Abstract
Haploinsufficiency for CHD7, an ATP-dependent nucleosome remodeling factor, is the leading cause of CHARGE syndrome. While congenital heart defects (CHDs) are major clinical features of CHARGE syndrome, affecting >75% of patients, it remains unclear whether CHD7 can directly regulate cardiogenic genes in embryos. Our complementary yeast two-hybrid and biochemical assays reveal that CHD7 is a novel interaction partner of canonical BMP signaling pathway nuclear mediators, SMAD1/5/8, in the embryonic heart. Moreover, CHD7 associates in a BMP-dependent manner with the enhancers of a critical cardiac transcription factor, Nkx2.5, that contain functional SMAD1-binding elements. Both the active epigenetic signature of Nkx2.5 regulatory elements and its proper expression in cardiomyocytes require CHD7. Finally, inactivation of Chd7 in mice impairs multiple BMP signaling-regulated cardiogenic processes. Our results thus support the model that CHD7 is recruited by SMAD1/5/8 to the enhancers of BMP-targeted cardiogenic genes to epigenetically regulate their expression. Impaired BMP activities in embryonic hearts may thus have a major contribution to CHDs in CHARGE syndrome.
INTRODUCTION
CHARGE (Coloboma of the eye, Heart defects, Atresia of the choanae, severe Retardation of growth/development, Genital abnormalities and Ear abnormalities) is a complex genetic disorder with an incidence of 1/10 000 (1–5). Congenital heart defects (CHDs) affect >75% of CHARGE patients and are the major cause of early morbidity. Tetralogy of Fallot, double-outlet-right-ventricle, patent ductus arteriosus, chamber septation abnormalities and cleft atrioventricular (AV) valves are the most commonly observed CHDs in CHARGE (1–5).
Haploinsufficiency for the chromodomain helicase DNA-binding 7 (CHD7) gene is the leading cause of CHARGE (6), accounting for ∼70% of all cases (2). CHD7 encodes a 2997-amino acid (aa) protein containing two chromodomains (aa 799–864), an ATPase/helicase domain (aa 964–1165), a helicase C-terminal domain (aa 1320–1404), a SLIDE domain (aa 1540–1848), a SANT domain (aa 1962–2028) and two BRK domains (aa 2564–2613). CHD7 is highly conserved among vertebrates, with mCHD7 being 95% identical and 97% similar to hCHD7 (7). As predicted from sequence analysis, CHD7 is capable of remodeling nucleosomes using energy from ATP hydrolysis (8). CHD7 can interact with histone H3 methylated at lysine 4 (H3K4me) through its chromodomains and is highly enriched in a subset of active enhancer elements with high levels of H3K4me (9,10). As CHD7 does not contain any known sequence-specific DNA-binding domain, a key question to address is how this chromatin remodeling factor is targeted to specific enhancers. Chd7 homozygous mutant mice die at ∼E10.5, whereas heterozygosity for Chd7 leads to multiple CHARGE-like phenotypes, including cardiovascular abnormalities (7,11). CHD7 can regulate neurogenesis (12–14), neural crest cell activation (15,16), middle and inner ear development (17–19) and great vessel morphogenesis (20). However, it remains unclear whether CHD7 can directly regulate cardiogenic genes in embryos.
BMP signaling plays complex roles during heart development (21). BMP signals are transduced through heterodimeric complexes of serine/threonine kinase receptors. The ligand/receptor complex at cell membranes phosphorylates intracellular BMP-activated SMADs (R-SMADs), SMAD1/5/8, which then associate with SMAD4 and translocate into the nucleus to regulate transcription of target genes (22). Our current study shows that CHD7 interacts with BMP R-SMADs to modulate the expression of critical cardiogenic transcription factors, such as Nkx2.5. The complex epigenetic regulatory mechanisms of heart development have only started to be revealed and have received a great deal of attention recently (23,24). We provide convincing evidence supporting CHD7 as a critical epigenetic regulator of cardiogenesis.
RESULTS
Identification of CHD7 as a novel SMAD1 interaction partner
We screened an embryonic mouse heart yeast two-hybrid library (25) using Smad1 as bait and obtained two identical clones of Chd7 (Fig. 1A). The two clones aligned with bp7230–8266 of Chd7 mRNA, which encodes aa 2301–2646 of mCHD7 (corresponding to aa 2312–2657 of hCHD7). Direct interaction between the C-terminal region of mCHD7 and SMAD1 was revealed by in vitro GST pull-down assays (Fig. 1B), and their interaction in mammalian cells was shown with mammalian cell GST pull-down assays (Supplementary Material, Fig. S1). An interaction between full-length CHD7 and HA-tagged SMAD1 was confirmed with co-immunoprecipitation (co-IP) experiments in COS cells (Fig. 1C). SMAD1 interacts with CHD7 through its MH2 domain (Fig. 1C). The CHD7–SMAD1 interaction was only detectable when a plasmid encoding constitutively active ALK6 was co-transfected, indicating that active BMP signaling is required for their interaction. We confirmed the interaction between endogenous CHD7 and SMAD1 with co-IP experiments in NkL-TAg immortal cardiomyocytes (26) and in mouse embryonic hearts (Fig. 1D and E). CHD7 also interacts with the other two BMP R-SMADs, SMAD5 and SMAD8 (Supplementary Material, Fig. S2). We therefore concluded that CHD7 is a novel interaction partner of BMP R-SMADs in embryonic hearts.
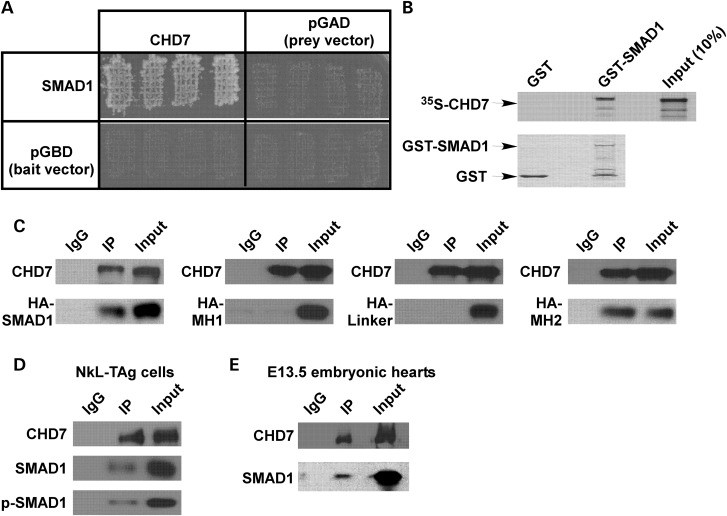
Figure 1.
CHD7 interacts with SMAD1. (A) AH109 yeast cells harboring different plasmids were grown on selective plates. Growth indicates positive interaction between the bait and the prey. SMAD1 and the C-terminal region of mCHD7 (aa 2301–2646) interact with each other, but do not interact with the bait or prey vector. (B) The C-terminal region of CHD7 that was in vitro-translated and labeled with 35S-Met was incubated with bacterial-expressed GST or GST-SMAD1. 35S-labeled CHD7 was co-purified with GST-SMAD1 but not with GST alone (top panel). Bottom panel shows Coomassie blue staining of SDS–PAGE gel. Arrows indicate the GST and GST-SMAD1 bands. (C) COS cells were transfected with plasmids encoding HA-SMAD1, HA-MH1, HA-Linker or HA-MH2. IP was performed using a CHD7 antibody. In this experiment, we took advantage of the fact that COS cells endogenously express CHD7. After IP, western analysis was performed using an HA antibody to test whether the HA-tagged protein was co-purified with CHD7 (bottom panels). Western analysis using a CHD7 antibody showed successful IP of CHD7 (top panels). Pre-immune IgG was used as a negative control. (D) To test whether endogenous SMAD1 and CHD7 interact with each other, NkL-TAg cell extracts were used for IP assay with a CHD7 antibody. A SMAD1 antibody was used to detect SMAD1 by western analysis. NkL-TAg cells were treated with 100 ng/ml of BMP4 for 4 h before co-IP. Western analysis using an anti-p-SMAD1 antibody confirmed that CHD7 interacts with active SMAD1. (E) Co-IP analysis was performed using E13.5 embryonic heart protein extracts and a CHD7 antibody. SMAD1 was co-immunoprecipitated with CHD7. Pre-immune IgG was used as a negative control.
