Abstract
The Undiagnosed Diseases Program at the National Institutes of Health uses High Throughput Sequencing (HTS) to diagnose rare and novel diseases. HTS techniques generate large numbers of DNA sequence variants, which must be analyzed and filtered to find candidates for disease causation. Despite the publication of an increasing number of successful exome-based projects, there has been little formal discussion of the analytic steps applied to HTS variant lists. We present the results of our experience with over 30 families for whom HTS sequencing was used in an attempt to find clinical diagnoses. For each family, exome sequence was augmented with high-density SNP-array data. We present a discussion of the theory and practical application of each analytic step and provide example data to illustrate our approach. The paper is designed to provide an analytic roadmap for variant analysis, thereby enabling a wide range of researchers and clinical genetics practitioners to perform direct analysis of HTS data for their patients and projects.
Keywords: genomics, next generation sequencing, exome, molecular diagnosis
INTRODUCTION
The NIH Undiagnosed Diseases Program (UDP) is designed to evaluate medical syndromes that have been refractory to diagnosis despite extensive assessment [Gahl, et al., 2011; Gahl and Tifft, 2011]. Once accepted, participants undergo in-depth medical evaluation at the NIH Clinical Center. Of the individuals or families seen at the NIH, 10% – 20% are diagnosed with a known condition based on clinical evaluation. The remaining participants become candidates for research studies designed to detect ultra-rare or new diseases that would be difficult, if not impossible, to diagnose using conventional means.
High-throughput sequencing (HTS) has emerged as a powerful tool to study undiagnosed diseases. Many recent publications describe new genes discovered by whole exome sequencing [Bilguvar, et al., 2010; Bonnefond, et al., 2010; Choi, et al., 2009; Erlich, et al., 2011; Hoischen, et al., 2010; Kalay, et al., 2011; Klein, et al., 2011; Krawitz, et al., 2010; Lalonde, et al., 2010; Ng, et al., 2010a; Ng, et al., 2010b; Puente, et al., 2011; Simpson, et al., 2011; Sobreira, et al., 2010; Walsh, et al., 2010; Wei, et al., 2011; Worthey, et al., 2011], and additional publications report genes identified by related techniques [Brkanac, et al., 2009; Johnston, et al., 2010; Kahrizi, et al., 2011; Lupski, et al., 2010; Nikopoulos, et al., 2010; Rehman, et al., 2010; Rios, et al., 2010; Summerer, et al., 2010; Volpi, et al., 2010].
HTS methods produce a list of genotype calls numbering on the order of 104 per exome, 105 for the combined exomes of a small family, and 106 per genome. The genotype list contains common polymorphisms, rare variants and false positives. In the early stages of analysis, variants are prioritized and filtered to produce a subset of potentially disease-causing candidate variants. Filtering is based on factors such as population frequency, segregation according to a proposed genetic model, and predicted consequences for gene function. In addition, many of the published HTS diagnostic successes to date have made use of clues that were present before sequencing commenced. Examples include linkage data [Rehman, et al., 2010], regions of homozygosity [Walsh, et al., 2010], the presence of non-physiologic metabolites [Rios, et al., 2010], and clinical similarity to known syndromes.
Application of HTS techniques to the UDP participant cohort is challenging due to the paucity of pre-sequencing clues. Many families have apparently unique syndromes and no history of consanguinity. The available family members often comprise a pedigree that is too small for traditional linkage methods. The nature of the cases has driven the development of methods to maximize the information obtained from small families and/or individuals. Using both previously described and novel techniques, we have found disease-causing mutations in 5 of 30 families to which HTS methods have been applied. A number of additional families have generated highly suggestive candidates that are undergoing functional validation.
In this paper we describe the step by step process used to analyze DNA sequence variants produced by HTS for our UDP participants. We provide a composite/artificial set of exome data to assist with the implementation of our techniques at other sites where similar clinical work is being performed. For each step we provide a discussion of the rationale behind our approach, a description of how to carry out the analysis with the example data set, and a brief discussion of the tools available for similar analyses. It is our intention to describe an approach that small and medium sized centers can use with their own patients, using next generation sequencing (NGS) data obtained by collaboration or from commercial sources.
METHODS
Supp. Table S1 provides a beginning-to-end outline of the major steps involved in exome sequencing. Most of the discussion in this paper focuses on the “Variant Filtering and Analysis” step in that table. The table can be used to provide some context for the following discussion.
Starting Dataset Acquisition, Annotation and Characteristics
Rationale
The starting point for our analysis is a list of annotated DNA sequence variants—the candidate list. As the analysis proceeds, groups of variants will be tentatively removed from the candidate list until there are few enough variants that each may be scrutinized on an individual basis.
The starting candidate list is the product of the following generalized steps: data acquisition (generating sequence short reads from DNA); alignment (matching the short reads to a pre-existing reference genome) [Lin, et al., 2011; Miller, et al., 2010; Schatz, et al., 2010]; base calling (determination of the best-guess for the genotype, or other sequence feature, at each aligned position) [Ledergerber and Dessimoz, 2011]; and annotation. These steps have been reviewed elsewhere [McKenna, et al., 2010]. The term annotation, as used here, requires special mention. Annotation involves multiple procedures used to gather and record information about each detected sequence variant. Examples include, but are not limited to; the alignment of the variant to a specific base position in a known gene; the assessment of the variant’s potential to disrupt gene function (“pathogenicity”); and the presence of the variant in databases such as dbSNP. Many annotations can be accomplished with free, publicly available tools such as the Genome Analysis Toolkit (GATK) [McKenna, et al., 2010], SeattleSeq (http://gvs.gs.washington.edu/SeattleSeqAnnotation/index.jsp) and the Galaxy website [Giardine, et al., 2005; Goecks, et al., 2010; Taylor, et al., 2007]. A few types of annotation are generated using custom programs developed at individual sequencing centers. For smaller sites lacking the bioinformatics resources of the large centers, the performance of some annotation procedures may be negotiated with a collaborating academic sequencing center or commercial vendor. In any case, a commitment to ongoing communication between the sequencing center and the researcher should be a pre-requisite for any collaboration.
For the purposes of this paper, a specified set of annotations will be assumed to have been performed before candidate list analysis begins. A few annotations are performed by software that is not yet freely available in a stand-alone form. While those annotations are not absolutely necessary, omission will result in a longer final candidate list. As an alternative, we have developed of a suite of Linux-based software scripts called VAR-MD and report this in a separate publication [Sincan, et al, 2012]. VAR-MD will provide the variant annotations used in this paper starting with a basic set of genotype calls. It will also automate many of the analytic procedures described below. Overlapping functionality is also available in the VAAST program, a recently released tool that can automate some annotation and candidate list manipulation tasks [Yandell, et al., 2011].
