Abstract
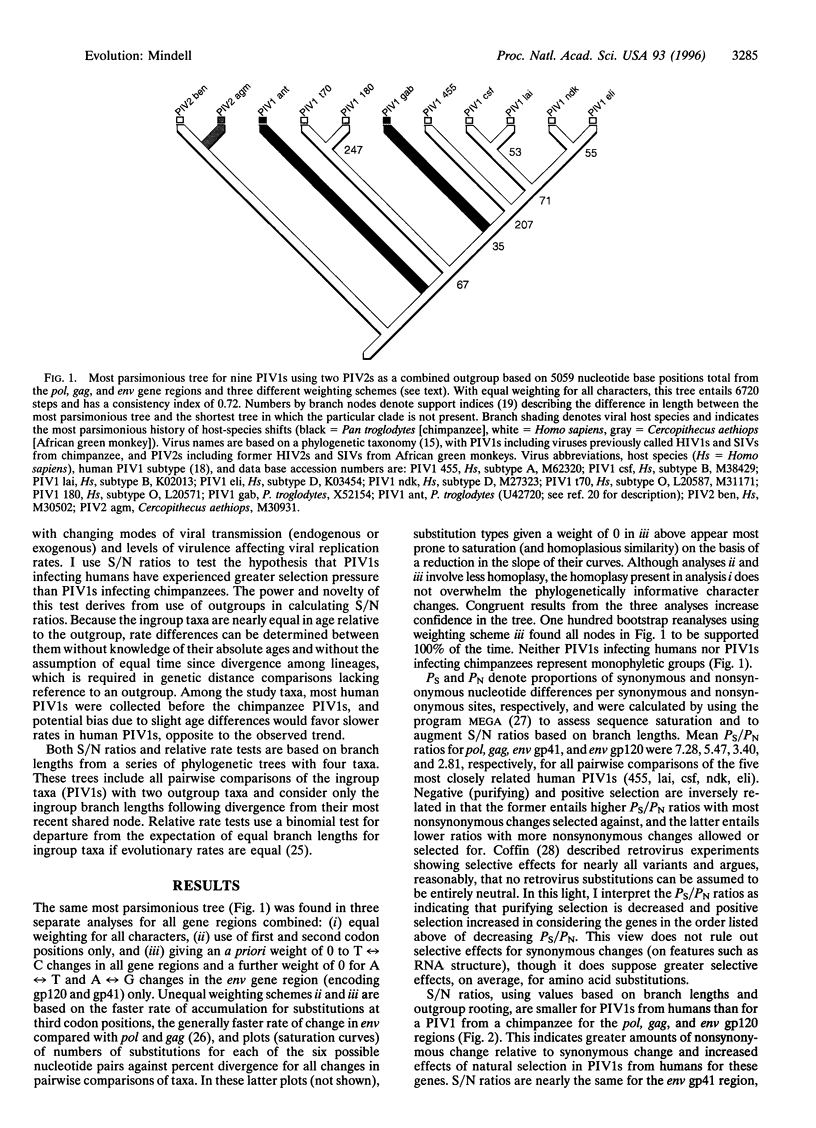
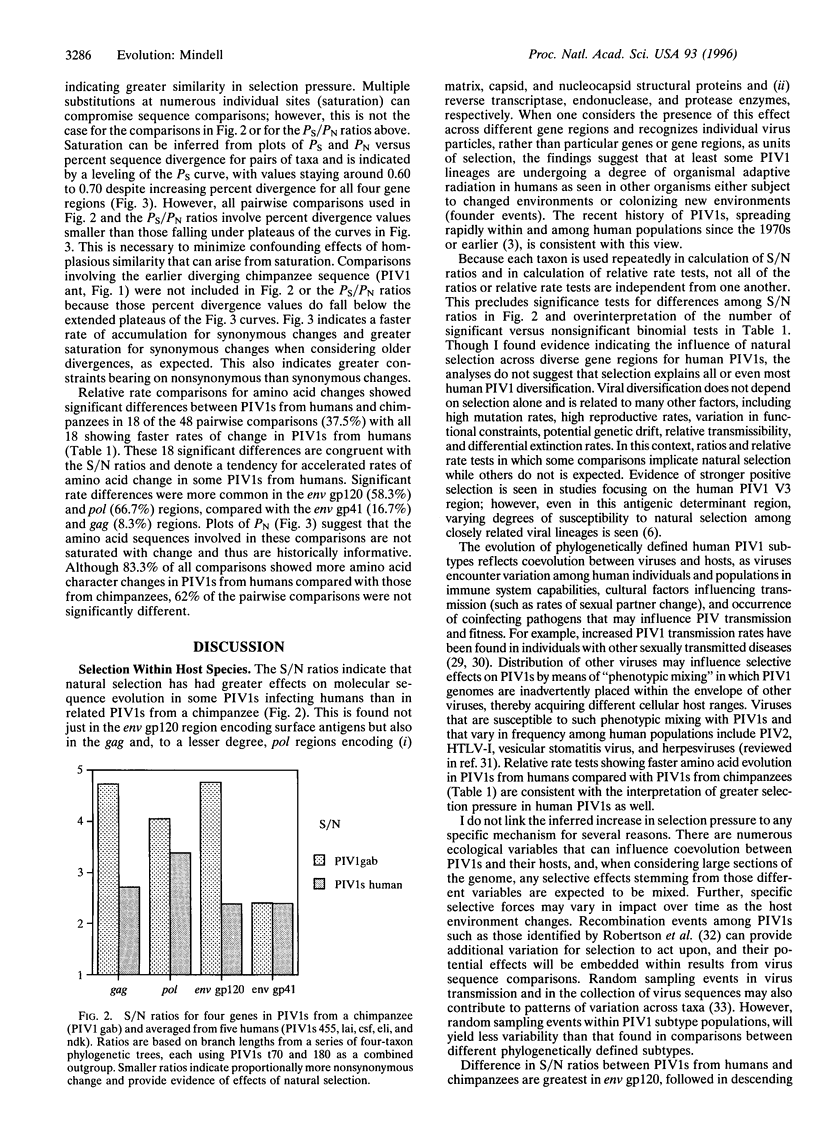
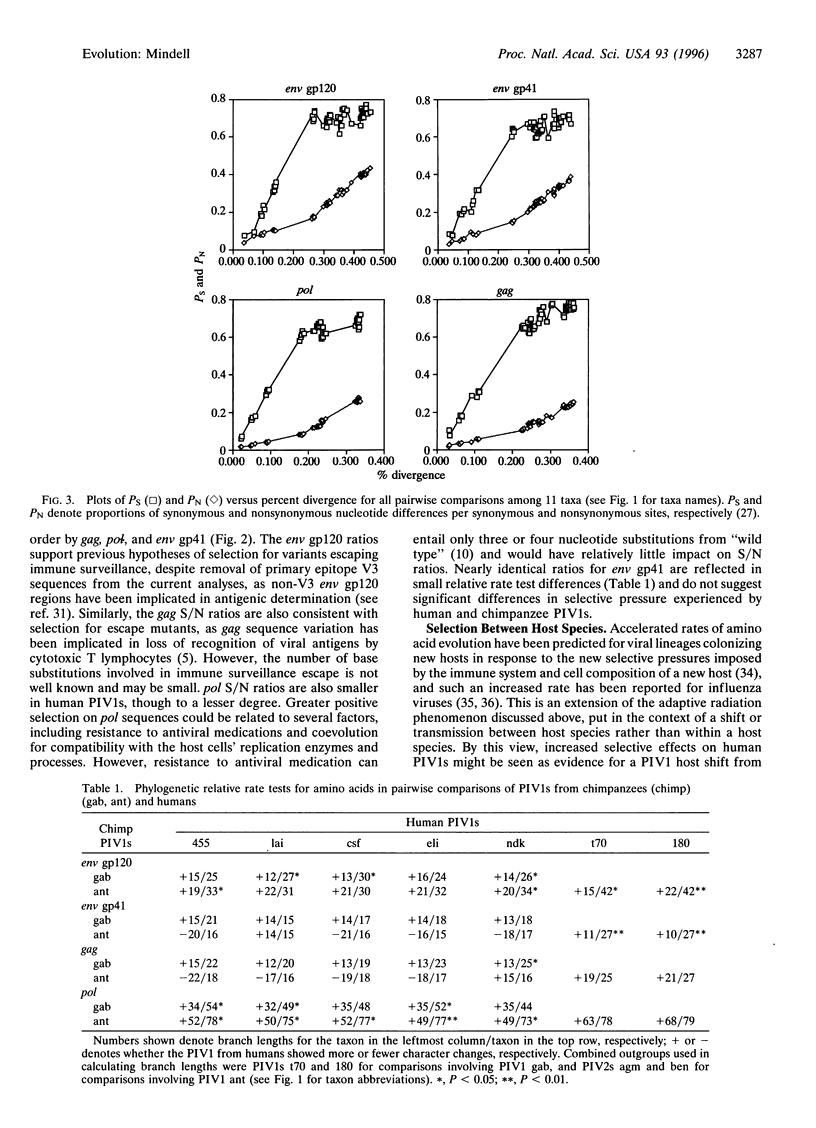
Evolutionary theory predicts the recent spread of primate immunodeficiency viruses (PIVs) to new human populations to be accompanied by positive selection in response to new host environments and/or by random genetic drift. I assess evidence for positive selection in human and chimpanzee PIVs type I (PIV1s), using ratios of synonymous to nonsynonymous nucleotide change based on branch lengths and outgroup rooting. Ratios are smaller for PIV1s from humans than for PIV1 from a chimpanzee for the pol, gag, and env glycoprotein 120 (gp120) regions, indicating greater effects of positive selection in PIV1s from humans. Parsimony-based relative rate tests for amino acid changes showed significant differences between PIV1s from humans and chimpanzees in 18 of 48 pairwise comparisons, with all 18 showing faster rates of change in PIV1s from humans. This study indicates that in some instances, the recent evolution of human PIV1s follows a speciational pattern, in which increased diversification of taxa is correlated with greater amounts of character change appearing and being maintained through time. This extends the generality of the speciational pattern to a group of organisms (viruses) having the fastest known rates of anagenetic change for nucleotide characters and indicates that comprehensive understanding of PIV1 evolution requires consideration of both anagenetic change within viral lineages and the relative historical success of different viral clades. Phylogenetic analyses show that neither PIV1s infecting humans nor those infecting chimpanzees represent monophyletic groups and suggest multiple host-species shifts for PIV1s.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boucher C. A., O'Sullivan E., Mulder J. W., Ramautarsing C., Kellam P., Darby G., Lange J. M., Goudsmit J., Larder B. A. Ordered appearance of zidovudine resistance mutations during treatment of 18 human immunodeficiency virus-positive subjects. J Infect Dis. 1992 Jan;165(1):105–110. doi: 10.1093/infdis/165.1.105. [DOI] [PubMed] [Google Scholar]
