Abstract
Objective:
To identify the genetic cause of axonal hereditary motor and sensory neuropathy (HMSN2) with infrequent giant axons.
Methods:
We studied 11 members of a previously described HMSN2 family with infrequent giant axons and variable cardiomyopathy. Whole-exome sequencing (WES) was performed on 2 affected persons and 1 unaffected person. Sanger sequencing was utilized to confirm the identified novel variant tracking with the affected status. Linkage analysis and haplotype mapping were obtained to confirm the causal nature of the identified variant. Cotransfection of HEK293 cells and co-immunoprecipitation assay were performed to assess the impact of the identified mutant protein in the implicated ubiquitin ligase pathway.
Results:
Giant axons with neurofilament accumulations were found in 3 affected persons who had undergone nerve biopsy evaluations. Six novel variants were identified by WES, but only DCAF8 p.R317C tracked with affected status within the family. Linkage and haplotype analysis using microsatellite markers supported this variant as causal. The mutation is within the DCAF8 WD repeat region critical for its binding to DDB1. Functional analysis shows DCAF8 p.R317C reduces the association of DCAF8 and DDB1, which is important in Cul4-ubiquitin E3 function.
Conclusions:
Our results indicate that DCAF8 p.R317C mutation is responsible for this specific variety of HMSN2 with infrequent giant axons and mild cardiomyopathy. This mutation results in decreased DDB1–DCAF8 association, leading to an E3 ubiquitin ligase defect that is likely associated with neurofilament degradation.
The hereditary motor and sensory neuropathies (HMSNs), also known as Charcot-Marie-Tooth, are caused by diverse genetic mutations.1 We previously reported a large German HMSN2 kinship with unique features, i.e., infrequent axonal swellings with neurofilament accumulations in sural nerve biopsies performed on 3 patients with the most severe neurologic phenotypes.2 These 3 severely affected patients also had mild cardiomyopathy. Subsequently, several HMSN2 cases with giant axons were reported, and genetic cause has been associated with mutations of NEFL (HMSN2E) and SH3TC2 (HMSN4C).3–5 A recent study also linked BAG3 mutation with giant axons in patients with prominent distal myofibrillar myopathy, cardiomyopathy, and mild sensory neuropathy.6 Despite sharing some similar characteristics, these disorders are different from the originally described giant axonal neuropathy, which is autosomal recessively inherited and caused by mutations of GAN (gigaxonin).7,8 The disorder described in this report is autosomal dominantly inherited; affected persons have no kinky hair or progressive CNS involvement as occurs with GAN mutations. In addition, their phenotypic onset usually occurred after the second decade, whereas patients with GAN mutations present phenotypes during toddler years and often die by the third decade. GAN protein has been recognized as an adaptor for CUL3-E3 ubiquitin ligase and is important in the proteasomal degradation pathway involved in clearing neuronal neurofilaments.7
In this report, we describe a novel genetic cause for HMSN2 with neurofilament accumulations and infrequent giant axons. The causal mutation also occurs in a protein important in the E3 ubiquitin ligase pathway, namely DDB1 and CUL4 associated factor 8 (DCAF8).
METHODS
Standard protocol approvals and patient consents.
This study was reviewed and approved by the Mayo Institutional Review Board. Written informed consent was obtained from all persons participating in this research study. Eleven members of this previously described HMSN2 kindred2 participated; 6 were affected and 5 unaffected (figure 1).
