This lecture honors the memory of Robert Wartenberg, who was born in Lithuania, trained in Germany, and in 1935 moved to the United States, where he started his second career as a scholar and teacher at the University of California, San Francisco.1
Fifty years after the description of the first bona fide mitochondrial disease2 and 25 years after the report of the first pathogenic mutations in mitochondrial DNA (mtDNA),3,4 it seems appropriate to take stock of the extraordinary, and ongoing, progress in mitochondrial neurology.
To start at the beginning, we have to go back about 2 billion years, when primordial eukaryotic cells were invaded by proteobacteria that had adapted to an increasingly oxygen-rich atmosphere and were on their way to becoming the permanent endosymbionts that we call mitochondria.5 The bacteria brought into the host cells their DNA, which explains why all eukaryotic cells have 2 types of DNA, their original nuclear DNA (nDNA) plus mtDNA, which is a relic—but not a fossil—of the bacterial symbionts.
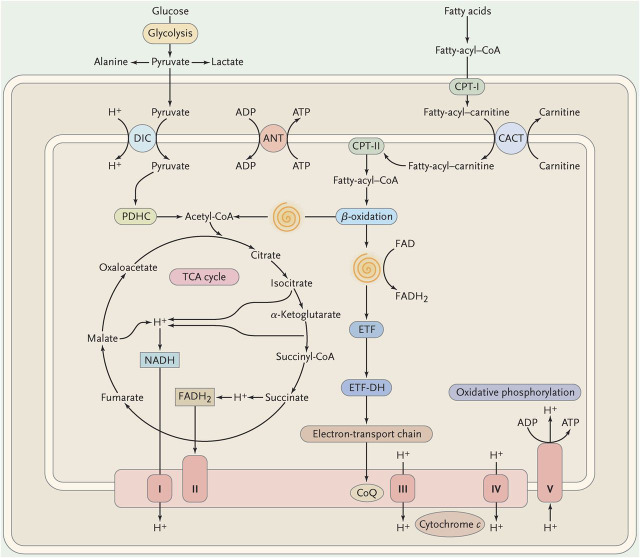
Although mitochondria have innumerable functions subserved by about 1,700 nDNA-encoded proteins, arguably the most important role of mitochondria is the provision of energy in the form of adenosine triphosphate (ATP). Figure 1 is a vastly oversimplified scheme of mitochondrial metabolism showing the points of entry of metabolic fuels (carbohydrates through pyruvate transport, fatty acids through the carnitine-acylcarnitine shuttle). The intramitochondrial oxidation of pyruvate by the pyruvate dehydrogenase complex and of fatty acyl-CoAs by the β-oxidation spirals results in a common compound, acetyl-CoA, which is then further oxidized in the Krebs cycle. Electrons derived from dehydrogenases in the Krebs cycle and in the β-oxidation pathway flow down the electron carrier chain, composed of 4 protein complexes (I to IV) and 2 small carriers (coenzyme Q10 [C0Q10] and cytochrome c). Concomitant with this horizontal flow of electrons, there is a vertical flow of protons from the mitochondrial matrix through the inner mitochondrial membrane (IMM) into the intermembrane space. This creates an electrochemical proton gradient that is used to drive complex V (ATP synthase), a magnificent rotary motor that uses the reverse flow of protons to convert ADP to ATP. I have highlighted in red the electron transport chain and the ATP synthase (together known as respiratory chain) for 2 main reasons: 1) this is the “business end” of mitochondrial metabolism, where ATP is generated; and 2) this is the only metabolic pathway in the cell that depends not only on nDNA but also on mtDNA.
Figure 1. Schematic view of mitochondrial metabolism.
The spirals represent the cyclic reactions of the β-oxidation pathway resulting in the formation of acetyl-coenzyme A (CoA) and the reduction of flavoprotein. ADP = adenosine diphosphate; ATP = adenosine triphosphate; ANT = adenine nucleotide translocase; CACT = carnitine-acylcarnitine translocase; CoQ = coenzyme Q; CPT = carnitine palmitoyltransferase; DIC = dicarboxylate carrier; ETF = electron-transfer flavoprotein; ETF-DH = electron-transfer flavoprotein dehydrogenase; FAD = flavin adenine-dinucleotide; FADH = reduced flavin adenine-dinucleotide; NADH = reduced nicotinamide adenine dinucleotide; PDHC = pyruvate dehydrogenase complex; TCA = tricarboxylic acid; I = complex I; II = complex II; III = complex III; IV = complex IV; V = complex V. (Modified with permission from DiMauro and Schon, New England Journal of Medicine 2003;348:2656–2668.)
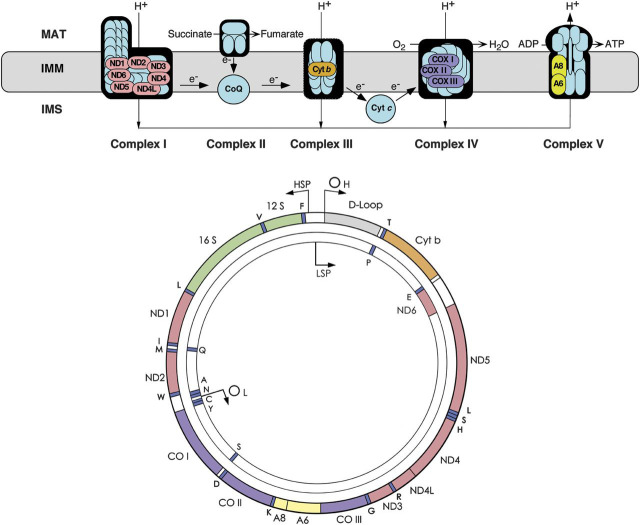
The interdependence of the respiratory chain and mtDNA is shown in figure 2, where the 13 protein-coding genes of mtDNA are color-coordinated with their products, which are all subunits of the respiratory chain, 7 in complex I, 1 in complex III, 3 in complex IV, and 2 in complex V. Notably, however, most proteins of the respiratory chain, including all 4 subunits of complex II, are encoded by nDNA.
Figure 2. The respiratory chain (top) and the mitochondrial DNA (bottom).