Our candidate lists are provided by our collaborators at the NIH Intramural Sequencing Center (NISC) in the form of tab-delimited text files with one variant per line. The included annotations and potential data sources are outlined in Table 1. The NISC methodology used to generate the exome data in this paper is outlined in Supp. Methods S1. A wide variety of computer programs can be used to view and manipulate a candidate list. We use a java program called VarSifter, developed by Jamie Teer at NISC (Teer, et al., 2012, available at http://research.nhgri.nih.gov/software/VarSifter/). Our candidate list, with accompanying annotations, is in a text-file format readable by VarSifter. The VarSifter file format, including information that is common to all similar files, is detailed in Supp. Methods S2. Alternately, many candidate list manipulations can be carried out using the Galaxy website [Blankenberg, et al., 2010; Goecks, et al., 2010], GATK, and/or a spreadsheet like Microsoft Excel (Microsoft Corporation, Renton, WA). Commercial solutions are available, and some offer alignment and/or annotation functionality as well, e.g., Nextgene (State College, PA) and the tools provided with sequence data generated by Knome (Cambridge, MA).
Table 1.
Example Candidate List Annotations
| Item Name | Annotation Sources* | Implementation Notes |
|---|---|---|
| Identifier (unique for each variant in candidate list) | Sequencing/Assembling/Genotyping Facility**, GATK | |
| Chromosome Number | Sequencing/Assembling/Genotyping Facility**, GATK | |
| Variant Position Within Chromosome | Sequencing/Assembling/Genotyping Facility**, GATK | Positions are given in the context of a specific reference genome, e.g. NCBI hg18/build36 |
| Reference allele | Sequencing/Asembling/Genotyping Facility**, GATK | |
| Variant allele | Sequencing/Asembling/Genotyping Facility**, GATK | |
| Variant type (exon, intron, etc.) | Annovar, SeattleSeq, GATK, VAAST | |
| Gene name | Annovar, SeattleSeq, GATK, VAAST | |
| Transcript | Annovar, SeattleSeq, GATK, VAAST | |
| Strand | Annovar, SeattleSeq, GATK, VAAST | |
| Reference Amino Acid | Annovar, SeattleSeq, GATK, VAAST | |
| Variant Amino Acid | Annovar, SeattleSeq, GATK, VAAST | |
| Amino Acid Position | Annovar, SeattleSeq, GATK, VAAST | |
| Pathogenicity Score | Galaxy, GATK, PolyPhen, many others | NISC provides “CDPred” score |
| Coverage | Samtools, Bed tools, GATK | |
| Quality Measure | Samtools, GATK | NISC provides MPG and MPG/coverage scores. Quality scores should be calibrated to a specific sequencing center/source |
| Mendelian Consistency for various genetic models | Manual inspection with spreadsheet, VAR-MD, VAAST | NISC provides annotation with in-house software |
| Compound Heterozygote Pairing for Autosomal Recessive Genetic Model | Manual inspection with spreadsheet | NISC provides annotation with in-house software |
These are incomplete lists. A broad and rapidly expanding list of tools is available.
Often a collaborating sequencing facility can provide some or all of the annotations listed here. Most of the annotations can be carried out separately if needed. However, synergistic benefits can accrue if assembling and genotyping are performed by the same team.
Genome-sequencing will eventually become standard for many HTS applications. Until that time, however, the addition of genome-wide data from a high-density SNP array has the potential to add critical additional information to an HTS project, particularly in the case of exome analysis. We also obtain SNP array data for every HTS project. We use the Illumina platform and the associated analysis program Genome Studio (Illumina, San Diego, CA). Other types of SNP arrays would be equally suitable.
The guiding principle behind our filtering procedure is that an HTS variant-analysis process must be flexible enough to allow adjustment of all analytic parameters. Those performing the analysis must understand the rational, procedures and assumptions inherent in each step.
Procedure
The files used in the following analyses are available in one of two places. An example data set and interval post-processing results are located at ftp://ftp.nhgri.nih.gov/pub/NIHUDP/ADAMS_METHODS/. The example dataset compexome_30_unfiltered.vs is an exome candidate list created and modified from several projects to protect individual patient data. Each included project involves a family with a similar structure: four individuals including two parents and two full sibs. One sibling is affected with a disorder that appears to be early-onset, severe and likely to be highly penetrant at an early age. There is no history of consanguinity. High-density SNP arrays have been run for each family member. Individual variations are all biologically derived and there is one verified positive finding in the dataset. The positive finding was found in a family for which the affected child had a childhood-onset neurodegenerative disorder. A number of consistent known diseases, including some lysosomal storage diseases, had been ruled out by specific clinical testing. The story of the original exome-based diagnosis for that family is in press in a separate publication [Pierson, et al., 2011].
Genotyping Quality Measurement
Rationale
HTS technology and methods are evolving rapidly. In addition to falling prices, aspects of the laboratory techniques used for data generation change every few months. Interpretation of an HTS candidate list requires an understanding of the genotyping-quality issues associated with the specific techniques used to acquire the data. Excellent reviews of HTS quality assessment are available [Teer, et al., 2010]. Quality for a given project should be assessed by, or with the group who performed the data acquisition. Only that group can provide historical data about their experience with the specific techniques they use. Key issues include variant-call quality near the ends of sequence reads and assemblies, quality of insertion/deletion variant calling and assessment of pre-sequencing laboratory-work.
The average depth of HTS short reads in a sequence alignment is a frequently reported metric of variant-call quality. Coverage for an entire HTS project can be reported in different ways such as “average coverage per base” or “percent of bases covered to depth n”. An example of one potential pitfall of using coverage as the sole measure of variant-call quality is the compression misalignment. In a compression, reads from two highly similar regions, e.g., a gene and matching pesudogene, are aligned to the same position on the reference sequence. The two slightly-different sequences create apparent non-reference genotype calls where they differ, and simultaneously create an area of falsely-reassuring deep coverage.
Procedure
Quality assessment metrics for our data were developed by NISC and include a Bayesian statistic for each base call (the Most Probable Genotype or MPG score) and a ratio of the MPG score to the coverage for any given variant [Teer, et al., 2010]. The latter makes intuitive sense. The quality score should increase in proportion to the coverage. A deeply-covered variant with an inadequately high quality score may indicate a false-positive genotype call. For the example dataset, variants have been included if at least one family member exceeds a lower-cutoff for quality. The lower cutoffs for the MPG and MPG/coverage were empirically derived and set at MPG = 10 and MPG/coverage = 0.5.
Candidate List Filtering: Variant Type
Rationale
Each analyst must define a starting point with regard to assumptions about the nature of the DNA change(s) affecting their gene of interest. Our usual starting assumptions have failed in some cases, and proven successful in others. Failure to find a convincing candidate simply prompts an additional pass through the data with different assumptions.
Procedure
As a first pass, we will guess that the disease-causing variation, or variations, involves coding sequence or a canonical splice site. We will further postulate that it will be a typical pathogenic variant, e.g., a missense change versus a less common type such as a synonymous splice modifier. After loading compexome_30_unfiltered.vs into VarSifter, the number of variant positions displayed is 116,837—a typically large number for a family of four. The following variant types are selected: insertions/deletions, missense mutations, nonsense mutations, and canonical splice-site mutations. Selecting those variants and applying the filter reduces the number of variants to 14,338 (compexome_31_pathogenic_variants.vs). The mechanism by which filtering occurs is straightforward. VarSifter uses one column of the candidate list file (“type”) to look up the annotated mutation type. Any mutation types not included in the filter are removed from the current view. To relax the criteria, intronic and other mutation categories may be added, followed by re-filtering of the original data.
