Abstract
Several limitations of current epigenomic technology preclude their use in many experimental and clinical settings. Here we describe Assay for Transposase Accessible Chromatin using sequencing (ATAC-seq)— based on direct in vitro transposition of sequencing adapters into native chromatin – as a rapid and sensitive method for integrative epigenomic analysis. ATAC-seq captures open chromatin sites using a simple 2-step protocol from 500 to 50,000 cells, and reveals the interplay between genomic locations of open chromatin, DNA binding proteins, individual nucleosomes, and higher-order compaction at regulatory regions with nucleotide resolution. We discover classes of DNA binding factor that strictly avoid, can tolerate, or tend to overlap with nucleosomes. Using ATAC-seq, we measured and interpreted the serial daily epigenomes of resting human T cells from a proband via standard blood draws, demonstrating the feasibility of reading personal epigenomes in clinical timescales for monitoring health and disease.
Introduction
Eukaryotic genomes are hierarchically packaged into chromatin1, and the nature of this packaging plays a central role in gene regulation2,3. Major insights into the epigenetic information encoded within the nucleoprotein structure of chromatin have come from high-throughput, genome-wide methods for separately assaying the chromatin accessibility (“open chromatin”)4,5, nucleosome positioning6–8, and transcription factor (TF) occupancy9. While powerful, published protocols for existing methods require millions of cells as starting material, complex and time-consuming sample preparations, and cannot simultaneously probe the interplay of nucleosome positioning, chromatin accessibility, and TF binding. These limitations are problematic in three major ways: First, current methods can average over and “drown out” heterogeneity in cellular populations. Second, cells must often be grown ex vivo to obtain sufficient biomaterials, perturbing the in vivo context and modulating the epigenetic state in unknown ways. Third, input requirements often prevent application of these assays to well-defined clinical samples, precluding generation of “personal epigenomes” in diagnostic timescales.
Here we report a robust and sensitive method for epigenomic profiling that can provide a multi-dimensional portrait of gene regulation. We use ATAC-seq to identify regions of open chromatin, identify nucleosome-bound and nucleosome-free positions in regulatory regions, and infer the positions of DNA binding proteins using “footprinting” in a B-cell line. Finally we demonstrate that this method is compatible with clinical timescales and standard blood draws by observing the open chromatin landscape of a healthy volunteer.
Results
ATAC-seq probes chromatin accessibility with transposons
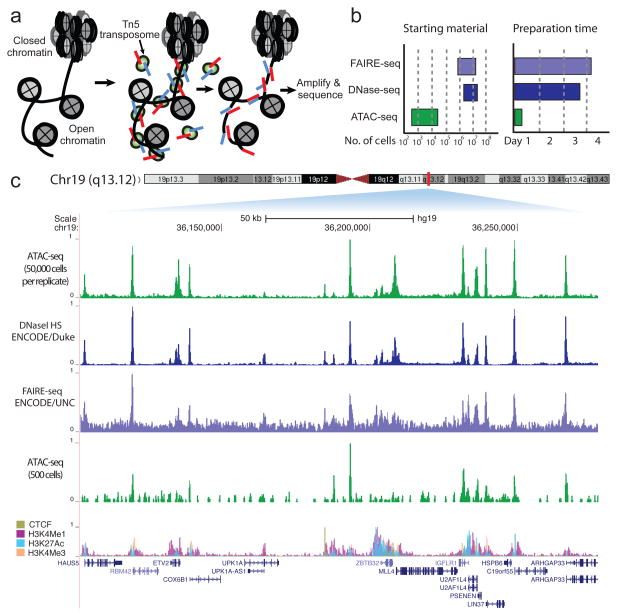
Hyperactive Tn5 transposase10,11, loaded in vitro with adapters for high-throughput DNA sequencing, can simultaneously fragment and tag a genome with sequencing adapters (previously described as “tagmentation”11). Because transposons have been shown to integrate into active regulatory elements in vivo12, we hypothesized that transposition by purified Tn5, a prokaryotic transposase, on small numbers of unfixed eukaryotic nuclei would interrogate regions of accessible chromatin. Here we describe Assay for Transposase Accessible Chromatin followed by high-throughput sequencing (ATAC-seq). ATAC-seq uses Tn5 transposase to integrate its adapter payload into regions of accessible chromatin, whereas steric hindrance less accessible chromatin makes transposition less probable. Therefore, amplifiable DNA fragments suitable for high-throughput sequencing are preferentially generated at locations of open chromatin (Fig 1a). The entire assay and library construction can be carried out in a simple two-step process involving Tn5 insertion and PCR. In contrast, published DNase- and FAIRE-seq protocols for assaying chromatin accessibility involve multi-step protocols and many potentially loss-prone steps, such as adapter ligation, gel purification, and crosslink reversal. For instance, a published DNase-seq protocol calls for approximately 44 steps, and two overnight incubations, while published FAIRE-seq protocols require two overnight incubations carried out over at least 3 days13,14. Furthermore, these protocols require 1–50 million cells (FAIRE) or 50 million cells (DNase-seq)13,14, perhaps because of these complex workflows (Fig 1b). In comparison to established methods, ATAC-seq enables rapid and efficient library generation because assay and library preparation are carried out in a single enzymatic step.