- Burns D. P., Desrosiers R. C. Selection of genetic variants of simian immunodeficiency virus in persistently infected rhesus monkeys. J Virol. 1991 Apr;65(4):1843–1854. doi: 10.1128/jvi.65.4.1843-1854.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichutek K., Norley S., Linde R., Kreuz W., Gahr M., Löwer J., von Wangenheim G., Kurth R. Lack of HIV-1 V3 region sequence diversity in two haemophiliac patients infected with a putative biologic clone of HIV-1. AIDS. 1991 Oct;5(10):1185–1187. doi: 10.1097/00002030-199110000-00005. [DOI] [PubMed] [Google Scholar]
- Coffin J. M. Genetic diversity and evolution of retroviruses. Curr Top Microbiol Immunol. 1992;176:143–164. doi: 10.1007/978-3-642-77011-1_10. [DOI] [PubMed] [Google Scholar]
- Eernisse D. J. DNA Translator and Aligner: HyperCard utilities to aid phylogenetic analysis of molecules. Comput Appl Biosci. 1992 Apr;8(2):177–184. doi: 10.1093/bioinformatics/8.2.177. [DOI] [PubMed] [Google Scholar]
- Gammelin M., Altmüller A., Reinhardt U., Mandler J., Harley V. R., Hudson P. J., Fitch W. M., Scholtissek C. Phylogenetic analysis of nucleoproteins suggests that human influenza A viruses emerged from a 19th-century avian ancestor. Mol Biol Evol. 1990 Mar;7(2):194–200. doi: 10.1093/oxfordjournals.molbev.a040594. [DOI] [PubMed] [Google Scholar]
- Gorman O. T., Bean W. J., Kawaoka Y., Donatelli I., Guo Y. J., Webster R. G. Evolution of influenza A virus nucleoprotein genes: implications for the origins of H1N1 human and classical swine viruses. J Virol. 1991 Jul;65(7):3704–3714. doi: 10.1128/jvi.65.7.3704-3714.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., Eldredge N. Punctuated equilibrium comes of age. Nature. 1993 Nov 18;366(6452):223–227. doi: 10.1038/366223a0. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Bleasby A. J., Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992 Apr;8(2):189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Holmes E. C., Zhang L. Q., Robertson P., Cleland A., Harvey E., Simmonds P., Leigh Brown A. J. The molecular epidemiology of human immunodeficiency virus type 1 in Edinburgh. J Infect Dis. 1995 Jan;171(1):45–53. doi: 10.1093/infdis/171.1.45. [DOI] [PubMed] [Google Scholar]
