Abstract
Neuronal ceroid lipofuscinosis (NCL) comprises ∼13 genetically distinct lysosomal disorders primarily affecting the central nervous system. Here we report successful reprograming of patient fibroblasts into induced pluripotent stem cells (iPSCs) for the two most common NCL subtypes: classic late-infantile NCL, caused by TPP1(CLN2) mutation, and juvenile NCL, caused by CLN3 mutation. CLN2/TPP1- and CLN3-iPSCs displayed overlapping but distinct biochemical and morphological abnormalities within the endosomal–lysosomal system. In neuronal derivatives, further abnormalities were observed in mitochondria, Golgi and endoplasmic reticulum. While lysosomal storage was undetectable in iPSCs, progressive disease subtype-specific storage material was evident upon neural differentiation and was rescued by reintroducing the non-mutated NCL proteins. In proof-of-concept studies, we further documented differential effects of potential small molecule TPP1 activity inducers. Fenofibrate and gemfibrozil, previously reported to induce TPP1 activity in control cells, failed to increase TPP1 activity in patient iPSC-derived neural progenitor cells. Conversely, nonsense suppression by PTC124 resulted in both an increase of TPP1 activity and attenuation of neuropathology in patient iPSC-derived neural progenitor cells. This study therefore documents the high value of this powerful new set of tools for improved drug screening and for investigating early mechanisms driving NCL pathogenesis.
INTRODUCTION
The neuronal ceroid lipofuscinosis (NCL) disorders (sometimes referred to as Batten disease) are a group of at least 13 distinct lysosomal diseases with overlapping clinical features including progressive motor and cognitive decline, pigmentary retinal degeneration and visual loss in most cases, seizures, movement disorder and eventual premature death (1). Collectively, the NCLs represent the major Mendelian cause of neurodegeneration among children, affecting ∼1–4 in 100 000 live births in most geographic regions (2). Of the distinct NCL subtypes, the most common are classic late-infantile NCL (CLN2), caused by loss-of-function mutations in TPP1, which encodes the soluble lysosomal enzyme tripeptidyl peptidase-1 (TPP1), and juvenile NCL (CLN3), caused by loss-of-function mutations in CLN3, which encodes a transmembrane endosomal/lysosomal protein (CLN3 or battenin) with orthologs from yeast to mammals (2). The histopathologic features common to the NCLs include lysosomal accumulation of autofluorescent, electron-dense material composed of oxidized lipids and undigested proteins, in particular, subunit c of the mitochondrial ATP synthase complex (3). These findings suggest the differing genetic underpinnings of NCL subtypes converge on a final common pathway that may involve incomplete clearance of mitochondria targeted to lysosomes (4).
While TPP1 functions as a lysosomal enzyme, its in vivo substrates are unknown, though subunit c is likely one of them (4). How TPP1 deficiency ultimately results in neuronal cell dysfunction is unknown. Similarly, though cloned more than a decade ago, the primary CLN3 protein function remains unresolved (for a recent review 5). An expanding CLN3 protein interaction network (6–11) links the protein to regulators of endocytic pathway trafficking and the cytoskeleton, consistent with research using lower mammalian and other eukaryotic NCL models suggesting that defects in endocytic trafficking, autophagy, lipid processing and cytoskeletal organization are major components of NCL pathogenesis, particularly for juvenile NCL (CLN3) (1,12–15). The yeast ortholog of CLN3, Btn1p, has been shown to be important for vacuolar pH homeostasis and amino acid transport (16–19), and to regulate transport of a subset of proteins between the vacuole (the yeast equivalent of the lysosome) and the trans-Golgi network (20, 21) and a similar role for CLN5 in retrograde trafficking has been demonstrated in HeLa cells (22).
While single-cell and other organism models offer useful guiding principles, it remains unknown whether the observations made in them are valid in human neurons or recapitulate critical aspects of the human disease. For this reason, the derivation of induced pluripotent stem cells (iPSCs) from patients and subsequent differentiation to specific cell types of interest has emerged as a powerful tool for human disease modeling and therapeutic screening (23, 24). Although subject to the limitations of any in vitro model system, iPSCs and their derivatives from patients with Mendelian disorders provide the precise genetic lesion and genetic background for testing mechanistic hypotheses and potential therapies in a genotype-specific manner.
Here we provide the first report of successful reprograming of patient fibroblasts with mutations in CLN2/TPP1 and CLN3. We have interrogated the earliest disease events in the human iPSCs at the pluripotent stage and through neuronal differentiation, and demonstrate distinct, but also overlapping, early-stage pathology in multiple subcellular compartments. We also report on assays and proof-of-concept studies that establish a new platform for improved genotype-directed NCL therapy development and drug screening in patient neurons.
RESULTS
Derivation and characterization of iPSCs from late-infantile and juvenile NCL patient fibroblasts
Fibroblasts from two late-infantile NCL patients (CLN2; hereafter designated TPP1 lines), four juvenile NCL patients (homozygous or compound heterozygous for CLN3 mutations; hereafter designated CLN3 lines) and two unaffected individuals (normal TPP1 and CLN3 alleles, or heterozygous for the common CLN3 ∼1-kb deletion) were reprogramed using the standard four-factor retroviral reprograming method (OCT4, SOX2, MYC and KLF4) (25–27). One additional iPSC line previously established by the same method was also used as a control in these studies (27). Multiple iPSC lines from each of the patient fibroblast lines were isolated and expanded for further characterization, as summarized in Supplementary Material, Table S1.
The pluripotent state of iPSC clones was confirmed by positive staining for alkaline phosphatase and the pluripotency markers SSEA4, OCT4 and TRA-1–60 (Supplementary Material, Fig. S1A). Silencing of exogenous transgenes was verified by qRT–PCR using freshly infected donor fibroblasts as a positive control, and by the loss of GFP reporter signal from the MIG retroviral vectors (see Materials and Methods). Viral transgenes were efficiently silenced in all patient-derived iPSC clones (Supplementary Material, Fig. S1B and data not shown). Furthermore, all iPSC clones could be induced to form embryoid bodies which gave rise to cells from all three germ layers in vitro: meso-, endo- and ectoderm (Supplementary Material, Fig. S1C, left panel). PCR analysis of embryoid bodies confirmed expression of genes specific for differentiation (Supplementary Material, Fig. S1C, right panel). Thus, iPSCs derived from TPP1 and CLN3 patients, and unaffected individuals, are pluripotent. The TPP1 and CLN3 mutation status of the iPSC lines was also confirmed by PCR (Supplementary Material, Fig. S2A and B). As observed in brain tissue and cerebellar cells derived from the Cln3Δex7/8 knockin model, which bears the common ∼1-kb CLN3 deletion mutation found in most juvenile NCL patients, and wild-type littermates (28), multiple CLN3 transcripts in addition to the major ∼1.8-kb form were present in control iPSCs and neural progenitor cells (NPCs). Cells homozygous for the common CLN3 deletion, however, showed multiple, distinct smaller transcripts, with a major form at ∼1.4 kb (Supplementary Material, Fig. S2C). 5′-UTR and 3′-UTR-anchored RT–PCR and cloning of CLN3 cDNA from CLN3IVS13/E15 fibroblasts and iPSCs showed stable transcripts bearing the point mutation on the c.1247 A>G allele, as well as multiple aberrant splice products surrounding the intron 13 (IVS13) mutation, confirming the pathogenicity of the c.1056 + 3 A>C mutation (Supplementary Material, Fig. S2C and Table S2).
The late endosomal–lysosomal compartment is disrupted in CLN3 iPSCs
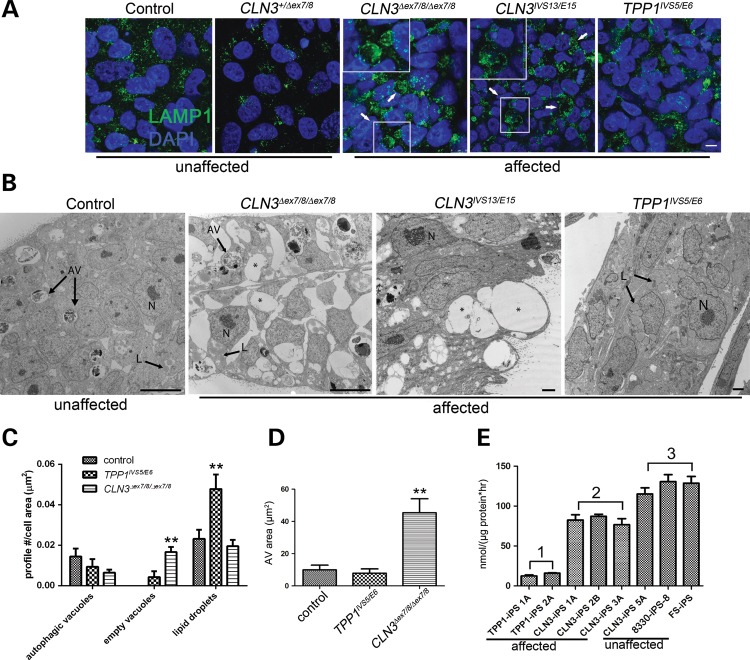
Prior evidence exists for abnormalities in the lysosomal, mitochondrial and Golgi compartments in models of NCL, particularly juvenile NCL (for example 15, 21). However, it remains controversial what of these defects is primary in the disease pathogenesis, and the primary defects probably differ depending on the gene mutation. This motivated us to survey the membrane organelles in our set of unaffected and affected NCL iPSCs. While immunostaining for Golgi and mitochondrial marker proteins (GM130 and GRP75, respectively) was unrevealing of clear genotypic differences (data not shown), immunofluorescence for the lysosomal transmembrane protein LAMP1 highlighted frequent, unusually large ring-like structures in CLN3 patient iPSCs, which were not observed in control and TPP1 patient iPSCs (Fig. 1A). Similarly, we observed numerous apparently empty vacuolar structures (up to 20 µm in diameter) by transmission electron microscopy (TEM) (Fig. 1B), although definitively determining whether these correspond to the LAMP1-labeled ring-like structures at the immunofluorescence level would require further studies. Notably, despite the observed differences in LAMP1 immunofluorescence, total LAMP1 levels, detected by immunoblotting of cell lysates, were not significantly different as compared with unaffected cell lines (Supplementary Material, Fig. S3A).
Figure 1.
TPP1 and CLN3 patient iPSCs show distinct pathology in lysosomal and autophagic compartments. (A) iPSC cells derived from control (unaffected) and patient (affected) fibroblasts were grown on Matrigel-coated cover slips and immunostained for lysosomal vesicles with LAMP1. Arrows and insets demonstrate enlarged LAMP1-positive vacuoles in two different representative CLN3 iPSC lines. Scale bar: 10 µm. (B) Representative TEM images of control (unaffected) and patient-derived (affected) iPSCs. N, nucleus; AV, degradative autophagic vacuoles; L, lipid droplets; asterisks, empty vacuoles. Scale bars: 10 nm (control and CLN3Δex7/8/Δex7/8), 2 nm (CLN3IVS13/E15 and TPP1IVS5/E6). (C) Quantification of structures demonstrated in (B). No significant difference was observed in the autophagic vacuole profile count per cell area (P > 0.05). Significantly more empty vacuoles were observed in CLN3Δex7/8/Δex7/8 cells (**P < 0.01), and significantly more cytoplasmic lipid droplets were observed in TPP1IVS5/E6 cells (**P < 0.01). Data are shown as mean ± SD. Similar qualitative patterns were observed in at least two additional iPSC lines each from TPP1 and CLN3-affected patients. (D) AV area measured for at least 20 representative degradative autophagic vacuoles per cell line, which were defined as described in Materials and Methods. **P < 0.01. (E) TPP1 enzyme activity in patient iPSC lines (affected), specifically TPP1 lines (Group 1) and CLN3 lines (Group 2), compared with control (unaffected) lines (Group 3; note that CLN3-iPS 5A is from a phenotypically normal donor heterozygous for the common CLN3Δex7/8 deletion). P < 0.01, Group 1 versus Groups 2 and 3. P < 0.05, Group 2 versus Group 3. Data are shown as mean ± SD of three technical replicates per line normalized to total protein.
