Abstract
A familial form of Amyotrophic lateral sclerosis (ALS8) is caused by a point mutation (P56S) in the vesicle-associated membrane protein associated protein B (VapB). Human VapB and Drosophila Vap-33-1 (Vap) are homologous type II transmembrane proteins that are localized to the ER. However, the precise consequences of the defects associated with the P56S mutation in the endoplasmic reticulum (ER) and its role in the pathology of ALS are not well understood. Here we show that Vap is required for ER protein quality control (ERQC). Loss of Vap in flies shows various ERQC associated defects, including protein accumulation, ER expansion, and ER stress. We also show that wild type Vap, but not the ALS8 mutant Vap, interacts with a lipid-binding protein, Oxysterol binding protein (Osbp), and that Vap is required for the proper localization of Osbp to the ER. Restoring the expression of Osbp in the ER suppresses the defects associated with loss of Vap and the ALS8 mutant Vap. Hence, we propose that the ALS8 mutation impairs the interaction of Vap with Osbp, resulting in hypomorphic defects that might contribute to the pathology of ALS8.
INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by preferential loss of motor neurons. Approximately 90% of all ALS cases occur sporadically, whereas the remaining 10% are inherited (1,2). Although mutations in almost 20 genes have now been shown to cause ALS (3), their proposed functions appear quite divergent, lacking any obvious link that would hint towards a specific molecular pathway (4).
ALS8 is an autosomal dominant form of ALS caused by a point mutation (P56S) in the gene encoding the VapB protein (5). Human vapB is evolutionarily conserved, with homologs in numerous species (6), including Drosophila Vap-33-1 or vap. Vaps contain an amino (N)-terminal domain, called the major sperm protein (MSP) domain (7,8) and a transmembrane domain that anchors the protein in the ER (9–11). Studies with Drosophila and Caenorhabditis elegans demonstrate that Vap has non-autonomous functions. Indeed, the MSP domain of Vap is cleaved and secreted from neurons (12,13). The cleaved MSP acts as a ligand for growth cone guidance receptors expressed on muscle surfaces and affects mitochondrial dynamics in the muscles. However, Vaps also have autonomous functions as they are ER associated proteins. They have been shown to function in glucose transport (14), neurite extension (15), the development of the neuromuscular junctions (16) and ER-to-Golgi protein trafficking (17). Importantly, Vaps have been implicated in the regulation of phospholipid biosynthetic proteins (18,19). Vaps interact with proteins containing two phenylalanines in an acidic tract (FFAT)-motif (20), which include lipid-binding proteins like Oxysterol binding protein (Osbp) (21) and ceramide transfer protein (Cert) (22). Studies with cultured cells indicate that the Vap/Osbp interaction is required for sphingomyelin (SM) biosynthesis in response to 25-hydroxycholesterol (18,23,24). Hence, Vap seems to be required for Osbp function in the ER or in ER–Golgi membrane contact sites (18,17).
The ER is the site where newly synthesized proteins are folded and modified. Protein folding in the ER is monitored by a stringent ER quality control (ERQC) system that only permits properly folded proteins to traffic to the Golgi (25–27). The accumulation of misfolded proteins in the ER, caused by alterations in ER homeostasis, initiates ER stress and attempts to resolve the protein-folding defects (28,29). Interestingly, ER stress has been observed in human sporadic ALS patients (30) and in SOD1 transgenic mice (31,32). Overexpression of the ALS8 mutant Vap has been shown to cause ER stress in flies (12) and mice (33). However, the role of Vap in ER stress and ER biology remains to be determined.
To determine the function of Vap, we characterized the loss of function phenotype associated with the loss of Vap in null mutant and transgenic flies expressing the ALS8 mutant Vap at physiological levels in the appropriate tissues. We found that the ALS8 mutation causes the Vap protein to be less active and we show that Vap is required for ER protein homeostasis. Loss of Vap causes defects in ERQC, resulting in protein accumulation and ER stress. Loss of the Vap protein also causes Osbp to be mislocalized from ER to Golgi, and restoring expression of Osbp in the ER partially suppresses the defects in vap null and ALS8 transgenic flies. We propose that loss of Vap contributes to ER stress and that this stress might play a role in the pathology of ALS.
RESULTS
Vap is required for ER proteostasis
Vap is localized to the ER and overexpression of the ALS8 mutant isoform causes ER stress in flies (12). However, the precise role of wild type (WT) of Vap (VapWT) in the ER remains to be determined. We therefore examined whether Vap is required for ER proteostasis. The ER is integral to maintaining protein homeostasis (proteostasis), as protein-folding of transmembrane and secreted proteins occurs under the supervision of ERQC (34). The ERQC is able to identify misfolded proteins, retrotranslocate the misfolded proteins and promote their degradation. ERQC overload induces ER stress, which restores proteostasis by halting protein translation, and by activating signaling pathways that lead to an increased production of molecular chaperones, that facilitate protein folding (35). The ERQC is especially important for membrane proteins, which are prone to aggregation due to their inherent tendency to assemble in oligomers (36).
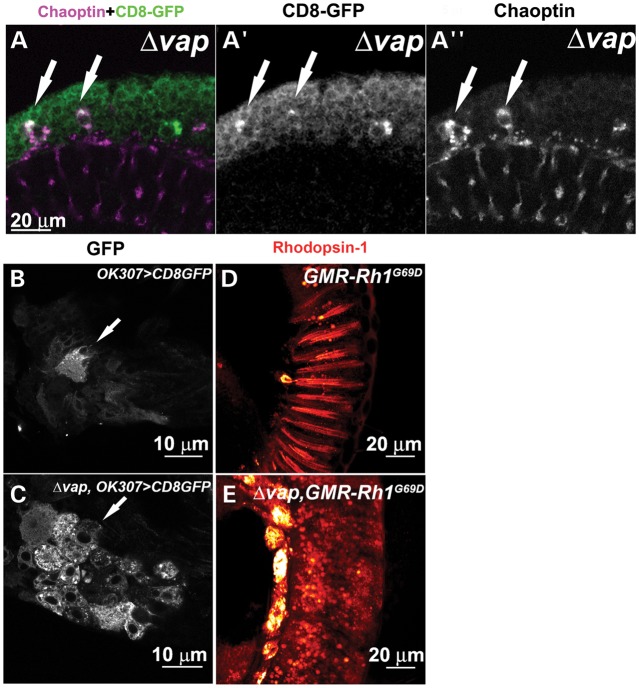
To determine whether the Vap functions in ER proteostasis, we examined for membrane protein accumulation in the neurons of the vap null mutant (16). We first stained adult brains of control and vap null mutant flies with an antibody for Chaoptin, a membrane protein that is expressed in photoreceptor cells (37). Some neurons of the adult cortex of control flies express low levels of Chaoptin, a feature that has not been previously reported (Fig. 1A, arrows). However, in cortical neurons of vap mutants, Chaoptin accumulates in punctae in the cytoplasm of the cell body (Fig. 1B, B′ and B″, arrows). In addition, we find that other membrane proteins such as Robo-1 (38) and N-cadherin (39) also accumulate in the cytoplasm of neurons of vap null mutants (Fig. 1D and data not shown), but not in control flies (Fig. 1C). These accumulated proteins are very closely associated with the ER membrane, as shown in neurons of the adult brain that are co-stained with anti-Chaoptin and anti-Boca antibody, an ER marker (40) (Fig. 1E, E′ and E″ and Supplementary movie 1). Interestingly, Chaoptin accumulates in only 64.6 ± 8.4 neurons per adult brain of vap null mutants (N = 3), and hence affect a small subpopulation of neurons (Fig. 1). To test whether a specific neuronal population is affected in the vap mutants we expressed a membrane anchored GFP (CD8-GFP) protein (41) driven by a neuronal GAL4 driver (OK307-GAL4) (42) and stained adult cortical neurons with anti-Chaoptin and anti-GFP antibodies (Fig. 2A, A′ and A″). Although CD8-GFP is expressed in all cortical neurons (Fig. 2A and A′), only a selective subset of neurons accumulate both Chaoptin and CD8-GFP (Fig. 2A, A′ and A″, arrows). Moreover, CD8-GFP also accumulates in adult motor neurons of vap mutants (Fig. 2C) whereas CD8-GFP is barely expressed in the control (Fig. 2B), suggesting that the adult motor neurons are affected in the vap null. Furthermore, although CD8-GFP accumulates in the cytoplasm of vap null adult flies, we did not observe this in mutant larvae (data not shown), indicating that protein accumulations are stage- and possibly age-dependent and that Vap might be required for ER proteostasis.
Figure 1.
Loss of Vap causes accumulation of membrane proteins in the adult neurons. (A and B) Neurons in the brain of 1-day-old adult flies stained with anti-Chaoptin and anti-Elav antibodies. In control flies, Chaoptin labels individual synapses in the lamina (dots) as previously published (102). However, it is also expressed in few cortical neurons of the brain at low levels (A, arrows). In vap null mutant brains, Chaoptin accumulates in the cytoplasm of the neurons of the cortex (B, B’ and B", arrows). A precise excision of P{Mae-UAS.6.11} in vap was used as a control in A-A” (see Materials and Methods). (C and D) Staining of cortical neurons in the adult brain of 1-day-old flies with anti-Robo-1 and anti-Elav. In vap null mutants, Robo-1 accumulates in the cytoplasm of a subset of neurons (arrows, D). (E) Staining of affected neurons in the adult brain of the vap null mutant. Accumulated Chaoptin protein is very closely associated with Boca, an ER marker (indicated by * in E″).
Figure 2.
Loss of Vap causes accumulation of membrane proteins in the ER of neurons. (A) Staining of cortical neurons in the adult brain of 1-day-old flies expressing the membrane anchored GFP (CD8-GFP) with anti-Chaoptin and anti-GFP antibody. In the vap null mutant neurons, some neurons accumulate both CD8-GFP (arrows, A′) and Chaoptin (arrows, A″). (B and C) Motor neurons in the second thoracic segment of 1-day-old adult flies. CD8-GFP, a membrane protein, is expressed in control flies and the vap null mutant flies with OK307-GAL4 line, a driver with expression in motor neurons (42). CD8-GFP accumulates in the vap null mutant (C, arrow), but not in the WT flies (B, arrow). (D and E) Retina of 1-day-old adult flies stained with anti-Rhodopsin 1. The mutant Rhodopsin (Rh1G69D), a substrate of ERQC, is expressed in control and the vap null mutant eyes with GMR promoter. Rh1G69D mislocalizes and accumulates in punctae of the vap null mutant (E), but not in the control flies (D).
A mutation in Rhodopsin-1(Rh-1G69D) has been characterized as an ERQC substrate (43). To determine whether Vap is required for ER proteostasis, we expressed the mutant Rh-1 in the eye of control and vap null mutants using the GMR-GAL4 driver (44) and stained eyes with Rh-1 antibody (45). As shown in Figure 2D and E, mutant Rh-1 aberrantly accumulates in punctae in the eyes of vap null mutants and the levels of Rh-1 are upregulated in the vap mutants when compared with control flies, suggesting defects in ER proteostasis in the vap null mutants.