- Holmes E. C., Zhang L. Q., Simmonds P., Ludlam C. A., Brown A. J. Convergent and divergent sequence evolution in the surface envelope glycoprotein of human immunodeficiency virus type 1 within a single infected patient. Proc Natl Acad Sci U S A. 1992 Jun 1;89(11):4835–4839. doi: 10.1073/pnas.89.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A. L., Nei M. Nucleotide substitution at major histocompatibility complex class II loci: evidence for overdominant selection. Proc Natl Acad Sci U S A. 1989 Feb;86(3):958–962. doi: 10.1073/pnas.86.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter D. J., Maggwa B. N., Mati J. K., Tukei P. M., Mbugua S. Sexual behavior, sexually transmitted diseases, male circumcision and risk of HIV infection among women in Nairobi, Kenya. AIDS. 1994 Jan;8(1):93–99. doi: 10.1097/00002030-199401000-00014. [DOI] [PubMed] [Google Scholar]
- Kawaoka Y., Webster R. G. Molecular mechanism of acquisition of virulence in influenza virus in nature. Microb Pathog. 1988 Nov;5(5):311–318. doi: 10.1016/0882-4010(88)90032-0. [DOI] [PubMed] [Google Scholar]
- Lenski R. E., Travisano M. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc Natl Acad Sci U S A. 1994 Jul 19;91(15):6808–6814. doi: 10.1073/pnas.91.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louwagie J., McCutchan F. E., Peeters M., Brennan T. P., Sanders-Buell E., Eddy G. A., van der Groen G., Fransen K., Gershy-Damet G. M., Deleys R. Phylogenetic analysis of gag genes from 70 international HIV-1 isolates provides evidence for multiple genotypes. AIDS. 1993 Jun;7(6):769–780. doi: 10.1097/00002030-199306000-00003. [DOI] [PubMed] [Google Scholar]
- Louwagie J., McCutchan F., Van der Groen G., Peeters M., Fransen K., Piot P., Gershy-Damet G. M., Roelants G., Van Heuverswyn H., Eddy G. Genetic comparison of HIV-1 isolates from Africa, Europe, and North America. AIDS Res Hum Retroviruses. 1992 Aug;8(8):1467–1469. doi: 10.1089/aid.1992.8.1467. [DOI] [PubMed] [Google Scholar]
- McClure M. A., Johnson M. S., Feng D. F., Doolittle R. F. Sequence comparisons of retroviral proteins: relative rates of change and general phylogeny. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2469–2473. doi: 10.1073/pnas.85.8.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers G. Tenth anniversary perspectives on AIDS. HIV: between past and future. AIDS Res Hum Retroviruses. 1994 Nov;10(11):1317–1324. doi: 10.1089/aid.1994.10.1317. [DOI] [PubMed] [Google Scholar]