We also probed the NCL iPSCs for disease characteristic storage material using an established subunit c antibody (29) and by ultrastructural examination. Autofluorescence was not detected above background levels present in unaffected iPSC lines (data not shown). Similarly, no disease-related subunit c accumulation was detected in any of the patient iPSC lines (Supplementary Material, Fig. S3B and data not shown). Moreover, we did not observe abnormal storage material by TEM analysis of the iPSC lines (Fig. 1). In all iPSC lines examined, autophagic compartments were readily observed. The major autophagic structures observed were interpreted as late/degradative autophagic vacuoles and autolysosomes (‘AV’ in Fig. 1) containing a single limiting membrane and cellular constituents at various stages of degradation, based on established morphologic criteria as outlined in Materials and Methods. Double-membrane-bound initial autophagosomes containing morphologically intact organelles were not readily observed. Although the average number of AV per cell, normalized to total cell area, did not differ significantly between control, CLN3 and TPP1 lines, the average area of the identified AV in CLN3 patient lines was significantly larger than those observed in control or TPP1 lines (Fig. 1C and D). Intriguingly, the TPP1 patient lines also showed abundant cytoplasmic lipid droplets but few empty vacuoles, and AVs were not enlarged (Fig. 1B–D). Finally, a fluorometric assay for TPP1 enzyme activity in iPSCs demonstrated robust activity in control cells and markedly reduced levels in the TPP1 patient cells as expected. Interestingly, intermediate TPP1 levels were found in all CLN3-affected patient iPSC lines examined (∼60% of control levels, Fig. 1E).
Generation of neural progenitor cells from control and patient-specific iPSCs
To gain further insight into NCL pathophysiology in vulnerable cell populations such as neurons, we generated and characterized NPCs from iPSCs. NPCs expressing the expected neural progenitor markers Nestin, SOX1, SOX2 and Musashi were generated by first differentiating the iPSCs to embryoid bodies, followed by dissociation and isolation and expansion of neural rosettes (see Materials and Methods and Supplementary Material, Fig. S4A). To further confirm the neural nature of the generated NPCs and therefore the loss of pluripotency, we performed qRT–PCR analysis comparing NPC marker gene expression levels to those of iPSCs. As expected, pluripotency genes such as NANOG and OCT4 were downregulated within the NPC populations whereas genes characteristic for NPCs such as PAX6, SOX1 and MUSASHI1 were upregulated compared with iPSCs (Supplementary Material, Fig. S4B). We additionally compared gene expression levels of NPC lines to those of embryoid bodies. Genes for meso- and endoderm (GATA4 and T, respectively) as well as for epidermis (KRT18) and neural crest (PAX3) were markedly downregulated in comparison to embryoid bodies; CDX2, which is prominently expressed in extra-embryonic tissue, yielded no amplification (Supplementary Material, Fig. S4C).
Neural precursor cells derived from CLN3 and TPP1 patient iPSCs display NCL-like characteristics
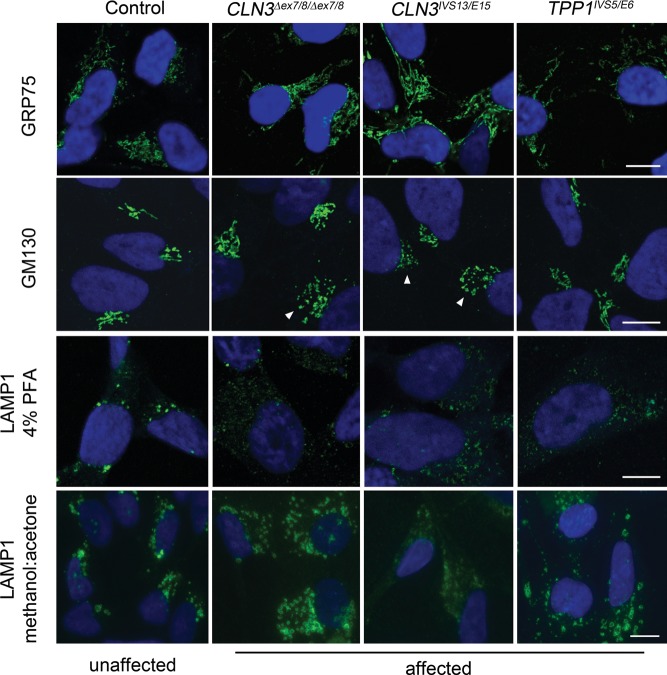
We next sought to further evaluate organellar pathology (mitochondria, Golgi, late endosomes/lysosomes) in the CLN3 and TPP1 patient iPSC-derived NPC lines. Immunostaining for the mitochondrial marker GRP75 showed a variable staining pattern in the different NPC lines and therefore genotypic differences at the immunoflourescence level were not immediately evident for this marker (Fig. 2). Immunostaining for the cis-Golgi marker GM130, though also somewhat variable in the different NPC lines, consistently appeared more vesiculated and less organized in all CLN3 patient NPC lines compared with control or TPP1 lines (Fig. 2). Image analysis of the GM130 immunostained structures revealed a larger proportion of Golgi structures with a high circularity score in the CLN3 NPCs, compared with the proportion of structures with a high circularity score in control NPCs, consistent with increased vesiculation. In control NPCs, 38 ± 2% of GM130 structures analyzed had a circularity score between 0.9 and 1 using ImageJ. The percentage of identified GM130 structures with a circularity score between 0.9 and 1 in CLN3 NPCs was 46 ± 1% for CLN3Δex7/8/Δex7/8 NPCs (P < 0.01 versus controls) and 52 ± 2% for CLN3IVS13/E15 NPCs (P < 0.01 versus controls). The GM130 staining pattern in TPP1 patient NPCs was not easily distinguishable from control NPCs (TPP1IVS5/E6 = 37 ± 1%, P > 0.5 versus controls).
Figure 2.
TPP1 and CLN3 patient NPCs display abnormal Golgi and endosomal/lysosomal marker staining patterns. Control (unaffected) and patient (affected) NPCs were fixed and stained for GRP75 (mitochondria), GM130 (cis-Golgi), or LAMP1 (late endosomes/lysosomes). Representative images are shown. White arrowheads denote examples of CLN3 patient cells with a vesiculated GM130 staining pattern. Scale bars: 10 µm. PFA, paraformaldehyde.
LAMP1 immunostaining also revealed morphological alterations in CLN3 and TPP1 patient NPCs, as compared with control cells. Brightly stained puncta were observed mostly in the perinuclear region in control cells fixed with paraformaldehyde (PFA), while faintly labeled LAMP1 puncta were often dispersed into the cell periphery in CLN3 and TPP1 patient cells fixed with PFA (Fig. 2). LAMP1-positive structures were more readily observed when the NPCs were fixed using methanol/acetone (50%/50%), rather than PFA (Fig. 2). The improved LAMP1 antibody staining revealed numerous ring-like structures that extended into the cell periphery in both CLN3 and TPP1 patient NPCs (67 ± 12% and 78 ± 14% of the CLN3 and TPP1 NPC cells, respectively, displayed this pattern, compared with 35 ± 12% of control NPC cells displaying prominent peripheral LAMP1-positive vesicles; P < 0.01).
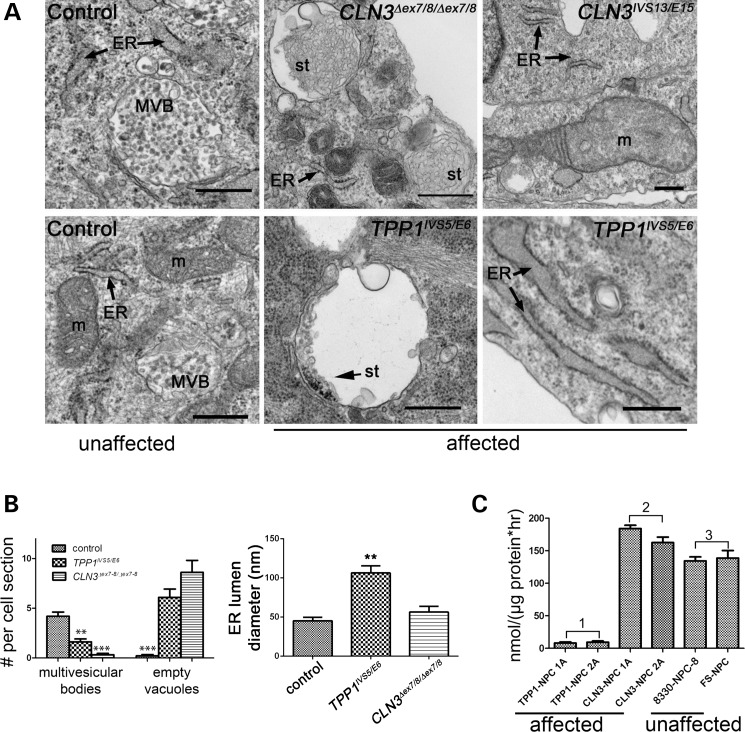
Significantly, by TEM we also observed membrane-bound structures partly filled with storage-like material in both CLN3 and TPP1 patient lines in ∼5% of cells (Fig. 3A). This storage-like material resembled curvilinear bodies but was not identical to the classic curvilinear storage material typically observed in clinically progressed NCL patients at the time of diagnosis. TEM of the TPP1 and CLN3 patient NPC lines also showed numerous ‘empty’ vacuolar structures ∼200–800 nm in diameter, similar to but smaller than those observed in CLN3 patient iPSCs (Fig. 3A and B). Correlated with the presence of the mostly empty vacuoles was a marked depletion of multivesicular bodies, a subset of secondary endosomes ∼100–600 nm in diameter containing intraluminal vesicles of ∼25–50 nm (Fig. 3B). This deficiency was especially pronounced in CLN3 patient NPCs. We also observed morphologic aberrations in ∼15–25% of mitochondria sampled in CLN3 patient NPC lines, including distended mitochondria, disruption of the mitochondrial internal architecture and loss of cristae. These mitochondrial abnormalities were not observed in control and TPP1 patient NPC lines (Fig. 3A). TPP1 patient NPCs most notably displayed pronounced dilation of the endoplasmic reticulum (ER) and a marked deficiency of TPP1 enzyme activity (Fig. 3B and C). Intriguingly, rather than the relative TPP1 enzyme deficiency observed at the iPSC stage, CLN3 patient NPCs showed slight but statistically significant elevations of TPP1 enzyme activity compared with control NPCs (Fig. 3C).
Figure 3.
NCL-like storage and organelle abnormalities are observed in ultrastructural studies of TPP1 and CLN3 patient NPCs. (A) Representative TEM images showing control (unaffected) and patient (affected) NPCs (represented genotypes are indicated) grown under subconfluent conditions. Abbreviations: ER, endoplasmic reticulum; MVB, multivesicular body; st, storage material; m, mitochondria. Scale bars: 500 nm. (B) Manual counts of multivesicular bodies and empty vacuoles, and ER lumen diameter measurements, from representative control versus TPP1 and CLN3-affected patient lines. Data are shown as mean ± SEM of counts in at least 20 cells per line. **P < 0.01; ***P < 0.001; nm, nanometers. (C) TPP1 enzyme activity in affected TPP1 lines (Group 1), affected CLN3 lines (Group 2) and unaffected control lines (Group 3). P < 0.001, Group 1 versus Groups 2 and 3. P < 0.05, Group 2 versus Group 3. Data are shown as mean ± SD of three technical replicates per line normalized to total protein.