Loss of Vap causes ER stress
Defects in ER proteostasis cause ER stress (28,29). To determine the ultrastructural features associated with the loss of Vap, we carried out transmission electron microscopy (TEM) of adult brains of WT and vap null mutant flies. These images revealed large intracellular vacuoles in a subset of neurons, often near the nuclei (Fig. 3A and B, 100% of the brains exhibit vacuoles in a small percentage of the neurons in three vap null mutants versus 0% in three brains of control flies). These vacuoles are surrounded by a single membrane and contain non-homogenous materials (Fig. 3B, asterisk). In addition, the vacuoles are contiguous with the nuclear membrane (Fig. 3C, arrows) and decorated with electron-dense ribosomes (Fig. 3D, arrow heads), indicating that they correspond to a vastly expanded ER. These ER expansions are typically observed in the presence of ER stress (46), which induces ire-1-mediated unconventional splicing of the Drosophila xbp1 mRNA (47). Upon expression of the ER stress sensor Xbp1-GFP (43,48) in control and vap mutant flies, we find that GFP is significantly upregulated in a subpopulation of mutant neurons (Fig. 4B and D), but not in control flies (Fig. 4A and C). Similarly, Bip, a chaperone and ER stress indicator (48), is upregulated in mutant brains relative to control flies (Fig. 4E and F), further suggesting that loss of Vap causes ER stress.
Figure 3.

Loss of Vap causes an aberrant ER expansion. (A–D) TEM analysis of neurons of control (A) and of vap null mutants (B–D). Large cytoplasmic vacuoles in the neurons of the vap null mutant (asterisk (*), B). The vacuole is contiguous with the nuclear membrane (arrow heads, C). The vacuole is also decorated with electron-dense ribosomes (arrows, D), suggesting that the vacuoles are aberrantly expanded ER. N: Nucleus. A precise excision line was used as a control.
Figure 4.
Loss of Vap causes ER stress. (A–D) An ER stress reporter Xbp1-GFP is significantly upregulated in the neurons of the 1-day-old vap null mutant adult brain (B and D), but not in the control flies (A and C). A precise excision line was used as a control. (E and F) Neurons of 1-day-old adult fly brains stained with anti-Bip, an ER stress marker. Bip is upregulated in the neurons of the vap null mutant (arrows in F). (G and H) Staining of the neurons in the cortex of the adult brain of control (G) and the vap null mutant (H) with Ubiquitin antibody. Ubiquitinated proteins accumulate in the cytoplasm of cortical neurons in adult vap null mutant (H and H′), but not in control adults (G and G′). (I) MARCM analysis of adult brains of y w, hsFLP, tub-GAL80, FRT19A/Δ vap, FRT19A; Act-GAL4, UAS-CD8:GFP flies. Ubiquitinated proteins accumulate only in GFP positive vap null mutant clones (I, arrows in I″), suggesting that accumulation of ubiquitinated proteins is associated with cellular autonomous defects. A membrane protein, CD8-GFP accumulates in the cytoplasm of vap null mutant clones (arrows in I′). Staining with GFP (I′), Ubiquitin (I″) and Elav (I‴) antibodies.
In summary, our data suggest that loss of Vap causes defects in ER proteostasis, resulting in accumulation of membrane proteins and ER stress in a subpopulation of neurons in the fruitfly brain.
Loss of Vap causes accumulation of ubiquitinated proteins
Continuous ER stress causes an impairment of the proteasome system and leads to accumulation of ubiquitinated proteins (49). Moreover, ubiquitin immunoreactive inclusions in lower motor neurons represent a characteristic pathological feature of ALS (50). We therefore assessed the presence of ubiquitinated proteins in adult brains using an ubiquitin antibody. We find that ubiquitinated proteins accumulate in subpopulations of cortical neurons of adult mutant flies (Fig. 4H and H′), but not in those of adult control flies (Fig. 4G and G′). We did not observe accumulations of ubiquitinated proteins in mutant larvae (data not shown). Hence, our data suggest that loss of Vap leads to accumulation of ubiquitinated proteins in a stage/age-dependent manner. As shown previously (12), Vap is cleaved and acts as a ligand for axon guidance receptors expressed in muscles. To determine whether the ERQC phenotypes are cell-autonomous or non-autonomous, we performed clonal analysis (51) and find that only vap null clones exhibit an accumulation of ubiquitinated proteins (average 2.3 Ubiquitin positive cells per 27.6 GFP positive cells per brain, N = 3 brains) (Fig. 4I, I′, and I″), suggesting that accumulation of ubiquitinated proteins is a cell-autonomous defect in vap mutant neurons. Hence, Vap has at least two independent functions: a cell-autonomous function in ER proteostasis and a non-cell-autonomous function as a secreted factor.
Vap is required for the proper localization of Osbp
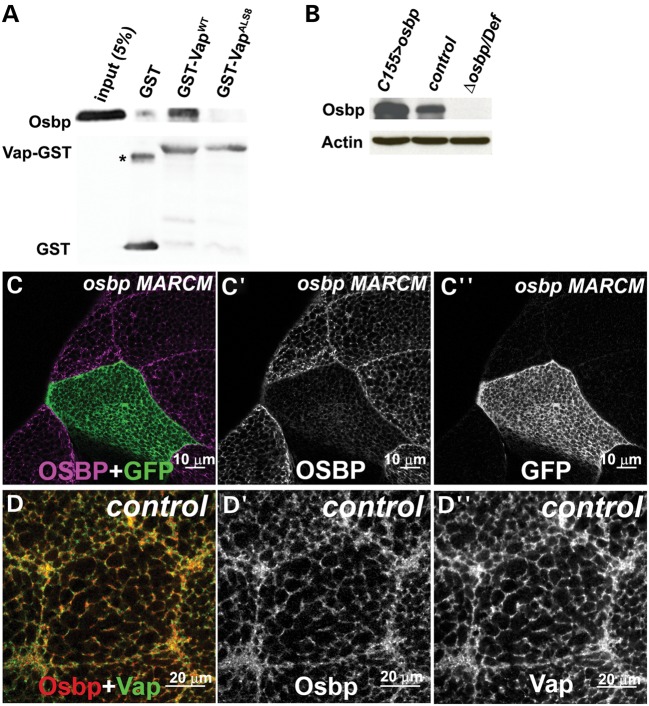
Neurons in which the ALS8 protein is overexpressed also exhibit ER stress, similar to what is observed in the vap null mutant ((12) and Fig. 4). We therefore examined if the ALS8 mutation causes loss of interactions with proteins that are required for its normal function. We performed a two-hybrid screen using a human adult brain cDNA library to identify proteins that are able to bind to the WT human VapB protein, but are unable to interact with mutant ALS8 human VapB protein (52). Among the positives we found two Osbp-related proteins: ORP3 (53–55) and OsbpL6 (56). Both interact with WT VapB, but are unable to interact with ALS8 VapB, as reported previously (17). The Drosophila Osbp homolog encoded by CG6708 contains a Vap binding site or FFAT motif (57,58). As shown in Figure 5A, Drosophila WT Vap, but not the ALS8 mutant version, can interact with Osbp in GST pull down assays, indicating that the interaction between Vap and Osbp is direct and evolutionarily conserved.
Figure 5.
Osbp colocalizes with Vap in the ER. (A) Vap and Osbp interaction in a GST pull-down assay. GST-VapWT binds Osbp, but GST-VapALS8 binding is severely reduced. GST-VapWT and GST-VapALS8 were expressed in bacteria and purified with glutathione–sepharose beads. Osbp protein was overexpressed and extracted from flies. The GST-Vap proteins linked to beads were then incubated with the lysate of these flies. The precipitate and input were analyzed with immunoblots using anti-Osbp antibody and anti-GST antibody. A GST only control was used to test specificity in binding of Vap GST and Osbp proteins. *: GST dimer. (B) Immunoblot analysis of proteins extracted from flies overexpressing Osbp, control (WT) and Δosbp over deficiency Df(3R)ED622, Δosbp/Df flies. Note the lack of Osbp protein expression in Δosbp/Df, showing the specificity of the Osbp antibody (GP89). (C) MARCM analysis of y w, hsFLP, tub-GAL4, UAS-GFP; FRT 82B, Δosbp/FRT 82B, y+, tub-GAL80 flies. Only GFP negative (WT) clones (C″) show Osbp expression (C′), suggesting the specificity of the guinea pig anti-Osbp (GP89) antibody. A portion of the salivary gland stained with anti-Osbp antibody (GP89). GFP marks the osbp null mutant cells (C and C″). (D) Immunostaining of a WT salivary gland cell with anti-Osbp (GP89) and anti-Vap antibody (Rb92). Osbp colocalizes with Vap in the ER.
The Osbp protein family is an evolutionarily conserved protein family whose function was initially linked to non-vesicular intracellular transport of sterols in yeast (59–61). However, several studies suggest that Osbps integrate sterol and SM metabolism (61–63), and control microtubule-dependent motility of endosomes/lysosomes, and exocytosis (60). Studies with cultured cells indicate that the Vap/Osbp interaction is required for SM biosynthesis in response to 25-hydroxycholesterol (18,23,24). Vap seems to be required for Osbp function in SM biosynthesis at ER–Golgi membrane contact sites (18). However, how Osbp is required for Vap function in the ER remains to be determined.
To determine the cellular distribution of Osbp, we generated antisera against Drosophila Osbp. To confirm the specificity of this antibody, we created null alleles of osbp (see Materials and Methods) and confirmed the deletion of the osbp locus with Southern blotting (data not shown), and the loss of Osbp protein expression with western blotting (Fig. 5B). Mosaic analyses (64) (Fig. 5C) show that mutant osbp clones marked by GFP lack Osbp, showing that the antibody is specific. To examine the cellular distribution of Osbp and Vap, we co-stained salivary glands with anti-Osbp and anti-Vap antibody. As shown in Figure 5D, Osbp colocalizes with Vap in the ER and the cell membrane.
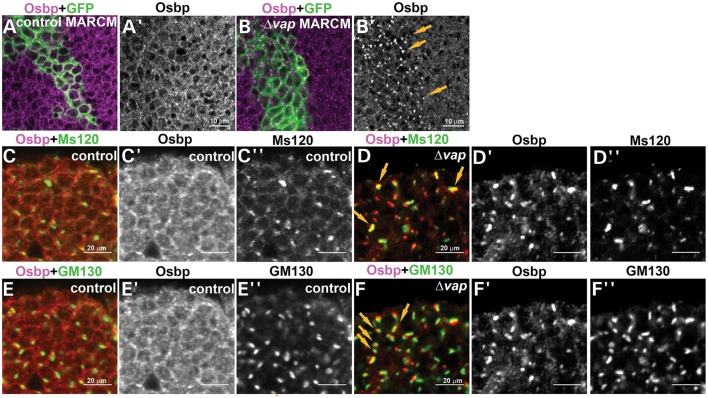
To determine whether Vap is required for the proper localization and function of Osbp, we examined the localization of Osbp in vap null mutant clones. We created vap mutant clones and labeled them with Osbp antibody. As shown in Figure 6, loss of vap (marked by GFP) leads to an aberrant distribution of Osbp and an accumulation of Osbp in punctae (arrows in Fig. 6B′). To determine the intracellular distribution of Osbp in vap null mutants, we stained neurons in control flies and vap null mutant flies with Osbp, Ms120 (medial-Golgi marker) (65) and GM130 (cis-Golgi marker) (66). In Drosophila, Golgi cisternae are stacked but not connected to form a ribbon, even during interphase (67). Ms120 protein and GM130 are always closely associated, but do not overlap (67). We found that Osbp is recruited to or is very closely associated with the Ms120 or GM130 positive punctae in vap null mutants (Fig. 6D and F, arrows), but not in control flies (Fig. 6C and E). We conclude that in the absence of vap, Osbp is mislocalized from ER to Golgi.