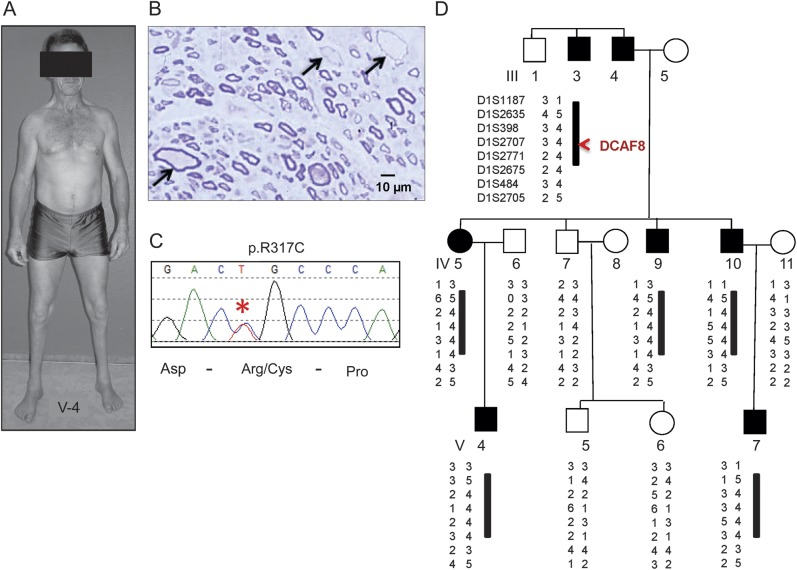
Figure 1. Genetic analysis and nerve biopsy in DCAF8 p.R317C family.
(A) The distribution of peroneal atrophy seen in patient V-4, which had not significantly worsened compared to an earlier photograph 28 years prior.2 (B) Sural nerve of patient IV-5 shows giant myelinated axons (arrows), also seen in 2 other biopsied persons with neurofilament accumulations.2 (C) Chromatograph of Sanger sequencing showing the DCAF8 p.R317C mutation (asterisk) identified by whole-exome sequencing. (D) The linkage analysis and a conserved haplotype at chromosome 1q23.2 supported the DCAF8 p.R317C variant as causal.
Kindred review.
Extensive kindred evaluation by neurologic examinations and nerve conduction studies indicated variable severity among affected persons. Needle EMG showed characteristics of chronic denervation with prolonged durations (up to 30 msec, normal <10 msec) and decreased firing with maximal contraction in affected muscles. The clinical presentations included gait abnormality, ankle weakness with prominent peroneal muscle atrophy (figure 1A), and hand weakness. Bony abnormalities such as pes cavus were present since infancy or childhood; hyporeflexia and areflexia were usually limited to the Achilles reflexes. For the 3 most severely affected patients (IV-5, IV-4, and V-4), neither the muscle action potential nor the nerve action potential could be elicited from their intrinsic foot muscles and their sural nerves, respectively. Sensory loss was limited to distal extremities and presented as either delayed cortical somatosensory evoked potential or desynchronization of sensory nerve action potential. Of 6 affected persons, 3 (IV-5, IV-10, and V-4) had undergone nerve biopsy evaluations. Their nerve biopsies revealed a reduction in the number of myelinated fibers and enlarged fibers with giant axons and thin myelin sheaths in patchy distribution (figure 1B).2 These giant axons had greatly increased numbers of misdirected neurofilaments; in addition, microtubules and mitochondria were displaced to the periphery of the axons. All 6 affected patients underwent cardiovascular assessment. Patients III-3, IV-9, and V-5 had normal ECGs and no signs of cardiac involvement. Patients IV-5, IV-10, and V-4, who had the most severe neurologic signs, all had low-grade systolic murmur; ECG and cardiac catheterization testing revealed mild cardiomyopathy. Since the initial report of this family, the clinical follow-up was available on all affected persons and none had overt neurologic progression. However, 2 affected patients died from cardiomyopathy-related problems. Patient IV-5 died at age 72 years with congestive heart failure and patient IV-10 died at age 68 years with cardiomyopathy-related problems. Patient III-3 died at age 76 years for reasons unrelated to cardiomyopathy. Patient IV-9 with most prominent distal peroneal atrophy is alive at age 52 years. Patient V-4 is 42 years old and still engaged in sport activities without symptomatic cardiomyopathy. His weakness remains predominantly of the below knee segments (figure in initial publication2 and figure 1A). Patient V-7, currently in his early 40s, was considered affected based on examination and mild pes cavus and has not had significant progression.
Genetic analysis.
