |
| BJH34 | M | 56 | 56 | 23.12 | 135/80 | Yes | No | No | No | 4.66 | 5.21 | 0.56 | 1.68 | 3.42 | Yes | No | No | No | No | No | No |
| BJH35 | M | 59 | 60 | 29.54 | 180/110 | Yes | Yes | Yes | Yes | 7.31 | 5.12 | 0.85 | 1.45 | 3.50 | Yes | Yes | No | No | No | Yes | Yes |
| BJH36 | M | 41 | 38 | 27.76 | 135/75 | No | No | No | Yes | 12.4 | 5.65 | 1.09 | 1.46 | 3.97 | Yes | Yes | No | No | No | No | No |
| BJH37 | M | 58 | 58 | 26.57 | 160/110 | No | Yes | Yes | No | 5.18 | 4.87 | 1.45 | 1.35 | 3.23 | Yes | Yes | Yes | No | No | Yes | Yes |
| BJH38 | M | 63 | 63 | 23.03 | 130/80 | No | No | No | No | 5.44 | 5.11 | 1.65 | 1.99 | 2.79 | Yes | Yes | No | No | No | No | No |
| BJH39 | M | 63 | 66 | 27.76 | 125/75 | Yes | No | No | Yes | 5.12 | 6.03 | 1.45 | 0.56 | 5.18 | Yes | Yes | No | No | No | No | Yes |
| BJH40 | M | 55 | 59 | 31.17 | 130/65 | Yes | No | No | Yes | 6.56 | 6.65 | 1.36 | 1.54 | 4.84 | Yes | Yes | No | No | No | No | No |
| BJH41 | M | Die | Die | 25.71 | 180/120 | Yes | Yes | Yes | No | 4.69 | 5.65 | 2.08 | 1.27 | 3.96 | Yes | Yes | No | Yes | No | Yes | No |
| BJH42 | M | 79 | 79 | 24.16 | 160/90 | No | Yes | No | No | 5.44 | 5.12 | 1.03 | 1.37 | 3.54 | Yes | No | No | Yes | No | No | No |
| BJH43 | F | 55 | 50 | 25.30 | 130/70 | No | No | No | No | 5.78 | 4.33 | 2.54 | 1.32 | 2.50 | Yes | Yes | No | No | No | No | Yes |
| BJH44 | F | 67 | 69 | 23.34 | 160/80 | No | Yes | No | No | 4.99 | 5.12 | 1.66 | 1.46 | 3.33 | Yes | Yes | No | No | No | Yes | No |
| BJH45 | F | 66 | 67 | 26.56 | 200/100 | No | Yes | Yes | No | 5.01 | 6.12 | 1.05 | 1.52 | 4.39 | Yes | Yes | Yes | No | Yes | No | No |
| BJH46 | M | 61 | 63 | 22.38 | 130/75 | No | No | No | No | 5.65 | 4.86 | 1.79 | 1.51 | 2.99 | Yes | Yes | No | No | No | No | No |
| BJH47 | M | 55 | 55 | 23.89 | 120/70 | Yes | No | No | No | 5.78 | 5.22 | 1.86 | 1.34 | 3.51 | Yes | Yes | No | No | No | No | No |
| BJH48 | M | 50 | 46 | 25.71 | 150/90 | No | Yes | No | No | 4.98 | 5.36 | 1.03 | 0.65 | 4.50 | Yes | NO | No | No | No | No | No |
| BJH49 | F | 59 | 57 | 27.03 | 160/100 | No | Yes | No | No | 4.68 | 4.69 | 1.05 | 1.23 | 3.25 | Yes | Yes | No | No | No | Yes | No |
| BJH50 | M | 55 | 60 | 31.64 | 125/70 | No | No | No | No | 5.23 | 5.32 | 1.21 | 1.62 | 3.46 | Yes | Yes | No | No | No | No | Yes |
| BJH51 | M | 56 | 58 | 22.02 | 130/70 | Yes | No | No | No | 5.42 | 6.03 | 1.31 | 1.25 | 4.52 | Yes | Yes | No | No | No | No | No |
| BJH52 | M | 60 | 61 | 27.18 | 200/120 | No | Yes | Yes | No | 5.44 | 5.32 | 1.54 | 1.32 | 3.69 | Yes | Yes | No | No | Yes | Yes | No |
| BJH53 | F | 73 | 73 | 26.11 | 220/160 | No | Yes | Yes | Yes | 7.65 | 4.96 | 0.85 | 1.54 | 3.25 | Yes | No | No | No | No | Yes | No |
| BJH54 | F | 62 | 66 | 30.48 | 140/90 | No | Yes | No | No | 5.14 | 5.77 | 1.06 | 1.65 | 3.91 | Yes | Yes | No | No | No | No | No |
| BJH55 | M | 49 | 53 | 25.06 | 230/180 | Yes | Yes | Yes | No | 5.41 | 4.99 | 0.99 | 1.24 | 3.55 | Yes | Yes | No | No | No | Yes | No |
| BJH56 | M | 70 | 73 | 24.03 | 190/100 | Yes | Yes | Yes | No | 4.78 | 4.68 | 1.06 | 1.32 | 3.15 | Yes | Yes | No | No | No | Yes | No |
| BJH57 | M | 69 | 69 | 26.33 | 170/100 | Yes | Yes | Yes | Yes | 14.1 | 5.32 | 1.54 | 1.62 | 3.39 | Yes | No | No | No | No | No | No |
| BJH58 | M | 50 | 48 | 26.30 | 180/110 | Yes | Yes | Yes | No | 4.99 | 5.24 | 1.75 | 1.33 | 3.56 | Yes | Yes | No | No | No | Yes | No |