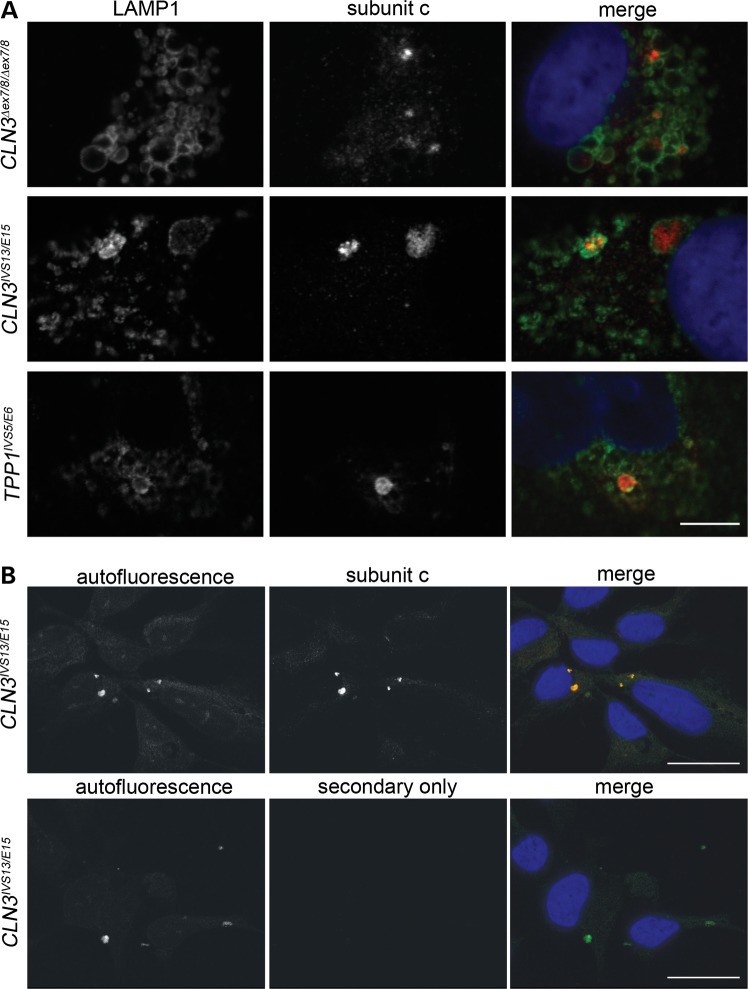
The observation of storage-like material by TEM prompted us to further examine the NCL patient NPCs by subunit c immunostaining and autofluorescence analyses. Indeed, we observed cytoplasmic, subunit c-positive accumulations in the main cell body of all TPP1 and CLN3 patient NPC lines examined. These accumulations of subunit c were localized to LAMP1-positive compartments (Fig. 4A). Moreover, the subunit c-positive accumulations co-localized with autofluorescent deposits (Fig. 4B). Thus, the features of the storage material observed in the NCL patient NPCs were highly consistent with what has been described in other in vitro and in vivo NCL model systems and in patient autopsy material (for example 2,15,28,29).
Figure 4.
Characterization of NCL-like storage material in TPP1 and CLN3 patient NPCs. (A) Representative immunofluorescence images are shown of LAMP1 and subunit c co-immunostained NCL patient NPCs. Scale bar: 10 µm. (B) Representative images of subunit c staining and autofluorescence analysis of NCL patient NPCs are shown (a representative CLN3IVS13/E15 line is shown, and results are typical of that observed in other patient lines). ‘Secondary only’ indicates results obtained when primary antibody was omitted. Scale bars: 20 µm.
Mature TPP1 and CLN3 patient neurons show progressive organellar pathology and characteristic NCL storage material
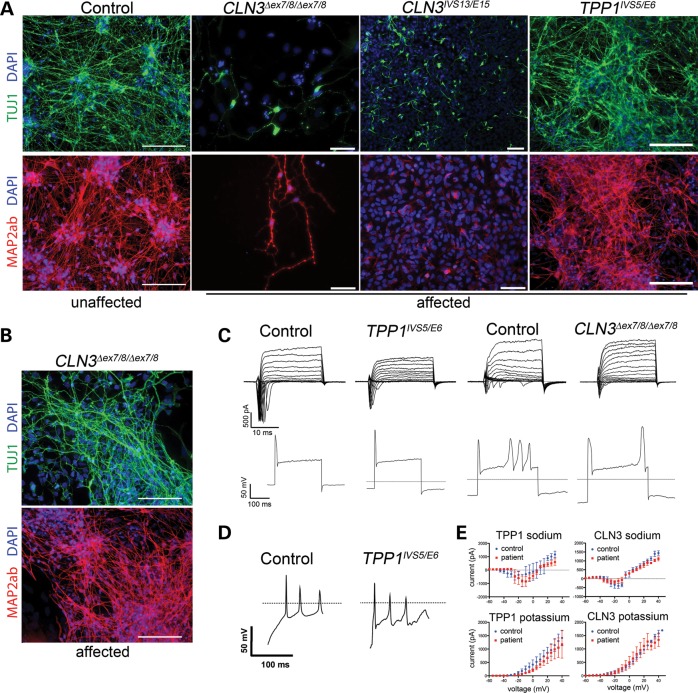
Mature neurons derived from TPP1 and CLN3 patients are of great interest because they represent the primary cell type that undergoes progressive degeneration during the NCL disease process. We therefore investigated the neuronal differentiation potential of TPP1 and CLN3 patient NPCs using a standard protocol involving withdrawal of mitogens and addition of cyclic AMP to NPC cultures grown as monolayers for at least 2 weeks. While robust neuronal differentiation was achieved using this method for control and TPP1 patient NPC cultures, this approach resulted in a poor yield of immature and mature neurons in CLN3 patient NPC cultures, which tended to detach from the plates over the differentiation period and displayed only stunted neuritic outgrowths in the remaining cells (Fig. 5A). Extending differentiation to 3 or 4 weeks did not increase the number of neurons derived from CLN3 patient NPCs (data not shown). The failure of CLN3 patient NPC lines to efficiently differentiate from monolayer cultures was independent of passage number and reproducible across cell lines from multiple donor patients, and prompted us to also examine growth. In BrdU incorporation assays, we consistently observed a considerable deficiency in the CLN3 patient NPCs' recovery after plating in comparison to control and TPP1 patient NPCs, though no significant differences in growth rate per se were observed over the subsequent 24-h period (Supplementary Material, Fig. S5A, B). TUNEL assay 24 h postreplating revealed that there was no increase in apoptosis in still-attached TPP1 and CLN3 cells compared with control NPCs (Supplementary Material, Fig. 5C).
Figure 5.
Differentiation potential of TPP1 and CLN3 patient iPSCs and NPCs. (A) NPC monolayer cultures derived from control (unaffected), CLN3-affected patient (CLN3Δex7/8/Δex7/8 and CLN3IVS13/E15) and TPP1-affected patient (TPP1IVS5/E6) iPSCs were differentiated for 14 days and stained for markers of immature neurons (TUJ1, green) and mature neurons (MAP2ab, red). Representative images from one line per genotype are shown. Cell nuclei are counterstained with DAPI (blue). Scale bars: 100 µm. (B) CLN3-affected patient neurons, differentiated via iPSC-derived neuralized embryoid bodies (EB), are shown. Cultures were differentiated for 14 days and stained for markers of immature (TUJ1, green) as well as mature neurons (MAP2ab, red). Representative images from a CLN3 patient iPSC line are shown. Cell nuclei are counterstained with DAPI (blue). Scale bars: 100 µm. (C–E) Electrophysiological properties of neurons from control or patient-derived iPSCs. (C) Representative traces from voltage-clamp recordings are depicted for each genotype. Two different control (unaffected) examples are shown, representing neurons derived using the monolayer differentiation method (left) and the neuralized EB differentiation method (right), to allow for the best comparisons to the TPP1 and CLN3-affected patient neuron traces, respectively. Representative images from current-clamp recordings show the ability to fire action potentials in response to depolarizing stimuli. (D) Spontaneous firing of action potentials seen in control (unaffected) and TPP1-affected patient-derived neurons. (E) Current–voltage relationships are shown.
An alternative neuronal differentiation approach that promotes cell–cell contact in neuralized embryoid bodies was therefore also tested (30). Using this approach, CLN3 patient iPSCs successfully gave rise to TUJ1 (bIII tubulin)-positive immature and MAP2ab-positive mature neurons (Fig. 5B), to an extent comparable with that from control iPSCs (not shown), as well as from control and TPP1-deficient NPC lines grown as monolayer cultures and differentiated for 14 days (Fig. 5A). Further, electrophysiological analysis showed the expression of sodium channels and the capability of firing spontaneous and depolarization-dependent action potentials, thus confirming the acquisition of properties of mature and functional neurons from both differentiation methods (Fig. 5C–E, Supplementary Material, Table S4). No significant differences were observed in the electrophysiological properties of TPP1 or CLN3 patient neurons, compared with the respective control neurons, though sodium currents were significantly larger after monolayer differentiation in general (Fig. 5E; Supplementary Material, Table S4).
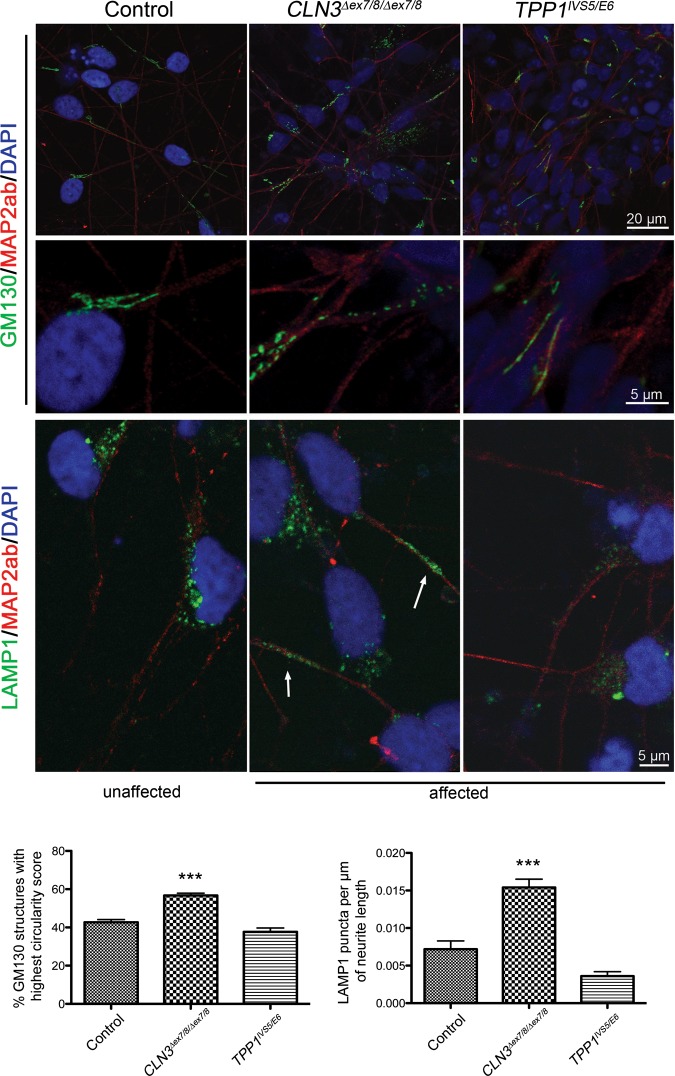
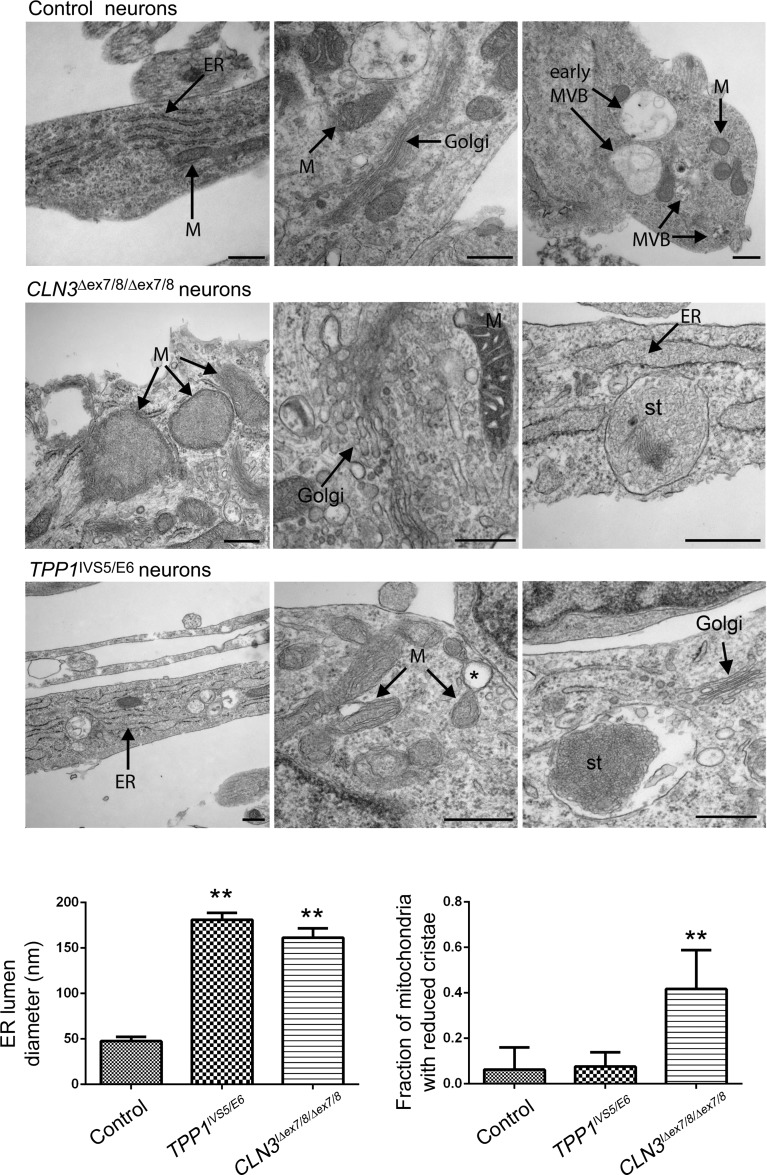
We next evaluated differentiated TPP1 and CLN3 neurons for organellar pathology and the presence of storage material. Similar to our observation in NPC cultures, GM130 staining in the patient lines revealed dispersion of normally compact, juxtanuclear Golgi structures and a significantly increased percentage of GM130 labeled structures with a high circularity score as compared with control or TPP neurons (Fig. 6). A markedly vesicular Golgi network with disorganized cisternal stacking and disrupted intercisternal bridging was also observed by TEM (Fig. 7). In contrast to control neurons where LAMP1-positive puncta were mostly localized to the perinuclear region, LAMP1 staining was dispersed throughout much of the cell bodies and often into primary and secondary neurites in CLN3 patient neurons (Fig. 6). Quantification revealed an ∼2-fold increase of neurite-localized LAMP1 puncta in the CLN3 neurons versus control neurons (Fig. 6). Clear alterations in LAMP1 immunostaining were not immediately evident in the TPP1 neurons, versus the control neurons (Fig. 6). GRP75 immunostaining to label the mitochondrial network was again variable, as it was in the NPCs, and therefore did not highlight obvious genotypic differences (Supplementary Material, Fig. S6). However, the mitochondrial dysmorphology seen in CLN3 patient NPCs at the TEM level was even more pronounced in mature neurons, with ∼50% of the mitochondria displaying reduced cristae (Fig. 7). Though no major significant changes in mitochondrial cristae density were observed in TPP1 patient neurons, occasional mitochondria in these cells were noted to display expansion of intermembrane spaces and vacuolation (Fig. 7), similar to the pattern described in SOD1 mouse models of amyotrophic lateral sclerosis (31). The ER in TPP1 patient neurons was globally dilated, and a subset of the ER in CLN3 patient neurons was also significantly dilated (Fig. 7). Neurons also showed numerous intracytoplasmic, membrane-bound structures containing storage material of curvilinear morphology in TPP1 patient neurons and mixed curvilinear and fingerprint morphologies in CLN3 patient neurons (Fig. 7). It is noteworthy that the curvilinear profile predominates in TPP1 patient biopsy samples, while the fingerprint profile predominates in CLN3 patient biopsy samples (2), indicating that the iPSC model system recapitulates diagnostic features of NCL. The empty vacuoles present in the cytoplasm at the NPC stage were not as evident in mature neurons, while mature MVBs remained reduced in the CLN3 patient neurons.