Whole-exome sequencing (WES) was performed in patients II-3, IV-5, and IV-7. SureSelect All-Exon Kits V3 (50 Mb) (Agilent Technologies, Santa Clara, CA) was used for exome capture and HiSeq2000 (Illumina, San Diego, CA) for 101 bp paired-end sequencing. Data were analyzed using in-house developed pipeline. Briefly, the reads were aligned to reference genome build 37.1 using Novoalign (v2.07.13) followed by realignment, recalibration, and variant calling using Genome Analysis Toolkit (GATK)9 (v1.226). The called variants were filtered and annotated using the latest Targeted RE-sequencing Annotation Tool (TREAT),10 incorporating dbSNP135, 1KGenome, and ESP6500 databases. Nonsynonymous variants were evaluated by PolyPhen-2, SIFT, Mutation Taster, and Genomic Evolutionary Rate Profiling (GERP) score. A custom-designed TaqMan genotyping assay (Life Technologies, Carlsbad, CA) was utilized to screen DCAF8-R317C in 1,536 controls with European ancestry. Sanger sequencing was utilized to confirm the DCAF8-R317C tracking with disease in the family and to screen the DCAF8 coding exons in 108 additional HMSN2 probands.
Linkage haplotype analysis.
High-definition mapping at chromosome 1q23.2 was carried out using 8 sequence-tagged site (STS) markers. Family structure and marker values were confirmed using the Pedcheck program.11 Multipoint linkage was calculated using the package Merlin,12 assuming a rare susceptibility allele (frequency 0.001) and an autosomal dominant mode of inheritance. The penetrance was assumed to be 0.001 for noncarriers and 0.9 for carriers. Marker allele frequencies were estimated from our data and equal recombination fractions for males and females were assumed. An estimate of the maximum attainable logarithm of odds (LOD) score possible with this pedigree was obtained using FastSLINK.13
Co-immunoprecipitation assay.
Site-directed mutagenesis of DCAF8-R317C was carried out using the QuikChange kit (Stratagene, La Jolla, CA). HEK293T cells were cotransfected with wild-type or mutant Flag-tagged DCAF and hemagglutinin (HA)-tagged DDB1 using FugeneHD (Promega, Madison, WI). Forty-eight hours posttransfection, cells were lysed using IP Lysis buffer (Pierce, Rockford, IL). Lysates were cleared by centrifugation and affinity purification was performed using Magnetic HA-Tag IP/Co-IP kit (Pierce). The eluted samples were analyzed by Western blot with anti-Flag antibody (A8592; Sigma-Aldrich, St. Louis, MO). The efficiency of affinity purification of DDB1 was monitored using anti- HA antibody (ab1190, Abcam, Cambridge, UK). The comparable transfection efficiency of FLAG-DCAF8 was checked using cell lysates with anti-Flag antibody (A8592; Sigma). The equal loading was checked by bicinchoninic acid protein assay (Pierce) and the expression of glyceraldehyde 3-phosphate dehydrogenase. In addition, a DCAF8 mutant construct R314H, initially utilized to demonstrate the critical role of WDxR motif in DCAF8-DDB1 association,8 was used as a positive control.
RESULTS
Of 6 affected persons in this HMSN2 kinship, the 3 most severely affected (IV-5, IV-10, and V-4) had undergone nerve biopsy evaluations, and giant axons with neurofilament accumulations were found in patchy distribution (figure 1B).2 We first performed Sanger sequencing of MFN2, GAN, and NEFL, but no mutation was identified. We then utilized WES in 2 affected patients (III-3, IV-5) and 1 unaffected patient (IV-7) (figure 1). The HiSeq2000 generated 13–15 Gb of sequencing data per sample and achieved an average sequencing depth of 125–150X with high-quality genotype calls (phred score14 > 30). Approximately 95% of targeted regions were covered by 10X sequencing depth in all 3 exomes. The alignment and variant calls were carried out using an in-house developed pipeline incorporating dbSNP135, HAPMAP, 1KGenome, and the National Heart, Lung and Blood Institute's Exome Sequencing Project (ESP6500) for in-depth novel variant filtering.10 While conducting novel variant analysis across the whole exome, we specifically interrogated all variants in the exons of known causal genes for HMSN and distal hereditary motor neuropathy (dHMN) and did not find any known pathogenic or novel variants in these causal genes. The gene list for HMSN and dHMN was generated based on recent review.1 We also ensured the absence of mutation and full coverage in the BAG3 gene, which has been shown to cause giant axonal neuropathy with cardiomyopathy. Our bioinformatics analysis identified 6 novel nonsynonymous variants existing only in affected patients III-3 and IV-5 but not in unaffected patient IV-7. Sanger sequencing was used to verify all the variants and to check whether variants tracked with the affected status within the family. Among these 6 variants, only DCAF8 p.R317C tracked with disease in the family (figure 1, C and D). This variant was absent in >17,000 control chromosomes from available databases (ESP6500, 1KGenome, dbSNP135, and HAPMAP). To further confirm the mutation is not present in normal controls, we designed a custom TaqMan genotyping assay (Life Technologies) to screen DCAF8 p.R317C in our 1,536 controls with European ancestry and no known neuropathy history and confirmed that DCAF8 p.R317C is absent in the 1,536 control samples.