CHD7 binds to enhancer regions of Nkx2.5 in a BMP-dependent manner
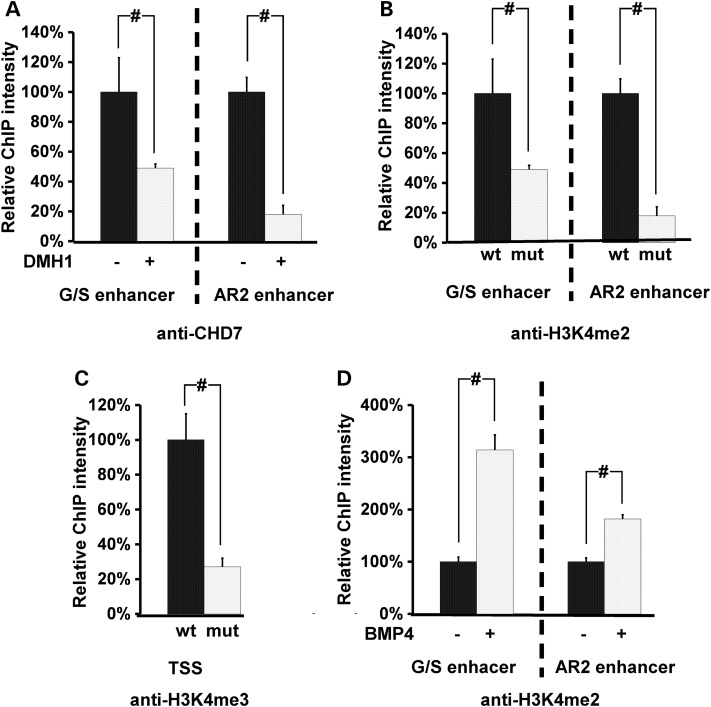
Nkx2.5 is a well-studied critical cardiogenic transcription factor whose cardiac expression is directly regulated by BMP R-SMADs (27–30). The immortal NkL-TAg cardiomyocyte cell line can respond to BMP stimulation to upregulate NKX2.5 expression (Supplementary Material, Fig. S3) and was thus used as the in vitro cell culture system to address the functional significance of the SMAD1–CHD7 interaction. We tested whether CHD7 is associated with Nkx2.5 enhancers by chromatin immunoprecipitation (ChIP) followed by quantitative PCR (qPCR) in NkL-TAg cells (26). We examined the G/S enhancer (28) and the AR2 enhancer (27, 29), both of which contain functional BMP R-SMAD-binding sites. We also examined the partial AR1 enhancer that contains GATA-binding sites (31). CHD7 was associated with G/S and AR2 enhancers, and the association was significantly increased upon BMP stimulation (Fig. 2A–C). The association with the partial AR1 enhancer was not altered by exogenous BMP4, consistent with the absence of SMAD-binding sites within this region. Treatment of cells with a BMP-specific blocker, DMH1 (32), inhibited the effect of BMP stimulation on CHD7 association with the two enhancers (Fig. 2D). Furthermore, ectopic overexpression of the SMAD1 MH2 domain significantly reduced BMP-induced CHD7 interaction with the Nkx2.5 enhancers (Fig. 2E). The MH2 domain of SMAD1 can interact with CHD7 but lacks the DNA-binding domain and therefore is expected to act in a dominant-negative manner to interfere with CHD7's interaction with DNA.
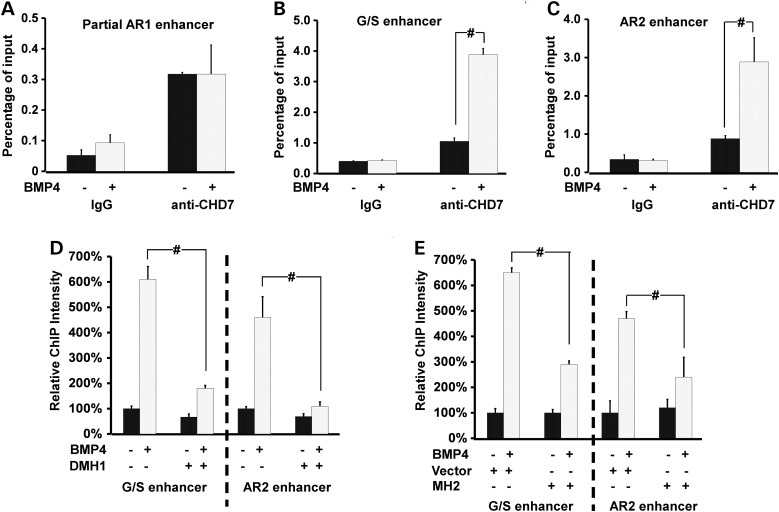
Figure 2.
CHD7 interacts with Nkx2.5 enhancers in a BMP-dependent fashion. (A–C) ChIP-qPCR analysis was performed on NkL-TAg cells treated with 100 ng/ml of BMP4 using a CHD7 antibody. The association of CHD7 with the partial AR1 genomic DNA was not altered (A), whereas the association with G/S (B) and AR2 (C) was significantly increased by BMP4 treatment. The Y-axis represents percentage of co-immunoprecipitated DNA over input. (D) Treatment of NkL-TAg cells with a BMP-specific inhibitor, DMH1 (8 uM), inhibited BMP4-enhanced CHD7 association with G/S and AR2 enhancers. The co-immunoprecipitated DNA from cells without BMP4 and DMH1 treatment was set at 100%. (E) NkL-TAg cells harboring a plasmid expressing the MH2 domain of SMAD1 or an empty vector (negative control) were treated with BMP4 followed by ChIP-qPCR using a CHD7 antibody. The co-immunoprecipitated DNA from cells harboring the vector and without BMP stimulation was set at 100%. Data were averaged from 3 to 5 independent experiments and were presented as mean ± standard error (SEM). # represents P < 0.01, student t-test.
Chd7 regulates expression of Nkx2.5 in NkL-TAg cells
We next determined the role of Chd7 on the expression of NKX2.5 in NkL-TAg cardiomyocytes. Knocking-down CHD7 expression using a published short-hairpin RNA (shRNA) construct (15) abolished BMP4-stimulated upregulation of NKX2.5 (Fig. 3A). Similar results were obtained using another shRNA construct. Therefore, Chd7 is required for BMP-induced upregulation of NKX2.5 in the cardiomyocyte cell line. We next tested whether the chromodomains and ATPase/helicase domain are required for CHD7 to promote the expression of NKX2.5. The genomic DNA fragment harboring G/S and AR2 enhancers of Nkx2.5 was fused with a SV40 minimum promoter and cloned into the pREP4-luc reporter vector. pREP4-luc is an Epstein–Barr virus-based episomal vector that undergoes chromatinization after transfection into mammalian cells and is thus particularly suitable for studying epigenetic regulation of enhancers (16,33). Overexpression of full-length CHD7 significantly increased the reporter activity upon BMP4 stimulation (Fig. 3B and C). Remarkably, ectopic expression of CHD7 lacking either of the chromodomains or the ATPase/helicase domain reduced the reporter activity. These truncated proteins presumably act in a dominant-negative fashion, preventing endogenous wild-type CHD7 from interacting with BMP R-SMADs and binding the Nkx2.5 enhancers. Our data suggest that CHD7's histone-binding and nucleosome-remodeling activities are required to promote Nkx2.5 expression.
Figure 3.