Figure 6.
Osbp is mislocalized to the Golgi in vap null mutant cells. (A and B) MARCM analysis shows that Osbp accumulates in cytoplasmic punctae of vap null mutant cells (B and arrows in B′) but not in WT cells (A and A′). (C and F) Osbp accumulates in the Golgi of vap null mutant cells. Immunostaining of the 1-day-old adult brain of WT (control) (C and E) and vap null mutant neurons (D and F) with anti-Osbp (C′–F′), anti-Ms120 (C″ and D″) and anti-GM130 (E″ and F″) antibodies, Golgi markers. Osbp barely colocalizes with the Golgi markers, Ms120 KDa protein (C–C″) and GM130 (E–E″) in the WT control, whereas Osbp colocalizes or is closely associated with Ms120 KDa protein (arrows, D–D′) and GM130 (arrows, F–F″) in the vap null mutant neurons.
ALS8 mutation causes a partial loss of function of Vap
We previously showed that overexpression of ALS8 mutant Vap (VapALS8) in flies affects the secretion of the MSP amino terminal domain of VapALS8, causing a loss of function of Vap. Furthermore, expression of VapALS8 causes an ER stress when overexpressed in neurons. However, overexpression of VapALS8 also rescues the lethality associated with the loss of Vap. The data suggest that VapALS8 is a partial loss of function mutation, and that overexpression may mask or create some of the associated cell biological defects (12) (68). To obtain the proper tissue specificity and express VapWT and VapALS8 proteins at physiological levels, we created transgenic flies carrying a genomic fragment of vap using P-element mediated transformation with and without a site-specific attB docking sites, to attenuate positional effects of transgenes (69). These constructs were transformed using classical P-element mediated transformation or phi-C31 mediated transformation into the VK31 and VK33 docking sites (70). We confirmed with western blots that both the VapWT and VapALS8 proteins encoded by these transgenes are expressed at levels that are comparable with endogenous Vap (Supplementary Material, Fig. S1A).
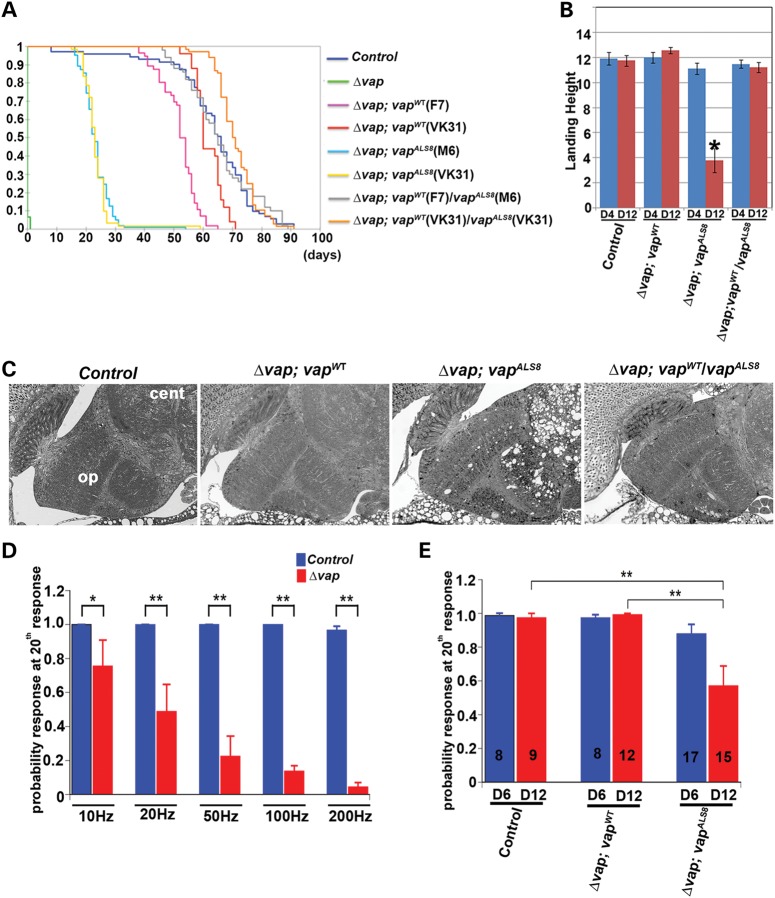
We first tested if the genomic vapWT transgene can rescue the lethality associated with vap null mutants (Δvap). Loss of vap causes pupal or pharate adult lethality with occasional adult escapers (Fig. 7A). All tested genomic vapWT transgenes (four lines) rescue the lethality associated with loss of vap and significantly restore the lifespan of the vap null flies (Fig. 7A and Supplementary Material, Fig. S1B; P < 0.001). Intriguingly, 8 out of 11 P-element vapALS8 transgenic lines also rescued the lethality, suggesting that the VapALS8 protein retains some WT protein function (Fig. 7A, Supplementary Material, Fig. S1B and data not shown). However, all vapALS8 rescued flies exhibit a severely reduced lifespan when compared with vapWT rescued flies (Fig. 7A and Supplementary Material, Fig. S1B, compare Δvap; vapWT (F7), average life span is 52 days (N = 56) versus Δvap; vapALS8 (M6), 23 days (N = 102; P < 0.001). Furthermore, a single copy WT vap (Δvap; vapWT/vapALS8) compensates for the defects associated with vapALS8 (compare the lifespan of Δvap; vapALS8 (M6), 23 days and Δvap ;vapWT(F7) /vapALS8 (M6), 65 days (N = 50); P < 0.001), suggesting that the ALS8 mutation only causes a partial loss-of-function.
Figure 7.
ALS8 mutation causes a partial loss of function of VAP and progressive defects in adult fly brain. (A) Longevity of adult flies. A Kaplan–Meier curve was used to show the probability of members of each genotype to exceed a certain lifespan. The vapALS8 transgene can rescue the lethality associated with loss of vap, but vapALS8 is not as active as the vapWT transgene. A copy of vapWT can compensate for the defects associated with vapALS8. Genotypes shown: Δvap (vap null mutant), Δvap; vapWT (vap null mutant carrying genomic vapWT, line F7 and line VK31), Δvap; vapALS8 (vap null mutant carrying genomic vapALS8, line M6 and line VK31), Δvap; vapWT/ vap ALS8 (vap null mutant carrying genomic vapWT and vapALS8). Line F7 and M6 are P-element mediated transgenes. VK31 lines are site-specific integrated transgenes. A one-way independent groups ANOVA and post-hoc Tukey HSD tests were used to show a significant difference in lifespan between Δvap; vapWT and Δvap; vapALS8 flies. Longevity: Δvap (pharate lethal, n = 46), control (precise excision line, average 64 days, n = 70), Δvap; vapWT (F7) (average 52 days, n = 56), Δvap; vapWT (VK31) (average 62 days, n = 25), Δvap; vapALS8 (M6) (average 23 days, * P < 0.0001, n = 102, compared with control), Δvap; vapALS8 (VK31) (average14 days, * P < 0.0001 compared with control), Δvap; vapWT(F7)/vap ALS8 (M6) (average 65 days, n = 50) and Δvap; vapWT/vap ALS8 (VK31) (average 67 days, n = 68). (B) Flight test. vapWT can suppress the flight defects associated with vapALS8. Δvap; vapALS8 (VK31) flies exhibit flight defects at day 12 (* P < 0.001 compared with control), but not at day 4 after eclosion. However, Δvap; vapWT/vapALS8 flies do not show flight defects. Flies were individually dropped into a plastic cylinder, and the height at which they landed was recorded. The shorter the distance from the bottom to their landing point, the worse their ability to fly. Numbers tested: control (n = 24), Δvap; vapWT (n = 28), Δvap; vapALS8(n = 24), and Δvap; vapWT/vapALS8 (n = 27). Error bars represent SEM. (C) vapALS8 causes adult brain degeneration. Brain section of 12-day-old adult flies. Δvap; vapALS8 (VK31), but not Δvap; vapWT (VK31) flies show vacuolation of brains in the optic lobe and central lobe. A copy of vapWT suppresses the defects associated with vapALS8. op: optic lobe and cen: central lobe. (D) Loss of Vap causes functional defects in motor neurons in adult flies. In the vap null mutant 1-day-old flies, the TTM muscles are unable to follow a 200 Hz stimulus, but they can almost respond accurately when stimulated at 10 Hz. (D) shows the difference between WT control (n = 16) and vap null mutants (n = 9) at the 20th pulse when stimulated at 10 Hz (control P = 1; Δvap P = 0.76 ± 0.15; mean ± SEM), 20 Hz (control P = 1; Δvap P = 0.49 ± 0.16), 50 Hz (control P = 1; Δvap P = 0.23 ± 0.12), 100 Hz (control P = 1; Δvap P = 0.14 ± 0.03), 200 Hz (control P = 0.97 ± 0.02; Δvap P = 0.04 ± 0.03). n: the number of flies used. * P < 0.05 and ** P < 0.005. (E) vapALS8 causes functional defects in motor neurons in adult flies. Comparison of 6- and 12-day-old flies that are responding to high frequency nerve stimulation (200 Hz) between control, Δvap; vapWT (VK31) and Δvap; vapALS8 (VK31) at 20th pulse. There is no difference in 6- and 12-day-old control flies (6 day P = 0.99 ± 0.013, n = 8; 14 day: P = 0.98 ± 0.025, n = 9) as well as Δvap; vapWT (6 day: 0.98 ± 0.017, n = 8; 12 day: 0.99 ± 0.010, n = 12). However, there is a significant reduction in the ability to follow a 200 Hz stimulus in Δvap; vapALS8 (6 day: P = 0.87 ± 0.057, n = 17; 12 day: P = 0.57 ± 0.116, n = 15. Control flies are those that have a precise excision of P{Mae-UAS.6.11}.
To avoid possible positional effects of the different transgenes, we also compared the ability of vapWT and vapALS8 transgenes inserted in the same docking site (VK31 line) to rescue the vap\xEF\x80null associated phenotypes. We confirmed that vapALS8 rescued flies indeed have a severely reduced lifespan when compared with vapWT rescued flies (Fig. 7A, Δvap; vapALS8, average longevity 14 days (n = 61) versus Δvap; vapWT, average longevity 67 days (n = 25); P < 0.001), showing that positional effects are not responsible for the observed differences in life span.
To assess the physiological consequences of the incomplete rescue, we examined flight ability (Fig. 7B), brain pathology (Fig. 7C), and the electrophysiological properties of adult neurons using the giant fiber (GF) responses of Δvap;vapALS8 (VK31) transgenic flies (Fig. 7D and E). Interestingly, vapALS8 rescued flies exhibit a progressive flight defect (Fig. 7B) that worsens with age and a single copy of WT vap suppresses the defects associated with vapALS8 (Fig. 7B, compare Δvap; vapALS8 and Δvap/vapWT; vapALS8), once more suggesting that the defects caused by the ALS8 mutation are due to a partial loss of function. Hence, vapALS8 causes defects in flight ability in an age-dependent manner.