To increase the causative certainty of this alteration, we performed high-definition linkage mapping at 1q23.2 where DCAF8 localizes. We identified 9 STS markers spanning a 3.3-Mb region at chromosome 1q23.2 (chr1:158895175–162210226) and carried out microsatellite analysis. We applied FastSLINK13 to estimate the maximum attainable LOD score possible with this pedigree. Our microsatellite linkage analysis obtained a LOD score of 1.72, which is the maximum possible LOD score based on this pedigree. Haplotype analysis also confirmed the conserved segment where DCAF8 resides (between D12S707 and D12S771) as tracking with the affected status (figure 1D). The R317 residue is located within a highly conserved region (figure 2A) of DCAF8 and the alteration of R317C is predicted to be damaging by SIFT and disease-causing by Mutation Taster. The high fidelity of DCAF8 is further supported by the extremely rare occurrence of any missense variant in this gene. Based on the ESP6500 database, all DCAF8 nonsynonymous variants have minor allele frequency of <0.0009.
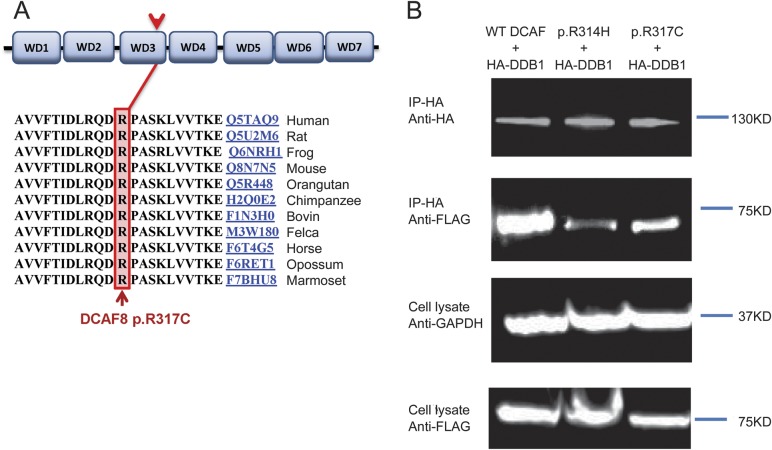
Figure 2. The conserved location of DCAF8 p.R317C and functional analysis.
(A) The identified p.R317C mutation occurs within a highly conserved region in the third WD repeat of DCAF8. (B) In vivo binding of DDB1 with wild-type (WT; left lane) or mutant p.R317C (right lane) and p.R314H (middle lane) DCAF8 proteins. Expression plasmids coding for Flag-tagged DCAF8 (pCDNA1-DCAF8) and hemagglutinin (HA)-tagged DDB1 (pGLUE-DDB1) were cotransfected into HEK293 cells. The association between DDB1 and the WT or mutant DCAF8 proteins was evaluated by affinity pull-down of DDB1 using anti-HA beads followed by Western blotting using anti-Flag antibodies (second panel). The efficiency of affinity purification of DDB1 and the transfection efficiency of Flag-DCAF8 were monitored as shown in the top panel and third panel, respectively. The equal loading was checked using anti-GAPDH (bottom panel). Flag-DCAF8-R314H was used as positive control. The images shown are representative of 3 independent experiments. GADPH = glyceraldehyde 3-phosphate dehydrogenase.