Figure 6.
Golgi and late endosome/lysosome markers analysis of TPP1 and CLN3 patient neurons. Monolayer differentiated TPP1-affected neurons or iPSC-derived neuralized embryoid body differentiated CLN3-affected neurons were immunostained for the cis-Golgi marker GM130 and the lysosomal marker LAMP1. Representative images of control (unaffected) neurons stained for GM130 and LAMP1 are also shown. MAP2ab was used to mark neurons and DAPI was used to counterstain nuclei. Arrows show LAMP1-positive vesicles along neurites. Scale bar sizes are indicated. Bar graphs represent mean ± SEM values for % GM130 structures with a circularity score between 0.9 and 1, where 1 represents a perfect circle (left bar graph), and LAMP1 puncta per µm of neurite length. ***P < 0.001.
Figure 7.
TPP1 and CLN3 patient neurons show disease subtype-specific storage material and progressed non-lysosomal pathology. Representative EM images are shown for control (unaffected), TPP1 and CLN3 (affected) patient neurons, 14 days after NPC differentiation or iPSC-derived neuralized embryoid body differentiation in the case of CLN3-affected patient neurons. ER, endoplasmic reticulum; MVB, multivesicular body; M, mitochondria; st, storage material. Scale bars: 500 nm. The single asterisk depicts an example of a mitochondrial vacuolation in a TPP1 patient neuron. Bar graphs represent mean ± SEM values for ER lumen diameter measurements (left bar graph) and the fraction of mitochondria with reduced cristae (right bar graph). **P < 0.01.
CLN3 and TPP1 protein overexpression rescues neuropathological events in patient NPCs
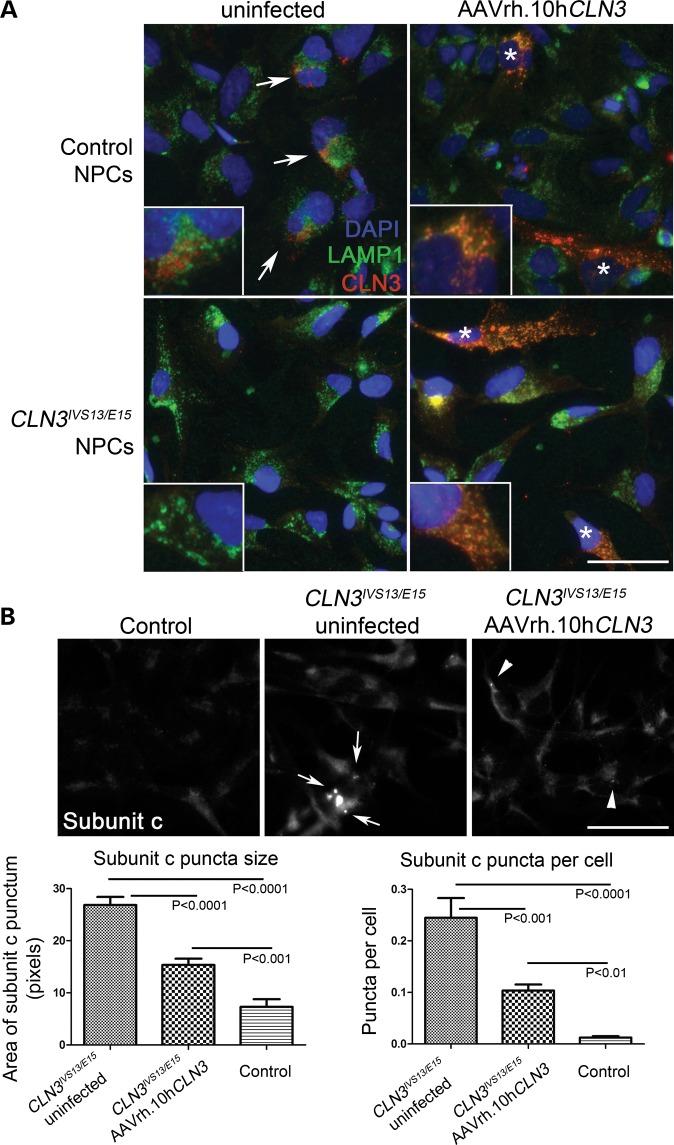
To test the utility of these patient-specific neuronal cell culture models as screening tools for agents that modulate NCL-related phenotypes, we first sought to reintroduce the wild-type proteins. Using an AAVrh.10 virus bearing wild-type human CLN3 cDNA, which was recently tested in vivo and shown to partially rescue neuropathological phenotypes in Cln3Δex7/8 mice (Sondhi, Scott, Chen, Hackett, Wong, Kubiak, Nelvagal, Cotman, Cooper and Crystal, accepted manuscript, Hum Gene Therapy), we infected both control and CLN3 patient NPCs, and analyzed the extent of expression. In infected control NPC cultures, using an established CLN3 antibody (batp1; (28)), strong CLN3 staining was observed in ∼10–20% of cells (see Materials and Methods for multiplicity of infection (MOI) details), in a pattern that was observed to extensively but not entirely overlap with the LAMP1 signal (Fig. 8A; Supplementary Material, Fig. S7). Infection of the CLN3 patient NPCs, followed by anti-CLN3 immunostaining yielded similar results to those in control NPCs (Fig. 8A; Supplementary Material, Fig. S7). Notably, the CLN3 antibody also weakly labeled uninfected control NPCs displaying vesicular staining with partial LAMP1 overlap, consistent with previous studies using this antibody (15, 28) (see arrows in Fig. 8A). As expected, the batp1 signal was weaker in the uninfected CLN3 patient versus control NPCs. The incomplete overlap with LAMP1 in the uninfected cells may reflect a difference in trafficking of the endogenous versus overexpressed protein. Indeed, differential localization of CLN3 protein based on its expression level has been previously reported (20).
Figure 8.
Overexpression of non-mutated CLN3 rescues the accumulation of storage material in patient NPCs. (A) Representative control (unaffected) and affected CLN3 patient NPC lines were grown as usual or infected with AAVrh.10hCLN3 for 72 h at an MOI of 3 × 105, then methanol:acetone fixed and co-stained for LAMP1 (green) and human CLN3 (batp1 antibody, red). Nuclei were counterstained with DAPI (blue). Representative images are shown, with the white arrows indicating examples of the signal observed in uninfected control NPCs, and white asterisks (*) indicating cells highly expressing CLN3 as detected with the batp1 antibody. The CLN3 signal was partly but not completely overlapping with LAMP1 signal in these cells, as indicated by the orange-yellow color in the merged image. Separated channels are provided in Supplementary Material, Figure S5. Scale bars: 50 µm. (B) Control and uninfected or infected CLN3IVS13/E15 NPCs were immunostained for subunit c to assess the extent of lysosomal storage. Representative images are shown. Scale bars: 50 µm. Subunit c storage deposits, indicated by white arrows and white arrowheads, were quantified as described in Materials and Methods. A significant reduction in subunit c puncta size and number was observed in the AAVrh.10hCLN3-infected CLN3 patient NPCs, as compared with the uninfected CLN3 patient NPCs. A significant >2-fold reduction in subunit c puncta size (P < 0.0001) and number of subunit c puncta per cell (P < 0.001) was observed by the treatment. However, consistent with the incomplete infection rate and expected lack of CLN3 cross-correction, the rescue did not bring the subunit c storage down to normal levels seen in control NPCs. Error bars represent SEM.
Next we sought to determine whether introduction of full-length, non-mutated CLN3 would improve the hallmark NCL lysosomal storage pathology in CLN3 patient cell lines. This was assessed by an established immunostaining assay against the major stored protein, subunit c of the mitochondrial ATP synthase (29), followed by image analysis. While abnormal subunit c storage deposits were readily detected in the uninfected CLN3 patient NPCs (white arrows, Fig. 8B), consistent with our previous observations shown in Figure 4, the subunit c puncta in the AAVrh.10hCLN3-infected cultures were both significantly smaller and less abundant (white arrowheads, Fig. 8B), indicating that the introduced non-mutated protein was able to partially correct the CLN3 deficiency in the patient NPCs. We were unable to directly correlate CLN3 expression and subunit c storage by co-immunostaining since both the CLN3 and subunit c antibodies were made in rabbit. Nevertheless, the partial correction of subunit c storage was overall consistent with the infection rate of the AAVrh.10hCLN3 vector in this system.