Candidate List Filtering: Population Frequency
Rationale
Filtering by population frequency is an attempt to remove common polymorphisms that are unlikely to be disease causing. It is conversely equivalent to the practice of reporting of negative results in a panel of normal controls when describing a new mutation. The disease-causing variant is implicitly assumed to be rare, high-penetrance, and responsible for a large phenotypic effect.
dbSNP [Sherry, et al., 2001] is highly-utilized public database of DNA sequence variations. Entries have a regular format, but are not curated and have non-required fields. Most of the HTS analysis papers to-date have used dbSNP entries as a filter to remove common variations. Unintentional generation of false-positive or false-negative filtering results can occur with inappropriate application of the dbSNP database. Many dbSNP entries lack population frequency information and/or derive from studies with few individuals. The dbSNP database is known to contain pathogenic mutations; it was never designed to exclude them. In a 2008 study, Won et al. demonstrated that 8% of the sequence variations in dbSNP (v.126) were also present in the Human Gene Mutation Database (HGMD)[Won, et al., 2008]. The HGMD (BIOBASE Biological Databases, Wolfenbüttel, Germany) is ostensibly a list of human disease-causing variations, although it is known to be only as good as the medical literature it collates. The HGMD/dbSNP overlap serves to illustrate the potential for misclassification of DNA sequence variants by using an unselected database.
The 1000 Genomes Project is increasingly providing an invaluable resource for identifying common DNA sequence variations. It is available as a subset of current versions of dbSNP or by itself from the 1000 Genomes website [Sudmant, et al., 2010]. 1000 Genomes variants are annotated with heterozygosity information allowing for the construction of filters with a specified lower limit of population heterozygosity. Determining an appropriate heterozygosity exclusion criterion requires an estimate of disease incidence. For ultra-rare conditions in Hardy-Weinberg equilibrium and with incidences on the order of 1:1,000,000, the expected heterozygosity in the population is 1/500 or 0.002 (0.2%). For a condition with an incidence of 1/10,000 it is 2%. It is preferable to set the criterion too high rather than too low as the latter will run the risk of excluding the disease-causing variation being searched for.
Procedure
Varsifter allows filtering using BED-formatted text files, the BED format providing a means to define arbitrary genomic intervals. Recent developments at the Galaxy website allow for the rapid construction of BED files with dbSNP data. Filters should be re-constructed with each dbSNP release as new data are added regularly. Table 2 shows the results of population frequency filtering with files for several different heterozygosity cutoffs including 0.5, 1, 2 and 5%. The 1% heterozygosity filter in Table 2 was applied using both dbSNP131 and dbSNP132 to highlight the fact that using updated filters is important to maximize the number of excluded variants. The dbSNP132 filter excluded 3000 more variants than the prior version. Each BED file is available at ftp://ftp.nhgri.nih.gov/pub/NIHUDP/ADAMS_METHODS/. For our filters, we use a subset of dbSNP that includes 1000 Genomes data and HapMap variants/polymorphisms that align uniquely to the genome. A method for constructing such filters using the Galaxy website is provided as Supp. Methods S3. Using the dbSNP132 1% filter, the example candidate list is reduced in size from 14,338 to 5,041 (compexome_32_DB132.vs). The population filtering threshold can be adjusted by creating files for various SNP heterozygosity cutoffs and substituting files as desired to adjust filtration.
Table 2.
Population Frequency Filtering of Candidate List from Exome Sequencing
| Filename | dbSNP version | Heterozygosity cutoff | Starting variants (full candidate variant list) | Post-filtering variants |
|---|---|---|---|---|
| dbSNP131_1percent.BED | 131 | 1% | 116,837 | 56,452 |
| dbSNP132_0.5percent.BED* | 132 | 0.5% | 116,837 | 53,514 |
| dbSNP132_1percent.BED* | 132 | 1% | 116,837 | 53,762 |
| dbSNP132_2percent.BED* | 132 | 2% | 116,837 | 54,267 |
| dbSNP132_5percent.BED* | 132 | 5% | 116,837 | 56,126 |
Only includes HapMap and 1000 Genomes Project data, and only uniquely mapping sites
Note that filtration numbers are all in the same order of magnitude, suggesting that the majority of the excluded SNPs are relatively common and appear in all of the filters. An additional ~3000 SNPs were filtered out by updating the filter from db131 to db132 highlighting the fact that the databases are significantly updated between releases.
Candidate List Filtering: Gene and Site Exclusion Lists
Rationale
Some sequence variants can often be excluded a priori during a first pass analysis. Two types of exclusion are explored in Fuentes Fajardo et al. [Fuentes Fajardo, et al, 2012]. Excluded genes contain multiple variants in every HTS-sequenced individual and are identified by restrospective analysis of accumulated exome data. These genes may fall into one of several categories: pseudogenes, groups of paralogs such as olfactory receptors, and/or chromosomal regions with biologically important hypervariability. An example of the last is the HLA region on chromosome 6. Additionally, individual base pairs can be excluded. Base-pair exclusions are made based on the meta-analysis of a collection of exomes, preferably from one sequencing center and set of related sequencing methods. Examples include sites that are always heterozygous (likely to be caused by alignment problems specific to a given alignment methodology) and sites that are always homozygous non-reference (sites where the reference sequence contains a minor allele).
Occasionally, certain projects will require the re-inclusion of typically excluded genes or sites. The analyst should be familiar with the contents of any exclusion lists employed, so that modifications can be made as needed.
Procedure
We use two exclusion lists developed using the techniques referred to in Fuentes Fajardo et al [Fuentes Fajardo et al, 2012]. The gene list is a text file with gene names, and the individual-base-pair list is a complemented BED file similar to the one used in the earlier population frequency filter. Application of the base pair exclusions reduces the candidate list from 5,041 to 3,752 (compexome_33_HWE_BEDfile_2.vs), and application of the gene list reduces the number further to 2,360 (compexome_34_Gene_Kill_List.vs). The BED file may be specific to our data acquisition methods, but the gene list should be useable by other centers. Both files are provided as gene exclusion_list.txt and base-pair_exclusion_list.txt, respectively.
Candidate List Filtering: Genotyping Quality Criteria
Rationale
Low-confidence genotype calls may be removed during the data acquisition and annotation process. Only highly-compelling criteria should prompt such variant removal. In the remaining cases, a quality score can be used to provide guidance to the candidate list analyst. Take as an example a case where three out of four family members have good quality data suggesting an important candidate variant. The variant may deserve consideration despite the fact that one family member has poor-quality data. Such variants are examples of what to revisit if an answer is not found during a first pass analysis. Genotyping quality scores, therefore, represent an additional variable that can be used to adjust filtration stringency.
Procedure
As mentioned previously, our collaborators at NISC use the MPG score and MPG score/coverage ratio to annotate variant quality. The VarSifter program allows specification of the number of family members in a pedigree who need to exceed a given cutoff for inclusion in the post-filtration list. For our example, we specify that all four family members need to have an MPG score of at least 10 and an MPG/coverage score of 0.5 or greater. The subsequent filtration reduces the number of variants from 2360 to 1469 (compexome_35_Quality_filters.vs).