Genes and corresponding gene products are similarly color-coded. ND denotes the subunits of NADH-coenzyme Q oxidoreductase (complex I); cyt b = cytochrome b; subunits of cytochrome c oxidase are labeled CO in the mtDNA scheme and COX in the respiratory chain rendition; A6 and A8 indicate subunits 6 and 8 of ATP synthase. The 22 tRNA genes are denoted by one-letter amino acid nomenclature; 12S and 16S denote ribosomal RNAs (rRNAs). OH and OL are the origin of heavy- and light-strand replication; HSP and LSP are the promoters of heavy- and light-strand transcription. ADP = adenosine diphosphate; ATP = adenosine triphosphate; IMM = inner mitochondrial membrane; IMS = intermembrane space; MAT = mitochondrial matrix.
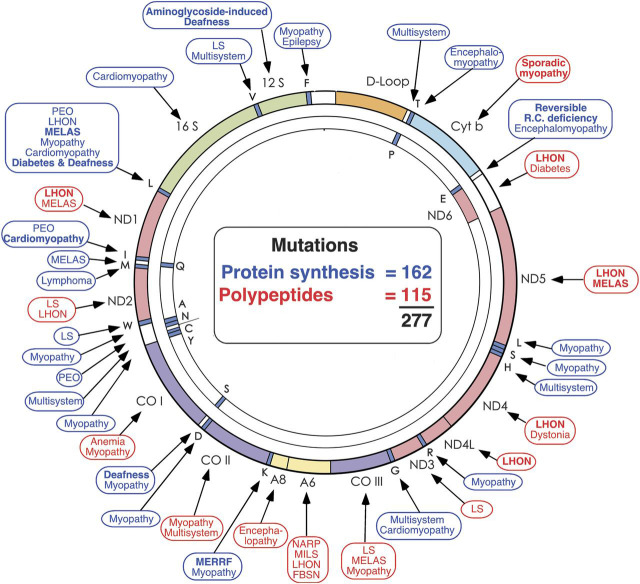
The mtDNA, also shown in figure 3, is a very small molecule (16,569 base pairs, a seemingly piddling amount when compared with the 3 billion base pairs of nDNA). Besides the 13 protein-coding genes, mtDNA has 2 ribosomal RNA (rRNA) genes and 22 transfer RNA (tRNA) genes that are interspersed among the rRNA and the protein-coding genes. Notably, there are no introns in mtDNA. Within the space of 25 years, the small mtDNA has become crowded with pathogenic point mutations (261 at last count, figure 3), to which one must add hundreds of large-scale deletions. Some point mutations affect protein-coding genes but even more affect protein synthesis as a whole. The boxes indicate the clinical pictures corresponding to mutations in different genes, often denoted by syndromic acronyms, including mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes (MELAS), myoclonus epilepsy and ragged-red fibers (MERRF), neuropathy, ataxia, retinitis pigmentosa (NARP), maternally inherited Leigh syndrome (MILS), Leber hereditary optic neuropathy (LHON), or familial bilateral striatal necrosis.
Figure 3. Morbidity map of the mitochondrial DNA as of 2013.
Disorders caused by mutations in protein-coding genes are shown in red; disorders caused by mutations in genes controlling protein synthesis are shown in blue. FBSN = familial bilateral striatal necrosis; LHON = Leber hereditary optic neuropathy; LS = Leigh syndrome; MELAS = mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes; MERRF = myoclonus epilepsy and ragged-red fibers; MILS = maternally inherited Leigh syndrome; NARP = neuropathy, ataxia, retinitis pigmentosa; PEO = progressive external ophthalmoplegia. (Modified with permission from DiMauro and Schon, New England Journal of Medicine 2003;348:2656-2668.)
The abundance of mtDNA-related disorders has called attention to mitochondrial genetics, which differs from mendelian genetics in 4 major ways. First, inheritance of mtDNA point mutations (single deletions are usually sporadic events) is maternal: at fertilization, all mitochondria—and all mtDNA—derive from the oocyte. Therefore, a pathogenic mtDNA mutation is transmitted from the mother to all her children, boys and girls, but only women will pass it on to their progeny. Second, mitochondrial genetics is population genetics because there are thousands of copies of mtDNA instead of the 2 alleles of each nuclear autosome. In most mtDNA-related diseases, mutated and wild-type mitochondrial genomes coexist, a situation known as heteroplasmy. Third, a logical corollary of heteroplasmy is that a critical number of mutated mtDNAs is needed for the respiratory chain to suffer and for the disease to manifest (threshold effect). This pathogenic threshold varies in different tissues according to their dependence on oxidative metabolism, explaining why brain and skeletal muscle are so often affected (“mitochondrial encephalomyopathies”), and why these disorders are of special interest to neurologists. The fourth concept, mitotic segregation, refers to the fact that the number of mutated mtDNAs (the mutation load) can change in some tissues from one generation of cell to the next: thus both genotype and clinical phenotype of mtDNA-related disorders may vary with time.6,7
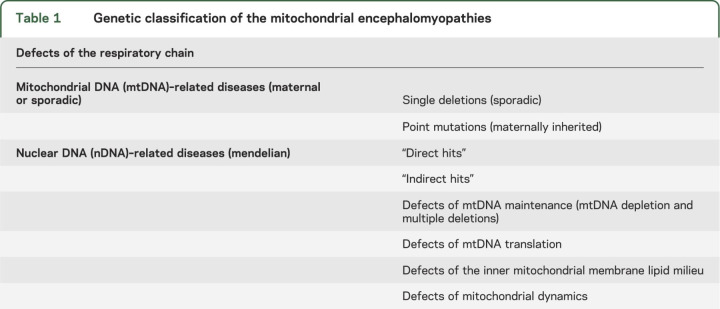
At this point, a genetic classification can be proposed (table 1), which divides the mitochondrial diseases into 2 major groups: 1) disorders due to mutations in mtDNA (these are usually inherited maternally but can also be sporadic); and 2) disorders due to mutations in nDNA, inherited as mendelian traits.
Table 1.