| BJH59 | F | 53 | 57 | 18.03 | 135/70 | No | No | No | No | 4.85 | 4.12 | 1.63 | 1.24 | 2.55 | Yes | Yes | No | No | No | No | No |
| BJH60 | M | 54 | 58 | 22.06 | 180/100 | Yes | Yes | Yes | No | 5.12 | 4.55 | 2.35 | 1.34 | 2.74 | Yes | Yes | No | No | No | Yes | No |
| BJH61 | M | 62 | 65 | 25.47 | 145/95 | Yes | No | No | No | 5.21 | 5.21 | 1.55 | 1.24 | 3.66 | Yes | No | No | No | No | No | No |
| BJH62 | F | 68 | 69 | 28.30 | 210/120 | No | Yes | Yes | No | 4.65 | 5.68 | 1.45 | 1.35 | 4.04 | Yes | Yes | No | No | No | Yes | No |
| BJH63 | M | 76 | 78 | 19.81 | 200/120 | No | Yes | Yes | Yes | 7.3 | 4.32 | 1.68 | 1.65 | 2.33 | Yes | Yes | No | No | No | Yes | Yes |
| BJH64 | F | 53 | 55 | 22.50 | 135/85 | No | No | No | No | 5.12 | 5.12 | 1.77 | 1.25 | 3.52 | Yes | No | Yes | No | Yes | No | No |
| BJH65 | M | 64 | 65 | 26.96 | 160/100 | Yes | Yes | No | No | 5.32 | 5.24 | 1.69 | 1.38 | 3.52 | Yes | Yes | No | No | No | Yes | No |
| BJH66 | M | 66 | 70 | 23.66 | 125/75 | Yes | No | No | No | 5.42 | 6.12 | 2.01 | 1.39 | 4.33 | Yes | Yes | No | No | No | No | No |
| BJH67 | M | 47 | 49 | 26.89 | 160/90 | Yes | Yes | No | No | 5.21 | 5.47 | 1.54 | 1.41 | 3.75 | Yes | Yes | No | No | No | Yes | No |
| BJH68 | M | 66 | 67 | 26.57 | 170/100 | No | Yes | Yes | No | 4.77 | 5.65 | 1.65 | 1.65 | 3.67 | Yes | Yes | No | No | No | Yes | No |
| BJH69 | F | 51 | 51 | 23.34 | 180/100 | No | Yes | Yes | No | 4.85 | 4.89 | 1.87 | 1.34 | 3.18 | Yes | Yes | No | No | No | Yes | Yes |
| BJH70 | M | 76 | 78 | 27.78 | 170/90 | Yes | Yes | Yes | No | 4.86 | 5.32 | 1.96 | 1.62 | 3.31 | Yes | NO | No | No | No | Yes | No |
| BJH71 | M | 43 | 46 | 25.95 | 138/85 | Yes | No | No | No | 5.21 | 5.01 | 2.03 | 1.02 | 3.58 | No | NO | No | No | No | No | No |
| BJH72 | M | 64 | 65 | 24.16 | 160/90 | Yes | Yes | No | No | 5.26 | 4.35 | 2.01 | 0.96 | 2.99 | Yes | Yes | No | No | No | No | No |
| BJH73 | M | 53 | 55 | 26.03 | 220/120 | Yes | Yes | Yes | No | 4.98 | 5.21 | 1.54 | 1.35 | 3.55 | Yes | Yes | No | No | No | Yes | No |
| BJH74 | M | 67 | 70 | 22.91 | 200/100 | Yes | Yes | Yes | No | 5.27 | 5.68 | 1.68 | 1.24 | 4.10 | Yes | Yes | Yes | No | No | Yes | No |
| BJH75 | F | 53 | 53 | 27.55 | 135/85 | No | No | No | No | 5.68 | 5.45 | 1.58 | 1.35 | 3.78 | Yes | Yes | No | No | No | No | Yes |
| BJH76 | F | 64 | 67 | 27.77 | 150/90 | No | Yes | No | No | 5.32 | 4.87 | 1.37 | 1.24 | 3.36 | Yes | NO | No | No | Yes | No | No |
| BJH77 | M | 69 | 70 | 27.68 | 200/100 | Yes | Yes | Yes | No | 5.45 | 6.03 | 1.65 | 1.26 | 4.44 | Yes | Yes | No | No | No | Yes | No |
| BJH78 | F | 72 | 72 | 22.58 | 180/100 | No | Yes | Yes | No | 4.89 | 5.31 | 1.54 | 1.25 | 3.75 | Yes | Yes | No | No | No | Yes | Yes |
| BJH79 | F | 67 | 69 | 26.49 | 160/90 | No | Yes | No | No | 4.68 | 4.78 | 1.03 | 1.35 | 3.22 | Yes | Yes | No | No | No | No | No |
| BJH80 | M | 54 | 55 | 27.44 | 130/70 | No | No | No | Yes | 7.8 | 5.12 | 0.65 | 1.25 | 3.74 | No | Yes | No | No | No | No | Yes |
AMI indicates acute myocardial infarction; BMI, body mass index; DBP, diastolic blood pressure; DM, diabetes mellitus; FBG, fasting blood glucose; HDL‐C, high‐density lipoprotein cholesterol; HT, hypertension; LDL‐C, low‐density lipoprotein cholesterol; LVH, left ventricular hypertrophy; SBP, systolic blood pressure; T2DM, diabetes mellitus type 2; TC, total cholesterol; TG, triglyceride.