To assess the morphological consequences of the Δvap flies rescued with vapALS8, we performed histological sections of adult brains that are 12 days old. As shown in Figure 7C, we observe significant histological differences in these brains when compared with those of controls. The adult brains of Δvap; vapWT flies did not show any obvious defects (Fig. 7C). In contrast, Δvap; vapALS8flies exhibit numerous vacuoles in the optic lobe and central brain (Fig. 7C), though appearing more frequently in the central lobe. Importantly, one copy of vapWT completely suppresses the defects associated with vapALS8. In summary, the defects that we observe show that the central nervous system (CNS) is affected in Δvap; vapALS8flies.
To assess neuronal function in the adults we recorded from the giant fiber system (GFS). The GFS is comprised of large neurons whose input consists of visual and mechanosensory inputs (71). Within the GFS, there are two types of motor neurons that are activated by a stimulus that induces escape behavior: the dorsal longitudinal flight motor and tergotrochanteral jump motor neurons. Their activation leads to contractions of the dorsal longitudinal muscles (DLMs) and tergotrochanteral muscles (TTMs) respectively (71,72). To stimulate the GFS, one impales the eyes with stimulation electrodes and records from the different muscles simultaneously (71,72). Following GF stimulation, we monitored responses of the DLM and TTM using different stimulation protocols (from 10 to 200 Hz with fixed 20 pulses, see Materials and Methods). The DLM and TTM responses accurately reflect the activity of the motor neurons (72,73), providing a read-out of motor neuron function in the adult fly and allowing us to assess an age-dependent demise of the function of these neurons. As shown in Figure 7D, control flies can maintain responses in TTM and DLM at the 20th pulse of a 200 Hz stimulation. In contrast, 1-day-old vap null mutants are unable to follow increasing stimulation rates, suggesting that loss of vap causes severe functional defects in the adult motor neuron system (Fig. 7D and Data not shown). However, the neuronal network must still be intact, as the flies still respond to a 10 Hz stimulation efficiently at day 1 when vap is lost.
To determine the rescuing effects of VapALS8 in adult motor neurons, we also performed GF recordings of Δvap; vapWT and Δvap; vapALS8 transgenic flies (Fig. 7E). Importantly, the vapWT transgene can fully rescue the defects in Δvap adult motor neurons (Fig. 7E and Data not shown). However, vapALS8 rescues the defects only partially (day 6) and exhibits a defect that progressively worsens (Δvap; vapALS8, 6 versus 12 days), showing that vapALS8 causes a progressive demise of the GFS. All the phenotypic data indicate that vapALS8 transgene behaves as a partial loss of function allele of vap. To determine if the vapALS8 transgene causes similar ERQC defects as observed in Δvap flies, we examined the accumulation of ubiquitinated proteins in Δvap; vapALS8 flies. We performed immunostaining of adult brains that are 1 day and 12 days old with a polyubiquitin antibody. As shown in Supplementary Material, Figure S3, we found that Δvap; vapALS8 exhibits cytoplasmic accumulations of ubiquitinated proteins in 12-day-old flies, but not in 1-day-old flies (Supplementary Material, Fig. S3C, C′, C″ and data not shown). In contrast, we did not observe accumulation of ubiquitinated proteins in control and vap null mutants carrying the genomic WT vap (Δvap; vapWT) (Supplementary Material, Fig. S3A and B). Importantly, the accumulation of ubiquitinated proteins in Δvap; vapALS8 does not overlap with the accumulation of the ubiquitinated mutant VapALS8 protein (Supplementary Material, Fig. S3 C, C and C″) (12). This suggests that mutant Vap ALS8 is responsible for defects in protein homeostasis in the ER, and results in the accumulation of ubiquitinated proteins. Lastly, we examined the ER stress in Δvap; vap ALS8. A consequence of ER stress is the cytoplasmic, unconventional splicing of the small intron of xbp1 (xbp1s) by the ER stress sensor Ire1 (74). Thus, we examined if vap ALS8 causes xbp1 splicing. We specifically amplified xbp1s transcripts by conventional PCR using a primer straddling the ER-sensitive intron that cannot hybridize with xbp1 unspliced transcripts (see Material and Methods). Consistent with the finding that vapALS8 induces xbp1 expression, flies expressing VapALS8 accumulate higher levels of xbp1 transcripts than Δvap; vapWT flies (Supplementary Material, Fig. S3D and E). Hence, we conclude that vap null mutants carrying a genomic vapALS8 exhibit the ERQC phenotypes. However, this phenotype is milder than the ERQC phenotype observed in vap null mutants.
Osbp genetically interacts with the ALS8 mutation
To determine whether loss of osbp shows similar defects as loss of vap, we tested the phenotype associated with loss of osbp (Δosbp) (75). Flies lacking osbp (Δosbp/Df(3R)ED622) are viable and male-sterile, in agreement with previously published observations (58). However, in contrast to loss of vap, loss of osbp does not lead to the accumulation of ubiquitinated proteins or ER stress (data not shown). These observations suggest that osbp may play a redundant role with other osbp paralogues in flies (58). To determine whether Osbp and its paralogue, CG1513 have redundant function, we created a null allele of CG1513 (see Material and Methods). Neither CG1513 nor osbp mutants (Δ CG1513/Df(2R)BSC298 and Δosbp/Df(3R)ED622) present obvious defects in viability (58), whereas double mutants lacking both osbp and CG1513 die during early larval stages (data not shown), indicating a redundant function between osbp and CG1513.
Since Osbp is mislocalized to the Golgi in vap null mutants, and if CG1513 has a redundant function, the protein encoded by CG1513 may also be mislocalized in the vap mutant. Indeed, loss of vap (marked by GFP) leads to an aberrant distribution of the CG1513 protein and an accumulation of CG1513 in punctae (arrows in Supplementary Material, Fig. S2A). Importantly, CG1513 accumulates in the same intracellular regions where Osbp accumulates in the vap null clone (Supplementary Material, Fig. S2B), suggesting that Vap mediates the function of Osbp and its paralogue, CG1513.
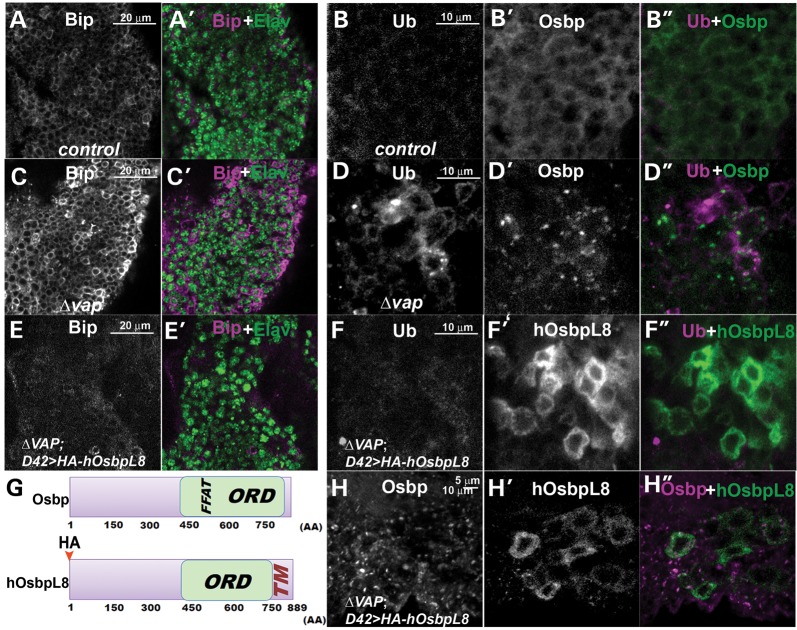
To determine if the mislocalization of the Osbps contribute to defects in the ER when Vap is lost, we created transgenic flies expressing human OsbpL8 (75). There are 16 Osbp homologues in humans and only four Osbp homologues (Osbp (CG6708), CG1513, CG3860 and CG5077) in flies. All Osbp homologues contain the Osbp domain, which includes a binding site for sterols or cholesterol (59–61). Importantly, each Osbp protein is also likely to have distinct functions. Some, but not all human and Drosophila Osbps have an FFAT motif and bind to Vap proteins, suggesting that Vap is required for the function of some Osbp proteins. Other Osbp proteins do not seem to require Vap for their function. Human OsbpL8 contains a conserved Osbp domain and an ER retention signal, but unlike Drosophila Osbp (CG6708), CG1513 or human Osbp6, it does not contain the FFAT binding motif (Fig. 8G) (75), suggesting that human OsbpL8 may be able to function in the ER in the absence of vap. We therefore expressed human OsbpL8 HA tagged protein in neurons of vap null mutants. As expected, human OsbpL8 localizes in the ER of neurons in the vap null mutant as shown by colocalization with an ER marker, Calnexin (76) (Supplementary Material, Fig. S4B, B’, and B”). However, Drosophila Osbp is still mislocalized in the vap null mutant (compare Fig. 8H and H’). Hence, human OsbpL8 localization, unlike Drosophila Osbp or CG1513, does not depend on Vap expression. Loss of Vap causes increased levels of Bip protein (compare Fig. 8C with A) and accumulation of ubiquitinated proteins (Fig. 8D compared with B) but the presence of human OsbpL8 in vap mutants strongly attenuates the upregulation of Bip and the accumulation of ubiquitinated proteins (Fig. 8E and F compared with 8C and D). In contrast, Drosophila Osbp does not rescue the defects associated with loss of Vap efficiently (data not shown), suggesting that restoring expression of Osbp in the ER is able to rescue the defects associated with loss of vap.
Figure 8.
Expression of human OsbpL8 suppresses the ER defects associated with loss of Vap. (A–F, H) Immunostaining of the adult brain of WT (control) (A and B), vap null mutant (C and D), vap null mutants expressing HA-hOsbpL8 with neuronal GAL4 (D42) driver (E, F and H). Expression of human OsbpL8 suppresses the upregulation of Bip and accumulation of ubiquitinated proteins caused by loss of Vap (compare E and F to C and D) without restoring the localization of Osbp (H). (G) Graphical overview of the Osbp and hOsbpL8 proteins. Human OsbpL8 contains a transmembrane domain, but neither a Vap binding FFAT motif nor a signal peptide. FFAT: Vap binding site, ORD: Osbp-related domain, TM: transmembrane domain. In UAS hosbpL8 transgene, the HA epitope tag was inserted at the amino terminus of human OsbpL8.