Chd7 promotes the expression of NKX2.5 in NkL-TAg cells. (A) NkL-TAg cells were transduced with a lentiviral shRNA construct against Chd7 (15) or a scramble shRNA. Cells were treated with 50 ng/ml of BMP4 followed by western analysis using antibodies against CHD7 and NKX2.5. Experiments were repeated three times. (B) NkL-TAg cells were transfected with a plasmid expressing an HA-tagged form of CHD7 or an empty vector. Forty-eight hours after transfection, western analysis was performed using an anti-HA antibody. Tubulin was used as a loading control. ‘del chromo 1’, ‘del chromo 2’ and ‘del ATPase’ refer to ‘deletion of Chromodomain 1’, ‘deletion of Chromodomain 2’ and ‘deletion of ATPase/helicase domain’, respectively. This experiment confirmed the expression of the transfected CHD7 constructs in NkL-TAg cells. Owing to the large size of the proteins (full-length CHD7: 336 kDa; del chromo 1: 335 kDa; del chromo 2: 335 kDa; del ATPase: 318 kDa), only subtle differences in band positions were observed. All constructs were sequenced to confirm that no mutation was introduced during subcloning. (C) The genomic DNA fragment harboring G/S and AR2 enhancers of Nkx2.5 was fused with a SV40 minimum promoter and cloned into the pREP4-luc reporter vector (an Epstein–Barr virus-based episomal vector that undergoes chromatinization after transfection into mammalian cells). The reporter construct was co-transfected with an empty vector or with a plasmid expressing full-length or a deletion construct of CHD7 into NkL-TAg cells, followed by BMP4 (100 ng/ml) or BSA (negative control) treatment for 48 h. Luciferase activity was then measured. The activity of cells transfected with empty vector and treated with BSA was arbitrarily set at 1.0. Data were averaged from three independent experiments with each experiment being performed in triplicate. Error bars represent SEM. # represents P < 0.01, Student's t-test.
Inactivation of Chd7 leads to multiple embryonic heart defects
To reveal the in vivo role of Chd7 during cardiogenesis, we purchased the Chd7tm1a(EUCOMM)Wtsi mice from IKMC. In this allele, a cassette containing an mRNA splice acceptor site, a lacZ reporter and a neomycin selection marker is inserted between exons 2 and 3 of Chd7. This cassette is predicted to stop transcription/translation after exon 2 of Chd7 mRNA. The transcript will only generate a truncated CHD7 protein lacking any known functional domain, and thus this allele is expected to be null (or strong hypomorphic). Chd7tm1a/+ mice were viable and some displayed head-bobbing and circling, a phenotype consistent with other Chd7 mutant mouse models (7,11). Cardiac expression of the lacZ reporter was detected at all stages examined (Supplementary Material, Fig. S4). LacZ was ubiquitously expressed in myocardial, endocardial and AV cushion mesenchymal cells at E9.5 (Supplementary Material, Fig. S5). The expression of CHD7 in various cardiac cell types was confirmed with an immunostaining study using a CHD7 antibody (Fig. 4A). No signal was detected in Chd7tm1a/tm1a embryos, confirming the specificity of the antibody and the absence of CHD7 in homozygous mutants (Fig. 4B). No apparent gross abnormality was observed in mutants at E9.5. At E10.5, all mutant embryos (living) displayed a growth arrest/delayed development defect (Fig. 4C). No living mutants were recovered after E10.5, consistent with a previous report (11). Histological studies showed no apparent myocardial defect in mutants at E9.5 (Fig. 4D and E); however, the number of AV cushion mesenchymal cells was reduced at this stage (Fig. 4D–E′). At E10.5, the mutant hearts displayed hypocellular myocardial wall and hypocellular AV cushion defects (Fig. 4F and G).
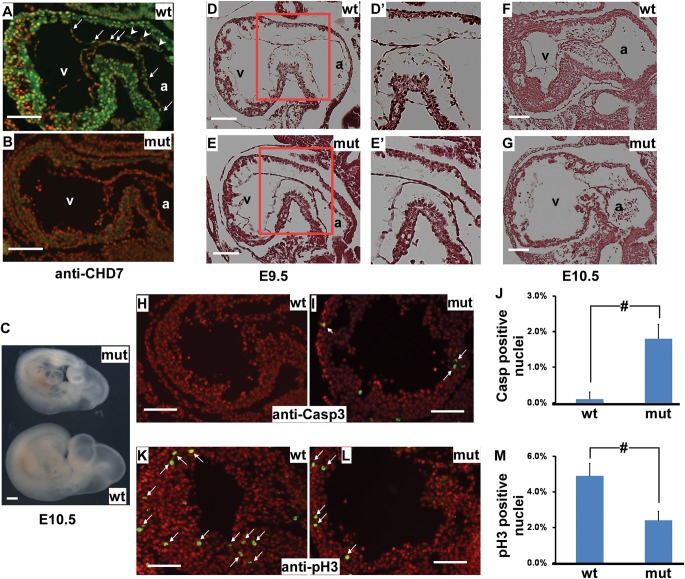
Figure 4.
Inactivation of Chd7 leads to multiple cardiogenic defects. Wild-type (A) and mutant (B) embryos at E9.5 were sagittally sectioned and stained with an anti-CHD7 antibody (green). Total nuclei were stained with DAPI (red). Arrows and arrow heads indicate examples of positively stained endocardial and mesenchymal cells, respectively. (C) Littermate mutant and wild-type embryos at E10.5 were isolated and photographed. Only living embryos (as judged from heart-beating) were used for further analysis throughout this study. (D–G) Sagittal sections of control and mutant embryos at E9.5 (D, E) and E10.5 (F, G) were HE-stained. D′ and E′ correspond to the boxed area of D and E, respectively. Fewer mesenchymal cells were present in mutant AV cushions. (H–J) Wild-type (H) and mutant (I) embryos at E10.0 (∼29–34 somites) were sagittally sectioned and stained with an antibody against cleaved Casp3 (an apoptosis marker, green). Total nuclei were stained with DAPI (red). Arrows indicate examples of positively stained nuclei. The percentage of Casp3-positive nuclei over total nuclei in ventricles was plotted (J). (K–M) Wild-type (K) and mutant (L) embryos at E10.0 were sagittally sectioned and stained with an antibody against phospho-histone H3 (pH3, a cell proliferation marker, green). Total nuclei were stained with DAPI (red). Arrows indicate examples of positive stained nuclei. The percentage of pH3-positive nuclei over total nuclei in ventricles was plotted (M). For J and M, data were averaged from three independent embryos of each genotype and were presented as mean ± SEM. a—atrium, v—ventricle, mut—mutant, Chd7tm1a/tm1a, wt—wild type, Chd7+/+, # represents P < 0.01, student t-test. The bar in panel C represents 0.4 mm. The bars in all other panels represent 0.1 mm.
To alleviate the concern that the observed cardiac defects are because of general growth delay, we only used living embryos at E10.0 (∼29–34 somites) for further cellular/molecular examination. At this stage, the gross growth delay was not apparent in mutants. Maintaining normal cell proliferation and survival are key roles of BMP signaling in embryonic hearts (34,35). At E10.0, we observed both significantly increased cell death and reduced cell proliferation in the myocardium of mutants (Fig. 4H–M), indicating that Chd7 is required for cardiomyocyte proliferation and survival.
Chd7 is required for AV cushion mesenchymal formation
BMP signaling is critical for mesenchymalization of AV cushions, [e.g. (36,37)], which are precursors of AV valves and AV septum. The hypocellular AV cushion defect was apparent at E9.5 (Fig. 4D–E′). We next performed ex vivo collagen gel analysis to quantitatively examine how inactivation of Chd7 affects cushion mesenchymal formation. As shown in Figure 5, the number of mesenchymal cells formed on mutant AV explants (E9.5) was reduced to <30% of the control level. The identity of the mesenchymal cells on the gel was confirmed with immunofluorescence (IF) studies using an antibody against α-smooth muscle actin (SMA), which is a commonly used cushion mesenchymal cell marker, [e.g. (38)]. Therefore, inactivation of Chd7 impairs multiple BMP-dependent cardiogenic processes, including myocardial cell proliferation and survival and AV cushion mesenchyme formation.