Mutational Analysis of 22 Mitochondrial tRNA Genes
DNA fragments spanning 22 tRNA genes were PCR‐amplified from genomic DNA of 80 affected Chinese subjects and 512 unaffected controls. Each fragment was purified and subsequently analyzed by DNA sequencing. Comparison of the resultant sequence with the Cambridge consensus sequence identified 16 (1 novel and 15 known) nucleotide changes in the 9 tRNA genes, as shown in Table 2. The novel variants were T7546C in tRNAAsp, whereas the known variants were T4386C in tRNAGln, A5592G and C5601T in tRNAAla, G5821A in tRNACys, A10005G and T10007C in tRNAGly, T10454C in tRNAArg, A12172G in tRNAHis, A14687G and A14693G in tRNAGlu, T15889C, and G15927A, G15928A, G15930A, and A15951G in tRNAThr.24 All the nucleotide changes were verified by sequence analysis of both strands and appeared to be homoplasmy. Among 80 subjects with coronary heart disease, 25 subjects carried 1 tRNA variant, whereas none of other 55 subjects harbored any mitochondrial tRNA variant.
Evaluation of Mitochondrial tRNA Variants
To identify putative deleterious mutation, these variants were further evaluated using the following 3 criteria: (1) present in <1% of the controls; (2) CI >75%, proposed by Ruiz‐Pesini and Wallace23; and (3) potential structural and functional alterations. First, we used the secondary structure of tRNAs to localize each variant with either a stem or a loop and to analyze if the base changes within stems altered the classic Watson‐Crick base pair. As shown in Figure 1, 9 variants were located at the loops, whereas 6 variants occurred in the stems of tRNAs. As shown in Table 4 and Figure 1, the A5592G variant in tRNAAla created a putative C‐G base‐pairing, whereas variants G5821A in tRNACys and T15889C, G15927A, G15928A, and A15951G in tRNAThr abolished putative base pairing(s). In addition, a phylogenetic analysis was performed by comparing the human tRNA nucleotide variants with those in 16 other vertebrates. As shown in Table 4, CI among the variants ranged from 12.5% (tRNAThr G15930A variant) to 100% (tRNAAla A5592G and tRNAGlu A14693G variants). In particular, the CI of 7 variants was >75%, the CI of 6 other variants was between 75% and 50%, and the CI of the remaining variants was <50%. These variants were then evaluated by examining the allelic frequency in 512 Han Chinese controls and 2704 control mtDNAs.25 The T7546C variant was absent in both this Chinese control population and the 2704 mtDNAs. Five variants were absent in this cohort of Chinese controls and <1% in the 2704 mtDNAs, whereas the allelic frequency of 10 other variants was >1% in this control population and/or in the 2704 mtDNAs. Based on these criteria, the tRNAAla A5592G and tRNAAsp T7546C mutations may have functional significance. In addition, our previous investigation showed that the tRNAThr G15927G mutation led to a failure in tRNA metabolism.26 Thus, these 3 tRNA variants are putative mutations associated with coronary heart disease. Furthermore, statistical analysis was carried out using Fisher's exact test to evaluate the difference in mitochondrial tRNA mutations between 80 subjects with CHD and 512 Chinese controls. The variants with P<0.05 were T15889C, G15927A, and G15930A, whereas the P value of 4 variants (A5592G, T7546C, A1005G, and T1007C) was 0.135. The higher P value of these 4 variants may be a result of the small sample size of subjects with CHD.
Figure 1.
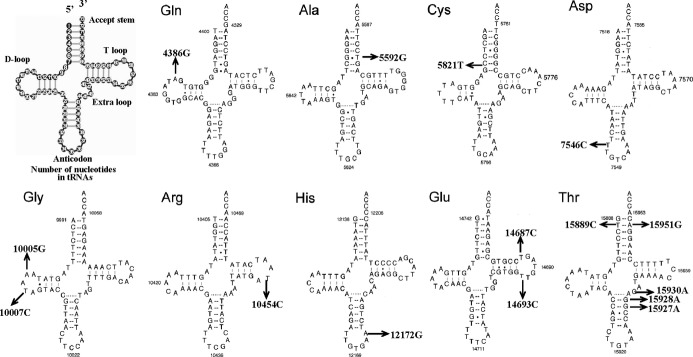
Mitochondrial tRNA variants in Chinese subjects with coronary heart disease. Cloverleaf structures of canonical tRNA and 9 mitochondrial tRNAs are shown. Circled numbers represent the nucleotide positions according to the conventional tRNA numbering system.17 Tertiary interactions between nucleotides are indicated by dotted lines. Arrows indicate the position of the tRNA mutations.
Table 4.