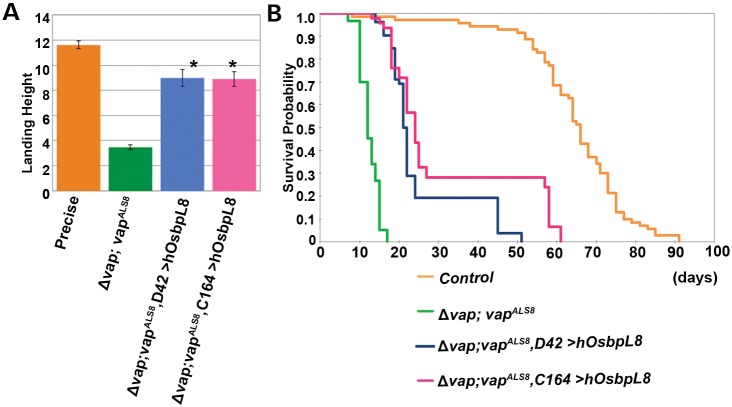
To further characterize the contribution of Osbp in the loss of vap, we examined whether expressing human OsbpL8 in neuronal tissues can rescue the flight defects and lethality associated with the hypomorphic vapALS8 mutation. As shown in Figure 9A and B, restoring expression of Osbp in the ER with human OsbpL8 is able to rescue the flight defects (Fig. 9A) and lethality (Fig. 9B) associated with the hypomorphic defects caused by the vapALS8 mutation. vapALS8 causes severe flight defects in 12-day-old flies and expression of human OsbpL8 in neurons with two different GAL4 drivers (D42-GAL4 and C164-GAL4) alleviates the defects associated with vapALS8 (Fig. 9A, compare Δvap; vapALS8 and Δvap; vapALS8; C164-GAL4 > UAS-human OsbpL8 or Δvap; vapALS8; D42-GAL4 > UAS-human OsbpL8; P < 0.0001). Moreover, human OsbpL8 expression in neurons also improves the longevity of Δvap; vapALS8 flies (Fig. 9B, compare life span of Δvap; vapALS8 (average 12.47 days, n = 150), Δvap; vapALS8; D42-GAL4 > UAS-human OsbpL8 (average 25.33 days, n = 52, P < 0.0001) and Δvap; vapALS8; C164-GAL4 > UAS-human OsbpL8, (average 31.87 days, n = 46, P < 0.0001), showing that restoring Osbp function in the ER of neuronal tissues partially rescues the defects associated with vapALS8.
Figure 9.
Neuronal expression of OsbpL8 enhances the longevity associated with vapALS8 transgenic flies. (A) Flight assay of Δvap; vapALS8 flies expressing human OsbpL8 in neurons. Δvap; vapALS8 flies exhibit flight defects at day 12 of adulthood (*P < 0.0001 compared with control). Neuron-specific expression (using D42 GAL4 and C164 GAL4) of OsbpL8 in Δvap; vapALS8 flies significantly abates the defects associated with Δvap; vapALS8. Flies tested: control (precise excision line, average 11.61 in, n = 59) Δvap; vapALS8 (average 3.47 in, n = 49), Δvap; vapALS8; D42-GAL4>UAS-human OsbpL8 (average 8.99 in, n = 45, *P < 0.0001 compared with vapALS8 mutant) and Δvap; vapALS8; C164-GAL4>UAS-human OsbpL8 (average 8.90 in, n = 39, *P < 0.0001 compared with vapALS8 mutant). Error bars represent SEM. (B) Longevities of Δvap; vapALS8 flies expressing human OsbpL8 in neurons. Human OsbpL8 was expressed using two different neuron-specific GAL4 drivers (D42-GAL4 and C164-GAL4). Expression of OsbpL8 in neurons of Δvap; vapALS8 flies significantly enhances the longevity associated with these flies. Longevity: control (precise excision line, average 63.69 days, n = 71), Δvap; vapALS8 (average 12.47 days, n = 150), Δvap; vapALS8; D42-GAL4>UAS-human OsbpL8 (average 25.33 days, n = 52, *P < 0.0001 compared with vapALS8 mutant) and Δvap; vapALS8; C164-GAL4>UAS-human OsbpL8 (average 31.87 days, n = 46, *P < 0.0001 compared with vapALS8 mutant).
DISCUSSION
Based on our findings, we propose that an impairment of the normal function of Vap in the ER may contribute to the pathology of ALS8. Vap is required for the proper localization of Osbp in the ER, and the interaction between Vap and Osbp function is apparently required for the implementation of the ERQC. Failure of Vap-Osbp to function causes a defect in ER proteostasis, resulting in protein accumulations in the ER and ER stress. These defects in the ER overload probably promote the Ubiquitin Proteasome System (UPS), which subsequently leads to an accumulation of ubiquitinated proteins. In the autosomal dominant form of ALS, ALS8, patients express both WT Vap and the ALS8 mutant Vap. As the ALS8 mutant Vap is not able to bind to Osbp, this loss of binding seems to result in the partial loss of function of Vap. This in turn may cause a gradual decrease in function of the ERQC. These defects may also lead to secondary defects that have been previously reported: namely the lack of the secretion of MSP (12,13).
Defects in ER proteostasis have been implicated in the pathology of ALS. The ER constantly requires maintenance of protein homeostasis, or proteostasis (76). To this end, the ER carries the burden of continuously modulating protein folding and degradation to avoid accumulation of misfolded proteins (34). An overloaded ERQC induces ER stress, and severe or prolonged ER stress leads to cell dysfunction and eventually cell death. Various proteins involved in ERQC have been identified, including VCP (77), Derlin-1 (78) and Calreticulin (79). Moreover, a familial form of ALS is associated with mutations in VCP (80) and mutations in SOD1 have been shown to inhibit the function of Derlin-1 (31) and reduce the levels of Calreticulin and trigger ER stress (81). In SOD1 mice, the ER stress seems to contribute to selective motor neuron degeneration (31,32), and ER stress has also been implicated in numerous patients with sporadic ALS (30). Together, these observations suggest that defects in ER proteostasis might be a common pathological feature of ALS. In many neurodegenerative diseases, only a subpopulation of neurons is initially targeted. However, in most cases the disease-causing proteins are ubiquitously expressed. Why some regions of the brain are more susceptible to defects remains unknown. We found specific defects in some subtypes of neurons in the vap null mutants. The defects are not limited to motor neurons. Interestingly, VapB levels decrease concomitantly with the disease's progression in the SOD1 mouse model (82), and sporadic ALS patients have been reported to have decreased levels of the VapB protein (82,83), suggesting that impaired VapB function may contribute to the pathogenesis of familial and sporadic forms of ALS. It is therefore likely that the molecular mechanism by which loss of Vap affects the cells is not restricted to ALS8.
An important question raised by our results is how VapB functions in protein homeostasis in the ER. The ERQC is involved in identifying aberrantly misfolded proteins, retrotranslocating the misfolded proteins and processing the degraded retrotranslocated proteins (34). These processes seem to be tightly linked. Indeed, many proteins function in multiple different steps in the ERQC. However, Vap is unlikely to function in chaperone-dependent refolding, since a molecular chaperone, Bip, is upregulated in vap null mutants. Moreover, Bip overexpression fails to rescue the ER stress in the vap null mutants (data not shown).
Restoring the levels of an Osbp that is not dependent on Vap binding suppresses the ER defects caused by loss of Vap, suggesting that this pathway is required for the execution of the ERQC. Mammalian Osbp and Osbp-related protein (Orp) constitute a large eukaryotic gene family characterized by a conserved C-terminal sterol binding domain (Fig. 8G) (56). This domain organization suggests that a primary function of Osbp/Orp is to sense cholesterol or oxysterols to control transport between target membranes (63,84,85). Differential localization of Osbp between organelles in response to exogenous and endogenous sterol ligands indicates that Osbps transfer cholesterol and/or oxysterols between these organelles. Although the ER membrane is cholesterol poor (3–6% of total lipids) (86), acute cholesterol depletion in culture medium impairs the mobility of membrane proteins and protein secretion from the ER in cultured cells (87). Hence, the defects in ER proteostasis might be due to decreased levels of cholesterol in the ER caused by loss of Osbp/Vap function. In addition, Osbps are also coupled to the activation of Cert and SM synthesis through increased activity of PI4KIIα, a cholesterol sensitive PI 4-kinase (63,88). This suggests that loss of Vap may also cause defects in ceramide transport from the ER, which may result in accumulation of ceramide and defects in ER proteostasis, consistent with the observation that ceramide accumulates in ER stress (89).
In this paper, we document the defects caused by mutant VapALS8 protein expressed under the control of its endogenous regulatory elements in a genomic rescue construct. Previous studies suggested that the VapALS8 protein causes dominant negative defects when the mutant protein is overexpressed in flies or cultured cells (33,90,91). However, overexpression of VapALS8 rescues the lethality associated with the vap null mutant (12) and the data presented here based on the vapALS8 genomic transgene strongly suggests that the defects caused by the ALS8 mutant protein are hypomorphic rather than dominant negative. Consistent with our findings, Kabashi et al. (2013), reported two mutations (A145 V and S160Δ) in the vapB gene in an ALS patients. They examined if WT and the mutant vap mRNA expression were able rescue the defects caused by loss of vapB in zebrafish and observed that these mutations (A145 V and S160Δ) partially fail to rescue (68). We therefore propose that the ALS8 mutation in patients is a partial loss of function mutation and that vapB is haploinsufficient in humans.
MATERIALS AND METHODS
Fly transgenes and strains
To generate UAS-osbp, the PCR fragment of the Drosophila EST clone (LD31802) containing the osbp cDNA was subcloned into pUAST and pUASTattB vector (92). To generate UAS-human OsbpL8, the PCR fragment of the human EST clone (MHS1010-98684599) containing OsbpL8 was subcloned into attB pUASTHA vector (gift from S. Yamamoto). Human OsbpL8 protein does not have a signal peptide. The HA epitope tag was inserted at the amino terminus of human OsbpL8. To generate the genomic WT vap construct, a PCR fragment was amplified with the following primers, 5′-GAATTCCTTGCTTGTGGACCCCGCTGGCTG-3′ and 5′-GGATACAATCTCTGCTGCTTAAATGGGATTT-3′. The genomic ALS8 mutant vap construct was created by chimeric PCR with primers containing a P58S mutation. The genomic fragment was subcloned into the pCasper 4 vector. To create site-specific transgenes (70), the attB sequence was inserted in the genomic WT or ALS8 mutant vap/pCasper 4 constructs. The attB constructs were injected into VK31 and VK33 attP docking sites (70).
The following strains were used:
y w, vapΔ31/FM7, Kr-GAL4, UAS-GFP [vapΔ31] was created by imprecise excision of the P-element present in yw, P{Mae-UAS.6.11},Vap-33-1GG01069 (93). Southern blotting and western blotting with the Vap (Rb92) antibody (12) shows that [vapΔ31] is a null allele. The stock containing the precise excision of y w, P{Mae-UAS.6.11},Vap-33-1GG01069 was used as a control. The osbp null mutant was created by screening for a deletion of the DNA between two piggyBac elements, PBac{WH}Osbpf00496 and PBac{RB}e04437 (94) using the method described by Golic and Golic (95). The CG1513 mutant was created by imprecise excision of the P-element in y1 w*; P{EP}CG1513G2193. Southern blotting and PCR shows the deletion of exons 5 and 6 which contain coding sequence of CG1513 (548–651 amino acids). The other strains used were yw; P{w[+mC] = UAS-mCD8::GFP.L}LL5(51) and w; P{w[+mW.hs] = GawB}D42 (D42-GAL4) (96), P{w+mW.hs = GawB}C164 (C164-GAL4) (97), GMR-Rh-1G69D (43), P{hsFLP}1, P{tubP-GAL80}LL1, w* P{neoFRT}19A; P{UAS-mCD8::GFP.L}LL5 (BDSC stock 5134), and yw, P{hsFLP}, P{tubP-GAL4}, P{w[+mC] = UAS-mCD8::GFP.L}; P{neoFRT}82B, P{tubP-GAL80}/ TM6B, Tb (Gift from Gary Struhl).