Candidate List Filtering: Family Structure
Rationale
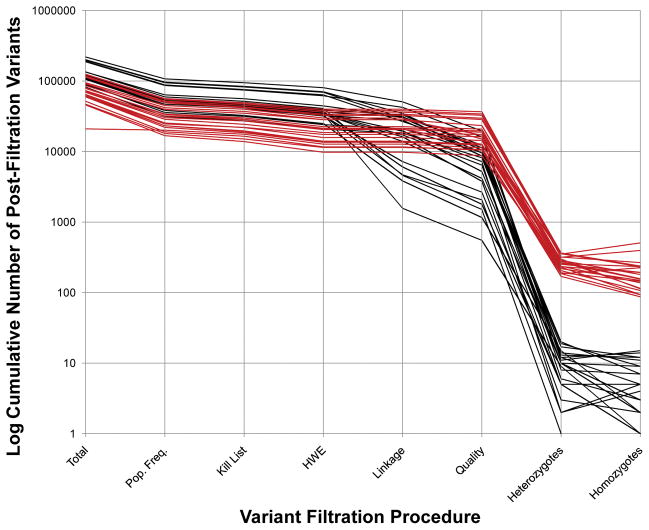
In the near-past, HTS data acquisition costs were frequently a limiting factor in experimental design. As costs drop, data acquisition feasibility is giving way to other design issues. One consideration is whether or not to sequence additional family members, beyond the proband. Added family members have the potential to directly and substantially decrease the number of candidate variations in an HTS project. Fig. 1 illustrates the effect of the incorporation of family data on final candidate list size.
Figure 1.
Cumulative Filtration of Exome Variant Lists from 22 Families. A set of 22 exome projects is displayed using two different analytical approaches: one uses all available family data (black) and the other uses only data from the proband (red). The y-axis is the log10 of the number of cumulatively filtered, residual variants. The x-axis shows filtration steps, which are sequential from left to right. The last two steps (homozygotes and heterozygotes) use the post-quality-filtration variants to filter and are not sequential. Note that the implementation of homozygote and heterozygote filters differs between single exome analyses and family based analyses. Mendelian segregation and phase information is not available in the case of single exome analysis. Homozygotes are not checked for inheritance from both parents. The “heterozygote” count is a tabulation of all pair-wise combinations of variants for those cases where more than one heterozygous variant is found in the same gene. Single exome projects start with fewer variants and end with a larger number of candidates for further study. See the text for a further explanation of the various filtration steps.
Added family members can be analyzed with concurrent SNP array analysis to provide recombination mapping (precise segregation-consistent chromosomal intervals) [Roach, et al., 2010], mosaicism detection [Gonzalez, et al., 2011; Markello, et al., 2011a] identification of regions of homozygosity, estimates of inbreeding coefficients, confirmation of parentage, uniparental disomy analysis and detection/interpretation of copy number variations. As an example, if a proband and a father share a single copy deletion, then the sequence of the corresponding maternal allele in the proband should be interrogated for possible complementary loss-of-function variations that might generate a phenotype when paired with the paternally-inherited deletion. If the same deletion is new to the proband, then a different set of mechanisms can be considered including haploinsufficiency or a complementary variant inherited from whichever parent contributed the non-deleted allele.
While recombination mapping can be performed using genome sequencing data, exome projects require the addition of genome-spanning high-density SNP array data. Construction of recombination maps using SNP data is described in an accompanying paper by Markello et al.[Markello, et al., 2011b]. Recombination mapping is analogous to traditional linkage analysis, which produces variable likelihood-based estimates of linkage between widely spaced markers. The close proximity of SNPs on a high-density SNP array means that the probability of a double-crossover event between a given pair of markers is small. Consequently, sites of recombination can be mapped in a “square wave” fashion, with regions of consistent and non-consistent segregation mapped to a precision on the order of a few kilobases. For exome candidate list analysis, regions that have segregated in a manner consistent with a given genetic model can be defined with a BED file. Variants outside the consistent regions are filtered out.
Consistent segregation can also be verified for individual variants [Choi, et al., 2009; Ng, et al., 2010b]. The group of variants filtered by recombination mapping overlaps but is not identical to the set of variants excluded by individual-variant segregation filtering. The difference probably represents variants in segregation-valid regions that are sequencing false positives. The stringency of segregation filtering is determined by the number of “errors” tolerated by the filter. For instance, consider the following situation. Given a postulated autosomal recessive model, a pattern of variation for a family of four includes a consistent proband, one consistent sib, one consistent parent, and a second parent with missing data (e.g., a local sequencing failure). Should this family be included or excluded? The rules used to answer that question will define the stringency of the filter.
Procedure
Genome Studio was used for the high-density SNP-array analyses including the straightforward visualization of copy number variants and the more complex detection of recombination sites using Boolean rule-sets. The methods for the latter types of analyses are provided in the papers referenced above.
We decided to obtain exome sequence on multiple family members. The decision was based on several factors: 1) There was no evidence of consanguinity or potential for homozygosity mapping based on previously obtained SNP array data; 2) There were no clinical findings to suggest a specific set of genes implicated in disease causation (that had not been excluded by clinical testing); and, 3) There was no linkage region or other mapping data to establish a genomic candidate region. We therefore chose the most powerful approach for agnostic screening of the exome and sequenced both parents and one unaffected sibling along with the proband.
Recombination mapping was carried out using the methods described in Markello et al [Markello, et al., 2011b]. The procedure involves using Genome Studio to apply a set of Boolean segregation rules to SNP array data. The resulting recombination map was defined in a BED-formatted file (Linkage File.txt). The BED file was applied using VarSifter and reduced the candidate number from 1469 to 958 (compexome_36_linkage_regions.vs).
Our candidate list includes specific annotations regarding Mendelian consistency. Custom scripts use family-relationship data to test whether a given variant did or did not segregate in a biologically feasible manner and flag it as inconsistent if it did not. Furthermore, regions defined by gene boundaries are surveyed for pairs of variants that could make up a compound heterozygote set. Such variants are annotated with a column that lists the index number(s) of the complementing variant or variants (Nancy Hansen, unpublished data). The Mendelian consistency annotations may not be available in candidate lists from all sequencing centers. The VAR-MD [Sincan et al, 2012] and VAAST [Yandell, et al., 2011] programs can incorporate such information. However, once the variant list gets short enough, a spreadsheet can be used to sort the variants by gene name. Once sorted, the contents of individual loci can be inspected for Mendelian relationships.
For our candidate list, we postulated an autosomal recessive genetic model because both parents were unaffected. A new dominant model would also be appropriate for a potential subsequent analysis. The recessive inheritance could arise from homozygous or compound heterozygous mutations. Application of the appropriate filters with VarSifter results in 7 homozygote candidates (compexome_37a_homozygous_recessive.vs) and 94 compound heterozygote candidates (compexome_37b_compound_heterozygotes.vs).
Working With the Candidate List: Assessment of Individual Variants
Rationale
Inspection of the example files show that the candidate list is now small enough for each variant to be considered individually for goodness of fit with the clinical syndrome. Additional tools become useful at this stage. Individual variant positions should be looked up in any available databases of known genomic variants. Homozygous variant positions should be compared with the positions of any regions of homozygosity identified by SNP array analysis or other means. Homozygous variants should also be correlated with any single copy deletions, to see if the two might combine to cause an autosomal recessive disease.