The incidence of HMSN2 with giant axons is unknown. To date, only a few cases have been reported.3,4,6 We did not identify any cases with similar phenotype in our sample bank. To investigate whether DCAF8 mutation may occur in other HMSN2 cases, we screened all coding exons of DCAF8 in 108 HMSN2 probands without known genetic cause. Not only did we not identify any novel nonsynonymous variants within DCAF8, we also did not find any nonsynonymous variant among 108 probands. This result supports the highly conserved nature of the DCAF8 gene (figure 1C) and underscores the fact that DACF8 mutation is very specific for this phenotypic presentation.
To investigate the functional impact of DCAF8 p.R317C, we applied site-directed mutagenesis and generated a DCAF8 p.R317C expression vector. HEK293T cells were cotransfected with wild-type or mutant Flag-tagged DCAF and HA-tagged DDB1 using FugeneHD (Promega). The eluted samples were analyzed on Western blot with anti-Flag antibody (A8592; Sigma). The DCAF8 mutant construct R314H, which was introduced through mutagenesis and demonstrated the critical role of WDxR motif in DCAF8-DDB1 association,8 was used as a positive control. Our co-immunoprecipitation assay of DDB1 with wild-type and DCAF8 p.R317C proteins showed R317C mutant protein decreased DCAF8 binding to DDB1, indicating a negative impact on the E3 ubiquitin ligase substrate recruiting (figure 2B).
DISCUSSION
The genetic causes for the majority of HMSN2 cases remain unknown.15 Our discovery of this DCAF8 mutation expands the genetic causes of HMSN2 and should be especially considered with giant axons on nerve biopsy and in association with cardiomyopathy. The DCAF8 protein, also known as WDR42A, contains 7 WD repeats that form a β-propeller and interacts with DDB1 protein in the CUL4-DDB1 E3 ubiquitin ligase macromolecular complex (figure 2A). Recent studies have shown that DCAF8 functions as a substrate-recognition receptor for CUL4-DDB1 E3 ubiquitin ligase.8 The CUL4-DDB1 ubiquitin ligase regulates cell proliferation, DNA repair, and chromatin integrity through targeted ubiquitination of specific substrates. Previous studies focusing on structure-based analysis of DCAF8 recognized a relatively conserved WDxR motif among the WD40-containing DCAFs.8 The WDxR motif resides on the solvent-exposed surface of the WD40 propeller fold and is crucial for DCAF proteins binding to DDB1 in the E3 ubiquitin ligase complex. Reduced binding to DDB1 has been demonstrated through mutagenesis analysis introducing WDxR motif mutation DCAF8-R314H.8 The importance of the WDxR motif is emphasized by another WDxR motif mutation, R273H, in the WD40 β-propeller protein DDB2, which causes human xeroderma pigmentosum with neuropathy.8,16,17 The DCAF8 R317 locates right next to this conserved WDxR motif on the solvent-exposed surface of WD40 β-propeller. Switching hydrophilic arginine to hydrophobic cysteine is likely to have consequential impact on WD40 β-propeller interaction. Our co-immunoprecipitation assay of DDB1 with wild-type and DCAF8 p.R317C proteins showed R317C mutant protein decreased DCAF8 binding to DDB1, indicating a negative impact on the E3 ubiquitin ligase substrate recruiting (figure 2B). DACF8 is one of the important players in Cul4-E3 ubiquitin ligase and possibly involved in the neurofilament substrate degradation. However, the structure of DCAF8 has not been solved. How exactly R317C mutation affects the association with DDB1 remains to be elucidated. Future crystallographic studies of the DDB1-DCAF8 complex will illustrate the more precise effect of R317C mutation on the interaction of DDB1and DCAF8.