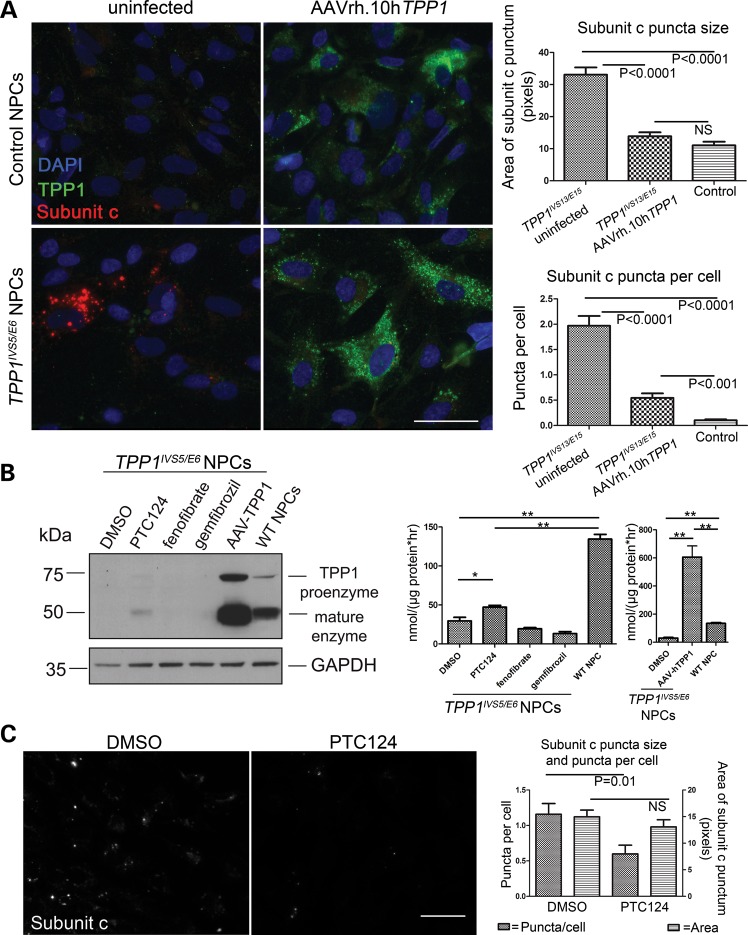
Infection of TPP1 patient NPCs with an AAVrh.10 virus containing wild-type human TPP1 cDNA (32) likewise caused marked elevations of TPP1 expression compared with that observed in uninfected NPCs, as assessed by immunostaining (Fig. 9A). Not surprisingly, given the potential for cross-correction with the TPP1 secreted enzyme, nearly all of the cells in the infected TPP1 patient NPC cultures showed elevated TPP1 expression. Co-immunostaining for TPP1 expression and subunit c storage, followed by image analysis, further demonstrated a clear dramatic reduction in storage material by overexpression of TPP1 (Fig. 9A).
Figure 9.
Rescue of TPP1 enzyme deficiency in patient NPCs by infection with AAVrh.10hTPP1 or by suppression of nonsense-mediated decay. (A) Representative control and patient NPC lines were grown as usual or infected with AAVrh.10hTPP1 for 72 h at an MOI of 3.6 × 105, then methanol:acetone fixed and co-stained for TPP1 (green) and subunit c (red). Nuclei were counterstained with DAPI (blue). Scale bars: 50 µm. Representative images are shown. AAVrh.10hTPP1-mediated TPP1 expression was evident in nearly all of the cells by bright punctate staining that was dramatically lower in uninfected control NPCs and virtually absent in uninfected patient NPCs. This result was consistent with a high infection rate and/or cross-correction by the secreted TPP1 enzyme. Subunit c storage was quantified as described in Materials and Methods. A significant reduction in subunit c puncta size (P < 0.0001) and subunit c puncta per cell (P < 0.0001) was observed in the AAVrh.10hTPP1-infected TPP1 patient NPCs, as compared with the uninfected TPP1 patient NPCs. Consistent with the expression analysis, we observed a nearly complete rescue of subunit c storage by the treatment, as compared with the control NPCs. The subunit c puncta size was completely normalized, while the number of puncta per cell was dramatically reduced. Error bars represent SEM. (B) Left panel: TPP1IVS5/E6 NPCs were treated with the indicated compounds (0.5% DMSO, 18 µm PTC124, 10 µm fenofibrate, 25 µm gemfibrozil) for 72 h or infected at an MOI of 100 000 with the AAVrh.10hTPP1 virus. Cell lysates were immunoblotted for TPP1 (Abcam 54685, 1:300) and GAPDH (Abcam 9485, 1:5000). Right panel: in parallel, lysates were assayed for TPP1 enzyme activity. Data are shown as mean ± SD of three technical replicates per line normalized to total protein. *P < 0.01; **P < 0.0001. The difference between vehicle- and PTC124-treated cells was reproducible across three experimental iterations. (C) The effect of PTC124, compared with DMSO vehicle, was tested on the subunit c storage in TPP1 patient NPCs. Representative micrograph images are shown of cells immunostained to detect subunit c storage, following 72 h of treatment with 0.5% DMSO or 18 µm PTC124. Quantification of the subunit c puncta per cell showed a small, statistically significant effect by PTC124 treatment (P = 0.01), but no significant effect (NS) was observed on the size of the subunit c puncta.
Patient NPCs are useful screening tools for identifying pharmacologic modulators of early-stage subcellular phenotypes and TPP1 activity
To further demonstrate a proof-of-principle that the herein developed TPP1 NPC lines and enzyme screening assay will be useful for drug screening, we tested the effect of three compounds expected or previously reported to enhance TPP1 expression and activity. The impact of the nonsense suppressor PTC124 (33) was tested, rationalizing that enhanced expression from the single nonsense allele in the compound heterozygous TPP1 patient lines might be achieved. Two lipid-lowering drugs were also tested, fenofibrate and gemfibrozil, given the report that these compounds could increase Tpp1/TPP1 transcript and protein levels in wild-type mouse primary neurons and in human cells (34). Treatment of TPP1 patient NPCs with 18 µm PTC124 caused a small but reproducibly significant increase in TPP1 protein level and enzyme activity in NPCs (Fig. 9B), consistent with recently reported results in TPP1 patient lymphoblasts (35). Fenofibrate and gemfibrozil, however, at reported doses (10 and 25 µm doses, respectively) (34), failed to increase protein levels or enzyme activity above background in the TPP1 patient NPCs (Fig. 9B). Moreover, commensurate with the increase in TPP1 activity achieved by PTC124 treatment, we also observed a significant impact on subunit c storage (Fig. 9C). The average number of subunit c puncta per cell was reduced ∼2-fold compared with DMSO control, although the average size of the subunit c puncta was not significantly changed by PTC124, possibly due to the limited degree of TPP1 upregulation in the patient NPCs. Nevertheless, together with our adeno-associated virus infection studies, these results demonstrate the importance of this new human neuronal model system for NCL drug testing and discovery.
DISCUSSION
Herein, we have established the first human neuronal cell models of late-infantile NCL (CLN2/TPP1) and juvenile NCL (CLN3) using cellular reprograming. We have demonstrated progressive organellar pathology, initially in the endocytic pathway with distinct genotype-specific features at the stem cell stage, and we have shown that these presymptomatic models capture the onset and progression of NCL-type storage material. Overexpression of non-mutated TPP1 or CLN3 protein rescued the neuronal NCL pathology phenotypes, proving the specificity of this iPSC model system and lending support for AAV-mediated gene therapy, currently in clinical testing (www.clinicaltrials.gov, NCT01161576). Finally, the value of these cell lines was further demonstrated through proof-of-principle drug treatment studies.
A number of eukaryotic models of NCL, such as deficiency yeast, knockout and knockin mice, and naturally occurring canine models, have significantly expanded our understanding of NCL pathophysiology (36). Together with the expanding genetic and clinical spectrum of NCL disorders, highlighted by recently discovered genes such as GRN (CLN11) (37), CTSF (CLN12) (38), and ATP13A2 (CLN13) (39), the model systems reported here also suggest cellular and molecular pathways that underlie shared features of NCL subtypes. Commonalities may further extend to other neurodegenerative diseases that have shared pathogenetic features, including parkinsonism, mitochondrial disorders, and frontotemporal dementia (40). However, little is known about the events preceding end-stage lysosomal storage in NCL patient neurons and whether they might be exploited as subtype-specific biomarkers of disease onset and progression.
To gain a better understanding of these presymptomatic events, we interrogated the organelle systems in the newly generated set of iPSCs and derivative NPCs. Abnormal pathology, reproducible across multiple clones from unrelated patients, was present as early as the pluripotent state, in the absence of appreciable lysosomal storage. Ultrastructural analysis of iPSCs nevertheless pointed to other evidence of impaired autophagic clearance, a process that has been demonstrated to be impacted by CLN3 mutation in a knockin mouse model (14) and more recently in CLN3 patient fibroblasts (41). In particular, CLN3 patient iPSCs showed marked aberrations of the endo/lysosomal compartment evidenced by large LAMP1-positive ring-like structures at the immunofluorescence level, cytoplasmic vacuolations at the EM level, and by reduced TPP1 activity levels. Together with an increase in the size of AVs, these observations may reflect a defect in lysosomal maturation, pH maintenance and/or in fusion of lysosomes with autophagosomes. Interestingly, these morphological changes identified in the iPSC system are reminiscent of the vacuolations seen in peripheral blood lymphocytes from CLN3 patients, a finding specific to this form of NCL (42), and in the epididymal clear cells of male mice homozygous for the common Cln3 deletion (43). By contrast, TPP1 patient iPSCs showed only the expected TPP1 enzyme deficiency and a marked accumulation of intracellular lipid droplets. In aggregate, these results suggest that TPP1 and CLN3 deficiency affect the endosomal/lysosomal system in different ways and that these effects precede the appearance of lysosomal storage and obvious abnormalities in other subcellular organelles.
We also demonstrated that disease status does not affect the potential of these iPSC lines to differentiate into the three major germ layers and into functional neurons, though we observed that CLN3-patient-iPSC differentiation into neurons was sensitive to the differentiation method. NPCs derived from TPP1 and CLN3 iPSCs, which were neuronally committed but not terminally differentiated, showed overlapping as well as distinct pathology again in the endocytic pathway but also in additional intracellular compartments. CLN3 NPCs showed a particularly dramatic deficit of well-defined MVBs, a subset of endosomes that fuse with autophagosomes to form amphisomes, which play an important role in membrane protein degradation. The formation of the 25–50-nm intraluminal vesicles (ILVs) that characterize MVBs is coordinated by at least four distinct complexes of the ESCRT (endosomal sorting complex required for transport) machinery. Notably, downregulation of Hrs, a component of the ESCRT-0 complex that sorts ubiquitinated cargo into endosomes and nascent MVBs (44), impairs MVB biogenesis and produces enlarged, ‘empty’ LAMP1-positive vesicles (45), in much the same pattern observed in CLN3 patient iPSCs and NPCs. Neuron-specific knockout of Hrs in mice causes accumulation of ubiquitinated proteins, including p62, and progressive neurodegeneration, especially in the CA3 region of the hippocampus (46).
Degradative processing by the ESCRT machinery is closely coordinated with endosome-to-trans-Golgi cargo recycling by the retromer complex, of interest in NCL biology given the recently reported roles of CLN5 and the CLN3 yeast ortholog btn1 in retrograde trafficking (20,22). Disrupting the latter process by, for instance, overexpressing a phosphomimicking form of the SNARE component Sed5, caused dispersion and vesiculation of the Golgi complex, in a pattern similar to that observed in CLN3 patient NPCs and neurons and in an iPSC-derived neuronal model of another lysosomal storage disease, mucopolysaccharidosis type IIIB (47). It will therefore be of interest to determine whether CLN3 has a sorting role at the nexus of MVB biogenesis and retrograde trafficking.
Mitochondrial abnormalities have also previously been observed in cerebellar cell lines from a knockin murine Cln3 model (15). The mitochondrial phenotypes noted in this and previously reported CLN3 model systems are similar to those that have been observed in the setting of defective mitophagy, a form of autophagy that specifically targets damaged mitochondria for degradation and has been extensively studied in iPSC-derived neurons from patients deficient in PARK2, the gene most commonly mutated in juvenile-onset, autosomal recessive forms of parkinsonism (48).
The phenotypes we have observed in NCL patient neurons, as well as TPP1 enzyme activity measurements, will be powerful screening tools for interventions that modify pathology in human cells and in a neuron-specific manner. The observation that TPP1 activity was decreased in CLN3 patient iPSCs to ∼60% the normal level, but that conversely TPP1 activity was increased in the differentiated CLN3 patient NPCs relative to normal levels, may reflect a compensatory response early in the disease state or a cell-type-specific response to the CLN3 deficiency. Notably, increases in TPP1 activity have also been reported in Cln3 mouse knockout brain tissue (49) and in CLN3 patient brain (50,51). Thus, following abnormal TPP1 levels in CLN3-iPSCs and NPCs may serve as an important screening tool to identify specific early-stage CLN3 disease modifiers. Indeed, we have demonstrated the suitability of the TPP1 enzyme assay for detecting treatment-induced changes by showing that TPP1 activity could be increased by both AAV-mediated TPP1 overexpression and by pharmacological treatment with the nonsense suppressor PTC124. The latter was in accordance with the recent report by Miller and colleagues (35). In our study, we were also able to partially rescue the lysosomal storage. Whether increasing PTC124 dosage or using a longer chronic treatment time further increases TPP1 activity levels enough to more significantly impact neuronal cell pathology will require further testing. However, even small differences in TPP1 endogenous function achieved through drug treatment, like those documented here, may be beneficial, particularly in combination with other treatments such as gene therapy.