Procedure
In our example, each homozygous variant is associated with a dbSNP “rs” number, providing an additional source of information. Individual variants may require in depth research. For example, among the homozygotes is a p.A34E mutation in the PPT2 gene, dbSNP number rs3096696. The coverage is low for the mother and the proband at 14 reads (compare with other variations in the list with coverage in the 50 to 200 range). Inspection of the dbSNP record reveals that the variation has been seen in homozygous form in 19% of 39 cell lines derived from persons of Caucasoid, African-American or American Indian ethnicities, 28 out of 39 of whom had known consanguinity. The SNPs were reported by a researcher at the Fred Hutchinson Cancer Research Center and contact information is available. Inspection of the Online Mendelian Inheritance in Man website (OMIM, http://www.ncbi.nlm.nih.gov/omim) shows that PPT2 (MIM# 603298) has a known mouse model with a neurological phenotype. In addition to the dbSNP record, the laboratory that performed the HTS data acquisition should be able to inspect the raw alignment data to see if the variant is in an area consistent with genotyping errors. For the example case, similar research was able to deprioritize all of the homozygous variations.
The compound heterozygote list has 94 individual variants. Compound heterozygotes must have at least two pathogenic, trans-oriented mutations to satisfy an autosomal recessive model. A study of the specifics of the annotation of our candidate list provides an example of how knowledge about each step of data production is critical for interpretation.
First, for our list, family-based Mendelian-consistency annotation is carried out before the final quality-based variant exclusions are decided. As a result, some variants are removed from the dataset after compound heterozygosity variant pairings are established.
Second, an individual variant can be inherited in a manner consistent with a compound heterozygote model, but never have had a second mutation to complement it.
Third, and as a corollary to the second item, multiple variants at one locus may not be pairable if they all occur on the same allele.
Fourth, a pair of trans-oriented variants at a given site may have one good candidate and one poor candidate (poor quality, low pathogencity prediction and/or known benign changes based on literature or other information).
As a result of these four factors, the list of compound heterozygotes includes numerous variants that are annotated as consistent with compound heterozygous inheritance, but can be excluded due to lack of a second, high quality, trans variant. As mentioned in a previous section, part of the NISC annotation pipeline attempts to find variant pairs that together would explain compound heterozygous inheritance. VarSifter will display consistent pairings, and the example data set reveals only 3 pairs of variants (out of the original 94 individual variants).
Working With the Candidate List: Pathogenicity Assessment
Rationale
Pathogenicty prediction estimates the effect a DNA variation will have on gene function. It is not unique to HTS and is frequently incorporated into the analysis of unknown sequence variants from other sources. Most of the available automated tools focus on the alteration of amino acids in coding regions. However, specialized tools are available to predict the affect of non-coding variants in splicing [Brunak, et al., 1991; Desmet, et al., 2009; Hebsgaard, et al., 1996; Pertea, et al., 2001] and regulatory regions [Venter and Warnich, 2009].
The criteria used to assess the pathogenicity of missense mutations include intraspecies conservation, information about protein structure (predicted and experimental), amino acid chemical similarity, coincidence with disease and functional assay. Many related software programs exist including Polyphen [Adzhubei, et al., 2010], SIFT [Ng and Henikoff, 2001], Panther [Mi, et al., 2005; Thomas, et al., 2003], SNAP [Bromberg and Rost, 2007; Bromberg and Rost, 2008; Bromberg, et al., 2008] and others. In general, pathogenicity prediction has false positive and false negative rates between 10% and 20% [Ng and Henikoff, 2006]. As a result of these substantial error rates, the predictions are primarily useful for prioritizing variation candidates and must be used with caution in assessing individual variants. When choosing a pathogenicity prediction software program, there are several features to consider beyond ease of use and convention. The optimal program would have the following characteristics: 1) the criteria by which individual predictions are made should be accessible to the user; 2) programs should provide results for a wide variety of regions, but should also reflect the paucity or abundance of information for a given site; and 3) the software should produce a reasonably variable numeric score to allow the prioritization of a long list of variants.
Procedure
The UDP data sets are analyzed with CDpred, a component of the NISC annotation pipeline [Johnston, et al., 2010]. CDPred estimates variant pathogenicity using alignment conservation data from the Conserved Domain Database [Marchler-Bauer, et al., 2003]. When conserved domain alignments cannot be made, the program defaults to a BLOSUM matrix based on empirically derived substitution frequencies [Henikoff and Henikoff, 1992]. Increasingly positive and negative integers indicate decreasing and increasing pathogenicity, respectively. Stop mutations and canonical splice site mutations are arbitrarily set at −30, a value more negative than that seen for any missense mutation.
The example data set contains three compound heterozygote pairs, and the CDPred scores for each can be inspected to get a sense of how severe the mutations are. One pair has positive CDPred scores, suggestive of relatively mild effect on gene function. The other two pairs have negative scores, which are more consistent with a disease-causing mutation. All of the mutations are missense, so there are no very low scores such as the −30 seen for a stop mutation. As mentioned, using pathogenicity prediction software for individual variants is risky, and for a list this small it would mainly be used to get a general sense of mutation severity. However, in less favorable cases, there may be a long list of variants, and sorting by pathogenicity is a useful way to focus on a subset of data for initial analysis.
Working With the Candidate List: Previously Reported Mutations
Rationale
Numerous software tools are available to search for associations between candidate variants and known clinical syndromes. Examples include the Online Mendelian Inheritance in Man (OMIM) website, Pubmed, the Human Gene Mutation Database [D.N. Cooper, 2011], disease-specific mutation repositories, and software such as Alamut (Interactive Biosoftware, Amsterdam, The Netherlands) that can collate information from multiple sources. Many sequence variants listed as pathogenic will not have been adequately characterized, so care must be taken when assigning disease causation.
Procedure
One of the compound heterozygotes genes is GLB1, and one of the variants in the pair, p.R201H, has been reported as being associated with GM1 gangliosidosis. That information supports the hypothesis that GLB1 is the disease-causing gene.
Working With the Candidate List: Incorporation of Pre-Existing Information
Rationale
Pre-existing knowledge about the biology or genetics of a particular project can be added at any stage of analysis. If there is strong evidence that the causal variation(s) will be present in a specific chromosomal region, a targeted capture technique may be preferable to exome capture. Targeted capture will genotype a wider range of potential non-coding regulatory sites and intronic sequence in the region of interest. In either case, the candidate list can be narrowed by creating a BED file that defines specific regions of interest. Alternatively, a list of genes located in a candidate region can be specified.
Erlich et al. [Erlich, et al., 2011] reported an approach by which a candidate gene list was narrowed using disease network analysis. The approach is suited for cases where the syndrome being studied shows genetic heterogeneity. Genes known to cause the syndrome are inspected for commonalities in physical structure, expression patterns, shared domains, etc. Identified shared characteristics are then searched for in genes with variants in the whole exome candidate list. Several software tools available for network analysis are reviewed in the Erlich paper.
Individual clinical hypotheses can be studied by making gene lists associated with specific syndromes. For example a syndrome that has characteristics consistent with a mitochondrial disease would make use of a gene list of all known nuclear-encoded mitochondrial genes.