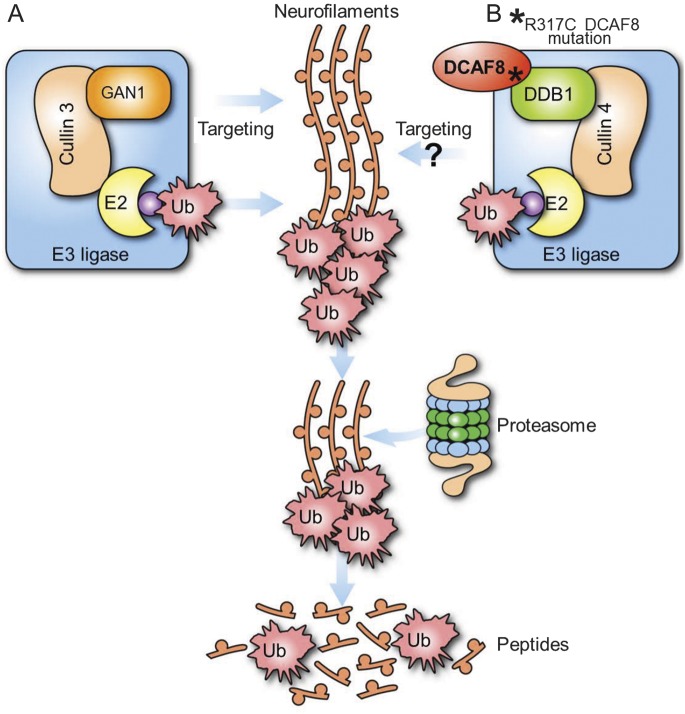
Giant axons and neurofilament accumulation on nerve biopsy were initially considered as the hallmarks of giant axonal neuropathy caused by biallelic mutations of GAN.18 Since then, giant axon and neurofilament accumulation have also been observed in HMSN2E caused by NEFL mutations and in HMSN4C caused by SH3TC2 mutations. GAN protein functions as an adaptor for CUL3-E3 ubiquitin ligase and is important in the proteasomal degradation pathway involved in clearing neuronal neurofilaments.7 SH3TC2 protein colocalizes with RAB11 on endosomal vesicles and is an adapter molecule involved in endocytic recycling.19 Recent study also discovered infrequent giant axons with neurofilament accumulations in myofibrillar myopathy patients with axonal neuropathy resulting from heterozygous BAG3 mutations6 and in early-onset axonal neuropathy patients by recessive mutations of TRIM2. BAG3 mediates degradation of misfolded proteins via autophagosome20; TRIM2 has been shown to be important in neurofilament ubiquitination.21 In addition, LRSAM1, which encodes a multidomain protein possessing E3 ubiquitin ligase activity, was found to be causal for axonal neuropathy in both dominant and recessive forms.22 Taken together, it is evident that the ubiquitination and protein degradation pathway plays a central role in the pathogenesis of giant axon and neurofilament accumulation.18 While the pathogenesis of DCAF8 p.R317C mutation remains to be discovered, it is plausible to speculate that DCAF8 mutation leads to impaired ubiquitination targeting of neurofilament in CUL4-E3 ubiquitin ligase–associated degradation pathway (figure 3).7 This study provides additional evidence supporting that the defects in E3 ubiquitin ligase lead to giant axon disorder and further emphasizes the imperative role of ubiquitin ligase–regulated proteasomal degradation pathway in maintaining the integrity of the nervous system and possibly cardiomyocytes.
Figure 3. E3 ubiquitin (Ub) ligase pathway in neurofilament regulation.
(A) Mutations in GAN lead to axonal accumulations in giant axonal neuropathy. GAN specifically directs Cul3-E3 complex to target neurofilaments. (B) The defective interaction of DCAF8 by the p.R317C mutation may lead to abnormal E3 ubiquitination. The pathologic observation of neurofilament accumulations with giant axons in affected patients' nerve biopsies suggests DCAF8 may directly target neurofilaments for ubiquitination.
ACKNOWLEDGMENT
The authors thank Dr. Stephane Angers at University of Toronto, Ontario, Canada for providing the expression vectors of DCAF8, DCAF8-R314H, and DDB1.
GLOSSARY
- dHMN
distal hereditary motor neuropathy
- HA
hemagglutinin
- HMSN
hereditary motor and sensory neuropathy
- LOD
logarithm of odds
- STS
sequence-tagged site
- WES
whole-exome sequencing
AUTHOR CONTRIBUTIONS
C.J.K.: study concept and design; study supervision; acquisition, analysis, and interpretation of data; drafting and revising the manuscript. P.J.D., Y.W., C.B., G.M.: acquisition, analysis, and interpretation of data; editing the manuscript. P.V., H.H.G.: patient acquisition, analysis and interpretation of data, editing the manuscript. K.Z., M.-V.B., X.D., S.M., E.J.A.: acquisition, analysis, and interpretation of data.