Variants in the Mitochondrial tRNA Genes in 80 Chinese Subjects With Coronary Heart Disease and 512 Controls
| Genes | Position | Replacement | Conservation Index (%)* | WC Base Pairs* | Number of 80 Patients (%) | Number of 512 Controls (%) | χ2 P Value | Number of 2704 mtDNAs* (%) | χ2 P Value |
|---|---|---|---|---|---|---|---|---|---|
| tRNAGln | 4386 | T to C | 75 | 1 (1.25) | 4 (0.78) | 0.517 | 51 (1.89) | 1.000 | |
| tRNAAla | 5592 | A to G | 100 | C‐G↑ | 1 (1.25) | 0 (0) | 0.135 | 3 (0.11) | 0.110 |
| 5601 | C to T | 50 | 2 (2.5) | 15 (2.9) | 0.709 | 37 (1.39) | 0.309 | ||
| tRNACys | 5821 | G to A | 62.5 | C‐G↓ | 1 (1.25) | 12 (2.3) | 1.000 | 14 (0.52) | 0.355 |
| tRNAAsp | 7546 | T to C | 100 | 1 (1.25) | 0 (0) | 0.135 | 0 (0) | 0.029 | |
| tRNAGly | 10005 | A to G | 87.5 | 1 (1.25) | 0 (0) | 0.135 | 3 (0.11) | 0.110 | |
| 10007 | T to C | 43.8 | 1 (1.25) | 0 (0) | 0.135 | 4 (0.15) | 0.136 | ||
| tRNAArg | 10454 | T to C | 50 | 1 (1.25) | 4 (0.78) | 0.517 | 11 (0.4) | 0.269 | |
| tRNAHis | 12172 | A to G | 93.8 | 1 (1.25) | 5 (0.98) | 0.583 | 31 (1.15) | 0.609 | |
| tRNAGlu | 14687 | A to G | 93.8 | 1 (1.25) | 1 (0.20) | 0.252 | 22 (0.81) | 0.490 | |
| 14693 | A to G | 100 | 2 (2.5) | 7 (1.37) | 0.349 | 10 (0.37) | 0.045 | ||
| tRNAThr | 15889 | T to C | 18.8 | U‐A↓ | 2 (2.5) | 1 (0.20) | 0.049 | 3 (0.11) | 0.008 |
| 15927 | G to A | 68.8 | G‐C↓ | 4 (5) | 7 (1.37) | 0.048 | 44 (1.62) | 0.047 | |
| 15928 | G to A | 68.8 | G‐C↓ | 1 (1.25) | 2 (0.39) | 0.354 | 132 (4.88) | 0.181 | |
| 15930 | G to A | 12.5 | 4 (5) | 5 (0.98) | 0.023 | 37 (1.39) | 0.029 | ||
| 15951 | A to G | 56.25 | A‐U↓ | 1 (1.25) | 2 (0.39) | 0.354 | 22 (0.81) | 0.490 |
The conservation index (CI) was then defined as the percentage of the human nucleotide variants with 16 other vertebrates that had the wild‐type nucleotide at that position.
Classic Watson–Crick (WC) base pair: created (↑) or abolished (↓).
Clinical and Genetic Characterization of 7 Chinese Subjects Carrying 1 of the Putative Mutations Associated With Coronary Heart Disease
Ten probands and other members in these families carrying 1 of the putative mutations underwent physical examinations and laboratory assessments of cardiovascular disease risk factors. Three probands, including subject BJH16 carrying the T7546C mutation and subjects BJH22 and BJH41 carrying the G15927A mutation, did not exhibit a family history of coronary heart disease. By contrast, 3 subjects had a family history of coronary heart disease. As shown in Figure 2, the pedigree of BJH24 carrying the A5592G mutation and the pedigrees of BJH15 and BJH45 harboring the G15927A mutation exhibited maternal transmission of coronary heart disease. In particular, 7 of 11 matrilineal relatives in the pedigree BJH24 and 6 of 17 matrilineal relatives in the pedigree BJH 45 suffered from coronary heart disease, whereas none of the affected fathers' offspring in these 2 families had clinical abnormalities. In the pedigree BJH15, 10 of 13 matrilineal relatives exhibited coronary heart disease, whereas all affected fathers with CHD, except subject II‐3 who married affected subject II‐4, never transmitted the trait to their offspring. These features are the maternal transmission of coronary heart disease in these 3 families.
Figure 2.
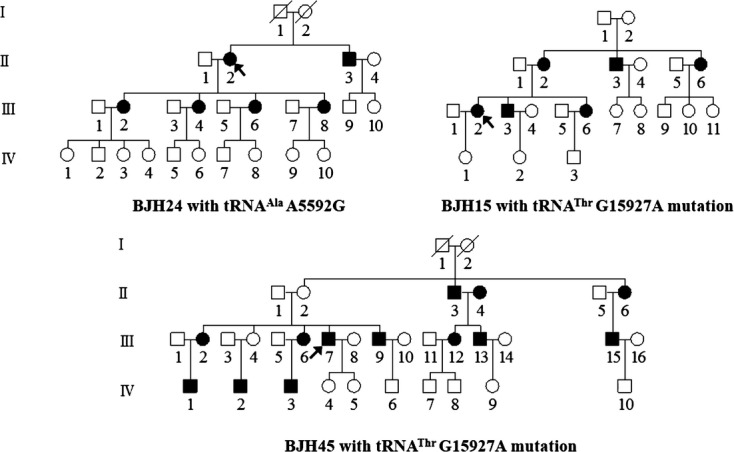
Three Han Chinese pedigrees with coronary heart disease. Affected individuals are indicated by filled symbols. An arrow denotes probands.
Mutational Analysis of Mitochondrial Genomes
To assess the contribution that mtDNA variants or haplogroups make toward the phenotypic expression of these putative mtDNA mutations in these Chinese pedigrees, we performed PCR amplification of fragments spanning the entire mtDNA and subsequent DNA sequence analysis in 2 probands carrying the G15927A mutation and 1 proband carrying the A5592G mutation. The sequence results from these Chinese subjects were aligned with the updated consensus Cambridge sequence.12 As shown in Table 5, these probands exhibited distinct sets of mtDNA polymorphisms. These included 27 variants in the D‐loop region, 5 known variants in the 12S rRNA gene, 2 known variants in the 16S rRNA gene, the known tRNAGln A5592G and tRNAThr G15927A mutations, and the known NC7 9‐bp deletion, as well as 29 (2 novel/27 known) silent variants and 11 known missense mutations in the polypeptide‐encoding genes.24 The mitochondrial genomes of subjects BJH15 and BJH45 belonged to the eastern Asian haplogroup B5b, whereas the mtDNA of subject BJH24 resided at haplogroup D4b.27 These variants in RNAs and polypeptides were further evaluated by phylogenetic analysis of these variants and sequences from 16 other organisms including mouse,28 bovine,29 and Xenopus laevis.30 Only the known CO1 G6969A (I356V) variant of subject BJH45 showed the high conservation in these species, proposed by Ruiz‐Pesini and Wallace,23 and had <1% frequency of 2706 mtDNAs. These data suggest that the CO1 G6962A(L356G) variant may have a role in the phenotypic manifestation of the G15927A mutation. By contrast, none of other variants showed both evolutionary conservation and <1% frequency of 2706 mtDNAs.