Genetic classification of the mitochondrial encephalomyopathies
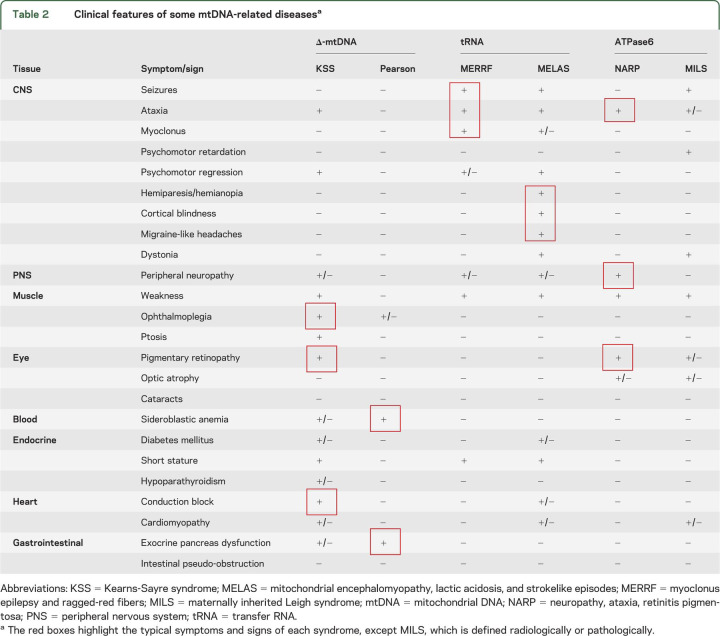
Mutations in mtDNA include single large-scale deletions (i.e., one patient harbors only one type of rearrangement) and point mutations. Table 2 is a compilation of the clinical features that accompany 6 mtDNA-related disorders: 2 (Kearns-Sayre syndrome [KSS] and Pearson syndrome) associated with single deletions; 2 (MELAS and MERRF) associated with point mutations in different tRNAs; and 2 (NARP and MILS) associated with one and the same mutation in a protein-coding gene, ATPase 6. Two points emerge from this table. The first is that—as mitochondria and mtDNA are ubiquitous—every tissue in the body can be affected, explaining the frequent multisystemic nature of mtDNA-related diseases. The second point is that—despite the morass of clinical presentations—some mtDNA mutations result in well-defined and clinically recognizable syndromes, such as the ones highlighted in the table.
Table 2.
Clinical features of some mtDNA-related diseasesa
It would be tedious to review all mtDNA-related diseases and I have chosen MELAS as one example because it is relatively common and typically very severe. MELAS is most often due to a mutation (m. 3243A > G) in tRNALeu (UUR) and is characterized by early-onset strokes that do not conform to the distribution of major brain vessels and commonly affect the occipital or parietal lobe, resulting in hemianopia, hemiparesis, or cortical blindness. The strokes are often preceded by seizures or migraine and are typically transient but recurrent, such that their cumulative effects are devastating and life is foreshortened. The lactic acidosis is well documented by 1H magnetic resonance spectroscopy of the brain, showing prominent lactate peaks both in the ventricles (CSF) and in the parenchyma.
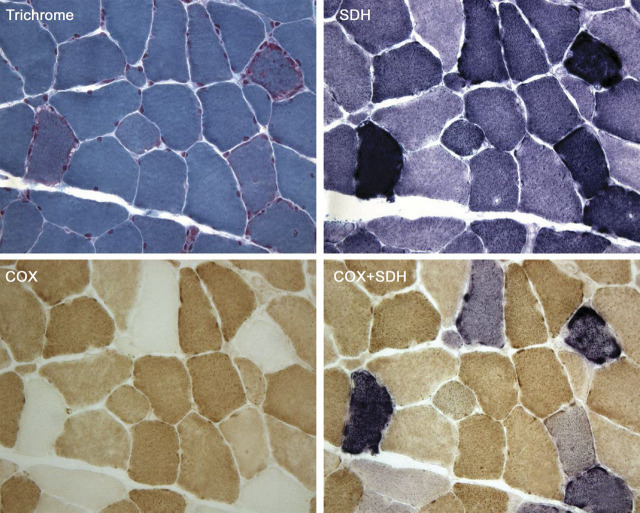
I will return to MELAS in discussing other aspects of mtDNA-related disorders. First, however, I will stress the importance of the muscle biopsy as a diagnostic gold standard for these diseases. Figure 4 shows serial cross-sections of muscle from a patient with MERRF stained with 4 histochemical methods: the modified Gomori trichrome, succinate dehydrogenase (SDH), cytochrome c oxidase (COX), and the combination of SDH and COX. The trichrome shows mitochondrial proliferation in classic ragged-red fibers.8 These are revealed even better by the SDH stain (“ragged-blue” fibers) because SDH (complex II of the respiratory chain), which is entirely encoded by nDNA, is unruffled by mutations in mtDNA and is a good marker of mitochondrial abundance. Conversely, COX, whose 3 catalytic subunits are encoded by mtDNA, is a good marker of respiratory chain function, as shown by the COX negativity of the ragged-red/blue fibers. Finally, my colleague Eduardo Bonilla made the astute observation that, when he superimposed the COX and SDH stain, the COX stain prevailed in normal fibers, which stained brown, whereas in COX-negative or even slightly COX-deficient fibers, the blue SDH stain shone through. Notably, all 4 stains show a checkerboard pattern, which is a telltale sign of heteroplasmy: in the elongated syncytial muscle fibers, the mutation load is not uniform along the length of the fiber, such that adjacent segmental sections may have widely different amounts of mutated mtDNAs.
Figure 4. Serial cross-sections of a muscle biopsy from a patient with myoclonus epilepsy and ragged-red fibers stained with different histochemical methods.
The modified Gomori trichrome shows classic ragged-red fibers, which stain intensely with the succinate dehydrogenase (SDH) stain (“ragged-blue”) and appear pale with the cytochrome c oxidase (COX) stain. The same fibers appear more or less intensely blue when the SDH and COX stain are superimposed.