Procedure
We have developed and/or adapted a number of disease gene lists for conditions known to exhibit significant genetic heterogeneity. For example, many of the UDP participants have medical syndromes that could be caused by mitochondrial disease. However, there are a large number of nuclear-encoded mitochondrial genes, and testing them by individual Sanger sequencing is time consuming and expensive. Exome sequencing provides an alternate approach to such diseases, and can be used to sequence all of the genes of interest within the limits of exome sequencing coverage characteristics. The exome candidate list is then examined by looking only at genes known to be associated with mitochondrial disease.
Unfortunately, in the example case, the clinical phenotype did not match any of those conditions. Inspection of the high-density SNP arrays did not show any genomic lesions that would provide a candidate locus or basis for targeted capture. However, the clinical presentation did have features consistent with a neurological lysosomal storage disease (LSD), including progressive symptoms.
Working With the Candidate List: Variant Validation
Rationale
Once variants are detected by an HTS technique, it is standard practice to validate candidates of interest using Sanger sequencing. For our sequencing collaborators, approximately 90% of HTS detected variants will validate. For variants that are not well supported by previous work, functional analysis must be the final determinant of the pathogenic role of a DNA sequence variant.
Procedure
Given a clinical syndrome suggestive of an LSD,and knowing that GLB1 causes GM1 gangliosidosis (a type of LSD), we performed in-house Sanger sequencing followed by CLIA laboratory sequencing to verify the HTS sequencing variants detected in the GLB1 gene. We then repeated prior clinical enzymatic testing that had ruled out GM1 gangliosidosis. It turned out that the prior negative testing had been a false-negative result and that the beta-galactosidase (GLB1 gene product) enzymatic-activity level was indeed reduced to a level consistent with disease [Pierson, et al., 2011].
DISCUSSION
We have presented a framework for the analysis of HTS data, starting with a large list of annotated DNA sequence variants, and ending with a small list of high-value candidates for confirmation or research follow-up. We have further provided an example data set that includes a validated result. During the introduction, we reported that we have found unequivocal disease-causing mutations in 5 of 30 families, plus several other compelling candidate DNA variants. Our record illustrates that many HTS projects do not generate a clear-cut answer during the early stages of analysis. In some cases, relaxing filtering constraints will expand the list to include an obvious candidate. In other cases, the result is a list of weak candidates that require significant laboratory work to definitively exclude or confirm.
When considering HTS failure, it is important to consider the types of variants not detected by the methodology. The default/first-pass scheme outlined for our example will not detect pathogenic synonymous mutations, non-cannonical splice site mutations, or common mutations that cause disease in certain circumstances (e.g., the common MTHFR c.677C>T mutation in the setting of folate deficiency). For our first-pass analysis, such variants are missed because of high filtration stringency. In other cases, the data acquisition methodology fails to genotype the variant of interest. Examples include regions not included in the capture design, and regions that are not well sequenced despite being captured. The latter is often due to local sequence characteristics including repetitive DNA elements and/or low sequence complexity. The analyst must know how the capture technique is designed (what would be captured under optimal conditions), how the capture methods perform in practice (what regions are actually captured by a given lab, chemistry and procedure), and how well individual regions are sequenced once captured.
Failure of HTS projects may be caused by incorrect hypotheses regarding the genetic mode of inheritance. In our example, the pedigree was consistent with several genetic/segregation mechanisms including autosomal recessive and new dominant. We do not discuss more complex models such as multi-allelic inheritance. Filtering based on a multi-allelic model can be attempted for individual hypotheses by viewing variants only from genes belonging to a specific pathway. Automated analysis is also possible, but requires bioinformatics procedures that are outside the scope of this article.
The provided example dataset included exome sequence for a small family. Other potential datasets include whole genomes, exomes from single individuals and custom capture of a genomic candidate region. Different specialized methods, and/or additional analyses, would be better suited to detect gene-gene interactions and epigenetic phenomena [Feng, et al., 2011]. The techniques outlined for our example have variable application to other datasets. Some elements, such as population frequency and kill-list filtering, are applicable to single exomes and subsets of whole genome sequence.
For single exomes, some filters will not be usable due to the lack of family structure data. However, with sufficient other clues, such as candidate regions or gene lists, the remaining filters may be adequate to find the causative variant(s). A special case of single exome sequencing is the simultaneous clinical testing of a large set of genes. An example of a disease for which such a technique might be advantageous is spinocerebellar ataxia (SCA). SCA is a neurologic syndrome comprising multiple overlapping diseases caused by multiple different genes [Matilla-Duenas, et al., 2010]. Given the current ~$1000 cost for an exome, it is enticing to consider using whole exome sequencing to screen the relevant genes for variants instead of paying the tens-of-thousands of dollars needed to screen the same genes using commercial clinical sequencing. However, several caveats need to be considered. First, several of the SCA’s are caused by repeat expansions, a type of genetic lesion that is not reliably detected by current HTS methods, especially exome sequencing. Second, exome data from any given lab must be carefully studied to determine how well the SCA genes are captured, covered and genotyped. Third, while exome sequencing is a convenient way to survey a group of genes for variants, it is more difficult to determine how well regaining regions have excluded variants. In other words, the pattern of false negative results is less well understood for HTS than for Sanger sequencing. Dias et al. looked at coverage for a group of genes known to be associated with inherited neurologic disease [Dias, et al., 2012]. Surprisingly, although most positions were well covered/genotyped in most individuals, there were no genes that were covered adequately in all individuals. Furthermore, the pattern of “missed” sites was distributed among the sequenced individuals rather than being concentrated in a few “bad reads.” These data suggest that we have work to do in understanding the false negative characteristics of exome sequencing.
Whole genome data will eventually replace exome data as prices fall and the significance of conserved intra-genic chromosomal regions are characterized [Margulies and Birney, 2008]. Genomic DNA will provide the genome-wide information we currently obtain from high-density SNP arrays. In addition, there is the potential for overall greater coverage of genes of interest, particularly non-coding regions that are missed by current exome HTS. Methods to filter conserved, non-coding regions of the genome are being developed, but are not yet widely available
HTS represents a fundamental advance in how genomic sequence data are measured, and has fundamental implications for both research and clinical work. We believe that interpretation of exome data must involve clinicians and scientists familiar with the subjects of the study and should not rely solely on bioinformatics specialists. Using this model, individual researchers need to be empowered to work directly with exome data. An understanding of the fundamental underpinnings of HTS data manipulation and processing will provide the means for that empowerment.
Supplementary Material
Acknowledgments
Supported by the Intramural division of the National Human Genome Research Institute, the National Institute of Neurological Disorders and Stroke, the NIH Clinical Center, and the NIH Office of the Director
We thank our patients and their families, who are partners in the pursuits of the NIH UDP. The clinical work to which we apply these methods would not be possible without the outstanding clinical nurse practitioners, research nurses, genetic counselors, consultants and other providers with whom we work. We appreciate the excellent technical skills of Roxanne Fischer and Richard Hess.