In summary, our initial findings in the first iPSC-based NCL models have validated observations in lower organism models and we have extended the knowledge surrounding the development of these phenotypes in presymptomatic stages. Our studies also demonstrate the utility of a human-based disease model for measuring both protein activity and the neuropathologic outcome of putative treatments. The opportunity to identify modifiers of the earliest disease events offers the best chance of developing treatments that will prevent the devastating symptoms of NCL. Importantly, we have also documented genotype–phenotype cell biological and biochemical relationships that, with expanded research and a broader genetic panel of patient-derived iPSCs, will provide key new insights into the pathways leading to highly similar end-stage pathologies involving lysosomal storage and neurodegeneration.
MATERIALS AND METHODS
Human fibroblast culture
For generating iPSC lines at the University of Dresden, CLN3-FB3, CLN3-FB4 and CLN3-FB5, and FS fibroblast lines were established from skin biopsies taken from CLN3 patients, an unaffected donor heterozygous for the CLN3 common deletion and an unaffected control, respectively. CLN2/TPP1 fibroblasts (TPP1-FB2) were kindly provided by Dr. Robert Steinfeld (52). All procedures were in accordance with the Helsinki convention and approved by the Ethical Committee of the University of Dresden (EK45022009). For generating iPSC lines through the Harvard iPS Cell Core, TPP1-FB1 and CLN3-FB2 lines were obtained from the NCL Biorepository at Massachusetts General Hospital, and CLN3-FB1 was kindly provided by Dr Nanbert Zhong (New York State Institute for Basic Research in Developmental Disabilities). A complete summary of the cell lines used in this study is also provided in Supplementary Material, Table S1. The use of these cell lines for this study was reviewed and approved by the Institutional Review Board of the Massachusetts General Hospital.
Methods for establishing fibroblast lines are provided in Supplemental Material.
Generation and expansion of iPSCs
The generation and characterization of control iPSC line 8330-iPS-8 was recently described (27). Fibroblast lines CLN3-FB1, CLN3-FB2 and TPP1-FB1 were reprogramed as previously described, using GFP as a marker of retroviral infection (27). Silencing of retroviral expression in iPSC clones derived from these fibroblast lines was monitored by loss of GFP expression.
Patient fibroblasts CLN3-FB3, CLN3-FB4, CLN3-FB5 and TPP1-FB2 were reprogramed using pMX-based retroviral vectors encoding the human cDNAs of OCT4, SOX2, KLF4 and cMYC (pMX vectors). Vectors were co-transfected with packaging-defective helper plasmids into 293T cells using Fugene 6 transfection reagent (Roche). Forty-eight hours later, virus supernatants were collected as described previously (53) and (54). Fibroblasts were plated at a density of 50 000 cells/well on 0.1% gelatin-coated 6-well plates and infected three times with a viral cocktail containing vectors expressing OCT4:SOX2:KLF4:cMYC in a 2:1:1:1 ratio in presence of 6 µg/ml protamine sulfate (Sigma-Aldrich) and 5 ng/ml FGF2 (Peprotech). Infected fibroblasts were plated onto mitomycin C (MMC, Tocris) inactivated CF-1 mouse embryonic fibroblasts (in-lab preparation) at a density of 900 cells/cm2 in fibroblast media. The next day media was exchanged to ES medium containing 78% knockout DMEM, 20% knockout serum replacement, 1% non-essential amino acids, 1% penicillin/streptomycin/glutamine and 50 µm β-mercaptoethanol (all from Invitrogen) supplemented with 5 ng/ml FGF2 and 1 mm valproic acid (Sigma-Aldrich). Media was changed every day to the same conditions. iPSC-like clusters started to appear at Day 7 postinfection, were manually picked 14 days postinfection and plated onto CF-1 feeder cells in regular ES-media containing 5 ng/ml FGF2. Stable clones were routinely passaged onto MMC-treated CF-1 feeder cells (Globalstem) using 1 mg/ml collagenase type IV (Invitrogen) and addition of 10 µm Y-27632 (Ascent Scientific) for the first 48 h after passaging. Media change with addition of fresh FGF2 was performed every day. Stable clones were analyzed by qRT–PCR for silencing of viral transgenes prior to further experimental procedures.
Methods used for pluripotency characterization and karyotyping are provided in Supplemental Material. Note that unless indicated otherwise, all phenotyping experiments were carried out on at least two-independent unaffected (control) iPSC lines and at least two-independent-affected patient lines for each genetic form of NCL. Data shown, unless indicated otherwise, are representative for phenotype observations consistent for all examined clones and for at least two-independent experiments.
Derivation of NPCs and neurons from iPSC clones
iPSCs were grown under standard conditions until they became confluent, at which point all cleaned colonies were detached with collagenase type IV (2 mg/ml) and plated into a 6-cm petri dish in ES media containing 5 µm Y-27632, 10 µm SB431542 and 1 µm dorsomorphin. The next day, embryoid bodies (EBs) were collected in a 15-ml Falcon tube by gravity flow. Media was aspirated and collected EBs were resuspended in fresh ES media containing 10 µm SB431542 and 1 µm dorsomorphin. Media was changed by this manner the next 2 days. On Day 4 of EB formation, collected EBs were resuspended in N2 media containing DMEM/F12, 1% penicillin/streptomycin and 1:75 N2 supplement, and plated onto MatrigelTM-coated plates Daily media change was performed the following 4 days. After 4 days neural rosettes were picked. Collected rosettes were plated into a 6-cm petri dish in DMEM/F12 containing 1% N2 supplement, 1% penicillin/streptomycin/glutamine, 2 µg/ml Heparin and 10 ng/ml FGF2. On Day 4, neurospheres were collected and resuspended in 0.25% trypsin/EDTA to dissociate neurospheres. Single cells were plated onto poly-l-ornithine/laminin-coated plates in DMEM/F12 containing 2% B27 supplement, 1% penicillin/streptomycin/glutamine and 20 ng/ml of each EGF and FGF2. NPC media consisted of 97% DMEM/F12, 2% B27 supplement and 1% penicillin/streptomycin. Media containing fresh growth factors was replaced every 3rd day and NPCs were routinely split when confluent at a 1:2 ratio using 0.05% trypsin/EDTA (Mediatech, Inc.) followed by inhibition with equal amounts of trypsin inhibitor.
For final differentiation into mature neurons, control and TPP1 patient NPCs were plated at a density of 40 000 cells/cm2 onto poly-l-ornithine/laminin-coated plates in NPC media with EGF and FGF2. The next day, media was changed to differentiation media containing DMEM/F12:Neurobasal 1:1, 1% B27 supplement without vitamin A (Invitrogen), 1% penicillin/streptomycin/glutamine, 0.5% N2 supplement and 0.5 mm cAMP (Sigma-Aldrich). Media change was performed twice a week for at least 14 days. CLN3 patient neurons were obtained by differentiation of freshly prepared neurospheres, generated as described above.
Electrophysiology
Cells were investigated 3–5 weeks after differentiation using standard whole-cell patch-clamp technique at room temperature. Recordings were made in the whole-cell voltage-clamp or current-clamp mode and data were recorded using an Axopatch 200B amplifier and Iso2 data acquisition software (Axon Instruments, Union City, CA, USA) essentially as described previously (55,56). Extracellular solution contained (in mmol l−1): 142 NaCl, 8.1 KCl, 1 CaCl2, 6 MgCl2, 10 HEPES, 10 d-glucose (pH 7.4). Pipette solution contained (in mmol l−1): 153 KCl, 1 MgCl2, 5 EGTA, 10 HEPES. Using these solutions, borosilicate pipettes had resistances of 4–8 MΩ. Seal resistances in the whole-cell mode were between 0.1 and 1 GΩ. Data were analyzed using Iso2, Prism4 and Microsoft Excel 97. Resting membrane potentials (RMP) were determined immediately after gaining whole-cell access. Action potentials (APs) were elicited by applying increasing depolarizing current pulses (5 pA current steps). The afterhyperpolarization (AHP) amplitude was measured from peak to beginning of plateau reached during the current injection and AP duration was measured at half amplitude.
Organelle markers immunostaining
Assessment of organelles in iPSCs, NPCs and neurons was performed using mouse anti-GM130 (1:200, BD Transduction Laboratories), mouse anti-GRP75 (1:200, Abcam) and mouse anti-LAMP1 (1:100, Santa Cruz). Lysosomal storage was evaluated with rabbit anti-subunit c (1:200; (29)).
After incubation with primary antibodies, samples were rinsed several times with PBS and incubated for 1 h at room temperature with Alexa-Fluor®-conjugated secondary antibodies (Invitrogen) diluted 1:500 in blocking solution. After several rinses, coverslips were mounted with ProLong® Gold antifade reagent with DAPI (Invitrogen). Nail polish-sealed coverslips were imaged on a Leica SP5 AOBS scanning laser confocal microscope (Leica Microsystems), or on a Zeiss epifluorescence system equipped with digital image capture. ‘No antibody’ and ‘secondary-only’ controls were included in these experiments, and imaging settings were optimized to ensure autofluorescence and non-specific signal from secondary antibody did not contribute to the immunofluorescence images.
For quantifying Golgi vesiculation, captured images containing ∼40–70 cells per image were analyzed in ImageJ. Golgi structures labeled by GM130 were segmented from thresholded images and the degree of circularity was obtained. Structures were binned into 10 groups by circularity score, and the percentage of structures in the top bin (0.9–1 circularity score, meaning the most circular group) was determined from three technical replicates. Statistical significance was determined by one-way ANOVA and Bonferroni multiple comparisons post hoc analysis, using Graphpad Prism.
To further assess the extent of the altered LAMP1 staining pattern in the patient NPCs, 100–150 cells per genotype were scored as displaying LAMP1-positive structures that extended into the cell periphery or as displaying predominantly perinuclear localized LAMP1-positive structures, a method that has been previously described for measuring LAMP1 distribution (57). The relative percentages of the two LAMP1 staining pattern categories within the field of image capture for each genotype were averaged (3–5 fields averaged from two representative experiments). One-way ANOVA and Bonferroni multiple comparisons post hoc analysis was performed to determine statistical significance, using Graphpad Prism. For quantifying LAMP1 vesicles in neurites in the PFA-fixed neurons immunostained for LAMP1, neurites (MAP2ab-positive) were first selected from thresholded images using ImageJ, and the number of LAMP1 puncta were subsequently determined from the selected neurite areas using Cell Profiler. In total, ∼400–800 neurites were analyzed per genotype. The statistical significance of genotypic differences in the mean number of LAMP1 puncta per µm of neurite length was determined using one-way ANOVA and post hoc Bonferroni multiple comparisons tests.
Electron microscopy analysis
Tissue culture specimens in confluent 6-well or 10-cm plates were fixed directly with gentle rocking for 15 min at room temperature with a glutaraldehyde fixative (2.5% glutaraldehyde, 2% PFA, 0.025% calcium chloride, in a 0.1 m sodium cacodylate buffer, pH 7.4), gently scraped and pelleted at 500g, and washed twice with cacodylate buffer. To make a cell block, the cells were centrifuged and resuspended in 60°C warm agar (2%) in a warm water bath to keep the agar fluid. The cells were then centrifuged again and the agar was allowed to solidify in an ice water bath. The tip of the centrifuge tube containing the tissue was cutoff resulting in an agar block with the cells embedded within it and routinely processed in a Leica Lynx automatic tissue processor. Briefly, the tissue was postfixed in osmium tetroxide, stained en bloc with uranyl acetate, dehydrated in graded ethanol solutions, infiltrated with propylene oxide/Epon mixtures, embedded in pure Epon and polymerized over night at 60°C. One-micron sections were cut, stained with toluidine blue and examined by light microscopy. Representative areas were chosen for electron microscopic study and the Epon blocks trimmed accordingly. Thin sections were cut with an LKB 8801 ultramicrotome and diamond knife, stained with lead citrate and examined in an FEI Morgagni transmission electron microscope. Images were captured with an AMT digital CCD camera.