Table 5.
mtDNA Variants in 3 Han Chinese Probands With Coronary Heart Disease
| Gene | Position | Replacement | Conservation (H/B/M/X)* | CRS* | BJH15 | BJH45 | BJH24 | Previously Reported |
|---|---|---|---|---|---|---|---|---|
| D‐loop | 73 | A to G | A | G | G | G | Yes | |
| 103 | G to A | G | A | A | Yes | |||
| 152 | T to C | T | C | Yes | ||||
| 189 | A to G | A | G | Yes | ||||
| 199 | T to C | T | C | Yes | ||||
| 204 | T to C | T | C | Yes | ||||
| 263 | A to G | A | G | G | G | Yes | ||
| 310 | T to CT/CTC/CTCC | T | CTC | CT | TC | Yes | ||
| 315 | C to CC | C | CC | Yes | ||||
| 481 | C to T | C | T | Yes | ||||
| 489 | T to G | T | G | Yes | ||||
| 514 | C to Del | C | Del C | Del C | Del C | Yes | ||
| 515 | A to Del | A | Del A | Del A | Del A | Yes | ||
| 16111 | C to T | C | T | Yes | ||||
| 16140 | T to C | T | C | Yes | ||||
| 16183 | A to C | A | C | Yes | ||||
| 16189 | T to C | T | C | C | C | Yes | ||
| 16193 | C to CC | C | CC | Yes | ||||
| 16223 | C to T | C | T | Yes | ||||
| 16234 | C to T | C | T | Yes | ||||
| 16243 | T to C | T | C | Yes | ||||
| 16344 | C to T | C | T | Yes | ||||
| 16362 | T to C | T | C | Yes | ||||
| 16463 | A to G | A | G | Yes | ||||
| 16519 | T to C | T | C | Yes | ||||
| 16569 | T to C | T | C | Yes | ||||
| 12S rRNA | 709 | G toA | G/G/A/‐ | G | A | A | Yes | |
| 750 | A to G | A/A/A/‐ | A | G | G | Yes | ||
| 1382 | A to C | A/A/A/G | A | C | Yes | |||
| 1438 | A to G | A/A/A/G | A | G | G | G | Yes | |
| 1598 | G toA | G/A/T/T | G | A | A | Yes | ||
| 16S rRNA | 2626 | T to C | T/T/A/G | T | C | Yes | ||
| 2706 | A to G | A/G/A/A | A | G | G | G | Yes | |
| ND1 | 4161 | C to T | C | T | Yes | |||
| ND2 | 4769 | A to G | A | G | G | Yes | ||
| 4883 | C to T | C | T | Yes | ||||
| 4895 | A to G | A | G | Yes | ||||
| 5178 | C to A (Leu to Met) | L/T/T/T | C | A | Yes | |||
| tRNAAla | 5592 | A to G | A/A/A/A | A | G | Yes | ||
| CO1 | 6962 | G to A (Leu to Gly) | L/L/L/L | G | A | Yes | ||
| 7028 | C to T | C | T | T | Yes | |||
| CO2 | 8020 | G to A | G | A | Yes | |||
| NC7 | 8271‐79 | 9‐bp del | C | 9‐bp Del | 9‐bp Del | Yes | ||
| ATP8 | 8414 | C to T (Leu to Phe) | L/F/M/W | C | T | Yes | ||
| ATP6 | 8584 | G to A (Ala to Thr) | A/V/V/I | G | A | A | Yes | |
| 8701 | A to G (Thr to Ala) | T/S/L/Q | A | G | Yes | |||
| 8828 | C to T | C | T | No | ||||
| 8829 | C to T | C | T | Yes | ||||
| 8856 | G to A | G | A | Yes | ||||
| 8860 | A to G (Thr to Ala) | T/A/A/T | A | G | G | G | Yes | |
| 8964 | C to T | C | T | Yes | ||||
| CO3 | 9296 | C to T | C | T | Yes | |||
| 9540 | T to C | T | C | Yes | ||||
| 9824 | T to C | T | C | Yes | ||||
| 9950 | T to C | T | C | Yes | ||||
| ND3 | 10398 | A to G (Thr to Ala) | T/T/T/A | A | G | G | G | Yes |
| 10400 | C to T | C | T | Yes | ||||
| ND4 | 10873 | T to C | T | C | Yes | |||
| 11101 | A to G | A | G | Yes | ||||
| 11719 | G to A | G | A | A | A | Yes | ||
| ND5 | 12361 | A to G | A | G | G | Yes | ||
| 12705 | C to T | C | T | Yes | ||||
| 13959 | C to T | C | T | No | ||||
| ND6 | 14221 | T to C | T | C | Yes | |||
| 14668 | C to T | C | T | Yes | ||||
| Cytb | 14766 | C to T (Thr to Ile) | T/S/T/S | C | T | T | T | Yes |
| 14783 | T to C | T | C | Yes | ||||
| 15043 | G to A | G | A | Yes | ||||
| 15223 | C to T | C | T | T | Yes | |||
| 15301 | G to A | G | A | Yes | ||||
| 15326 | A to G (Thr to Ala) | T/M/I/I | A | G | G | G | Yes | |
| 15508 | C to T | C | T | T | Yes | |||
| 15662 | A to G (Ile to Val) | I/L/F/L | A | G | G | Yes | ||
| 15850 | T to C | T | C | Yes | ||||
| 15851 | A to G (Ile to Val) | I/A/S/M | A | G | G | Yes | ||
| tRNAThr | 15927 | G to A | G/G/G/G | G | A | A | Yes |
CRS indicates Cambridge reference sequence.5
Conservation of amino acid for polypepides or nucleotide for RNAs in human (H), bovine (B), mouse (M), and Xenopus laevis (X).