Antibody generation and immunostaining
Domain of the Osbp protein (60–400 amino acids) and the CG1513 protein (125–421 amino acids) were expressed using the GST-fusion protein system. A polyclonal guinea pig Osbp antibody (GP89) and mouse CG1513 antibody (Ms54) were raised against the fusion proteins at Cocalico Biologicals (Reamstown, PA). Larvae and adult brains were fixed in 4% paraformaldehyde for 20 min and washed with PBS containing 0.2% Triton X-100. The following antibody dilutions were used: guinea pig anti-Osbp (GP89), 1:2000; mouse anti-CG1513 (Ms54), 1:300; rabbit anti-Vap (Rb92) (12), 1:1000; mouse monoclonal anti-Chaoptin (37), 1:200; Elav (7E8A10) (98), 1:200; guinea pig anti-Boca (40), 1:500; guinea pig anti-Bip (Ryoo et al., 2007) and mouse anti-Ubiquitin (FK1) (BIOMOL), 1:100, anti-Robo1 (99), 1:200, anti-N-Cadherin (39), 1:200 and anti-Rhodospin-1 (4C5), 1:200. Anti-HA antibody (clone 3F10, Roche) is used as 1:200 dilution to detect HA-hOSBPL8. Secondary antibodies conjugated to Cy3 or Alexa 488 (Jackson ImmunoResearch, Molecular Probes) were used at 1:1000. Images were captured with a Zeiss LSM510 confocal microscope and processed with ImageJ.
Protein chemistry
For immunoblotting, proteins from adult heads were extracted with Urea buffer [8 m urea, 4% SDS, 0.125 m Tris–HCl (pH 6.8), 12 mm EDTA, 3% β-mercaptoethanol, proteinase inhibitors (Roche), and 0.002% bromophenol blue]. Anti-Vap antibody (Rb92,1:5000) and mouse anti-Actin (MP Biomedicals) (1:10 000) was used for immunoblotting.
GST pull-down assays
The domain N-terminal to the transmembrane domain of the WT and ALS8 Vap protein (1–248 amino acids) was expressed using the GST-fusion protein system (12). Osbp protein is expressed in flies with UAS-osbp and tubulin-GAL4 and lysates were collected with TNT buffer (1% Triton, 50 mm Tris (pH8), 150 mm NaCl, 1 mm EDTA, proteinase inhibitors, 1 mm PMSF). The lysates were incubated with glutathione–Sepharose-bound WT or ALS8 Vap. The resulting precipitates were analyzed by western blotting with anti-Osbp antibodies (GP89, 1:5000) and anti-GST antibody (Sigma, 1:20 000).
Yeast two-hybrid assays
WT and ALS8 human Vap protein (1–222 amino acids) were subcloned into the pBD vector (52). Human OsbpL8 and Orp3 were subcloned into pAD vector (52). All interaction assays were performed by co-transfecting the two vectors encoding the hybrid proteins into the yeast strain Mav203 and plating on selective media lacking histidine. βGAL assays were performed as described previously (52).
TEM
TEM was performed as previously described (100).
Flight assay
Flight assays were performed as previously described (42,52).
Giant fiber recording
GF recordings were performed with a protocol modified from (71). Briefly, flies were anesthetized on ice, transferred to a petri dish filled with soft dental wax, and the fly wings and legs were mounted in wax, ventral side down, using forceps. For stimulations and recordings from the TTM and DLM, five electrolytically sharpened tungsten electrodes were used: two for stimulating the GF, one as a reference electrode, and two for recording from the TTM and DLM, respectively. To activate the GF, two sharp tungsten electrodes were inserted into each eye and voltage stimulation was applied at different frequency stimulations. GF-DLM and GF-TTM responses were measured through two electrodes implanted in the DLM and TTM. Prior to applying high frequency stimulation on the GF, low frequency stimulations at 0.5 Hz were applied after placing the two recording electrodes in TTM and DLM to ensure that the electrodes are recording from the proper muscles (the latency of responses for TTM: 0.8 ms and for DLM: 1.2 ms (71). High frequency train stimulations of 20 pulses were delivered to the GF at 10, 20, 50, 100 and 200 Hz in random order. Ten times repetitive stimulations were applied for each particular frequency train, interspersed with 5 min rests between two trains of stimuli. 0.5 Hz stimulations were used again after high frequency stimulation to confirm that electrodes were still in the proper muscle. Stimuli of the crossing electrodes were fixed at a duration of 0.1 ms at 8–15 V of amplitudes through a stimulus isolation unit (model DS2A, Digitimer Ltd, UK) and the frequency of train stimuli was controlled by AxoGraph acquisition software (AxoGraph Scientific). A microelectrode amplifier (Model 1800, A-M system) was used for all recordings. Digidata 1322A (Axon Instruments) was used for data acquisition. The probability of responses, under particular frequency of GF stimulation, due to a particular stimulus (nth, n = 1, 2,…, 20), was calculated from the proportion of successful responses (out of 10 of repetitive, note: not 20; also see S10-C) for both TTM and DLM pathways. The difference of probability of responses between control and mutant (P-value) for particular stimuli were calculated by a t-test (SigmaPlot 10; Systat Software, Inc.).
Reverse transcription polymerase chain reaction
RNA isolated from day 3 old adult fly heads (Qiagen RNeasy Mini Kit) was subjected to reverse transcription PCR (RT-PCR) using One Step RT-PCR kit (Qiagen). To detect Drosophila XBP1s, the spliced form of XBP-1 implicated in ER stress, we amplified a fragment of 148 bp using primers xbp-1F2 5′-ACCAACCTTGGATCTGCCG-3′ and xbp-1R2 5′-CGCCAAGCATGTCTTGTAGA-3 (Life Technologies), where the 3′ end of the reverse primer straddles the ER-sensitive intron (101). In all the reactions, a 482 bp fragment from Actin5C was amplified as internal control using the primers actinF 5′-CTGGACTTCGAGCAGGAGAT-3′ and actinR 5′-GGTGGCTTGGATGCTTAGAA-3′ (Life Technologies). PCR products were checked on 4% Metaphor agarose (Lonza) and 2% agarose gels.
SUPPLEMENTARY MATERIAL
FUNDING
Confocal microscopy at BCM was supported by National Institutes of Health (5P30HD024064-23) to the BCM Intellectual and Developmental Disabilities Research Center (IDDRC). H.T. was supported by grants from the ALS Association and the Canadian Institutes of Health Research (MOP-115153).
Supplementary Material
ACKNOWLEDGEMENTS
We thank X. Huang for Osbp strains, H. Steller and H. Ryoo for anti-Hsc3 antibodies, K. Basler for pUAS attB vector, Gary Struhl for 3R MARCM stock, S. Yamamoto for the attB pUAST HA vector and M. Vidal for vectors for Y2H. We thank C. Haueter for TEM analysis. We thank J. Petrov for technical assistance, H. Pan for injections and N. Giagtzoglou and B. Kamal for comments on the manuscript. H.J.B. is an Investigator of the HHMI.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Kiernan M.C., Vucic S., Cheah B.C., Turner M.R., Eisen A., Hardiman O., Burrell J.R., Zoing M.C. Amyotrophic lateral sclerosis. Lancet. 2011;377:942–955. doi: 10.1016/S0140-6736(10)61156-7. [DOI] [PubMed] [Google Scholar]
- 2.Pasinelli P., Brown R.H. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat. Rev. Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 3.Ferraiuolo L., Kirby J., Grierson A.J., Sendtner M., Shaw P.J. Molecular pathways of motor neuron injury in amyotrophic lateral sclerosis. Nat. Rev. Neurol. 2011;7:616–630. doi: 10.1038/nrneurol.2011.152. [DOI] [PubMed] [Google Scholar]
- 4.Al-Chalabi A., Jones A., Troakes C., King A., Al-Sarraj S., van den Berg L.H. The genetics and neuropathology of amyotrophic lateral sclerosis. Acta Neuropathol. 2012;124:339–352. doi: 10.1007/s00401-012-1022-4. [DOI] [PubMed] [Google Scholar]
- 5.Nishimura A.L., Mitne-Neto M., Silva H.C., Richieri-Costa A., Middleton S., Cascio D., Kok F., Oliveira J.R., Gillingwater T., Webb J., et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am. J. Hum. Genet. 2004;75:822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lev S., Ben Halevy D., Peretti D., Dahan N. The VAP protein family: from cellular functions to motor neuron disease. Trends Cell Biol. 2008;18:282–290. doi: 10.1016/j.tcb.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Nishimura Y., Hayashi M., Inada H., Tanaka T. Molecular cloning and characterization of mammalian homologues of vesicle-associated membrane protein-associated (VAMP-associated) proteins. Biochem. Biophys. Res. Commun. 1999;254:21–26. doi: 10.1006/bbrc.1998.9876. [DOI] [PubMed] [Google Scholar]
- 8.Weir M.L., Klip A., Trimble W.S. Identification of a human homologue of the vesicle-associated membrane protein (VAMP)-associated protein of 33 kDa (VAP-33): a broadly expressed protein that binds to VAMP. Biochem. J. 1998;333:247–251. doi: 10.1042/bj3330247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soussan L., Burakov D., Daniels M.P., Toister-Achituv M., Porat A., Yarden Y., Elazar Z. ERG30, a VAP-33-related protein, functions in protein transport mediated by COPI vesicles. J. Cell. Biol. 1999;146:301–311. doi: 10.1083/jcb.146.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skehel P.A., Fabian-Fine R., Kandel E.R. Mouse VAP33 is associated with the endoplasmic reticulum and microtubules. Proc. Natl. Acad. Sci. USA. 2000;97:1101–1106. doi: 10.1073/pnas.97.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaiser S.E., Brickner J.H., Reilein A.R., Fenn T.D., Walter P., Brunger A.T. Structural basis of FFAT motif-mediated ER targeting. Structure. 2005;13:1035–1045. doi: 10.1016/j.str.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Tsuda H., Han S.M., Yang Y., Tong C., Lin Y.Q., Mohan K., Haueter C., Zoghbi A., Harati Y., Kwan J., et al. The amyotrophic lateral sclerosis 8 protein VAPB is cleaved, secreted, and acts as a ligand for Eph receptors. Cell. 2008;133:963–977. doi: 10.1016/j.cell.2008.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han S.M., Tsuda H., Yang Y., Vibbert J., Cottee P., Lee S.J., Winek J., Haueter C., Bellen H.J., Miller M.A. Secreted VAPB/ALS8 major sperm protein domains modulate mitochondrial localization and morphology via growth cone guidance receptors. Dev. Cell. 2012 doi: 10.1016/j.devcel.2011.12.009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster L.J., Weir M.L., Lim D.Y., Liu Z., Trimble W.S., Klip A. A functional role for VAP-33 in insulin-stimulated GLUT4 traffic. Traffic. 2000;1:512–521. doi: 10.1034/j.1600-0854.2000.010609.x. [DOI] [PubMed] [Google Scholar]