AVs, empty vacuoles, lipid droplets and multivesicular bodies were scored in at least 20 cell profiles per cell line. AVs were scored as described (58). In particular, late AVs and autolysosomes were collectively defined as containing one limiting membrane and containing electron-dense cytoplasmic material and/or organelles at various stages of degradation. AV area was calculated in ImageJ by defining regions of interest. ER lumen diameter was measured in at least 20-cell profiles per cell line, using ImageJ. Fraction of mitochondria displaying reduced cristae was determined using ImageJ. Mitochondria were identified as having reduced cristae if the visible cristae crossed <75% of the length of the major axis. Statistical significance was determined using a Student's t-test.
TPP1 enzyme activity assay
Cells from one confluent well of a 6-well plate were washed with PBS, scraped into acetate buffer supplemented with 10 µm of the protease inhibitors pepstatin A and E64 (Sigma) and homogenized with 10 passes through a 27-G syringe. TPP1 activity was measured fluorometrically using the Ala-Ala-Phe-7-amido-4-methylcoumarin substrate (AAF-MCA) as described previously, with some modifications (59–61). Approximately 20 µg of total protein from the scraped cells was incubated in 150 µl acetate buffer (pH 4.0) containing a final concentration of 62.5 µm AAF-MCA for 20 h at 37°C. The reaction was stopped by the addition of 100 µl 0.5 m EDTA (pH 12.0). Fluorescence was measured using a Molecular Devices SpectraMax M2 spectrophotometer with an extinction wavelength of 355 nm and an emission wavelength of 460 nm.
Rescue experiments
AAVrh.10-CAG-hCLN2 (32) (AAVrh.10hTPP1 in this manuscript) and AAVrh.10-CAG-hCLN3 (AAVrh.10hCLN3 in this manuscript), consisting of a replication-defective AAV2 genome containing the respective human TPP1 and CLN3 transgenes driven by a CAG promoter, were packaged in a capsid of rhesus macaque-derived serotype AAVrh.10. Virus produced in 293T cells was purified by iodixanol density gradients and QHP affinity chromatography. The purified flow-through was spin-concentrated, and viral titer was assessed by a TaqMan assay. Patient TPP1 and CLN3 NPCs were transduced with the respective virus for 72 h at MOIs ranging from 105 to 5 × 105. Percent expressing cells for the AAVrh.10hTPP1 vector reached 100%, though whether this was due to viral infection or cross-correction was not clear. Percent expressing cells for the AAVrh.10hCLN3 vector maximally reached ∼20%, as analyzed by batp1 immunostaining to detect CLN3 expression. We are unaware whether the rh.10 serotype adeno-associated virus has previously been tested in iPSCs or iPSC-derived neurons, but we note that other serotypes of AAV have been reported to infect iPSCs at similar rates to those observed in our iPSC-derived NPCs (e.g. (62,63) and were consistent with in vitro infection rates for the AAVrh.10hTPP1 and AAVrh.10hCLN3 vectors in other cell systems (data not shown).
Gemfibrozil and fenofibrate were obtained from Sigma, and PTC124 was obtained from Selleck Chemicals. Compounds were solubilized in DMSO and used at the doses described in the text.
Lysates from uninfected and infected cultures were solubilized with 0.2% Triton X-100 and immunoblotted for TPP1 (Abcam 54685, 1:300) and GAPDH (Abcam 9485, 1:5000). Alternatively, immunocytochemistry on methanol:acetone fixed cells (ice cold 1:1 methanol:acetone for 10 min, followed by air drying and rehydration with tris buffered saline, pH 7.4) was performed to probe for TPP1 or CLN3 expression and subcellular localization, using the TPP1 antibody (Abcam 54685, 1:200) or batp1 (40) (1:400), with co-staining using antibodies against subunit c (29) (1:100) or LAMP1 (Santa Cruz, 1:200). To quantify subunit c storage, images were collected from 10 fields of view using the 20× objective on a Zeiss epifluorescence system equipped with digital image capture. Each image had >40 cells counted (range: 41–215). Using CellProfiler (64), puncta were identified from the subunit c image and related to the closest nucleus. Per field averages were calculated for the area of each punctum (‘subunit c puncta size’ in Figs 8 and 9) and for the number of puncta for each nucleus (‘subunit c puncta per cell’ in Figs 8 and 9). These were subsequently averaged and plotted using GraphPad Prism.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported in part by the National Institutes of Health: National Institute of Neurological Disorders & Stroke (grant number R01NS073813 to S.L.C.; grant numbers R01NS061848 U54NS065768 to R.G.C. and D.S.; and grant number K08NS079359 to J.F.S.); the Batten Disease Support and Research Association (to J.F.S., S.B. and S.L.C.); the National Institute of Mental Health (grant number R33MH087896 to S.J.H.); The National Contest for Life (NCL Stiftung; to A.S. and A.H.); the Tau Consortium (to S.J.H.); and by a Massachusetts General Hospital ECOR Tosteson Award (to J.F.S.); and by Catherine's Hope for a Cure (to K.B.S. and S.L.C.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the patients and their families for donating the skin biopsies. We also thank Drs. Chad Cowan and Laurence Daheron of the Harvard Stem Cell Institute-Harvard iPS Core for helpful advice and assistance in generating iPSC lines. Rosemary Barone, and the MGH Neurogenetics DNA Diagnostic Laboratory under the direction of Winnie Xin, assisted in genotype confirmation, and Jeff Brown assisted in CLN3 gene expression studies.
Conflict of Interest statement: The authors declare the following: J.F.S. is currently an employee of Biogen Idec and X.L. is currently an employee of Biocrea GmbH. S.L.C. and K.B.S. are members of the Batten Disease Support and Research Association Advisory Board.
REFERENCES
- 1.Jalanko A., Braulke T. Neuronal ceroid lipofuscinoses. Biochim. Biophys. Acta. 2009;1793:697–709. doi: 10.1016/j.bbamcr.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Mole S.E., Williams R.E., Goebel H.H. The Neuronal Ceroid Lipofuscinoses (Batten Disease) Oxford: Oxford University Press; 2011. [Google Scholar]
- 3.Palmer D.N., Fearnley I.M., Medd S.M., Walker J.E., Martinus R.D., Bayliss S.L., Hall N.A., Lake B.D., Wolfe L.S., Jolly R.D. Lysosomal storage of the DCCD reactive proteolipid subunit of mitochondrial ATP synthase in human and ovine ceroid lipofuscinoses. Adv. Exp. Med. Biol. 1989;266:211–222. doi: 10.1007/978-1-4899-5339-1_15. discussion 223. [DOI] [PubMed] [Google Scholar]
- 4.Kominami A.E. What are the requirements for lysosomal degradation of subunit c of mitochondrial ATPase? IUBMB Life. 2002;54:89–90. doi: 10.1080/15216540214307. [DOI] [PubMed] [Google Scholar]
- 5.Cotman S.L., Staropoli J.F. The juvenile Batten disease protein, CLN3, and its role in regulating anterograde and retrograde post-Golgi trafficking. Clin. Lipidol. 2012;7:79–91. doi: 10.2217/clp.11.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang J.W., Choi H., Kim H.J., Jo D.G., Jeon Y.J., Noh J.Y., Park W.J., Jung Y.K. Neuronal vulnerability of CLN3 deletion to calcium-induced cytotoxicity is mediated by calsenilin. Hum. Mol. Genet. 2007;16:317–326. doi: 10.1093/hmg/ddl466. [DOI] [PubMed] [Google Scholar]
- 7.Uusi-Rauva K., Luiro K., Tanhuanpaa K., Kopra O., Martin-Vasallo P., Kyttala A., Jalanko A. Novel interactions of CLN3 protein link Batten disease to dysregulation of fodrin-Na+, K+ ATPase complex. Exp. Cell Res. 2008;314:2895–2905. doi: 10.1016/j.yexcr.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 8.Lyly A., von Schantz C., Heine C., Schmiedt M.L., Sipila T., Jalanko A., Kyttala A. Novel interactions of CLN5 support molecular networking between neuronal ceroid lipofuscinosis proteins. BMC Cell Biol. 2009;10:83. doi: 10.1186/1471-2121-10-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Getty A.L., Benedict J.W., Pearce D.A. A novel interaction of CLN3 with nonmuscle myosin-IIB and defects in cell motility of Cln3(−/−) cells. Exp. Cell Res. 2011;317:51–69. doi: 10.1016/j.yexcr.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uusi-Rauva K., Kyttala A., van der Kant R., Vesa J., Tanhuanpaa K., Neefjes J., Olkkonen V.M., Jalanko A. Neuronal ceroid lipofuscinosis protein CLN3 interacts with motor proteins and modifies location of late endosomal compartments. Cell. Mol. Life Sci. 2012;69:2075–2089. doi: 10.1007/s00018-011-0913-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scifo E., Szwajda A., Debski J., Uusi-Rauva K., Kesti T., Dadlez M., Gingras A.C., Tyynela J., Baumann M.H., Jalanko A., et al. Drafting the CLN3 protein interactome in SH-SY5Y human neuroblastoma cells: a label-free quantitative proteomics approach. J. Proteome Res. 2013;12:2101–2115. doi: 10.1021/pr301125k. [DOI] [PubMed] [Google Scholar]
- 12.Cooper J.D., Russell C., Mitchison H.M. Progress towards understanding disease mechanisms in small vertebrate models of neuronal ceroid lipofuscinosis. Biochim. Biophys. Acta. 2006;1762:873–889. doi: 10.1016/j.bbadis.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Phillips S.N., Muzaffar N., Codlin S., Korey C.A., Taschner P.E., de Voer G., Mole S.E., Pearce D.A. Characterizing pathogenic processes in Batten disease: use of small eukaryotic model systems. Biochim. Biophys. Acta. 2006;1762:906–919. doi: 10.1016/j.bbadis.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Cao Y., Espinola J.A., Fossale E., Massey A.C., Cuervo A.M., MacDonald M.E., Cotman S.L. Autophagy is disrupted in a knock-in mouse model of juvenile neuronal ceroid lipofuscinosis. J. Biol. Chem. 2006;281:20483–20493. doi: 10.1074/jbc.M602180200. [DOI] [PubMed] [Google Scholar]
- 15.Fossale E., Wolf P., Espinola J.A., Lubicz-Nawrocka T., Teed A.M., Gao H., Rigamonti D., Cattaneo E., MacDonald M.E., Cotman S.L. Membrane trafficking and mitochondrial abnormalities precede subunit c deposition in a cerebellar cell model of juvenile neuronal ceroid lipofuscinosis. BMC Neurosci. 2004;5:57. doi: 10.1186/1471-2202-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearce D.A., Carr C.J., Das B., Sherman F. Phenotypic reversal of the btn1 defects in yeast by chloroquine: a yeast model for Batten disease. Proc. Natl. Acad. Sci. USA. 1999;96:11341–11345. doi: 10.1073/pnas.96.20.11341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearce D.A., Ferea T., Nosel S.A., Das B., Sherman F. Action of BTN1, the yeast orthologue of the gene mutated in Batten disease. Nat. Genet. 1999;22:55–58. doi: 10.1038/8861. [DOI] [PubMed] [Google Scholar]
- 18.Kim Y., Ramirez-Montealegre D., Pearce D.A. A role in vacuolar arginine transport for yeast Btn1p and for human CLN3, the protein defective in Batten disease. Proc. Natl. Acad. Sci. USA. 2003;100:15458–15462. doi: 10.1073/pnas.2136651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Codlin S., Haines R.L., Burden J.J., Mole S.E. Btn1 affects cytokinesis and cell-wall deposition by independent mechanisms, one of which is linked to dysregulation of vacuole pH. J. Cell Sci. 2008;121:2860–2870. doi: 10.1242/jcs.030122. [DOI] [PubMed] [Google Scholar]