Mitochondrial genetics does explain some important features of mtDNA-related diseases. For example, different degrees of heteroplasmy (70% vs 90%) readily explain how the same mutation (m. 8993T > G) in ATPase 6 can cause 2 different syndromes, often in members of the same family: a relatively benign and late-onset disorder with neuropathy, ataxia, and retinitis pigmentosa (NARP) or a devastating neurodegenerative disease of infancy or childhood with the neuroradiologic and neuropathologic features of Leigh syndrome (MILS).9,10
In 2000, I coauthored a review titled “Mutations in mtDNA: Are we scraping the bottom of the barrel?”11 It was clear then—as it is now—that we are still far from a full understanding of mtDNA-related diseases.12 New pathogenic mutations are still being added to the already-crowded mtDNA morbidity map (figure 3). Thanks to super-sophisticated next-generation sequencing, the concept that healthy individuals are homoplasmically normal (i.e., harbor only wild-type mtDNAs) has been replaced by the concept of universal heteroplasmy.13 The individual vulnerability, tissue selectivity, and occasional reversibility of homoplasmic pathogenic mtDNA mutations remains puzzling.14,15 The role of epigenetics in mtDNA-related diseases is unclear,16 although at least one epigenetic factor, lack of protective exposure to estrogens, has been considered responsible for the marked prevalence of affected men in LHON.17
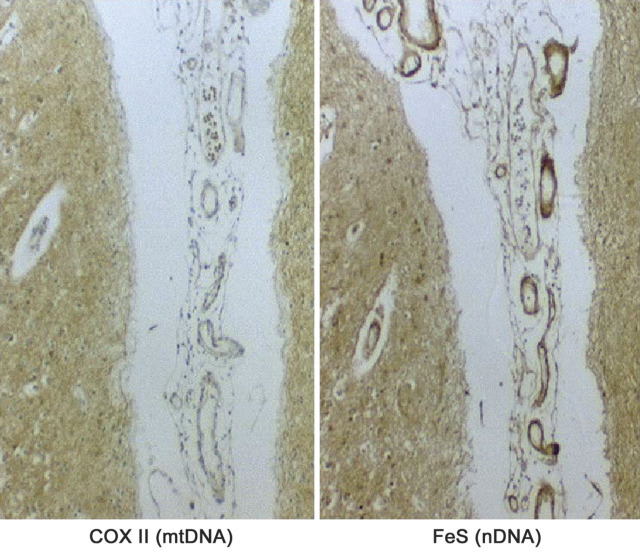
Even though, as mentioned above, the rules of mitochondrial genetics explain some features of mtDNA-related diseases, the pathogenesis of “typical” heteroplasmic diseases remains a conundrum. Simply put, why is KSS different from MELAS and MELAS different from MERRF? All 3 conditions result from impaired mitochondrial protein synthesis and ATP production: it would be logical to expect phenotypic homogeneity rather than syndromic individuality, but—despite occasional overlapping presentations—this is not the case. One possible explanation is that different mutations may have different spatial distribution, especially in the brain. This concept was borne out by immunohistochemical studies conducted by Eduardo Bonilla and Kurenai Tanji. To use MELAS again as one example, figure 5 shows how, in subpial arterioles, the expression of COX II, a mtDNA-encoded protein, was markedly decreased compared to that of the nDNA-encoded iron-sulfur Rieske protein (FeS), evidence that the m3243A > G MELAS mutation must be abundant in these vessels,18 as also shown by others.19 These findings make sense because MELAS is, ultimately, a mitochondrial micro- and macroangiopathy.20 However, the distinct spatial distribution of different mutations begs the question of what “directs” them to different areas of the brain.
Figure 5. Serial sections from the brain cortex of a patient with mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes showing COX II immunodeficiency and normal immunoreaction for the FeS protein in pial and intracortical arterioles.
(Reprinted with permission from Tanji et al., Semin Cell Dev Biol 2001;12:429-439.18)
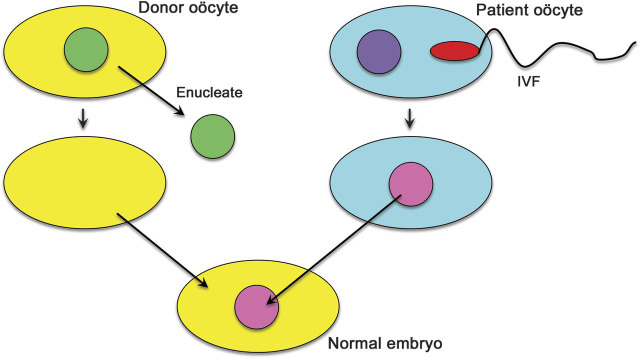
A provocative question is whether mtDNA-related disorders, especially those, like typical MELAS, due to point mutations in tRNA genes, are preventable. My colleague Darryl De Vivo and I faced this dilemma when the asymptomatic sister of our first patient with MELAS—a girl who had died at 10 years of age after multiple strokes21—was about to get married and asked us “How can I be sure not to have an affected child?” While there is no foolproof prenatal diagnosis for m3243A > G MELAS, there is a way to prevent mtDNA from getting into the fetus. An oocyte from the mutation carrier is fertilized in vitro with sperm from her husband. The pronucleus of the zygote is removed with as little as possible surrounding cytoplasm and is transferred into an enucleated oocyte from a normal donor (figure 6). After implantation, the resulting fetus will have the nuclear genes of the biological parents but the cytoplasm and the mitochondria from the donor, free of mtDNA mutations. In experiments with nonhuman primates using oocyte spindle-chromosomal complex transfer, the infants were healthy and devoid of maternal mtDNA.22 Experiments with fertilized or unfertilized but parthenogenically activated human oocytes are under way both in the United Kingdom23 and in the United States,24 and the results are very promising. This bodes well for implementation in humans, once lingering ethical questions have been resolved.
Figure 6. Schematic representation of the pronuclear transfer technique to prevent transmission of pathogenic mitochondrial DNA point mutations.
For explanation, see text.