Footnotes
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
References
- http://gvs.gs.washington.edu/SeattleSeqAnnotation/
- Online Mendelian Inheritance in Man, OMIM (TM) McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University; Baltimore, MD: National Center for Biotechnology Information, National Library of Medicine; Bethesda, MD: [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilguvar K, Ozturk AK, Louvi A, Kwan KY, Choi M, Tatli B, Yalnizoglu D, Tuysuz B, Caglayan AO, Gokben S, et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467(7312):207–10. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. 2010;Chapter 19(Unit 19):10, 1–21. doi: 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefond A, Durand E, Sand O, De Graeve F, Gallina S, Busiah K, Lobbens S, Simon A, Bellanne-Chantelot C, Letourneau L, et al. Molecular diagnosis of neonatal diabetes mellitus using next-generation sequencing of the whole exome. PLoS One. 2010;5(10):e13630. doi: 10.1371/journal.pone.0013630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brkanac Z, Spencer D, Shendure J, Robertson PD, Matsushita M, Vu T, Bird TD, Olson MV, Raskind WH. IFRD1 is a candidate gene for SMNA on chromosome 7q22-q23. Am J Hum Genet. 2009;84(5):692–7. doi: 10.1016/j.ajhg.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35(11):3823–35. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y, Rost B. Comprehensive in silico mutagenesis highlights functionally important residues in proteins. Bioinformatics. 2008;24(16):i207–12. doi: 10.1093/bioinformatics/btn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y, Yachdav G, Rost B. SNAP predicts effect of mutations on protein function. Bioinformatics. 2008;24(20):2397–8. doi: 10.1093/bioinformatics/btn435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunak S, Engelbrecht J, Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol. 1991;220(1):49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloglu A, Ozen S, Sanjad S, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106(45):19096–101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, EVB, Stenson PD, Phillips AD, Shaw K, Mort ME, Thomas NST. The Human Gene Mutation Database at the Institute of Medical Genetics in Cardiff 2011 [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37(9):e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias C, Sincan M, Rupps R, Briemberg H, Selby K, Mullikin J, Markello T, Adams D, Gahl WA, Boerkoel CF. Exome sequencing: diagnosis of genetically heterogeneous neuromuscular disorders. Hum Mutat. 2011;33:xxx–yyy. [Google Scholar]
- Erlich Y, Edvardson S, Hodges E, Zenvirt S, Thekkat P, Shaag A, Dor T, Hannon GJ, Elpeleg O. Exome sequencing and disease-network analysis of a single family implicate a mutation in KIF1A in hereditary spastic paraparesis. Genome Res. 2011;21(5):658–64. doi: 10.1101/gr.117143.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Rubbi L, Jacobsen SE, Pellegrini M. Determining DNA methylation profiles using sequencing. Methods Mol Biol. 2011;733:223–38. doi: 10.1007/978-1-61779-089-8_16. [DOI] [PubMed] [Google Scholar]
- Fuentes Fajardo KV, Adams D, Mason CE, Sincan M, Tifft C, Toro C, Boerkoel CF, Gahl W, Markello T NISC Comparative Sequencing Program. Detecting false positive signals in exome sequencing. Hum Mut. 2012;33:xxx–yyy. doi: 10.1002/humu.22033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahl WA, Markello TC, Toro C, Fajardo KF, Sincan M, Gill F, Carlson-Donohoe H, Gropman A, Pierson TM, Golas G, et al. The National Institutes of Health Undiagnosed Diseases Program: Insights into rare diseases. Genetics in medicine : official journal of the American College of Medical Genetics. 2011 doi: 10.1038/gim.0b013e318232a005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahl WA, Tifft CJ. The NIH Undiagnosed Diseases Program: lessons learned. JAMA : the journal of the American Medical Association. 2011;305(18):1904–5. doi: 10.1001/jama.2011.613. [DOI] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15(10):1451–5. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11(8):R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez JR, Rodriguez-Santiago B, Caceres A, Pique-Regi R, Rothman N, Chanock SJ, Armengol L, Perez-Jurado LA. A fast and accurate method to detect allelic genomic imbalances underlying mosaic rearrangements using SNP array data. BMC Bioinformatics. 2011;12(1):166. doi: 10.1186/1471-2105-12-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouze P, Brunak S. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 1996;24(17):3439–52. doi: 10.1093/nar/24.17.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992;89(22):10915–9. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoischen A, van Bon BW, Gilissen C, Arts P, van Lier B, Steehouwer M, de Vries P, de Reuver R, Wieskamp N, Mortier G, et al. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat Genet. 2010;42(6):483–5. doi: 10.1038/ng.581. [DOI] [PubMed] [Google Scholar]
- Johnston JJ, Teer JK, Cherukuri PF, Hansen NF, Loftus SK, Chong K, Mullikin JC, Biesecker LG. Massively parallel sequencing of exons on the X chromosome identifies RBM10 as the gene that causes a syndromic form of cleft palate. Am J Hum Genet. 2010;86(5):743–8. doi: 10.1016/j.ajhg.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahrizi K, Hu CH, Garshasbi M, Abedini SS, Ghadami S, Kariminejad R, Ullmann R, Chen W, Ropers HH, Kuss AW, et al. Next generation sequencing in a family with autosomal recessive Kahrizi syndrome (OMIM 612713) reveals a homozygous frameshift mutation in SRD5A3. Eur J Hum Genet. 2011;19(1):115–7. doi: 10.1038/ejhg.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalay E, Yigit G, Aslan Y, Brown KE, Pohl E, Bicknell LS, Kayserili H, Li Y, Tuysuz B, Nurnberg G, et al. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat Genet. 2011;43(1):23–6. doi: 10.1038/ng.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein CJ, Botuyan MV, Wu Y, Ward CJ, Nicholson GA, Hammans S, Hojo K, Yamanishi H, Karpf AR, Wallace DC, et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet. 2011 doi: 10.1038/ng.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawitz PM, Schweiger MR, Rodelsperger C, Marcelis C, Kolsch U, Meisel C, Stephani F, Kinoshita T, Murakami Y, Bauer S, et al. Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome. Nat Genet. 2010;42(10):827–9. doi: 10.1038/ng.653. [DOI] [PubMed] [Google Scholar]
- Lalonde E, Albrecht S, Ha KC, Jacob K, Bolduc N, Polychronakos C, Dechelotte P, Majewski J, Jabado N. Unexpected allelic heterogeneity and spectrum of mutations in Fowler syndrome revealed by next-generation exome sequencing. Hum Mutat. 2010;31(8):918–23. doi: 10.1002/humu.21293. [DOI] [PubMed] [Google Scholar]
- Ledergerber C, Dessimoz C. Base-calling for next-generation sequencing platforms. Brief Bioinform. 2011 doi: 10.1093/bib/bbq077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Li J, Shen H, Zhang L, Papasian CJ, Deng HW. Comparative Studies of de novo Assembly Tools for Next-generation Sequencing Technologies. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, Bainbridge M, Dinh H, Jing C, Wheeler DA, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362(13):1181–91. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, DeWeese-Scott C, Fedorova ND, Geer LY, He S, Hurwitz DI, Jackson JD, Jacobs AR, Lanczycki CJ, et al. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 2003;31(1):383–7. doi: 10.1093/nar/gkg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies EH, Birney E. Approaches to comparative sequence analysis: towards a functional view of vertebrate genomes. Nat Rev Genet. 2008;9(4):303–13. doi: 10.1038/nrg2185. [DOI] [PubMed] [Google Scholar]