STUDY FUNDING
NIH grant (NS065007, C.J.K.), Mayo Clinic Center for Individualized Medicine.
DISCLOSURE
The authors report no disclosures relevant to the manuscript. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Klein CJ, Duan X, Shy ME. Inherited neuropathies: clinical overview and update. Muscle Nerve 2013;48:604–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogel P, Gabriel M, Goebel HH, Dyck PJ. Hereditary motor sensory neuropathy type II with neurofilament accumulation: new finding or new disorder? Ann Neurol 1985;17:455–461 [DOI] [PubMed] [Google Scholar]
- 3.Lus G, Nelis E, Jordanova A, et al. Charcot-Marie-Tooth disease with giant axons: a clinicopathological and genetic entity. Neurology 2003;61:988–990 [DOI] [PubMed] [Google Scholar]
- 4.Fabrizi GM, Cavallaro T, Angiari C, et al. Giant axon and neurofilament accumulation in Charcot-Marie-Tooth disease type 2E. Neurology 2004;62:1429–1431 [DOI] [PubMed] [Google Scholar]
- 5.Azzedine H, Ravise N, Verny C, et al. Spine deformities in Charcot-Marie-Tooth 4C caused by SH3TC2 gene mutations. Neurology 2006;67:602–606 [DOI] [PubMed] [Google Scholar]
- 6.Jaffer F, Murphy SM, Scoto M, et al. BAG3 mutations: another cause of giant axonal neuropathy. J Peripher Nerv Syst 2012;17:210–216 [DOI] [PubMed] [Google Scholar]
- 7.Mahammad S, Murthy SN, Didonna A, et al. Giant axonal neuropathy-associated gigaxonin mutations impair intermediate filament protein degradation. J Clin Invest 2013;123:1964–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 2006;443:590–593 [DOI] [PubMed] [Google Scholar]
- 9.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asmann YW, Middha S, Hossain A, et al. TREAT: a bioinformatics tool for variant annotations and visualizations in targeted and exome sequencing data. Bioinformatics 2012;28:277–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 1998;63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 2002;30:97–101 [DOI] [PubMed] [Google Scholar]
- 13.Weeks DE, Ott J, Lathrop GM. SLINK: a general simulation program for linkage analysis. Am J Hum Genet 1990;47:A204 [Google Scholar]
- 14.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 1998;8:175–185 [DOI] [PubMed] [Google Scholar]
- 15.Saporta AS, Sottile SL, Miller LJ, Feely SM, Siskind CE, Shy ME. Charcot-Marie-Tooth disease subtypes and genetic testing strategies. Ann Neurol 2011;69:22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanda T, Oda M, Yonezawa M, et al. Peripheral neuropathy in xeroderma pigmentosum. Brain 1990;113:1025–1044 [DOI] [PubMed] [Google Scholar]
- 17.Wu F, Wang S, Xing J, Li M, Zheng C. Characterization of nuclear import and export signals determining the subcellular localization of WD repeat-containing protein 42A (WDR42A). FEBS Lett 2012;586:1079–1085 [DOI] [PubMed] [Google Scholar]
- 18.Bomont P, Cavalier L, Blondeau F, et al. The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat Genet 2000;26:370–374 [DOI] [PubMed] [Google Scholar]
- 19.Stendel C, Roos A, Kleine H, et al. SH3TC2, a protein mutant in Charcot-Marie-Tooth neuropathy, links peripheral nerve myelination to endosomal recycling. Brain 2010;133:2462–2474 [DOI] [PubMed] [Google Scholar]
- 20.Gamerdinger M, Kaya AM, Wolfrum U, Clement AM, Behl C. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep 2011;12:149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ylikallio E, Poyhonen R, Zimon M, et al. Deficiency of the E3 ubiquitin ligase TRIM2 in early-onset axonal neuropathy. Hum Mol Genet 2013;22:2975–2983 [DOI] [PubMed] [Google Scholar]
- 22.Weterman MA, Sorrentino V, Kasher PR, et al. A frameshift mutation in LRSAM1 is responsible for a dominant hereditary polyneuropathy. Hum Mol Genet 2012;21:358–370 [DOI] [PMC free article] [PubMed] [Google Scholar]