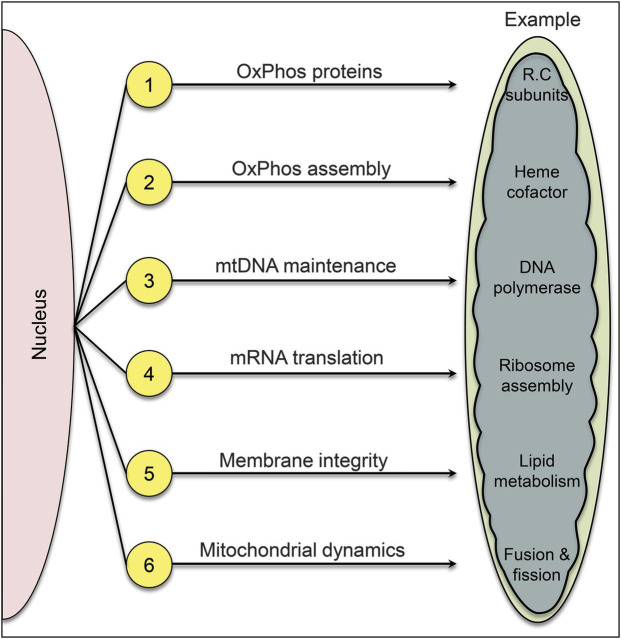
In the course of evolution, mitochondria have lost their bacterial independence and now rely on the nucleus for most of their vital functions. Therefore, mendelian mitochondrial disorders are many and varied (figure 7). There are at least 6 disease flavors, as pathogenic mutations can affect 1) genes encoding subunits of the respiratory chain (“direct hits”); 2) genes encoding assembly proteins of respiratory chain complexes (“indirect hits”); 3) genes needed for mtDNA integrity and replication (“mtDNA maintenance”); 4) genes needed for mtRNA translation; 5) genes controlling the phospholipid composition of the IMM; and 6) genes involved in mitochondrial dynamics.
Figure 7. Six different modalities of mendelian mitochondrial diseases.
1, “Direct hits”; 2, “indirect hits”; 3, defects of intergenomic communication resulting in mitochondrial DNA depletion and multiple mitochondrial DNA deletions; 4, defects of mRNA translation; 5, defects of the inner mitochondrial membrane phospholipid milieu; 6, defects of mitochondrial dynamics.
The advent of whole-exome sequencing and of the more narrowly targeted “mitoexome sequencing”25 has made it possible to sequence in one sweep the approximately 1,700 nuclear genes encoding mitochondrial proteins. Mitoexome sequencing has already enriched all 6 disease flavors with new variants,26–28 and it is conceivable that it may reveal new flavors, that is, alterations in as yet unsuspected mitochondrial functions.
It is not within the scope of this overview to describe the diverse clinical presentations in each disease category; rather, I will try to highlight some salient features of each group before considering in some detail the defects of the lipid milieu and the defects of mitochondrial dynamics.
In general, both direct and indirect hits of the respiratory chain affect infants or children and manifest as Leigh syndrome (LS), which reflects the ravages caused by impaired energy supply on the developing nervous system.29
Defects of mtDNA maintenance have been traditionally subdivided into 2 groups: 1) mtDNA depletion syndromes, manifesting early in life and presenting as myopathy, hepatocerebral disorder, or encephalomyopathy; and 2) multiple mtDNA deletions, usually associated with progressive external ophthalmoplegia (PEO)–plus.29–31 These disorders of intergenomic communication are usually caused either by defects of mtDNA replication (catalyzed by polymerase γ and by the helicase Twinkle [gene PEO1]) or by altered homeostasis of the intramitochondrial nucleoside pool, which is controlled by multiple genes.31 We have come to realize that mtDNA depletion and multiple mtDNA deletions often coexist in the same patient and are often caused by the same gene defect. Another peculiarity of these disorders is that, although they are unequivocally mendelian, they share many features of mitochondrial genetics, including heteroplasmy and the threshold effect. Accordingly, muscle biopsies also show a checkerboard pattern of ragged-blue and COX-negative fibers.
Reflecting the multiplicity of factors needed for mtRNA translation (initiation, elongation, and release factors, plus translational activators and specific tRNA base modifiers),32–34 the defects of mtRNA translation encompass a clinically heterogeneous and still rapidly expanding group of disorders. Most patients are infants or children, often with LS or LS-like disorders and with multiple respiratory chain enzyme defects.34,35
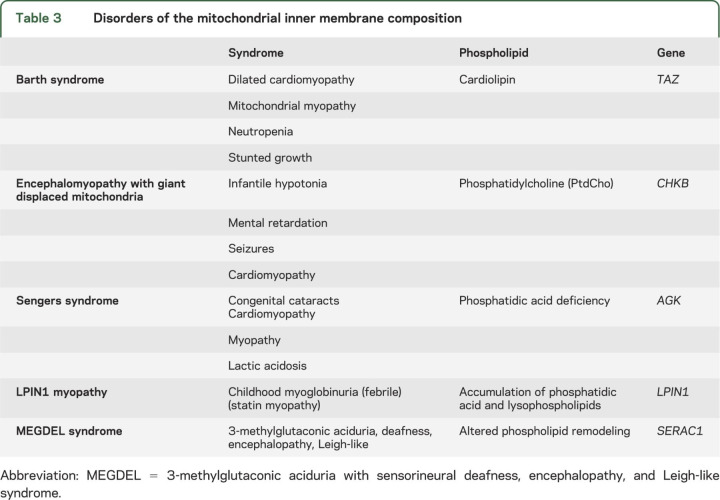
Until 2002, the only bona fide defect of the IMM lipid milieu was Barth syndrome, an X-linked mitochondrial myopathy and cardiopathy due to mutations in the tafazzin (TAZ) gene resulting in deficiency or abnormal composition of cardiolipin, the major phospholipid component of the IMM.36–38
In the past few years, 4 new entities have been described, affecting different phospholipids and clearly documenting that the lipid milieu is not just a scaffold holding the respiratory chain but an integral partner in its function (table 3). Sengers syndrome is also a mitochondrial myopathy and cardiopathy with the added clinical feature of congenital cataracts.39 When it was found that the adenine nucleotide translocator 1 (ANT1) was missing in muscle of patients but its gene was normal, the riddle was solved by whole-exome sequencing, which showed mutations in the AGK gene encoding acylglycerol kinase.27 AGK catalyzes the phosphorylation of diacyl- and monoacylglycerol to phosphatidic acid (PA) or lysophosphatidic acid (LPA), important intermediates in the synthesis of phospholipids. The loss of ANT1 is presumably secondary to the decreased levels of PA and LPA. As PA is also a precursor of cardiolipin, there are biochemical similarities with Barth syndrome that may explain the comparable clinical presentations.