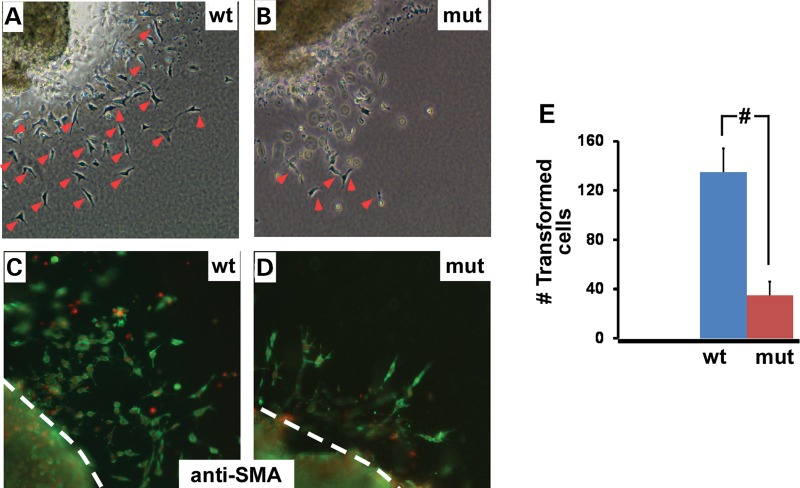
Figure 5.
Inactivation of Chd7 reduces AV cushion mesenchymal cell formation on collagen gel analysis. (A, B) AV explants from wild-type controls (A) and mutants (B) were cultured on type I collagen gel for 48 h. Pictures were taken with a phase-contrast microscope. Examples of mesenchymal cells are indicated with red arrow heads. (C, D) Wild-type AV explants (C) and mutant explants (D) were immunostained with an anti-SMA antibody (a cushion mesenchyme marker, green). Total nuclei were stained with DAPI (red). Dotted lines indicate the origin of the explants. (E) The number of mesenchymal cells formed was quantified and plotted. Data were averaged from at least seven independent explants for each genotype and were shown as mean ± SEM. mut—mutant, Chd7tm1a/tm1a, wt—wild type, Chd7+/+, # represents P < 0.01, student t-test.
Expression of BMP-targeted genes was down-regulated in mutant hearts
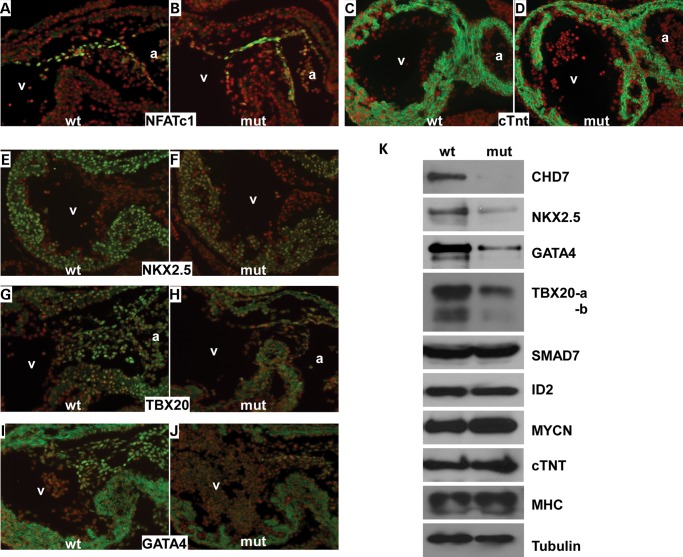
We next focused on examining the molecular defects in Chd7 mutant hearts. No altered expression of NFATc1 (an endocardial marker) or cTNT (a myocardial marker) was observed in mutants (Fig. 6A–D), suggesting that the general differentiation program of endocardial/ myocardial cells is not halted by inactivation of Chd7. The expression of known BMP-targeted, cardiogenic genes, including Nkx2.5, Gata4 and Tbx20, was visibly reduced in mutant sections (E10.0) (Fig. 6E–J). Reduced expression of these genes was confirmed with western analysis (Fig. 6K). Interestingly, the expression of ID2, MYCN and SMAD7, which are also BMP targets in developing hearts (35,39–41), was not altered in mutant hearts. These results suggest that CHD7 selectively regulates BMP target genes in mouse embryonic hearts.
Figure 6.
Chd7 regulates the expression of BMP downstream target genes in developing hearts. (A–J) Sagittal sections of wild-type and mutant embryos (E10.0) were immunostained with antibodies indicated (green). Total nuclei were stained with DAPI (red). (K) Total proteins were extracted from pooled embryonic hearts at E10.0 and subjected to western analysis using the indicated antibodies. Tubulin was used as the loading control. a—atrium, v—ventricle, mut—mutant, Chd7tm1a/tm1a, wt—wild type, Chd7+/+.
Chd7 is required for maintaining active epigenetic state of Nkx2.5 regulatory elements
We next tested whether the CHD7 interaction with Nkx2.5 enhancers relies on BMP signaling in embryonic hearts, as observed in NkL-TAg cells. Treatment of cultured embryonic hearts (E11.5) with DMH1 significantly decreased CHD7 association with G/S and AR2 enhancers (Fig. 7A), suggesting that BMP activity is essential for CHD7 interaction with Nkx2.5 enhancers in vivo. As an ATP-dependent nucleosome remodeling factor, we speculate that CHD7 modulates the epigenetic status of the regulatory elements of target genes. To test this idea, we performed ChIP-qPCR analyses to examine chromatin modifications in enhancers and the transcription start site (TSS) of Nkx2.5 in mutant embryos. Association of the Nkx2.5 enhancers and TSS with H3K4me2 and H3K4me3, respectively, was significantly reduced in mutant embryos (Fig. 7B and C). H3K4me2 is a marker for an open and active enhancer and H3K4me3 marks active transcription (42,43). We further showed that BMP4 stimulation significantly increased association of the G/S and AR2 enhancers with H3K4me2 (Fig. 7D). Our data therefore suggest that CHD7 is required for maintaining Nkx2.5 enhancers (G/S, AR2) and TSS in an active epigenetic state.
Figure 7.
Chd7 is required for maintaining the active epigenetic state of Nkx2.5 regulatory elements. (A) E11.5 embryonic hearts were cultured and treated with 8 uM DMH1 or DMSO alone for 4 h followed by ChIP-qPCR using a CHD7 antibody. The co-immunoprecipitated DNA from hearts without DMH1 treatment was set at 100%. Association of CHD7 with both the G/S and AR2 was significantly reduced by DMH1 treatment. (B, C) ChIP-qPCR analysis was performed on pooled wild-type and mutant embryos at E10.0 using the antibodies indicated. The co-immunoprecipitated DNA from wild-type embryos was set at 100%. (D) Wild-type embryonic hearts at E11.5 were treated with BMP4 (100 ng/ml) or BSA (negative control) for 4 h, followed by ChIP-qPCR analysis using an antibody against H3K4me2. The co-immunoprecipitated DNA from hearts treated with BSA was set at 100%. Data were averaged from at least three independent experiments and were presented as mean ± SEM. mut—mutant, Chd7tm1a/tm1a, wt—wild type, Chd7+/+, TSS—transcriptions start site, # represents P < 0.01, student t-test.
DISCUSSION
CHARGE syndrome is associated with multiple forms of CHDs (1–5), yet the underlying molecular mechanisms remain unknown. We reveal for the first time that CHD7 interacts with BMP R-SMADs to epigenetically regulate the expression of BMP-targeted, critical cardiogenic transcription factors in mouse embryos. Specifically, we show that Nkx2.5 is a direct downstream target of CHD7 in embryonic hearts. Considering the well-documented complexity of BMP signaling during cardiogenesis (21), we speculate that impaired BMP activity is a key contributing factor of CHDs in CHARGE patients. In addition to CHDs, CHARGE syndrome is characterized by abnormal development of multiple organs known to be regulated by BMP signaling, including eyes, ears and choanae. Therefore, the interaction between CHD7 and BMP R-SMADs likely also has roles in other clinical features of CHARGE.