- Markello TC, Carlson-Donohoe H, Sincan M, Adams D, Bodine DM, Farrar JE, Vlachos A, Lipton JM, Auerbach AD, Ostrander EA, et al. Sensitive Quantification of Mosaicism Using High Density SNP Arrays and the Cumulative Distribution Function. Molecular Genetics and Metabolism. 2011a doi: 10.1016/j.ymgme.2011.12.015. Accepted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markello TC, Han T, Carlson-Donohoe H, Ahaghotu C, Harper U, Jones M, Chandrasekharappa S, Anikster Y, Adams DR Program NCS and others. Recombination mapping using Boolean logic and high-density SNP genotyping for exome sequence filtering. Molecular Genetics and Metabolism. 2011b doi: 10.1016/j.ymgme.2011.12.014. Accepted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla-Duenas A, Sanchez I, Corral-Juan M, Davalos A, Alvarez R, Latorre P. Cellular and molecular pathways triggering neurodegeneration in the spinocerebellar ataxias. Cerebellum. 2010;9(2):148–66. doi: 10.1007/s12311-009-0144-2. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33(Database issue):D284–8. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, Koren S, Sutton G. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95(6):315–27. doi: 10.1016/j.ygeno.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–74. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010a;42(9):790–3. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010b;42(1):30–5. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikopoulos K, Gilissen C, Hoischen A, van Nouhuys CE, Boonstra FN, Blokland EA, Arts P, Wieskamp N, Strom TM, Ayuso C, et al. Next-generation sequencing of a 40 Mb linkage interval reveals TSPAN12 mutations in patients with familial exudative vitreoretinopathy. Am J Hum Genet. 2010;86(2):240–7. doi: 10.1016/j.ajhg.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Lin X, Salzberg SL. GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res. 2001;29(5):1185–90. doi: 10.1093/nar/29.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson TM, Adams DA, Markello T, Simeonov DR, Golas G, Yang S, Hansen NF, Cherukuri PF, Cruz P, et al. Program NCS. Exome sequencing as a diagnostic tool in a case of undiagnosed juvenile-onset GM1-gangliosidosis. Neurology. 2011 doi: 10.1212/WNL.0b013e31825f047a. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente XS, Quesada V, Osorio FG, Cabanillas R, Cadinanos J, Fraile JM, Ordonez GR, Puente DA, Gutierrez-Fernandez A, Fanjul-Fernandez M, et al. Exome Sequencing and Functional Analysis Identifies BANF1 Mutation as the Cause of a Hereditary Progeroid Syndrome. Am J Hum Genet. 2011;88(5):650–6. doi: 10.1016/j.ajhg.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman AU, Morell RJ, Belyantseva IA, Khan SY, Boger ET, Shahzad M, Ahmed ZM, Riazuddin S, Khan SN, Friedman TB. Targeted capture and next-generation sequencing identifies C9orf75, encoding taperin, as the mutated gene in nonsyndromic deafness DFNB79. Am J Hum Genet. 2010;86(3):378–88. doi: 10.1016/j.ajhg.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios J, Stein E, Shendure J, Hobbs HH, Cohen JC. Identification by whole-genome resequencing of gene defect responsible for severe hypercholesterolemia. Hum Mol Genet. 2010;19(22):4313–8. doi: 10.1093/hmg/ddq352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT, Rowen L, Pant KP, Goodman N, Bamshad M, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328(5978):636–9. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz MC, Delcher AL, Salzberg SL. Assembly of large genomes using second-generation sequencing. Genome Res. 2010;20(9):1165–73. doi: 10.1101/gr.101360.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson MA, Irving MD, Asilmaz E, Gray MJ, Dafou D, Elmslie FV, Mansour S, Holder SE, Brain CE, Burton BK, et al. Mutations in NOTCH2 cause Hajdu-Cheney syndrome, a disorder of severe and progressive bone loss. Nat Genet. 2011;43(4):303–5. doi: 10.1038/ng.779. [DOI] [PubMed] [Google Scholar]
- Sobreira NL, Cirulli ET, Avramopoulos D, Wohler E, Oswald GL, Stevens EL, Ge D, Shianna KV, Smith JP, Maia JM, et al. Whole-genome sequencing of a single proband together with linkage analysis identifies a Mendelian disease gene. PLoS Genet. 2010;6(6):e1000991. doi: 10.1371/journal.pgen.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Eichler EE. Diversity of human copy number variation and multicopy genes. Science. 2010;330(6004):641–6. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerer D, Schracke N, Wu H, Cheng Y, Bau S, Stahler CF, Stahler PF, Beier M. Targeted high throughput sequencing of a cancer-related exome subset by specific sequence capture with a fully automated microarray platform. Genomics. 2010;95(4):241–6. doi: 10.1016/j.ygeno.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Sincan M, Simeonov D, Adams D, Markello TC, Pierson T, Toro C, Gahl WA, Boerkoel CF. VAR-MD: A tool to analyze whole exome/genome variants in small human pedigrees with Mendelian inheritance. Hum Mutat. 2012;33:xxx–yyy. doi: 10.1002/humu.22034. [DOI] [PubMed] [Google Scholar]
- Taylor J, Schenck I, Blankenberg D, Nekrutenko A. Using galaxy to perform large-scale interactive data analyses. Curr Protoc Bioinformatics. 2007;Chapter 10(Unit 10):5. doi: 10.1002/0471250953.bi1005s19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teer JK, Bonnycastle LL, Chines PS, Hansen NF, Aoyama N, Swift AJ, Abaan HO, Albert TJ, Margulies EH, Green ED, et al. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res. 2010;20(10):1420–31. doi: 10.1101/gr.106716.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teer JK, Green ED, Mullikin JC, Biesecker LG. VarSifter: Visualizing and analyzing exome-scale sequence variation data on a desktop computer. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr711. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13(9):2129–41. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter M, Warnich L. In silico promoters: modelling of cis-regulatory context facilitates target predictio. J Cell Mol Med. 2009;13(2):270–8. doi: 10.1111/j.1582-4934.2008.00371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi L, Roversi G, Colombo EA, Leijsten N, Concolino D, Calabria A, Mencarelli MA, Fimiani M, Macciardi F, Pfundt R, et al. Targeted next-generation sequencing appoints c16orf57 as clericuzio-type poikiloderma with neutropenia gene. Am J Hum Genet. 2010;86(1):72–6. doi: 10.1016/j.ajhg.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, Shahin H, Elkan-Miller T, Lee MK, Thornton AM, Roeb W, Abu Rayyan A, Loulus S, Avraham KB, King MC, et al. Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of nonsyndromic hearing loss DFNB82. Am J Hum Genet. 2010;87(1):90–4. doi: 10.1016/j.ajhg.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, Davis S, Stemke-Hale K, Davies MA, Gershenwald JE, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43(5):442–6. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won HH, Kim HJ, Lee KA, Kim JW. Cataloging coding sequence variations in human genome databases. PLoS One. 2008;3(10):e3575. doi: 10.1371/journal.pone.0003575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthey EA, Mayer AN, Syverson GD, Helbling D, Bonacci BB, Decker B, Serpe JM, Dasu T, Tschannen MR, Veith RL, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13(3):255–62. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- Yandell M, Huff C, Hu H, Singleton M, Moore B, Xing J, Jorde LB, Reese MG. A probabilistic disease-gene finder for personal genomes. Genome research. 2011;21(9):1529–42. doi: 10.1101/gr.123158.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.