See the online mitochondrial genome database http://www.mitomap.org and http://www.genpat.uu.se/mtDB/
Discussion
In the present study, we performed a clinical, genetic, and molecular characterization of 80 Han Chinese subjects with coronary heart disease. Mutational analysis of mitochondrial tRNA genes identified 16 variants. These variants were further evaluated using the following criteria: (1) present in <1% of the controls; (2) evolutional conservation; (3) potential structural and functional alterations; (4) maternal transmission of coronary heart disease in matrilineal relatives carrying 1 of tRNA mutations. Of these variants, only tRNAGln A5592G and tRNAThr G15927A mutations were fitted with these criteria, suggesting that these mutations may be associated with coronary heart disease.
The first evidence was that the pedigrees of BJH24 carrying the A5592G mutation, BJH15, and BJH45 harboring the G15927A mutation exhibited maternal transmission of coronary heart disease. In particular, 7 of 11 matrilineal relatives in the pedigree BJH24, 6 of 17 matrilineal relatives in the pedigree BJH45, and 10 of 13 matrilineal relatives of pedigree BJH15 suffered from coronary heart disease, whereas none of affected fathers' offspring in these families had clinical abnormalities. The maternal transmissions of coronary heart disease in these 3 families strongly suggest that mutations in mitochondrial DNA are the molecular basis of this disorder in these families. Further sequence analysis of their mitochondrial genomes confirmed the presence of the homplasmic A5592G or G15927A mutations in all matrilineal relatives but not other members of these families. The absence of functionally significant mutations in mtDNAs of probands BJH15‐III‐2 and BJH24‐II‐2 indicates that mitochondrial backgrounds may not play an important role in the pathogenesis of coronary heart disease. On the other hand, the CO1 G6962A (L356G) variant may have a role in the phenotypic manifestation of the G15927A mutation in the pedigree of BJH45.
The T5592C mutation is localized at a highly conserved uridine (68U) on the acceptor stem of tRNAAla, where the position is important for the stability and identity of tRNA.17,31 The U‐to‐C transition at this position by the T5592C mutation is expected to create a highly conservative base pairing (5G‐68C) on the acceptor stem of this tRNA, alter the secondary structure of this tRNA, as in the case of the deafness‐associated tRNAHis T12201C mutation.32 Our previous investigation showed that the T12201C mutation destablized a highly conservative base pairing (5A‐68U) on the acceptor stem of this tRNA, leading to a failure in tRNA metabolism.32 In particular, ≈70% decrease in tRNAHis steady‐state level was observed in mutant cells carrying the T12201C mutation, compared with those of controls. These observations further supported the functional significance of the T5592C mutation in the pedigree BJH45 with coronary heart disease.
The homoplasmy G15927A mutation is at a highly conserved nucleotide (G42) in the anticodon stem of tRNAThr, where the position is important for the stability and identity of tRNA.31 The anticipated destabilization of base pairing (28C‐42G) by the G15927A mutation affects secondary structure and function of this tRNA, as in the cases of tRNAIle A4300G and tRNALeu(UUR) T3273C mutations.33–34 The G15927A mutation changed the conformation of tRNAThr, as suggested by slower electrophoretic mobility of mutated tRNA with respect to the wild‐type molecule. However, the aminoacylation level of the tRNAThr was not impaired, but the steady‐state level of tRNA was reduced 44% in lymphoblastoid cell lines derived from Chinese control subjects carrying the G15927A mutation.26 The alteration in tRNA metabolism by the G15927A mutation impaired mitochondrial translation and respiration, increasing the production of reactive oxygen species.35
The homoplasmic nature of the mtDNA A5592G and G15927A mutations hints at the mild nature of the mutations. These suggest that these mtDNA mutations are by themselves insufficient to produce a clinical phenotype19–20,36 but the inherited risk factors are necessary for the development of coronary heart disease. The nuclear modifier genes and environmental and epigenetic factors, as well as personal lifestyles, may also contribute to the development of coronary heart disease in these subjects carrying the mtDNA mutation.37–40 In particular, the tissue specificity of the tRNAAla A5592G or tRNAThr G15927A mutation in these Chinese families is likely attributed to tissue‐specific tRNA metabolism or the involvement of nuclear modifier genes.
In summary, our data provide evidence that mitochondrial genetic defects may lead to coronary heart disease. The mitochondrial tRNAAla A5592G and tRNAThr G15927A mutations altered the structure and function of their tRNAs, thereby causing mitochondrial dysfunctions and long‐standing increase of reactive oxygen species in cardiovascular cells. These 2 mutations may be the inherited risk factors for coronary heart disease. Thus, our findings may provide new insights into the understanding of the pathophysiology and valuable information for the management and treatment of coronary heart disease.
Sources of Funding
This work was supported by National Institutes of Health (NIH) grant RO1DC07696 from the National Institute on Deafness and Other Communication Disorders and a research grant (206‐08) from the Ministry of Science and Technology, Wenzhou City Government, China (to M.‐X.G.).
Disclosures
None.