CHD7 interacts with H3K4me through its chromodomain, and whole-genome studies revealed that CHD7 preferentially binds the H3K4 methylated enhancer regions (9,10). However, H3K4 methylation cannot be the sole determinant for the position of CHD7 along the chromatin, as CHD7 is only associated with a small subset of H3K4me-enriched enhancers. Based on our studies showing the novel SMAD1/5/8–CHD7 interaction and the association of CHD7 with Nkx2.5 enhancers, we propose that CHD7 is recruited to BMP-responsive enhancers by its interaction with SMAD1/5/8 through its C-terminal region (see model in Fig. 8). In addition, CHD7 also interacts with methylated histones through its N-terminal chromodomains. The combined interaction through both its N-terminal and C-terminal domains allow CHD7 to be loaded onto specific enhancers along chromosomes. In the case of Nkx2.5, CHD7 is required for sustaining the methylation of H3K4 at the G/S, AR2 and TSS to promote Nkx2.5 expression. As suggested from our reporter analysis, histone-binding and nucleosome-remodeling activities are required for CHD7 to promote Nkx2.5 expression. Moreover, whole-genome analysis revealed that CHD7 co-localizes with SMAD1 along chromosomes in ES cells to regulate gene expression (10). Therefore, our model may also be applied to other cell types.
Figure 8.
Model of CHD7 recruitment to the enhancer of a BMP/SMAD target gene. Upon BMP stimulation, the SMAD1/5/8-SMAD4 complex is transferred into nucleus, where the complex binds to the SMAD-binding elements in the enhancers of BMP/SMAD target genes. The C-terminus of CHD7 interacts with BMP R-SMADs, resulting in CHD7's recruitment to the enhancer. In addition, CHD7 also interacts with H3K4me histones through its N-terminal chromodomains. Once located on the enhancer, CHD7 acts in concert with other epigenetic regulators to modulate transcription of cardiogenic transcription factors downstream of BMP signaling cascades, such as Nkx2.5. CHD7 is required for maintaining the active epigenetic state of both enhancers and the TSS of Nkx2.5. Histone-binding and nucleosome-remodeling activities are essential functions of CHD7 that enhance the expression of Nkx2.5, as suggested from our reporter analysis (Fig. 3).
Inactivation of Chd7 impaired multiple BMP-dependent cardiogenic processes, including cardiomyocyte proliferation and survival and AV cushion mesenchymalization. The thin myocardial wall defect is expected to reduce cardiac contractility and is likely the major cause for embryonic lethality of mutants. The hypocellular AV cushion defect in mutants is unlikely secondary to the myocardial abnormalities, as no apparent myocardial wall defect was observed at E9.5 when the AV cushion defect was already apparent. The results from the collagen gel assay further support the direct role of Chd7 in AV cushion development. In this ex vivo explant assay, AV cushions were isolated and put on the gel. Formation of cushion mesenchymal cells on the collagen gel is thus relatively independent from other developmental processes of an embryo. At the molecular level, the expression of GATA4 and TBX20, two regulatory targets of BMP signaling important for AV cushion development, was reduced in mutant AV cushion cells. We conclude that Chd7 promotes the expression of BMP target genes in AV cushions to support normal valvuloseptal development. Significantly, abnormalities in the AV canal region are frequently observed in CHARGE patients (1,2,4). Our findings on the role of Chd7 during early stages of valvuloseptal development provide clues on the molecular etiology of these forms of CHDs in CHARGE.
Single mutations of BMP R-Smads do not cause obvious cardiac defects in mice (44–46), likely due to their redundant functions. However, conditional inactivation of the R-SMAD co-activator, Smad4, resulted in similar cardiac phenotypes to those observed in Chd7 mutants. Myocardial inactivation of Smad4 caused a thin myocardial wall defect (35) and endothelial inactivation caused hypocellular AV cushions (47,48). These data further support our conclusion that reduced BMP/SMAD activities contribute to the heart defects in Chd7 mutants.
It is noteworthy that not all BMP activities were disrupted in Chd7 mutant hearts. Initiation of trabeculation, which relies on Bmp10 (49), appeared normal in Chd7 mutants. This result suggests that Chd7 does not act as a general BMP signaling modifier, but rather, can selectively regulate BMP activities during cardiogenesis. Supporting this idea, the expression of some BMP target genes is not changed in Chd7 mutant hearts. For example, Chd7 inactivation has no effect on the expression of Mycn, which acts downstream of BMP signaling to promote trabeculation (50). For the genes whose expression does not rely on Chd7, it is possible that their enhancers recruit other ATP-dependent nucleosome remodeling factors, which compensate for the loss of Chd7. How CHD7 is differentially required for the normal expression of BMP target genes warrants further investigation.
In summary, we reveal that CHD7 interacts with BMP R-SMADs to epigenetically regulate the expression of BMP target genes such as Nkx2.5 in mouse embryonic hearts. Our results help understand how CHD7 regulates gene expression in a tissue-specific fashion and provide clues on the molecular mechanism for CHDs in CHARGE syndrome.
MATERIALS AND METHODS
Yeast two-hybrid screening
We used the MATCHMAKER Two-Hybrid System 3 (Clontech) to perform the yeast two-hybrid screening with detailed procedures previously described (51). Briefly, the Smad1 bait construct was co-transformed into AH109 yeast cells with an embryonic heart two-hybrid library (25). Cells were plated on selective medium lacking Trp, Leu, His and Ade. A positive interaction between the prey and bait allows yeast cells to grow. Candidate plasmids were purified and individually co-transformed with the Smad1 bait plasmid or empty bait vector into AH109 cells to re-test whether they could still grow on the selective medium. The candidates that only interacted with the Smad1 bait but not with the empty bait vector were sequenced. Two identical clones encoding aa 2301–2646 of mCHD7 were acquired.
Cell culture, short-hairpin RNA (shRNA) knock down and western analysis
The NkL-TAg cardiomyocyte cell line was kindly provided by Dr. Olson (UT Southwestern) and was cultured as previously described (26). shRNA lentiviral constructs against Chd7 were kindly provided by Dr. Wysocka (Stanford University). These constructs can efficiently and specifically knock down the expression of CHD7 as shown from a previous study (15). Lentivirus was packaged using the TRIPZ system purchased from Open Biosystems. NkL-TAg cells were transduced with the shRNA lentiviral constructs and selected with puromycin (5 ug/ml) for 14 days. A scramble shRNA construct was included as a negative control. For western analysis, proteins were extracted using 2× Laemmli buffer from cultured cells or embryos. The protein concentration was determined using the Dc Protein Assay kit purchased from BioRad. A total of 10–50 ug of protein was loaded onto SDS–PAGE gel, and western analysis was performed using the ECL kit from Millipore. Primary antibodies used in this study, and their concentrations in different applications are described later. Horseradish peroxidase-conjugated secondary antibodies (Invitrogen) were used to visualize the western signal.