Figure 1. ATAC-seq is a sensitive, accurate probe of open chromatin state.
(a) ATAC-seq reaction schematic. Transposase (green), loaded with sequencing adapters (red and blue), inserts only in regions of open chromatin (nucleosomes in grey) and generates sequencing library fragments that can be PCR amplified. (b) Approximate reported input material and sample preparation time requirements for genome-wide methods of open chromatin analysis. (c) A comparison of ATAC-seq to other open chromatin assays at a locus in GM12878 lymphoblastoid cells displaying high concordance. Lower ATAC-seq track was generated from 500 FACS-sorted cells.
Extensive analyses show that ATAC-seq provides accurate and sensitive measure of chromatin accessibility genome-wide. We carried out ATAC-seq on 50,000 and 500 unfixed nuclei isolated from GM12878 lymphoblastoid cell line (ENCODE Tier 115) for comparison and validation with chromatin accessibility data sets, including DNase-seq13 and FAIRE-seq16. At a locus previously highlighted by others5, (Fig. 1c), ATAC-seq has a signal-to-noise ratio similar to DNase-seq, which was generated from approximately 3 to 5 orders-of-magnitude more cells13,14. Peak intensities were highly reproducible between technical replicates (R=0.98), and highly correlated between ATAC-seq and DNase-seq (R=0.79 and R=0.83, Supplementary Fig. 1), and we note that the majority of reads within peaks come from intersections of DNase and ATAC-seq peaks (Supplementary Fig. 2). Comparing our data to DHSs identified in ENCODE DNase-seq data, receiver operating characteristic (ROC) curves demonstrate a similar sensitivity and specificity as DNase-seq (Supplementary Fig. 3). We also note that ATAC-seq peak intensities correlate well with markers of active chromatin and not with transposase sequence preference (Supplementary Fig. 4 and 5). Highly sensitive open chromatin detection is maintained even when using 5,000 or 500 human nuclei as starting material (Supplementary Fig. 3 and 6), although sensitivity is diminished for smaller numbers of input material, as can be seen in Fig 1c.
ATAC-seq insert sizes disclose nucleosome positions
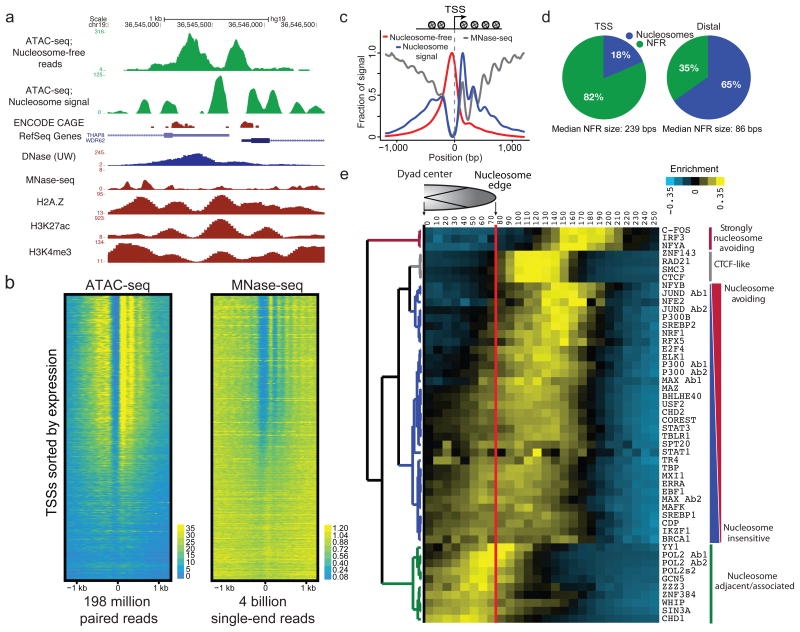
We found that ATAC-seq paired-end reads produce detailed information about nucleosome packing and positioning. The insert size distribution of sequenced fragments from human chromatin has clear periodicity of approximately 200 base pairs, suggesting many fragments are protected by integer multiples of nucleosomes (Fig 2a). This fragment size distribution also shows clear periodicity equal to the helical pitch of DNA11. By partitioning insert size distribution according to functional classes of chromatin as defined by previous models17, and normalizing to the global insert distribution (see Methods) we observe clear class-specific enrichments across this insert size distribution (Fig. 2b), demonstrating that these functional states of chromatin have an accessibility “fingerprint” that can be read out with ATAC-seq. These differential fragmentation patterns are consistent with the putative functional state of these classes, as CTCF-bound regions are enriched for short fragments of DNA, while transcription start sites are differentially depleted for mono-, di- and tri-nucleosome associated fragments. Transcribed and promoter flanking regions are enriched for longer multi-nucleosomal fragments, suggesting they may represent more compacted forms of chromatin. Finally, prior studies have shown that certain DNA sequences are refractory to nuclease digestion and released as large, multi-nucleosome-sized fragments18; subsequent studies showed that such fragments are condensed heterochromatin19. Indeed we found repressed regions are depleted for short fragments and enriched for phased multi-nucleosomal inserts, consistent with their expected inaccessible state. These data suggest that ATAC-seq reveals differentially accessible forms of chromatin, which have been long hypothesized to exist in vivo2,20,21.