Reference
- 1.Lopez AD, Mathers CD, Ezzati M, Jamison DT, Murray CJ. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet. 2006; 367:1747-1757 [DOI] [PubMed] [Google Scholar]
- 2.Zhang XH, Lu ZL, Liu L. Coronary heart disease in China. Heart. 2008; 94:1126-1131 [DOI] [PubMed] [Google Scholar]
- 3.Khot UN, Khot MB, Bajzer CT, Sapp SK, Ohman EM, Brener SJ, Ellis SG, Lincoff AM, Topol EJ. Prevalence of conventional risk factors in patients with coronary heart disease. JAMA. 2003; 290:898-904 [DOI] [PubMed] [Google Scholar]
- 4.Sing CF, Stengard JH, Kardia SL. Genes, environment, and cardiovascular disease. Arterioscler Thromb Vasc Biol. 2003; 23:1190-1196 [DOI] [PubMed] [Google Scholar]
- 5.Wilson PW, D'Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of coronary heart disease using risk factor categories. Circulation. 1998; 97:1837-1847 [DOI] [PubMed] [Google Scholar]
- 6.Wang Q. Molecular genetics of coronary artery disease. Curr Opin Cardiol. 2005; 20:182-188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peden JF, Farrall M. Thirty‐five common variants for coronary artery disease: the fruits of much collaborative labour. Hum Mol Genet. 2011; 20:R198-R205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang F, Xu CQ, He Q, Cai JP, Li XC, Wang D, Xiong X, Liao YH, Zeng QT, Yang YZ, Cheng X, Li C, Yang R, Wang CC, Wu G, Lu QL, Bai Y, Huang YF, Yin D, Yang Q, Wang XJ, Dai DP, Zhang RF, Wan J, Ren JH, Li SS, Zhao YY, Fu FF, Huang Y, Li QX, Shi SW, Lin N, Pan ZW, Li Y, Yu B, Wu YX, Ke YH, Lei J, Wang N, Luo CY, Ji LY, Gao LJ, Li L, Liu H, Huang EW, Cui J, Jia N, Ren X, Li H, Ke T, Zhang XQ, Liu JY, Liu MG, Xia H, Yang B, Shi LS, Xia YL, Tu X, Wang QK. Genome‐wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nat Genet. 2011; 43:345-349 [DOI] [PubMed] [Google Scholar]
- 9.Tregouet DA, Konig IR, Erdmann J, Munteanu A, Braund PS, Hall AS, Grosshennig A, Linsel‐Nitschke P, Perret C, DeSuremain M, Meitinger T, Wright BJ, Preuss M, Balmforth AJ, Ball SG, Meisinger C, Germain C, Evans A, Arveiler D, Luc G, Ruidavets JB, Morrison C, van der Harst P, Schreiber S, Neureuther K, Schafer A, Bugert P, El Mokhtari NE, Schrezenmeir J, Stark K, Rubin D, Wichmann HE, Hengstenberg C, Ouwehand W, Ziegler A, Tiret L, Thompson JR, Cambien F, Schunkert H, Samani NJ. Genome‐wide haplotype association study identifies the SLC22A3‐LPAL2‐LPA gene cluster as a risk locus for coronary artery disease. Nat Genet. 2009; 41:283-285 [DOI] [PubMed] [Google Scholar]
- 10.Mueller EE, Eder W, Ebner S, Schwaiger E, Santic D, Kreindl T, Stanger O, Paulweber B, Iglseder B, Oberkofler H, Maier R, Mayr JA, Krempler F, Weitgasser R, Patsch W, Sperl W, Kofler B. The mitochondrial T16189C polymorphism is associated with coronary artery disease in Middle European populations. PLoS One. 2011; 6:e16455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sobenin IA, Sazonova MA, Ivanova MM, Zhelankin AV, Myasoedova VA, Postnov AY, Nurbaev SD, Bobryshev YV, Orekhov AN. Mutation C3256T of mitochondrial genome in white blood cells: novel genetic marker of atherosclerosis and coronary heart disease. PLoS One. 2012; 7:e46573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999; 23:147. [DOI] [PubMed] [Google Scholar]
- 13.Guan MX, Enriquez JA, Fischel‐Ghodsian N, Puranam RS, Lin CP, Maw MA, Attardi G. The deafness‐associated mitochondrial DNA mutation at position 7445, which affects tRNASer(UCN) precursor processing, has long‐range effects on NADH dehydrogenase subunit ND6 gene expression. Mol Cell Biol. 1998; 18:5868-5879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ballinger SW. Mitochondrial dysfunction in cardiovascular disease. Free Radic Biol Med. 2005; 38:1278-1295 [DOI] [PubMed] [Google Scholar]
- 15.Liu C, Yang Q, Hwang SJ, Sun F, Johnson AD, Shirihai OS, Vasan RS, Levy D, Schwartz F. Association of genetic variation in the mitochondrial genome with blood pressure and metabolic traits. Hypertension. 2012; 60:949-956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marian AJ. Mitochondrial genetics and human systemic hypertension. Circ Res. 2011; 108:784-786 [DOI] [PubMed] [Google Scholar]
- 17.Suzuki T, Nagao A, Suzuki T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu Rev Genet. 2011; 45:299-329 [DOI] [PubMed] [Google Scholar]
- 18.Li R, Liu Y, Li Z, Yang L, Wang S, Guan MX. Failures in mitochondrial tRNAMet and tRNAGln metabolism caused by the novel 4401A>G mutation are involved in essential hypertension in a Han Chinese family. Hypertension. 2009; 54:329-337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Li R, Li Z, Wang XJ, Yang L, Wang S, Guan MX. Mitochondrial transfer RNAMet 4435A>G mutation is associated with maternally inherited hypertension in a Chinese pedigree. Hypertension. 2009; 53:1083-1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang S, Li R, Fettermann A, Li Z, Qian Y, Liu Y, Wang X, Zhou A, Mo JQ, Yang L, Jiang P, Taschner A, Rossmanith W, Guan MX. Maternally inherited essential hypertension is associated with the novel 4263A>G mutation in the mitochondrial tRNAIle gene in a large Han Chinese family. Circ Res. 2011; 108:862-870 [DOI] [PubMed] [Google Scholar]