Table 3.
Disorders of the mitochondrial inner membrane composition
A spontaneously mutant mouse strain with rostrocaudal muscular dystrophy and mutations in the gene encoding choline kinase beta (Chkb)40 suggested that the human homologous gene (CHKB) may be mutated in a form of human congenital muscular dystrophy with similar morphologic changes in muscle: giant mitochondria displaced to the periphery of the fibers.41,42
The acronym MEGDEL refers to a syndrome characterized by 3-methylglutaconic aciduria type IV, deafness, and LS-like encephalopathy, in which whole-exome sequencing revealed mutations in SERAC1.43
Finally, at least for now, recurrent muscle breakdown and myoglobinuria in children, a syndrome often occurring during febrile illnesses and rarely associated with known inborn errors of metabolism, has been ascribed to mutations in LPIN1, the gene that encodes the muscle-specific isoform of phosphatidic acid phosphatase.44
Increasing attention has been directed to a structure called mitochondria-associated membrane (MAM), which is where mitochondria and endoplasmic reticulum come into physical and functional association. MAM is involved in lipid exchange between the 2 organelles, cholesterol synthesis, calcium signaling, and mitochondrial dynamics.45 At least 2 of the 5 disorders of the lipid milieu listed in table 3 involve the MAM: the megaconial mitochondrial myopathy due to CHKB deficiency42 and MEGDEL, whose defective gene product SERAC1 is located in the MAM and controls the exchange of phospholipids. In addition, some forms of primary lateral sclerosis (defective gene ERLIN2)46 and of juvenile amyotrophic lateral sclerosis (defective gene SIGMAR1)47 have been associated with alterations of the lipid rafts that are part of the MAM, and there is increasing evidence that MAM dysfunction may play a central role in the pathogenesis of Alzheimer disease.48
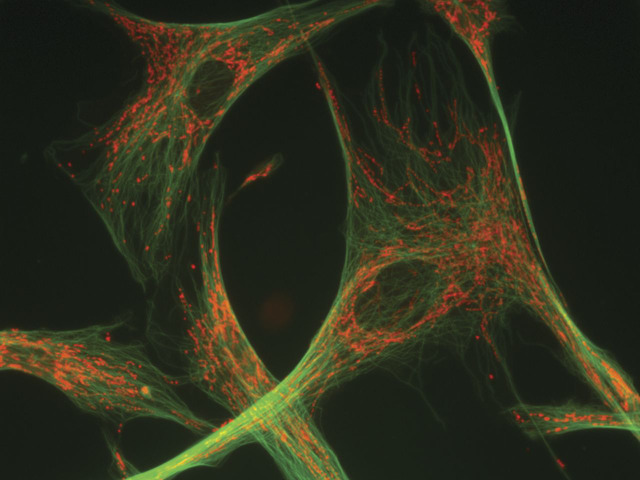
Mitochondria are highly dynamic organelles that move around the cell on microtubular tracks (figure 8) and, recalling their bacterial origins, go through repeated cycles of fusion and fission, resulting in extensive tubular structures or in discreet individual entities more akin to the conventional mitochondrial images. This restless activity is crucial, especially in neurons, to ensure adequate energy distribution throughout the cell, including axonal and dendritic processes, and to guarantee mitochondrial quality control through mitophagy, the autophagic elimination of damaged mitochondria.49,50
Figure 8. A neuron with mitochondria (stained in red with mitotracker) appended to microtubular “tracks” (stained in green).
Courtesy of Dr. Estela Area-Gómez.
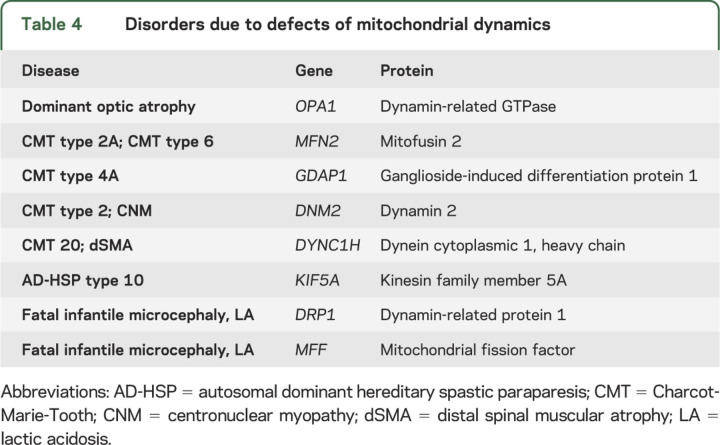
Defects of mitochondrial dynamics have exploded on us in the past decade and both the central and the peripheral nervous system are especially vulnerable. The clinical spectrum of these disorders includes dominant optic atrophy, various forms of Charcot-Marie-Tooth neuropathy and of autosomal dominant hereditary spastic paraplegia, a form of centronuclear myopathy, and microcephaly with optic atrophy (table 4).51,52
Table 4.
Disorders due to defects of mitochondrial dynamics
Importantly, mitochondrial fusion and fission are not independent functions; rather, they are closely integrated with energy metabolism and mtDNA maintenance, as documented by PEO-plus syndromes with multiple mtDNA deletions due to mutations in OPA1 and MFN2.53–56 It is also noteworthy that alterations of mitochondrial dynamics are involved, albeit not primarily, in the pathogenesis of neurodegenerative diseases.50,57
Considering the complexity of mtDNA-related disorders and the multiple categories of mendelian disorders listed in figure 7, it is apparent that the correct diagnosis of a suspected mitochondrial disease can be a formidable challenge. To help neurologists in this task and in fostering research in the field, we now have the North American Mitochondrial Disease Consortium (NAMDC) (http://rarediseasesnetwork.epi.usf.edu/namdc/), based at Columbia University but encompassing 18 centers of excellence in mitochondrial diseases.