Luciferase reporter analysis
The 3.2-kb genomic DNA fragment covering the G/S and AR2 enhancers of Nkx2.5 (bp5850 to bp2657 relative to the start codon) was PCR-amplified, fused with the SV40 minimum promoter and cloned into pREP4-luc (33) reporter vector (kindly provided by Dr. Zhao, NIH). pREP4-luc is an episomal reporter vector containing the Epstein–Barr virus replication origin (33). After transient transfection into mammalian cells, the plasmid undergoes chromatinization and is particularly useful for studying epigenetic regulation of enhancers, [i.e. (16,33)]. The plasmid expressing full-length hCHD7 (with 3×HA tag at the N-terminal end) was purchased from GeneCopoeia. The cDNA fragments (bp2269 to bp3535 relative to the start codon) of hCHD7 with a deletion of Chromodomain 1, Chromodomain 2 or the ATPase/helicase domain were synthesized by IDT, digested with AarI and AfeI and swapped with the AarI–AfeI fragment of full-length CHD7 to acquire constructs expressing mutated forms of CHD7. All constructs were confirmed with sequencing analysis. The reporter construct was co-transfected with a plasmid expressing full-length or various deletion forms of CHD7 into NkL-TAg cells using the Neon Electroporation Transfection System (Invitrogen). Cells were treated with 100 ng/ml of BMP4 or BSA for 48 h followed by measurement of luciferase activity using the Luciferase Assay kit (Promega). A plasmid that constitutively expresses a lacZ reporter was co-transfected for normalization of transfection efficiency. LacZ activity was measured using the Beta-Glo Assay System (Promega).
In vitro GST pull-down, mammalian cell GST pull-down and co-IP analysis
For the in vitro pull-down assay, the C-terminal region of mCHD7 (aa 2301–2646) was labeled with [35S]Met using the TNT-coupled transcription system (Promega) according to the manufacturer's protocol. GST or GST-SMAD1 proteins were purified using glutathione-conjugated agarose beads (Pharmacia) from BL21 bacterial following the manufacturer's instruction. Radio-labeled CHD7 was incubated with GST or GST-SMAD1 followed by extensive washing. Samples were heat-denatured and separated on SDS–PAGE gel. Gels were first stained with Coomassie blue to visualize GST and GST-SMAD1 proteins and then treated with Enhance (NEN) following the manufacturer's instructions. The treated gels were dried followed by auto-radiography. For mammalian cell GST pull-down, the DNA fragment from the prey construct encoding aa 2301–2646 of mCHD7 was cloned into pCMV-GST (52) to allow the expression of GST-CHD7 in mammalian cells. The empty GST vector or the GST-CHD7 plasmid was co-transfected with a plasmid expressing HA-SMAD1 into COS cells. The ALK6 (Q203D) construct encoding a constitutively active form of type I BMP receptor (kindly provided Dr. Attisano, University of Toronto) was co-transfected to activate the BMP intracellular signaling cascade. Cell lysates were subjected to GST-bead purification as described in the previous study (51). Western analysis was then performed to detect HA-SMAD1, GST and GST-CHD7. For co-IP experiments in COS cells, constructs expressing various HA-tagged proteins were transfected into COS cells. IP was performed using a CHD7 antibody followed by western analysis to detect HA-tagged proteins and CHD7. For co-IP in NkL-TAg cells and embryonic hearts, cells (treated with 100 ng/ml of BMP4 for 2 h) or hearts were lysed and lysates were subjected to IP using an anti-CHD7 antibody followed by western analysis to detect SMAD1, pSMAD1 and CHD7.
Chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) analysis
ChIP analysis was performed using the ab500 ChIP kit (Abcam) following the manufacturer's protocol. Briefly, NkL-TAg cells or embryo tissues were lysed using the lysis buffer provided in the kit. Lysates were treated with 1.1–1.5% formaldehyde to cross-link proteins to DNA. Genomic DNA was sheared using Bioruptor XL sonicator (Diagenode) for 10 min at the M2 intensity level to acquire optimal DNA fragment size of ∼500 bp. Samples were then subjected to IP using various antibodies followed by DNA purification. Pre-immune IgG was used as a negative control for all experiments. DNA samples were analyzed by qPCR using Roche LightCycler480 Real Time PCR machine (UAB Heflin Genomic Core). The relative DNA amount was calculated using the ΔΔCt method. Primer sequences for amplifying the G/S and AR2 enhancers were published previously (28). These sequences are G/S forward (5′ CAGTCTTGGGAGCTCAAGACTAACC), G/S backward (5′ CAGATCCCCAAGCTTACTAGC AACTAC), AR2 forward (5′ CTGCTCATCCATCAGCCAGACGAAGA) and AR2 backward: 5′ GAAAGATAAGCTGCAACTATCACCCGG). The primers for amplifying the partial AR1 are AR1 forward (5′ GGACAACAGCGGGGAAGG) and AR1 backward (5′ CGGCAAGATTTC CTCCGGG). The primers for amplifying the TSS are TSS_up (5′ GAGACTATCCCAAATGTGTCG) and TSS_dn (5′ GAAGAGGCGGGAGCAGGCTGG).
Animals
This study conforms to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication no. 85–23, revised 1996). All protocols were approved by the Institutional Animal Care and Use Committee at the University of Alabama at Birmingham. Chd7tm1a(EUCOMM)Wtsi/+ mice were purchased from International Knockout Mouse Consortium (IKMC) (http://www.knockoutmouse.org/martsearch/project/35711). In this allele, a cassette containing an mRNA splice acceptor site, a lacZ reporter and a neomycin selection marker is inserted between exons 2 and 3 of Chd7. This cassette is predicted to stop transcription/translation after exon 2 of Chd7 mRNA. The transcript will only generate a truncated CHD7 protein lacking any known functional domain, and thus this allele is expected to be null. Mice were maintained on the 129S6 background as suggested by the supplier. Genotyping was performed following the protocol provided by the supplier. Three primers were mixed in a PCR. The three primers are: Chd7_5P_F: 5′ TGCAGATGGGACGTTTTCAG; CAS_R1_Term: 5′ TCGTGGTATCGTTATGCGCC; Chd7_5P_R: 5′ CTGCAAGAACACAGGG CAAG. A 411-bp and a 610-bp product are generated from the wild-type and the mutant allele, respectively. PCR products from the founder mouse (originally purchased from IKMC) were sequenced to verify their identity.
Lacz staining, IF studies and ex vivo explant culture
LacZ staining on embryos or embryonic hearts was performed as described in the previous study (53). IF studies were performed on embryo sections using various primary antibodies. An Alexa Fluor 488-conjugated secondary antibody (green, Invitrogen) was used to visualize the signal. Samples were counter-stained with DAPI to visualize total nuclei. Slides were examined and photographed using a fluorescence microscope (EVOS, from AMG). To gain better contrast between antibody staining and nuclei staining, the blue color of DAPI staining was manually switched to the red color in Photoshop 11 (Adobe) without altering the relative signal intensity. Ex vivo explant culture was performed as previously described (37,47). Briefly, the AV canal regions from E9.5 embryos were isolated under a dissection microscope and placed onto a type I collagen gel. After a 48-h incubation, mesenchymal cells form and migrate away from the explant sites. Mesenchymal cells can be identified by staining with SMA or by their fibroblast-like morphology.