- 21.Patel MR, Dehmer GJ, Hirshfeld JW, Smith PK, Spertus JA. ACCF/SCAI/STS/AATS/AHA/ASNC 2009 Appropriateness Criteria for Coronary Revascularization: a report by the American College of Cardiology Foundation Appropriateness Criteria Task Force, Society for Cardiovascular Angiography and Interventions, Society of Thoracic Surgeons, American Association for Thoracic Surgery, American Heart Association, and the American Society of Nuclear Cardiology Endorsed by the American Society of Echocardiography, the Heart Failure Society of America, and the Society of Cardiovascular Computed Tomography. J Am Coll Cardiol. 2009; 53:530-553 [DOI] [PubMed] [Google Scholar]
- 22.Rieder MJ, Taylor SL, Tobe VO, Nickerson DA. Automating the identification of DNA variations using quality‐based fluorescence re‐sequencing: analysis of the human mitochondrial genome. Nucleic Acids Res. 1998; 26:967-973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruiz‐Pesini E, Wallace DC. Evidence for adaptive selection acting on the tRNA and rRNA genes of human mitochondrial DNA. Hum Mutat. 2006; 27:1072-1081 [DOI] [PubMed] [Google Scholar]
- 24.Ruiz‐Pesini E, Lott MT, Procaccio V, Poole JC, Brandon MC, Mishmar D, Yi C, Kreuziger J, Baldi P, Wallace DC. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007; 35:D823-D828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingman M, Gyllensten U. mtDB: human mitochondrial genome database, a resource for population genetics and medical sciences. Nucleic Acids Res. 2006; 34:D749-D751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Lu J, Zhu Y, Yang A, Yang L, Li R, Chen B, Qian Y, Tang X, Wang J, Zhang X, Guan MX. Mitochondrial tRNAThr G15927A mutation may modulate the phenotypic manifestation of ototoxic 12s rRNA A1555G mutation in four Chinese families. Pharmacogenet Genomics. 2008; 18:1059-1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong QP, Bandelt HJ, Sun C, Yao YG, Salas A, Achilli A, Wang CY, Zhong L, Zhu CL, Wu SF, Torroni A, Zhang YP. Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum Mol Genet. 2006; 15:2076-2086 [DOI] [PubMed] [Google Scholar]
- 28.Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981; 26:167-180 [DOI] [PubMed] [Google Scholar]
- 29.Gadaleta G, Pepe G, De Candia G, Quagliariello C, Sbisa E, Saccone C. The complete nucleotide sequence of the Rattus norvegicus mitochondrial genome: cryptic signals revealed by comparative analysis between vertebrates. J Mol Evol. 1989; 28:497-516 [DOI] [PubMed] [Google Scholar]
- 30.Roe BA, Ma DP, Wilson RK, Wong JF. The complete nucleotide sequence of the Xenopus laevis mitochondrial genome. J Biol Chem. 1985; 260:9759-9774 [PubMed] [Google Scholar]
- 31.Normanly J, Abelson J. tRNA identity. Annu Rev Biochem. 1989; 58:1029-1049 [DOI] [PubMed] [Google Scholar]
- 32.Yan X, Wang X, Wang Z, Sun S, Chen G, He Y, Mo JQ, Li R, Jiang P, Lin Q, Sun M, Li W, Bai Y, Zhang J, Zhu Y, Lu J, Yan Q, Li H, Guan MX. Maternally transmitted late‐onset non‐syndromic deafness is associated with the novel heteroplasmic T12201C mutation in the mitochondrial tRNAHis gene. J Med Genet. 2011; 48:682-690 [DOI] [PubMed] [Google Scholar]
- 33.Campos Y, Gamez J, Garcia A, Andreu AL, Rubio JC, Martin MA, del Hoyo P, Navarro C, Cervera C, Garesse R, Arenas J. A new mtDNA mutation in the tRNALeu(UUR) gene associated with ocular myopathy. Neuromuscul Disord. 2001; 11:477-480 [DOI] [PubMed] [Google Scholar]
- 34.Taylor RW, Giordano C, Davidson MM, d'Amati G, Bain H, Hayes CM, Leonard H, Barron MJ, Casali C, Santorelli FM, Hirano M, Lightowlers RN, DiMauro S, Turnbull DM. A homoplasmic mitochondrial transfer ribonucleic acid mutation as a cause of maternally inherited hypertrophic cardiomyopathy. J Am Coll Cardiol. 2003; 41:1786-1796 [DOI] [PubMed] [Google Scholar]
- 35.Jia Z, Wang X, Qin Y, Xue L, Jiang P, Meng Y, Shi S, Wang Y, Qin Mo J, Guan MX. Coronary heart disease is associated with a mutation in mitochondrial tRNA. Hum Mol Genet. 2013; 22:4064-4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu Q, Li R, Jiang P, Xue L, Lu Y, Song Y, Han J, Lu Z, Zhi S, Mo JQ, Guan MX. Mitochondrial tRNA mutations are associated with maternally inherited hypertension in two Han Chinese pedigrees. Hum Mutat. 2012; 33:1285-1293 [DOI] [PubMed] [Google Scholar]
- 37.Addabbo F, Montagnani M, Goligorsky MS. Mitochondria and reactive oxygen species. Hypertension. 2009; 53:885-892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Djousse L, Driver JA, Gaziano JM. Relation between modifiable lifestyle factors and lifetime risk of heart failure. JAMA. 2009; 302:394-400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galluzzi L, Kepp O, Trojel‐Hansen C, Kroemer G. Mitochondrial control of cellular life, stress, and death. Circ Res. 2012; 111:1198-1207 [DOI] [PubMed] [Google Scholar]
- 40.Vasan RS, Beiser A, Seshadri S, Larson MG, Kannel WB, D'Agostino RB, Levy D. Residual lifetime risk for developing hypertension in middle‐aged women and men: the Framingham Heart Study. JAMA. 2002; 287:1003-1010 [DOI] [PubMed] [Google Scholar]