Let me conclude with a quote from C. Miller Fisher: “If there were a dictum, it would be that it is all a matter of being in the right place at the right time, with the right mentor.”58 I owe my interest in mitochondrial diseases—indeed my whole academic career—to my mentor, Lewis P. Rowland. Thank you, Bud.
ACKNOWLEDGMENT
The author thanks Drs. Eric A. Schon and Michio Hirano for collaboration and Pablo Abreu for help with the figures.
STUDY FUNDING
Supported by grants from NICHD (P01HD032062), NINDS/NICHD (U54NS078059), and the Marriott Mitochondrial Disorder Clinical Research Fund (MMDCRF).
DISCLOSURE
The author reports no disclosures relevant to the manuscript. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Rowland LP. Molecular genetics, pseudogenetics, and clinical neurology. Neurology 1983;23:1179–1195. [DOI] [PubMed] [Google Scholar]
- 2.Luft R, Ikkos D, Palmieri G, Ernster L, Afzelius B. A case of severe hypermetabolism of nonthyroid origin with a defect in the maintenance of mitochondrial respiratory control: a correlated clinical, biochemical, and morphological study. J Clin Invest 1962;41:1776–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holt IJ, Harding AE, Morgan Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 1988;331:717–719. [DOI] [PubMed] [Google Scholar]
- 4.Wallace DC, Singh G, Lott MT, et al. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science 1988;242:1427–1430. [DOI] [PubMed] [Google Scholar]
- 5.Sagan L. On the origin of mitosing cells. J Theor Biol 1967;14:255–274. [DOI] [PubMed] [Google Scholar]
- 6.Larsson NG, Holme B, Kristiansson B. Progressive increase of the mutated mitochondrial DNA fraction in Kearns-Sayre syndrome. Pediatr Res 1990;28:131–136. [DOI] [PubMed] [Google Scholar]
- 7.McShane MA, Hammans SR, Sweeney M, et al. Pearson syndrome and mitochondrial encephalomyopathy in a patient with a deletion of mtDNA. Am J Hum Genet 1991;48:39–42. [PMC free article] [PubMed] [Google Scholar]
- 8.Engel WK, Cunningham CG. Rapid examination of muscle tissue: an improved trichrome stain method for fresh-frozen biopsy sections. Neurology 1963;13:919–923. [DOI] [PubMed] [Google Scholar]
- 9.Holt IJ, Harding AE, Petty RK, Morgan-Hughes JA. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am J Hum Genet 1990;46:428–433. [PMC free article] [PubMed] [Google Scholar]
- 10.Tatuch Y, Christodoulou J, Feigenbaum A, et al. Heteroplasmic mtDNA mutation (T > G) at 8993 can cause Leigh disease when the percentage of abnormal mtDNA is high. Am J Hum Genet 1992;50:852–858. [PMC free article] [PubMed] [Google Scholar]
- 11.DiMauro S, Andreu AL. Mutations in mtDNA: are we scraping the bottom of the barrel? Brain Pathol 2000;10:431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schon EA, DiMauro S, Hirano KI. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat Rev Genet 2012;13:878–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Payne BAJ, Wilson IJ, Yu-Wai-Man P, et al. Universal heteroplasmy of human mitochondrial DNA. Hum Mol Genet 2013;22:384–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McFarland R, Clark KM, Morris AAM, et al. Multiple neonatal deaths due to homoplasmic mitochondrial DNA mutation. Nat Genet 2002;30:145–146. [DOI] [PubMed] [Google Scholar]
- 15.Horvath R, Kemp JP, Tuppen HAL, et al. Molecular basis of infantile reversible cytochrome c oxidase deficiency. Brain 2009;132:3165–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chinnery PF, Elliott P, Hudson G, Samuels DC, Relton CL. Epigenetics, epidemiology and mitochondrial DNA diseases. Int J Epidemiol 2012;41:177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giordano C, Montopoli M, Perli E, et al. Oestrogens ameliorate mitochondrial dysfunction in Leber's hereditary optic neuropathy. Brain 2011;134:220–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanji K, Kunimatsu T, Vu TH, Bonilla E. Neuropathological features of mitochondrial disorders. Semin Cell Dev Biol 2001;12:429–439. [DOI] [PubMed] [Google Scholar]
- 19.Betts J, Jaros E, Perry RH, et al. Molecular neuropathology of MELAS: level of heteroplasmy in individual neurones and evidence of extensive vascular involvement. Neuropathol Appl Neurobiol 2006;32:359–373. [DOI] [PubMed] [Google Scholar]
- 20.Tay SHK, Nordli DR, Bonilla E, et al. Aortic rupture in mitochondrial encephalopathy, lactic acidosis, and stroke-like episodes. Arch Neurol 2006;63:281–283. [DOI] [PubMed] [Google Scholar]
- 21.Hirano M, Ricci E, Koenigsberger MR, et al. MELAS: an original case and clinical criteria for diagnosis. Neuromuscul Disord 1992;2:125–135. [DOI] [PubMed] [Google Scholar]
- 22.Tachibana M, Sparman M, Sritanaudomchai H, et al. Mitochondrial gene replacement in primate offspring and embryonic stem cells. Nature 2009;461:367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craven L, Elson JL, Irving L, et al. Mitochondrial DNA disease: new options for prevention. Hum Mol Genet 2011;20:R168–R174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paull D, Emmanuele V, Weiss KA, et al. Nuclear genome transfer in human oocytes eliminates mitochondrial DNA variants. Nature 2013;493:632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calvo S, Compton A, Hershman SG, et al. Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Sci Transl Med 2012;4:118ra110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ronchi D, Garone C, Bordoni A, et al. Next-generation sequencing discloses DGUOK mutations in adult patients with mtDNA multiple deletions. Brain 2012;135:3404–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayr JA, Haack TB, Graf E, et al. Lack of mitochondrial protein acylglycerol kinase causes Sengers syndrome. Am J Hum Genet 2012;90:314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kornblum C, Nicholls T, Haack TB, et al. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat Genet 2013;45:214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DiMauro S, Emmanuele V. The clinical spectrum of nuclear DNA-related mitochondrial disorders. In: Wong L-JC, ed. Mitochondrial Disorders Caused by Nuclear Genes. New York: Springer Science + Business Media; 2013:3–25. [Google Scholar]