Primary antibodies
The antibody against HA (HA.11, 1/1000 for western analysis) was purchased from Babco. The GST antibody (#27-4577, 1/5000 for western analysis) was purchased from Amersham. The anti-NFATc1 antibody (7A6, 1/50 for IF) was purchased from BD Pharmingen. Antibodies against cleaved CASP3 (#9661, 1/100 for IF), SMAD1 (#9743, 1/1000 for western analysis) and p-SMAD1 (#9511, 1/2000 for western analysis) were purchased from Cell Signaling. The anti-phospho-histone H3 antibody (#06-570, 1/500 For IF) was purchased from Upstate. Antibodies against SMA (1A4, 1/500 for IF), TBX20 (HPA008192, 1/100 for IF) and ID2 (HPA027612, 1/2000 for western analysis) were purchased from Sigma. Antibodies against Tubulin (E7, 1/5000 for western analysis), cardiac troponin T (CT3, 1/1000 for western analysis, 1/200 For IF) and cardiac myosin heavy chain (MF20, 1/2000 for western analysis, 1/750 for IF) were purchased from Iowa Hybridoma Bank. Antibodies against CHD7 (ab31824, 1/1000 for western analysis; 1/50 for IP and ChIP, 1/200 for IF), NKX2.5 (ab35842, 1/2000 for western analysis, 1/150 for IF), SMAD7 (ab76498, 1/2000 for western analysis), TBX20 (ab42468, 1/1000 for western analysis), MYCN (ab16898, 1/300 for western analysis), H3K4me2 (Y47, 10 ug/ml for ChIP) and H3K4me3 (ab8580, 10 ug/ml for ChIP) were purchased from Abcam. The anti-GATA4 antibody for western analysis (LS-b3074, 1/2000) was purchased from Lifespan, and the antibody for IF (G-4, 1/300) was purchased from Santa Cruz.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by a grant from National Institutes of Health (R01HL095783) awarded to K.J. C.H. was awarded with American Heart Association predoctoral fellowship (09PRE2261138).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Wysocka (Stanford), Olson (UT Southwestern), Zhao (NIH) and Attisano (University of Toronto) for providing regents, and Drs Scacheri (Case Western), Bouazoune and Kingston (Harvard) for helpful discussion.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Blake K.D., Prasad C. CHARGE syndrome. Orphanet. J. Rare Diseases. 2006;1:34. doi: 10.1186/1750-1172-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zentner G.E., Layman W.S., Martin D.M., Scacheri P.C. Molecular and phenotypic aspects of CHD7 mutation in CHARGE syndrome. Am. J. Med. Genet. A. 2010;152A:674–686. doi: 10.1002/ajmg.a.33323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergman J.E., Janssen N., Hoefsloot L.H., Jongmans M.C., Hofstra R.M., van Ravenswaaij-Arts C.M. CHD7 mutations and CHARGE syndrome: the clinical implications of an expanding phenotype. J. Med. Genet. 2011;48:334–342. doi: 10.1136/jmg.2010.087106. [DOI] [PubMed] [Google Scholar]
- 4.Sanlaville D., Verloes A. CHARGE syndrome: an update. Eur. J. Hum. Genet. 2007;15:389–399. doi: 10.1038/sj.ejhg.5201778. [DOI] [PubMed] [Google Scholar]
- 5.Wyse R.K., al-Mahdawi S., Burn J., Blake K. Congenital heart disease in CHARGE association. Pediatr. Cardiol. 1993;14:75–81. doi: 10.1007/BF00796983. [DOI] [PubMed] [Google Scholar]
- 6.Vissers L.E., van Ravenswaaij C.M., Admiraal R., Hurst J.A., de Vries B.B., Janssen I.M., van der Vliet W.A., Huys E.H., de Jong P.J., Hamel B.C., et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat. Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 7.Bosman E.A., Penn A.C., Ambrose J.C., Kettleborough R., Stemple D.L., Steel K.P. Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum. Mol. Genet. 2005;14:3463–3476. doi: 10.1093/hmg/ddi375. [DOI] [PubMed] [Google Scholar]
- 8.Bouazoune K., Kingston R.E. Chromatin remodeling by the CHD7 protein is impaired by mutations that cause human developmental disorders. Proc. Natl. Acad. Sci. U S A. 2012;109:19238–19243. doi: 10.1073/pnas.1213825109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnetz M.P., Bartels C.F., Shastri K., Balasubramanian D., Zentner G.E., Balaji R., Zhang X., Song L., Wang Z., Laframboise T., et al. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009;19:590–601. doi: 10.1101/gr.086983.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schnetz M.P., Handoko L., Akhtar-Zaidi B., Bartels C.F., Pereira C.F., Fisher A.G., Adams D.J., Flicek P., Crawford G.E., Laframboise T., et al. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS. Genet. 2010;6:e1001023. doi: 10.1371/journal.pgen.1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurd E.A., Capers P.L., Blauwkamp M.N., Adams M.E., Raphael Y., Poucher H.K., Martin D.M. Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm. Genome. 2007;18:94–104. doi: 10.1007/s00335-006-0107-6. [DOI] [PubMed] [Google Scholar]
- 12.Engelen E., Akinci U., Bryne J.C., Hou J., Gontan C., Moen M., Szumska D., Kockx C., van Ijcken W., Dekkers D.H., et al. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat. Genet. 2011;43:607–611. doi: 10.1038/ng.825. [DOI] [PubMed] [Google Scholar]
- 13.Adams M.E., Hurd E.A., Beyer L.A., Swiderski D.L., Raphael Y., Martin D.M. Defects in vestibular sensory epithelia and innervation in mice with loss of Chd7 function: implications for human CHARGE syndrome. J. Comp. Neurol. 2007;504:519–532. doi: 10.1002/cne.21460. [DOI] [PubMed] [Google Scholar]
- 14.Layman W.S., McEwen D.P., Beyer L.A., Lalani S.R., Fernbach S.D., Oh E., Swaroop A., Hegg C.C., Raphael Y., Martens J.R., et al. Defects in neural stem cell proliferation and olfaction in Chd7 deficient mice indicate a mechanism for hyposmia in human CHARGE syndrome. Hum. Mol. Genet. 2009;18:1909–1923. doi: 10.1093/hmg/ddp112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bajpai R., Chen D.A., Rada-lglesias A., Zhang J., Xiong Y., Helms J., Chang C.P., Zhao Y., Swigut T., Wysocka J. CHD7 cooperates with PBAF to control multipotent nerual crest formation. Nature. 2010;463:958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W., Xiong Y., Shang C., Twu K.Y., Hang C.T., Yang J., Han P., Lin C.Y., Lin C.J., Tsai F.C., et al. Brg1 governs distinct pathways to direct multiple aspects of mammalian neural crest cell development. Proc. Natl. Acad. Sci. U S A. 2013;110:1738–1743. doi: 10.1073/pnas.1218072110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurd E.A., Micucci J.A., Reamer E.N., Martin D.M. Delayed fusion and altered gene expression contribute to semicircular canal defects in Chd7 deficient mice. Mech. Dev. 2012;129:308–323. doi: 10.1016/j.mod.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurd E.A., Poucher H.K., Cheng K., Raphael Y., Martin D.M. The ATP-dependent chromatin remodeling enzyme CHD7 regulates pro-neural gene expression and neurogenesis in the inner ear. Development. 2010;137:3139–3150. doi: 10.1242/dev.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurd E.A., Adams M.E., Layman W.S., Swiderski D.L., Beyer L.A., Halsey K.E., Benson J.M., Gong T.W., Dolan D.F., Raphael Y., et al. Mature middle and inner ears express Chd7 and exhibit distinctive pathologies in a mouse model of CHARGE syndrome. Hear Res. 2011;282:184–195. doi: 10.1016/j.heares.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Randall V., McCue K., Roberts C., Kyriakopoulou V., Beddow S., Barrett A.N., Vitelli F., Prescott K., Shaw-Smith C., Devriendt K., et al. Great vessel development requires biallelic expression of Chd7 and Tbx1 in pharyngeal ectoderm in mice. J. Clin. Invest. 2009;119:3301–3310. doi: 10.1172/JCI37561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J., Greene S.B., Martin J.F. BMP signaling in congenital heart disease: new developments and future directions. Birth Defects Res. A Clin. Mol. Teratol. 2011;91:441–448. doi: 10.1002/bdra.20785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Attisano L., Wrana J.L. Signal transduction by the TGF-beta superfamily. Science. 2002;296:1646–1647. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- 23.Bruneau B.G. Chromatin remodeling in heart development. Curr. Opin. Genet. Dev. 2010;20:505–511. doi: 10.1016/j.gde.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 24.Chang C.P., Bruneau B.G. Epigenetics and cardiovascular development. Annu. Rev. Physiol. 2012;74:41–68. doi: 10.1146/annurev-physiol-020911-153242. [DOI] [PubMed] [Google Scholar]