- Peeters M., Fransen K., Delaporte E., Van den Haesevelde M., Gershy-Damet G. M., Kestens L., van der Groen G., Piot P. Isolation and characterization of a new chimpanzee lentivirus (simian immunodeficiency virus isolate cpz-ant) from a wild-captured chimpanzee. AIDS. 1992 May;6(5):447–451. doi: 10.1097/00002030-199205000-00002. [DOI] [PubMed] [Google Scholar]
- Phillips R. E., Rowland-Jones S., Nixon D. F., Gotch F. M., Edwards J. P., Ogunlesi A. O., Elvin J. G., Rothbard J. A., Bangham C. R., Rizza C. R. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature. 1991 Dec 12;354(6353):453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- Plummer F. A., Simonsen J. N., Cameron D. W., Ndinya-Achola J. O., Kreiss J. K., Gakinya M. N., Waiyaki P., Cheang M., Piot P., Ronald A. R. Cofactors in male-female sexual transmission of human immunodeficiency virus type 1. J Infect Dis. 1991 Feb;163(2):233–239. doi: 10.1093/infdis/163.2.233. [DOI] [PubMed] [Google Scholar]
- Robertson D. L., Sharp P. M., McCutchan F. E., Hahn B. H. Recombination in HIV-1. Nature. 1995 Mar 9;374(6518):124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- Seibert S. A., Howell C. Y., Hughes M. K., Hughes A. L. Natural selection on the gag, pol, and env genes of human immunodeficiency virus 1 (HIV-1). Mol Biol Evol. 1995 Sep;12(5):803–813. doi: 10.1093/oxfordjournals.molbev.a040257. [DOI] [PubMed] [Google Scholar]
- Shpaer E. G., Mullins J. I. Rates of amino acid change in the envelope protein correlate with pathogenicity of primate lentiviruses. J Mol Evol. 1993 Jul;37(1):57–65. doi: 10.1007/BF00170462. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Balfe P., Ludlam C. A., Bishop J. O., Brown A. J. Analysis of sequence diversity in hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J Virol. 1990 Dec;64(12):5840–5850. doi: 10.1128/jvi.64.12.5840-5850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer D. A., Holland J. J. Rapid evolution of RNA viruses. Annu Rev Microbiol. 1987;41:409–433. doi: 10.1146/annurev.mi.41.100187.002205. [DOI] [PubMed] [Google Scholar]