- 20.Kama R., Kanneganti V., Ungermann C., Gerst J.E. The yeast Batten disease orthologue Btn1 controls endosome-Golgi retrograde transport via SNARE assembly. J. Cell Biol. 2011;195:203–215. doi: 10.1083/jcb.201102115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Codlin S., Mole S.E. S. pombe btn1, the orthologue of the Batten disease gene CLN3, is required for vacuole protein sorting of Cpy1p and Golgi exit of Vps10p. J. Cell Sci. 2009;122:1163–1173. doi: 10.1242/jcs.038323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mamo A., Jules F., Dumaresq-Doiron K., Costantino S., Lefrancois S. The role of ceroid lipofuscinosis neuronal protein 5 (CLN5) in endosomal sorting. Mol. Cell. Biol. 2012;32:1855–1866. doi: 10.1128/MCB.06726-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Unternaehrer J.J., Daley G.Q. Induced pluripotent stem cells for modelling human diseases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011;366:2274–2285. doi: 10.1098/rstb.2011.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rai R., Li J.M., Zheng H., Lok G.T., Deng Y., Huen M.S., Chen J., Jin J., Chang S. The E3 ubiquitin ligase Rnf8 stabilizes Tpp1 to promote telomere end protection. Nat. Struct. Mol. Biol. 2011;18:1400–1407. doi: 10.1038/nsmb.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park I.H., Zhao R., West J.A., Yabuuchi A., Huo H., Ince T.A., Lerou P.H., Lensch M.W., Daley G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 27.Sheridan S.D., Theriault K.M., Reis S.A., Zhou F., Madison J.M., Daheron L., Loring J.F., Haggarty S.J. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS ONE. 2011;6:e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cotman S.L., Vrbanac V., Lebel L.A., Lee R.L., Johnson K.A., Donahue L.R., Teed A.M., Antonellis K., Bronson R.T., Lerner T.J., et al. Cln3(Deltaex7/8) knock-in mice with the common JNCL mutation exhibit progressive neurologic disease that begins before birth. Hum. Mol. Genet. 2002;11:2709–2721. doi: 10.1093/hmg/11.22.2709. [DOI] [PubMed] [Google Scholar]
- 29.Cao Y., Staropoli J.F., Biswas S., Espinola J.A., Macdonald M.E., Lee J.M., Cotman S.L. Distinct early molecular responses to mutations causing vLINCL and JNCL presage ATP synthase subunit C accumulation in cerebellar cells. PLoS ONE. 2011;6:e17118. doi: 10.1371/journal.pone.0017118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galli R., Gritti A., Bonfanti L., Vescovi A.L. Neural stem cells: an overview. Circ. Res. 2003;92:598–608. doi: 10.1161/01.RES.0000065580.02404.F4. [DOI] [PubMed] [Google Scholar]
- 31.Doi K., Nakano T., Kitayama M., Watanabe Y., Yasui K., Fukada Y., Morino S., Kaidoh T., Nakashima K., Inoue T. Mitochondrial changes in motor neurons of homozygotes of leucine 126 TT deletion SOD1 transgenic mice. Neuropathology. 2008;28:269–276. doi: 10.1111/j.1440-1789.2007.00876.x. [DOI] [PubMed] [Google Scholar]
- 32.Sondhi D., Hackett N.R., Peterson D.A., Stratton J., Baad M., Travis K.M., Wilson J.M., Crystal R.G. Enhanced survival of the LINCL mouse following CLN2 gene transfer using the rh.10 rhesus macaque-derived adeno-associated virus vector. Mol. Ther. 2007;15:481–491. doi: 10.1038/sj.mt.6300049. [DOI] [PubMed] [Google Scholar]
- 33.Welch E.M., Barton E.R., Zhuo J., Tomizawa Y., Friesen W.J., Trifillis P., Paushkin S., Patel M., Trotta C.R., Hwang S., et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 34.Ghosh A., Corbett G.T., Gonzalez F.J., Pahan K. Gemfibrozil and fenofibrate, FDA-approved lipid-lowering drugs, upregulate tripeptidyl-peptidase 1 in brain cells via peroxisome proliferator-activated receptor-a: implications for late infantile neuronal ceroid lipofuscinosis therapy. J. Biol. Chem. 2012;287:38922–38935. doi: 10.1074/jbc.M112.365148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller J.N., Chan C.H., Pearce D.A. The role of nonsense-mediated decay in neuronal ceroid lipofuscinosis. Hum. Mol. Genet. 2013;22:2723–2734. doi: 10.1093/hmg/ddt120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bond M., Holthaus S.M., Tammen I., Tear G., Russell C. Use of model organisms for the study of neuronal ceroid lipofuscinosis. Biochim. Biophys. Acta. 2013;1832:1842–1865. doi: 10.1016/j.bbadis.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Smith K.R., Damiano J., Franceschetti S., Carpenter S., Canafoglia L., Morbin M., Rossi G., Pareyson D., Mole S.E., Staropoli J.F., et al. Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am. J. Hum. Genet. 2012;90:1102–1107. doi: 10.1016/j.ajhg.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith K.R., Dahl H.H., Canafoglia L., Andermann E., Damiano J., Morbin M., Bruni A.C., Giaccone G., Cossette P., Saftig P., et al. Cathepsin F mutations cause Type B Kufs disease, an adult-onset neuronal ceroid lipofuscinosis. Hum. Mol. Genet. 2013;22:1417–1423. doi: 10.1093/hmg/dds558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bras J., Verloes A., Schneider S.A., Mole S.E., Guerreiro R.J. Mutation of the parkinsonism gene ATP13A2 causes neuronal ceroid-lipofuscinosis. Hum. Mol. Genet. 2012;21:2646–2650. doi: 10.1093/hmg/dds089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cotman S.L., Karaa A., Staropoli J.F., Sims K.B. Neuronal ceroid lipofuscinosis: impact of recent genetic advances and expansion of the clinicopathologic spectrum. Curr. Neurol. Neurosci. Rep. 2013;13:366. doi: 10.1007/s11910-013-0366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vidal-Donet J.M., Carcel-Trullols J., Casanova B., Aguado C., Knecht E. Alterations in ROS activity and lysosomal pH account for distinct patterns of macroautophagy in LINCL and JNCL fibroblasts. PLoS ONE. 2013;8:e55526. doi: 10.1371/journal.pone.0055526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson G., Smith V.V., Malone M., Sebire N.J. Blood film examination for vacuolated lymphocytes in the diagnosis of metabolic disorders; retrospective experience of more than 2,500 cases from a single centre. J. Clin. Pathol. 2005;58:1305–1310. doi: 10.1136/jcp.2005.027045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staropoli J.F., Haliw L., Biswas S., Garrett L., Holter S.M., Becker L., Skosyrski S., Da Silva-Buttkus P., Calzada-Wack J., Neff F., et al. Large-scale phenotyping of an accurate genetic mouse model of JNCL identifies novel early pathology outside the central nervous system. PLoS ONE. 2012;7:e38310. doi: 10.1371/journal.pone.0038310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Razi M., Futter C.E. Distinct roles for Tsg101 and Hrs in multivesicular body formation and inward vesiculation. Mol. Biol. Cell. 2006;17:3469–3483. doi: 10.1091/mbc.E05-11-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bache K.G., Brech A., Mehlum A., Stenmark H. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J. Cell Biol. 2003;162:435–442. doi: 10.1083/jcb.200302131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamai K., Toyoshima M., Tanaka N., Yamamoto N., Owada Y., Kiyonari H., Murata K., Ueno Y., Ono M., Shimosegawa T., et al. Loss of hrs in the central nervous system causes accumulation of ubiquitinated proteins and neurodegeneration. Am. J. Pathol. 2008;173:1806–1817. doi: 10.2353/ajpath.2008.080684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lemonnier T., Blanchard S., Toli D., Roy E., Bigou S., Froissart R., Rouvet I., Vitry S., Heard J.M., Bohl D. Modeling neuronal defects associated with a lysosomal disorder using patient-derived induced pluripotent stem cells. Hum. Mol. Genet. 2011;20:3653–3666. doi: 10.1093/hmg/ddr285. [DOI] [PubMed] [Google Scholar]
- 48.Imaizumi Y., Okada Y., Akamatsu W., Koike M., Kuzumaki N., Hayakawa H., Nihira T., Kobayashi T., Ohyama M., Sato S., et al. Mitochondrial dysfunction associated with increased oxidative stress and alpha-synuclein accumulation in PARK2 iPSC-derived neurons and postmortem brain tissue. Mol. Brain. 2012;5:35. doi: 10.1186/1756-6606-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitchison H.M., Bernard D.J., Greene N.D., Cooper J.D., Junaid M.A., Pullarkat R.K., de Vos N., Breuning M.H., Owens J.W., Mobley W.C., et al. Targeted disruption of the Cln3 gene provides a mouse model for Batten disease. The Batten Mouse Model Consortium [corrected] Neurobiol. Dis. 1999;6:321–334. doi: 10.1006/nbdi.1999.0267. [DOI] [PubMed] [Google Scholar]
- 50.Junaid M.A., Pullarkat R.K. Increased brain lysosomal pepstatin-insensitive proteinase activity in patients with neurodegenerative diseases. Neurosci. Lett. 1999;264:157–160. doi: 10.1016/s0304-3940(99)00095-6. [DOI] [PubMed] [Google Scholar]
- 51.Sleat D.E., Sohar I., Pullarkat P.S., Lobel P., Pullarkat R.K. Specific alterations in levels of mannose 6-phosphorylated glycoproteins in different neuronal ceroid lipofuscinoses. Biochem. J. 1998;334(Pt 3):547–551. doi: 10.1042/bj3340547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steinfeld R., Heim P., von Gregory H., Meyer K., Ullrich K., Goebel H.H., Kohlschutter A. Late infantile neuronal ceroid lipofuscinosis: quantitative description of the clinical course in patients with CLN2 mutations. Am. J. Med. Genet. 2002;112:347–354. doi: 10.1002/ajmg.10660. [DOI] [PubMed] [Google Scholar]
- 53.Zaehres H., Daley G.Q. Transgene expression and RNA interference in embryonic stem cells. Methods Enzymol. 2006;420:49–64. doi: 10.1016/S0076-6879(06)20004-1. [DOI] [PubMed] [Google Scholar]
- 54.Kim J.B., Greber B., Arauzo-Bravo M.J., Meyer J., Park K.I., Zaehres H., Scholer H.R. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461:649–643. doi: 10.1038/nature08436. [DOI] [PubMed] [Google Scholar]
- 55.Hermann A., Gastl R., Liebau S., Popa M.O., Fiedler J., Boehm B.O., Maisel M., Lerche H., Schwarz J., Brenner R., et al. Efficient generation of neural stem cell-like cells from adult human bone marrow stromal cells. J. Cell Sci. 2004;117:4411–4422. doi: 10.1242/jcs.01307. [DOI] [PubMed] [Google Scholar]
- 56.Hermann A., Maisel M., Wegner F., Liebau S., Kim D.W., Gerlach M., Schwarz J., Kim K.S., Storch A. Multipotent neural stem cells from the adult tegmentum with dopaminergic potential develop essential properties of functional neurons. Stem Cells. 2006;24:949–964. doi: 10.1634/stemcells.2005-0192. [DOI] [PubMed] [Google Scholar]
- 57.Korolchuk V.I., Saiki S., Lichtenberg M., Siddiqi F.H., Roberts E.A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F.M., et al. Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 2011;13:453–460. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klionsky D.J., Abdalla F.C., Abeliovich H., Abraham R.T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J.A., et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lukacs Z., Santavuori P., Keil A., Steinfeld R., Kohlschutter A. Rapid and simple assay for the determination of tripeptidyl peptidase and palmitoyl protein thioesterase activities in dried blood spots. Clin. Chem. 2003;49:509–511. doi: 10.1373/49.3.509. [DOI] [PubMed] [Google Scholar]
- 60.Ezaki J., Takeda-Ezaki M., Oda K., Kominami E. Characterization of endopeptidase activity of tripeptidyl peptidase-I/CLN2 protein which is deficient in classical late infantile neuronal ceroid lipofuscinosis. Biochem. Biophys. Res. Commun. 2000;268:904–908. doi: 10.1006/bbrc.2000.2207. [DOI] [PubMed] [Google Scholar]
- 61.Page A.E., Fuller K., Chambers T.J., Warburton M.J. Purification and characterization of a tripeptidyl peptidase I from human osteoclastomas: evidence for its role in bone resorption. Arch. Biochem. Biophys. 1993;306:354–359. doi: 10.1006/abbi.1993.1523. [DOI] [PubMed] [Google Scholar]
- 62.Asuri P., Bartel M.A., Vazin T., Jang J.H., Wong T.B., Schaffer D.V. Directed evolution of adeno-associated virus for enhanced gene delivery and gene targeting in human pluripotent stem cells. Mol. Ther. 2012;20:329–338. doi: 10.1038/mt.2011.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khan I.F., Hirata R.K., Wang P.R., Li Y., Kho J., Nelson A., Huo Y., Zavaljevski M., Ware C., Russell D.W. Engineering of human pluripotent stem cells by AAV-mediated gene targeting. Mol. Ther. 2010;18:1192–1199. doi: 10.1038/mt.2010.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.