- 30.Spinazzola A, Zeviani M. Disorders of nuclear-mitochondrial intergenomic signaling. Gene 2005;354:162–168. [DOI] [PubMed] [Google Scholar]
- 31.Suomalainen A, Isohanni P. Mitochondrial DNA depletion syndromes: many genes, common mechanisms. Neuromuscul Disord 2010;20:429–437. [DOI] [PubMed] [Google Scholar]
- 32.Chrzanowska-Lightowlers ZMA, Horvath R, Lightowlers RN. 175th ENMC International Workshop: mitochondrial protein synthesis in health and disease, 25-27th June, 2010, Naarden, the Netherlands. Neuromuscul Disord 2011;21:142–147. [DOI] [PubMed] [Google Scholar]
- 33.Christian BE, Spremulli LL. Mechanism of protein biosynthesis in mammalian mitochondria. Biochim Biophys Acta 2012;1819:1035–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobs HT, Turnbull DM. Nuclear genes and mitochondrial translation: a new class of genetic disease. Trends Genet 2005;21:312–314. [DOI] [PubMed] [Google Scholar]
- 35.Smits P, Smeitink JAM, van den Heuvel B. Mitochondrial translation and beyond: processes implicated in combined oxidative phosphorylation deficiencies. J Biomed Biotechnol 2010;2010:737385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barth PG, Scholte HR, Berden JA, et al. An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J Neurol Sci 1983;62:327–355. [DOI] [PubMed] [Google Scholar]
- 37.Schlame M, Ren M. Barth syndrome, a human disorder of cardiolipin metabolism. FEBS Lett 2006;580:5450–5455. [DOI] [PubMed] [Google Scholar]
- 38.Claypool SM, Koehler CM. The complexity of cardiolipin in health and disease. Trends Biochem Sci 2012;37:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sengers RC, Stadhouders AM, van Lakwijk Vondrovicova E, Kubat K, Ruitenbeek W. Hypertrophic cardiomyopathy associated with a mitochondrial myopathy of voluntary muscles and congenital cataract. Br Heart J 1985;54:543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sher RB, Aoyama C, Huebsch KA, et al. A rostrocaudal muscular dystrophy caused by a defect in choline kinase beta, the first enzyme in phosphatidylcholine biosynthesis. J Biol Chem 2006;281:4938–4948. [DOI] [PubMed] [Google Scholar]
- 41.Mitsuhashi S, Ohkuma A, Talim B, et al. A congenital muscular dystrophy with mitochondrial structural abnormalities caused by defective de novo phosphatidylcholine biosynthesis. Am J Hum Genet 2011;88:845–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gutierrez-Rios P, Kalra AA, Wilson JD, et al. Congenital megaconial myopathy due to a novel defect in the choline kinase beta (CHKB) gene. Arch Neurol 2012;69:657–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wortmann RL, Vaz FM, Gardeitchik T, et al. Mutation in the phospholipid remodeling gene SERAC1 impair mitochondrial function and intracellular cholesterol trafficking and cause dystonia and deafness. Nat Genet 2012;44:797–802. [DOI] [PubMed] [Google Scholar]
- 44.Zeharia A, Shaag A, Houtkooper RH, et al. Mutations in LPIN cause recurrent childhood myoglobinuria in childhood. Am J Hum Genet 2008;83:489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayashi T, Rizzuto R, Hajnoczky G, Su TP. MAM: more than just a housekeeper? Trends Cell Biol 2009;19:81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Al-Saif A, Bohlega S, Al-Mohanna F. Loss of ERLIN2 function leads to juvenile primary lateral sclerosis. Ann Neurol 2012;72:510–516. [DOI] [PubMed] [Google Scholar]
- 47.Bellzil VV, Rouleau GA. Endoplasmic reticulum lipid rafts and upper motor neuron degeneration. Ann Neurol 2012;72:479–480. [DOI] [PubMed] [Google Scholar]
- 48.Schon EA, Area Gomez E. Mitochondria-associated ER membranes in Alzheimer disease. Mol Cell Neurosci 2013;55:26–36. [DOI] [PubMed] [Google Scholar]
- 49.Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell 2006;125:1241–1252. [DOI] [PubMed] [Google Scholar]
- 50.Chen H, Chan DC. Mitochondrial dynamics—fusion, fission, movement, and mitophagy—in neurodegenerative diseases. Hum Mol Genet 2009;18:R169–R176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan DC. Mitochondrial dynamics in disease. N Engl J Med 2007;356:1707–1709. [DOI] [PubMed] [Google Scholar]
- 52.Milone M, Benarroch EE. Mitochondrial dynamics: general concepts and clinical implications. Neurology 2012;78:1612–1619. [DOI] [PubMed] [Google Scholar]
- 53.Amati-Bonneau P, Valentino ML, Reynier P, et al. OPA1 mutations induce mitochondrial DNA instability and optic atrophy "plus" phenotypes. Brain 2008;131:338–351. [DOI] [PubMed] [Google Scholar]
- 54.Hudson G, Amati-Bonneau P, Blakely E, et al. Mutation of OPA1 causes dominant optic atrophy with external ophthalmoplegia, ataxia, deafness and multiple mitochondrial DNA deletions: a novel disorder of mtDNA maintenance. Brain 2008;131:329–337. [DOI] [PubMed] [Google Scholar]
- 55.Rouzier C, Bannwarth S, Chaussenot A, et al. The MFN2 gene is responsible for mitochondrial DNA instability and optic atrophy "plus" phenotype. Brain 2012;136:23–34. [DOI] [PubMed] [Google Scholar]
- 56.Vielhaber S, Debska-Vielhaber G, Peeva V, et al. Mitofusin 2 mutations affect mitochondrial function by mitochondrial DNA depletion. Acta Neuropathol 2013;125:245–256. [DOI] [PubMed] [Google Scholar]
- 57.Schon EA, Przedborski S. Mitochondria in the next (neurode) generation. Neuron 2011;70:1033–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ropper AH. C. Miller Fisher (in memoriam). Ann Neurol 2012;72:1–3. [Google Scholar]