- 25.Debenedittis P., Harmelink C., Chen Y., Wang Q., Jiao K. Characterization of the novel interaction between muskelin and TBX20, a critical cardiogenic transcription factor. Biochem. Biophys. Res. Commun. 2011;409:338–343. doi: 10.1016/j.bbrc.2011.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rybkin I.I., Markham D.W., Yan Z., Bassel-Duby R., Williams R.S., Olson E.N. Conditional expression of SV40 T-antigen in mouse cardiomyocytes facilitates an inducible switch from proliferation to differentiation. J. Biol. Chem. 2003;278:15927–15934. doi: 10.1074/jbc.M213102200. [DOI] [PubMed] [Google Scholar]
- 27.Lien C.L., McAnally J., Richardson J.A., Olson E.N. Cardiac-specific activity of an Nkx2–5 enhancer requires an evolutionarily conserved Smad binding site. Dev. Biol. 2002;244:257–266. doi: 10.1006/dbio.2002.0603. [DOI] [PubMed] [Google Scholar]
- 28.Brown C.O., 3rd, Chi X., Garcia-Gras E., Shirai M., Feng X.H., Schwartz R.J. The cardiac determination factor, Nkx2–5, is activated by mutual cofactors GATA-4 and Smad1/4 via a novel upstream enhancer. J. Biol. Chem. 2004;279:10659–10669. doi: 10.1074/jbc.M301648200. [DOI] [PubMed] [Google Scholar]
- 29.Liberatore C.M., Searcy-Schrick R.D., Vincent E.B., Yutzey K.E. Nkx-2.5 gene induction in mice is mediated by a Smad consensus regulatory region. Dev. Biol. 2002;244:243–256. doi: 10.1006/dbio.2002.0604. [DOI] [PubMed] [Google Scholar]
- 30.Lee K.H., Evans S., Ruan T.Y., Lassar A.B. SMAD-mediated modulation of YY1 activity regulates the BMP response and cardiac-specific expression of a GATA4/5/6-dependent chick Nkx2.5 enhancer. Development. 2004;131:4709–4723. doi: 10.1242/dev.01344. [DOI] [PubMed] [Google Scholar]
- 31.Lien C.L., Wu C., Mercer B., Webb R., Richardson J.A., Olson E.N. Control of early cardiac-specific transcription of Nkx2–5 by a GATA-dependent enhancer. Development. 1999;126:75–84. doi: 10.1242/dev.126.1.75. [DOI] [PubMed] [Google Scholar]
- 32.Hao J., Ho J.N., Lewis J.A., Karim K.A., Daniels R.N., Gentry P.R., Hopkins C.R., Lindsley C.W., Hong C.C. In vivo structure–activity relationship study of dorsomorphin analogues identifies selective VEGF and BMP inhibitors. ACS. Chem. Biol. 2010;5:245–253. doi: 10.1021/cb9002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu R., Liu H., Chen X., Kirby M., Brown P.O., Zhao K. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001;106:309–318. doi: 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- 34.Gaussin V., Van de Putte T., Mishina Y., Hanks M.C., Zwijsen A., Huylebroeck D., Behringer R.R., Schneider M.D. Endocardial cushion and myocardial defects after cardiac myocyte-specific conditional deletion of the bone morphogenetic protein receptor ALK3. Proc. Natl. Acad. Sci. U S A. 2002;99:2878–2883. doi: 10.1073/pnas.042390499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song L., Yan W., Chen X., Deng C.X., Wang Q., Jiao K. Myocardial Smad4 is essential for cardiogenesis in mouse embryos. Circ. Res. 2007;101:277–285. doi: 10.1161/CIRCRESAHA.107.155630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma L., Lu M.F., Schwartz R.J., Martin J.F. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development. 2005;132:5601–5611. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- 37.Song L., Fassler R., Mishina Y., Jiao K., Baldwin H.S. Essential functions of Alk3 during AV cushion morphogenesis in mouse embryonic hearts. Dev. Biol. 2007;301:276–286. doi: 10.1016/j.ydbio.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 38.Sugi Y., Yamamura H., Okagawa H., Markwald R.R. Bone morphogenetic protein-2 can mediate myocardial regulation of atrioventricular cushion mesenchymal cell formation in mice. Dev. Biol. 2004;269:505–518. doi: 10.1016/j.ydbio.2004.01.045. [DOI] [PubMed] [Google Scholar]
- 39.Cai C.L., Zhou W., Yang L., Bu L., Qyang Y., Zhang X., Li X., Rosenfeld M.G., Chen J., Evans S. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development. 2005;132:2475–2487. doi: 10.1242/dev.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benchabane H., Wrana J.L. GATA- and Smad1-dependent enhancers in the Smad7 gene differentially interpret bone morphogenetic protein concentrations. Mol. Cell. Biol. 2003;23:6646–6661. doi: 10.1128/MCB.23.18.6646-6661.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Q., Chen H., Zheng D., Kuang C., Fang H., Zou B., Zhu W., Bu G., Jin T., Wang Z., et al. Smad7 is required for the development and function of the heart. J. Biol. Chem. 2009;284:292–300. doi: 10.1074/jbc.M807233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Justin N., De Marco V., Aasland R., Gamblin S.J. Reading, writing and editing methylated lysines on histone tails: new insights from recent structural studies. Curr. Opin. Struct. Biol. 2010;20:730–738. doi: 10.1016/j.sbi.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 43.Eissenberg J.C., Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev. Biol. 2010;339:240–249. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tremblay K.D., Dunn N.R., Robertson E.J. Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development. 2001;128:3609–3621. doi: 10.1242/dev.128.18.3609. [DOI] [PubMed] [Google Scholar]
- 45.Umans L., Cox L., Tjwa M., Bito V., Vermeire L., Laperre K., Sipido K., Moons L., Huylebroeck D., Zwijsen A. Inactivation of Smad5 in endothelial cells and smooth muscle cells demonstrates that Smad5 is required for cardiac homeostasis. Am. J. Pathol. 2007;170:1460–1472. doi: 10.2353/ajpath.2007.060839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hester M., Thompson J.C., Mills J., Liu Y., El-Hodiri H.M., Weinstein M. Smad1 and Smad8 function similarly in mammalian central nervous system development. Mol. Cell. Biol. 2005;25:4683–4692. doi: 10.1128/MCB.25.11.4683-4692.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song L., Zhao M., Wu B., Zhou B., Wang Q., Jiao K. Cell autonomous requirement of endocardial Smad4 during atrioventricular cushion development in mouse embryos. Dev. Dyn. 2011;240:211–220. doi: 10.1002/dvdy.22493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moskowitz I.P., Wang J., Peterson M.A., Pu W.T., Mackinnon A.C., Oxburgh L., Chu G.C., Sarkar M., Berul C., Smoot L., et al. Transcription factor genes Smad4 and Gata4 cooperatively regulate cardiac valve development. Proc. Natl. Acad. Sci. U S A. 2011;108:4006–4011. doi: 10.1073/pnas.1019025108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen H., Shi S., Acosta L., Li W., Lu J., Bao S., Chen Z., Yang Z., Schneider M.D., Chien K.R., et al. BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development. 2004;131:2219–2231. doi: 10.1242/dev.01094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harmelink C., Peng Y., Debenedittis P., Chen H., Shou W., Jiao K. Myocardial Mycn is essential for mouse ventricular wall morphogenesis. Dev. Biol. 2013;373:53–63. doi: 10.1016/j.ydbio.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiao K., Zhou Y., Hogan B.L. Identification of mZnf8, a mouse Kruppel-like transcriptional repressor, as a novel nuclear interaction partner of Smad1. Mol. Cell. Biol. 2002;22:7633–7644. doi: 10.1128/MCB.22.21.7633-7644.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsai R.Y., Reed R.R. Using a eukaryotic GST fusion vector for proteins difficult to express in E. coli. BioTechniques. 1997;23:794–796. doi: 10.2144/97235bm06. , 798, 800. [DOI] [PubMed] [Google Scholar]
- 53.Jiao K., Kulessa H., Tompkins K., Zhou Y., Batts L., Baldwin H.S., Hogan B.L. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 2003;17:2362–2367. doi: 10.1101/gad.1124803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.