- 15.Matsuzaki F., Shirane M., Matsumoto M., Nakayama K.I. Protrudin serves as an adaptor molecule that connects KIF5 and its cargoes in vesicular transport during process formation. Mol. Biol. Cell. 2011;22:4602–4620. doi: 10.1091/mbc.E11-01-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pennetta G., Hiesinger P.R., Fabian-Fine R., Meinertzhagen I.A., Bellen H.J. Drosophila VAP-33A directs bouton formation at neuromuscular junctions in a dosage-dependent manner. Neuron. 2002;35:291–306. doi: 10.1016/s0896-6273(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 17.Kuijpers M., Yu K.L., Teuling E., Akhmanova A., Jaarsma D., Hoogenraad C.C. The ALS8 protein VAPB interacts with the ER–Golgi recycling protein YIF1A and regulates membrane delivery into dendrites. EMBO J. 2013;32:2056–2072. doi: 10.1038/emboj.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peretti D., Dahan N., Shimoni E., Hirschberg K., Lev S. Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol. Biol. Cell. 2008;19:3871–3884. doi: 10.1091/mbc.E08-05-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forrest S., Chai A., Sanhueza M., Marescotti M., Parry K., Georgiev A., Sahota V., Mendez-Castro R., Pennetta G. Increased levels of phosphoinositides cause neurodegeneration in a Drosophila model of amyotrophic lateral sclerosis. Hum. Mol. Genet. 2013;22:2689–2704. doi: 10.1093/hmg/ddt118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mikitova V., Levine T.P. Analysis of the key elements of FFAT-like motifs identifies new proteins that potentially bind VAP on the ER, including two AKAPs and FAPP2. PloS One. 2012;7:e30455. doi: 10.1371/journal.pone.0030455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyles J.P., Ridgway N.D. VAMP-associated protein-A regulates partitioning of oxysterol-binding protein-related protein-9 between the endoplasmic reticulum and Golgi apparatus. Exp. Cell Res. 2004;297:533–547. doi: 10.1016/j.yexcr.2004.03.052. [DOI] [PubMed] [Google Scholar]
- 22.Kawano M., Kumagai K., Nishijima M., Hanada K. Efficient trafficking of ceramide from the endoplasmic reticulum to the Golgi apparatus requires a VAMP-associated protein-interacting FFAT motif of CERT. J. Biol. Chem. 2006;281:30279–30288. doi: 10.1074/jbc.M605032200. [DOI] [PubMed] [Google Scholar]
- 23.Perry R.J., Ridgway N.D. Oxysterol-binding protein and vesicle-associated membrane protein-associated protein are required for sterol-dependent activation of the ceramide transport protein. Mol. Biol. Cell. 2006;17:2604–2616. doi: 10.1091/mbc.E06-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagace T.A., Byers D.M., Cook H.W., Ridgway N.D. Chinese hamster ovary cells overexpressing the oxysterol binding protein (OSBP) display enhanced synthesis of sphingomyelin in response to 25-hydroxycholesterol. J. Lipid Res. 1999;40:109–116. [PubMed] [Google Scholar]
- 25.Balch W.E., Morimoto R.I., Dillin A., Kelly J.W. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 26.Braakman I., Bulleid N.J. Protein folding and modification in the mammalian endoplasmic reticulum. Annu. Rev. Biochem. 2011;80:71–99. doi: 10.1146/annurev-biochem-062209-093836. [DOI] [PubMed] [Google Scholar]
- 27.Araki K., Nagata K. Protein folding and quality control in the ER. Cold Spring Harb. Perspect. Biol. 2011;3:a007526. doi: 10.1101/cshperspect.a007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedlander R., Jarosch E., Urban J., Volkwein C., Sommer T. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat. Cell Biol. 2000;2:379–384. doi: 10.1038/35017001. [DOI] [PubMed] [Google Scholar]
- 29.Travers K.J., Patil C.K., Wodicka L., Lockhart D.J., Weissman J.S., Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 30.Atkin J.D., Farg M.A., Walker A.K., McLean C., Tomas D., Horne M.K. Endoplasmic reticulum stress and induction of the unfolded protein response in human sporadic amyotrophic lateral sclerosis. Neurobiol. Dis. 2008;30:400–407. doi: 10.1016/j.nbd.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 31.Nishitoh H., Kadowaki H., Nagai A., Maruyama T., Yokota T., Fukutomi H., Noguchi T., Matsuzawa A., Takeda K., Ichijo H. ALS-linked mutant SOD1 induces ER stress- and ASK1-dependent motor neuron death by targeting Derlin-1. Genes Dev. 2008;22:1451–1464. doi: 10.1101/gad.1640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saxena S., Cabuy E., Caroni P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat. Neurosci. 2009;12:627–636. doi: 10.1038/nn.2297. [DOI] [PubMed] [Google Scholar]
- 33.Aliaga L., Lai C., Yu J., Chub N., Shim H., Sun L., Xie C., Yang W.J., Lin X., O'Donovan M.J., et al. Amyotrophic lateral sclerosis-related VAPB P56S mutation differentially affects the function and survival of corticospinal and spinal motor neurons. Hum. Mol. Genet. 2013;22:4293–4305. doi: 10.1093/hmg/ddt279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vembar S.S., Brodsky J.L. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Bio. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernales S., Papa F.R., Walter P. Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- 36.Hurtley S.M., Helenius A. Protein oligomerization in the endoplasmic reticulum. Annu. Rev. Cell Biol. 1989;5:277–307. doi: 10.1146/annurev.cb.05.110189.001425. [DOI] [PubMed] [Google Scholar]
- 37.Van Vactor D., Jr., Krantz D.E., Reinke R., Zipursky S.L. Analysis of mutants in chaoptin, a photoreceptor cell-specific glycoprotein in Drosophila, reveals its role in cellular morphogenesis. Cell. 1988;52:281–290. doi: 10.1016/0092-8674(88)90517-x. [DOI] [PubMed] [Google Scholar]
- 38.Seeger M., Tear G., Ferres-Marco D., Goodman C.S. Mutations affecting growth cone guidance in Drosophila: genes necessary for guidance toward or away from the midline. Neuron. 1993;10:409–426. doi: 10.1016/0896-6273(93)90330-t. [DOI] [PubMed] [Google Scholar]
- 39.Iwai Y., Usui T., Hirano S., Steward R., Takeichi M., Uemura T. Axon patterning requires DN-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron. 1997;19:77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- 40.Culi J., Mann R.S. Boca, an endoplasmic reticulum protein required for wingless signaling and trafficking of LDL receptor family members in Drosophila. Cell. 2003;112:343–354. doi: 10.1016/s0092-8674(02)01279-5. [DOI] [PubMed] [Google Scholar]
- 41.Wong A.M., Wang J.W., Axel R. Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell. 2002;109:229–241. doi: 10.1016/s0092-8674(02)00707-9. [DOI] [PubMed] [Google Scholar]
- 42.Fayyazuddin A., Zaheer M.A., Hiesinger P.R., Bellen H.J. The nicotinic acetylcholine receptor Dalpha7 is required for an escape behavior in Drosophila. PLoS Biol. 2006;4:e63. doi: 10.1371/journal.pbio.0040063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang M.J., Ryoo H.D. Suppression of retinal degeneration in Drosophila by stimulation of ER-associated degradation. Proc. Natl. Acad. Sci. U.S A. 2009;106:17043–17048. doi: 10.1073/pnas.0905566106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hay B.A., Wolff T., Rubin G.M. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- 45.Colley N.J., Baker E.K., Stamnes M.A., Zuker C.S. The cyclophilin homolog ninaA is required in the secretory pathway. Cell. 1991;67:255–263. doi: 10.1016/0092-8674(91)90177-z. [DOI] [PubMed] [Google Scholar]
- 46.Schuck S., Prinz W.A., Thorn K.S., Voss C., Walter P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J. Cell Biol. 2009;187:525–536. doi: 10.1083/jcb.200907074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calfon M., Zeng H., Urano F., Till J.H., Hubbard S.R., Harding H.P., Clark S.G., Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 48.Ryoo H.D., Domingos P.M., Kang M.J., Steller H. Unfolded protein response in a Drosophila model for retinal degeneration. EMBO J. 2007;26:242–252. doi: 10.1038/sj.emboj.7601477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Menendez-Benito V., Verhoef L.G., Masucci M.G., Dantuma N.P. Endoplasmic reticulum stress compromises the ubiquitin–proteasome system. Hum. Mol. Genet. 2005;14:2787–2799. doi: 10.1093/hmg/ddi312. [DOI] [PubMed] [Google Scholar]
- 50.Leigh P.N., Whitwell H., Garofalo O., Buller J., Swash M., Martin J.E., Gallo J.M., Weller R.O., Anderton B.H. Ubiquitin-immunoreactive intraneuronal inclusions in amyotrophic lateral sclerosis. Morphology, distribution, and specificity. Brain. 1991;114:775–788. doi: 10.1093/brain/114.2.775. [DOI] [PubMed] [Google Scholar]
- 51.Lee T., Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 52.Rual J.F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 53.Gregorio-King C.C., Collier G.R., McMillan J.S., Waugh C.M., McLeod J.L., Collier F.M., Kirkland M.A. ORP-3, a human oxysterol-binding protein gene differentially expressed in hematopoietic cells. Blood. 2001;98:2279–2281. doi: 10.1182/blood.v98.7.2279. [DOI] [PubMed] [Google Scholar]
- 54.Lehto M., Hynynen R., Karjalainen K., Kuismanen E., Hyvarinen K., Olkkonen V.M. Targeting of OSBP-related protein 3 (ORP3) to endoplasmic reticulum and plasma membrane is controlled by multiple determinants. Exp. Cell. Res. 2005;310:445–462. doi: 10.1016/j.yexcr.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 55.Lehto M., Mayranpaa M.I., Pellinen T., Ihalmo P., Lehtonen S., Kovanen P.T., Groop P.H., Ivaska J., Olkkonen V.M. The R-Ras interaction partner ORP3 regulates cell adhesion. J. Cell Sci. 2008;121:695–705. doi: 10.1242/jcs.016964. [DOI] [PubMed] [Google Scholar]
- 56.Lehto M., Olkkonen V.M. The OSBP-related proteins: a novel protein family involved in vesicle transport, cellular lipid metabolism, and cell signalling. Biochim. Biophys. Acta. 2003;1631:1–11. doi: 10.1016/s1388-1981(02)00364-5. [DOI] [PubMed] [Google Scholar]
- 57.Alphey L., Jimenez J., Glover D. A Drosophila homologue of oxysterol binding protein (OSBP)—implications for the role of OSBP. Biochim. Biophys. Acta. 1998;1395:159–164. doi: 10.1016/s0167-4781(97)00159-0. [DOI] [PubMed] [Google Scholar]
- 58.Ma Z., Liu Z., Huang X. OSBP- and FAN-mediated sterol requirement for spermatogenesis in Drosophila. Development. 2010;137:3775–3784. doi: 10.1242/dev.049312. [DOI] [PubMed] [Google Scholar]
- 59.Ridgway N.D. Oxysterol-binding proteins. Subcell. Biochem. 2010;51:159–182. doi: 10.1007/978-90-481-8622-8_6. [DOI] [PubMed] [Google Scholar]
- 60.Raychaudhuri S., Prinz W.A. The diverse functions of oxysterol-binding proteins. Annu. Rev. Cell Dev. Biol. 2010;26:157–177. doi: 10.1146/annurev.cellbio.042308.113334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan D., Olkkonen V.M. Characteristics of oxysterol binding proteins. Int. Rev. Cytol. 2008;265:253–285. doi: 10.1016/S0074-7696(07)65007-4. [DOI] [PubMed] [Google Scholar]
- 62.Goto A., Liu X., Robinson C.A., Ridgway N.D. Multisite phosphorylation of oxysterol-binding protein regulates sterol binding and activation of sphingomyelin synthesis. Mol. Biol. Cell. 2012;23:3624–3635. doi: 10.1091/mbc.E12-04-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banerji S., Ngo M., Lane C.F., Robinson C.A., Minogue S., Ridgway N.D. Oxysterol binding protein-dependent activation of sphingomyelin synthesis in the golgi apparatus requires phosphatidylinositol 4-kinase IIalpha. Mol. Biol. Cell. 2010;21:4141–4150. doi: 10.1091/mbc.E10-05-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee T., Winter C., Marticke S.S., Lee A., Luo L. Essential roles of Drosophila RhoA in the regulation of neuroblast proliferation and dendritic but not axonal morphogenesis. Neuron. 2000;25:307–316. doi: 10.1016/s0896-6273(00)80896-x. [DOI] [PubMed] [Google Scholar]
- 65.Stanley H., Botas J., Malhotra V. The mechanism of Golgi segregation during mitosis is cell type-specific. Proc. Natl. Acad. Sci. USA. 1997;94:14467–14470. doi: 10.1073/pnas.94.26.14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barr F.A., Nakamura N., Warren G. Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. EMBO J. 1998;17:3258–3268. doi: 10.1093/emboj/17.12.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yano H., Yamamoto-Hino M., Abe M., Kuwahara R., Haraguchi S., Kusaka I., Awano W., Kinoshita-Toyoda A., Toyoda H., Goto S. Distinct functional units of the Golgi complex in Drosophila cells. Proc. Natl. Acad. Sci. USA. 2005;102:13467–13472. doi: 10.1073/pnas.0506681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kabashi E., El Oussini H., Bercier V., Gros-Louis F., Valdmanis P.N., McDearmid J., Mejier I.A., Dion P.A., Dupre N., Hollinger D., et al. Investigating the contribution of VAPB/ALS8 loss of function in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2013;22:2350–2360. doi: 10.1093/hmg/ddt080. [DOI] [PubMed] [Google Scholar]
- 69.Venken K.J., Simpson J.H., Bellen H.J. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron. 2011;72:202–230. doi: 10.1016/j.neuron.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Venken K.J., He Y., Hoskins R.A., Bellen H.J. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- 71.Tanouye M.A., Wyman R.J. Motor outputs of giant nerve fiber in Drosophila. J. Neurophysiol. 1980;44:405–421. doi: 10.1152/jn.1980.44.2.405. [DOI] [PubMed] [Google Scholar]
- 72.Pavlidis P., Tanouye M.A. Seizures and failures in the giant fiber pathway of Drosophila bang-sensitive paralytic mutants. J. Neurosci. 1995;15:5810–5819. doi: 10.1523/JNEUROSCI.15-08-05810.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koenig J.H., Ikeda K. Evidence for a presynaptic blockage of transmission in a temperature-sensitive mutant of Drosophila. J. Neurobiol. 1983;14:411–419. doi: 10.1002/neu.480140602. [DOI] [PubMed] [Google Scholar]
- 74.Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Bio. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 75.Parks A.L., Cook K.R., Belvin M., Dompe N.A., Fawcett R., Huppert K., Tan L.R., Winter C.G., Bogart K.P., Deal J.E., et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- 76.Brodsky J.L., McCracken A.A. ER protein quality control and proteasome-mediated protein degradation. Semin. Cell. Dev. Biol. 1999;10:507–513. doi: 10.1006/scdb.1999.0321. [DOI] [PubMed] [Google Scholar]
- 77.Latterich M., Frohlich K.U., Schekman R. Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell. 1995;82:885–893. doi: 10.1016/0092-8674(95)90268-6. [DOI] [PubMed] [Google Scholar]
- 78.Lilley B.N., Ploegh H.L. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- 79.Parlati F., Dominguez M., Bergeron J.J., Thomas D.Y. Saccharomyces cerevisiae CNE1 encodes an endoplasmic reticulum (ER) membrane protein with sequence similarity to calnexin and calreticulin and functions as a constituent of the ER quality control apparatus. J. Biol. Chem. 1995;270:244–253. doi: 10.1074/jbc.270.1.244. [DOI] [PubMed] [Google Scholar]
- 80.Johnson J.O., Mandrioli J., Benatar M., Abramzon Y., Van Deerlin V.M., Trojanowski J.Q., Gibbs J.R., Brunetti M., Gronka S., Wuu J., et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bernard-Marissal N., Moumen A., Sunyach C., Pellegrino C., Dudley K., Henderson C.E., Raoul C., Pettmann B. Reduced calreticulin levels link endoplasmic reticulum stress and Fas-triggered cell death in motoneurons vulnerable to ALS. J. Neurosci. 2012;32:4901–4912. doi: 10.1523/JNEUROSCI.5431-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Teuling E., Ahmed S., Haasdijk E., Demmers J., Steinmetz M.O., Akhmanova A., Jaarsma D., Hoogenraad C.C. Motor neuron disease-associated mutant vesicle-associated membrane protein-associated protein (VAP) B recruits wild-type VAPs into endoplasmic reticulum-derived tubular aggregates. J. Neurosci. 2007;27:9801–9815. doi: 10.1523/JNEUROSCI.2661-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Anagnostou G., Akbar M.T., Paul P., Angelinetta C., Steiner T.J., de Belleroche J. Vesicle associated membrane protein B (VAPB) is decreased in ALS spinal cord. Neurobiol. Aging. 2010;31:969–985. doi: 10.1016/j.neurobiolaging.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 84.Ngo M., Ridgway N.D. Oxysterol binding protein-related Protein 9 (ORP9) is a cholesterol transfer protein that regulates Golgi structure and function. Mol. Biol. Cell. 2009;20:1388–1399. doi: 10.1091/mbc.E08-09-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Suchanek M., Hynynen R., Wohlfahrt G., Lehto M., Johansson M., Saarinen H., Radzikowska A., Thiele C., Olkkonen V.M. The mammalian oxysterol-binding protein-related proteins (ORPs) bind 25-hydroxycholesterol in an evolutionarily conserved pocket. Biochem. J. 2007;405:473–480. doi: 10.1042/BJ20070176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lange Y. Disposition of intracellular cholesterol in human fibroblasts. J. Lipid Res. 1991;32:329–339. [PubMed] [Google Scholar]
- 87.Ridsdale A., Denis M., Gougeon P.Y., Ngsee J.K., Presley J.F., Zha X. Cholesterol is required for efficient endoplasmic reticulum-to-Golgi transport of secretory membrane proteins. Mol. Biol. Cell. 2006;17:1593–1605. doi: 10.1091/mbc.E05-02-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Waugh M.G., Minogue S., Chotai D., Berditchevski F., Hsuan J.J. Lipid and peptide control of phosphatidylinositol 4-kinase IIalpha activity on Golgi-endosomal rafts. J. Biol. Chem. 2006;281:3757–3763. doi: 10.1074/jbc.M506527200. [DOI] [PubMed] [Google Scholar]
- 89.Swanton C., Marani M., Pardo O., Warne P.H., Kelly G., Sahai E., Elustondo F., Chang J., Temple J., Ahmed A.A., et al. Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs. Cancer Cell. 2007;11:498–512. doi: 10.1016/j.ccr.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 90.Kanekura K., Nishimoto I., Aiso S., Matsuoka M. Characterization of amyotrophic lateral sclerosis-linked P56S mutation of vesicle-associated membrane protein-associated protein B (VAPB/ALS8) J. Biol. Chem. 2006;281:30223–30233. doi: 10.1074/jbc.M605049200. [DOI] [PubMed] [Google Scholar]
- 91.Ratnaparkhi A., Lawless G.M., Schweizer F.E., Golshani P., Jackson G.R. A Drosophila model of ALS: human ALS-associated mutation in VAP33A suggests a dominant negative mechanism. PLoS One. 2008;3:e2334. doi: 10.1371/journal.pone.0002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bischof J., Maeda R.K., Hediger M., Karch F., Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bellen H.J., Levis R.W., Liao G., He Y., Carlson J.W., Tsang G., Evans-Holm M., Hiesinger P.R., Schulze K.L., Rubin G.M., et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thibault S.T., Singer M.A., Miyazaki W.Y., Milash B., Dompe N.A., Singh C.M., Buchholz R., Demsky M., Fawcett R., Francis-Lang H.L., et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 95.Golic K.G., Golic M.M. Engineering the Drosophila genome: chromosome rearrangements by design. Genetics. 1996;144:1693–1711. doi: 10.1093/genetics/144.4.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yeh E., Gustafson K., Boulianne G.L. Green fluorescent protein as a vital marker and reporter of gene expression in Drosophila. Proc. Natl. Acad. Sci. USA. 1995;92:7036–7040. doi: 10.1073/pnas.92.15.7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Torroja L., Packard M., Gorczyca M., White K., Budnik V. The Drosophila beta-amyloid precursor protein homolog promotes synapse differentiation at the neuromuscular junction. J. Neurosci. 1999;19:7793–7803. doi: 10.1523/JNEUROSCI.19-18-07793.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.O'Neill E.M., Rebay I., Tjian R., Rubin G.M. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- 99.Kidd T., Brose K., Mitchell K.J., Fetter R.D., Tessier-Lavigne M., Goodman C.S., Tear G. Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell. 1998;92:205–215. doi: 10.1016/s0092-8674(00)80915-0. [DOI] [PubMed] [Google Scholar]
- 100.Verstreken P., Koh T.W., Schulze K.L., Zhai R.G., Hiesinger P.R., Zhou Y., Mehta S.Q., Cao Y., Roos J., Bellen H.J. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron. 2003;40:733–748. doi: 10.1016/s0896-6273(03)00644-5. [DOI] [PubMed] [Google Scholar]
- 101.Casas-Tinto S., Zhang Y., Sanchez-Garcia J., Gomez-Velazquez M., Rincon-Limas D.E., Fernandez-Funez P. The ER stress factor XBP1 s prevents amyloid-beta neurotoxicity. Hum. Mol. Genet. 2011;20:2144–2160. doi: 10.1093/hmg/ddr100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hiesinger P.R., Fayyazuddin A., Mehta S.Q., Rosenmund T., Schulze K.L., Zhai R.G., Verstreken P., Cao Y., Zhou Y., Kunz J., et al. The v-ATPase V0 subunit a1 is required for a late step in synaptic vesicle exocytosis in Drosophila. Cell. 2005;121:607–620. doi: 10.1016/j.cell.2005.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.