Figure 2. ATAC-seq provides genome-wide information on chromatin compaction.
(a) ATAC-seq fragment sizes generated from GM12878 nuclei (red) indicate chromatin-dependent periodicity with a spatial frequency consistent with nucleosomes, as well as a high frequency periodicity consistent with the pitch of the DNA helix for fragments less than 200 bp. (Inset) log-transformed histogram shows clear periodicity persists to 6 nucleosomes. (b) Normalized read enrichments for 7 classes of chromatin state previously defined17.
To explore nucleosome positioning within accessible chromatin in the GM12878 cell line, we partitioned our data into reads generated from putative nucleosome free regions of DNA, and reads likely derived from nucleosome associated DNA (see Methods and Supplementary Fig. 7). Using a simple heuristic that positively weights nucleosome associated fragments and negatively weights nucleosome free fragments (see Methods), we calculated a data track used to call nucleosome positions within regions of accessible chromatin22. An example locus (Fig. 3a) contains a putative bidirectional promoter with CAGE data showing two transcription start sites (TSS) separated by ~700bps. ATAC-seq reveals in fact two distinct nucleosome free regions, separated by a single well-positioned mononucleosome (Fig. 3a). Compared to MNase-seq23, ATAC-seq data is more amenable to detecting nucleosomes within putative regulatory regions, as the majority of reads are concentrated within accessible regions of chromatin (Fig. 3b). By averaging signal across all active TSSs, we note nucleosome-free fragments are enriched at a canonical nucleosome-free promoter region overlapping the TSS, while our nucleosome signal is enriched both upstream and downstream of the active TSS, and displays characteristic phasing of upstream and downstream nucleosomes6,7 (Fig. 3c). Because ATAC-seq reads are concentrated at regions of open chromatin, we see strong nucleosome signal at the +1 nucleosome, which decreases at the +2, +3 and +4 nucleosomes, in contrast, MNase-seq nucleosome signal increases at larger distances from the TSS likely due to over digestion of more accessible nucleosomes. Additionally, MNase-seq (4 billion reads) assays all nucleosomes, whereas reads generated from ATAC-seq (198 million paired reads) are concentrated at regulatory nucleosomes (Fig. 3b,c). Using our nucleosome calls, we further partitioned putative distal regulatory regions and TSSs into regions that were nucleosome free and regions that were predicted to be nucleosome bound. We note that TSSs were enriched for nucleosome free regions when compared to distal elements, which tend to remain nucleosome rich (Fig. 3d). These data suggest ATAC-seq can provide high-resolution readout of nucleosome associated and nucleosome free regions in regulatory elements genome wide.
Figure 3. ATAC-seq provides genome-wide information on nucleosome positioning in regulatory regions.
(a) An example locus containing two transcription start sites (TSSs) showing nucleosome free read track, calculated nucleosome track (Methods), as well as DNase, MNase, and H3K27ac, H3K4me3, and H2A.Z tracks for comparison. (b) ATAC-seq (198 million paired reads) and MNase-seq (4 billion single-end reads from ref 23) nucleosome signal shown for all active TSSs (n=64,836), TSSs are sorted by CAGE expression. (c) TSSs are enriched for nucleosome free fragments, and show phased nucleosomes similar to those seen by MNase-seq at the −2, −1, +1, +2, +3 and +4 positions. (d) Relative fraction of nucleosome associated vs. nucleosome free (NFR) bases in TSS and distal sites (see Methods). (e) Hierarchical clustering of DNA binding factor position with respect to the nearest nucleosome dyad within accessible chromatin reveals distinct classes of DNA binding factors. Factors strongly associated with nucleosomes are enriched for chromatin remodelers.
ATAC-seq reveals patterns of nucleosome-TF spacing
ATAC-seq high-resolution regulatory nucleosome maps can be used to understand the relationship between nucleosomes and DNA binding factors. Using ChIP-seq data, we plotted the position of a variety of DNA binding factors with respect to the dyad of the nearest nucleosome. Unsupervised hierarchical clustering (Figure 3e) revealed major classes of binding with respect to the proximal nucleosome, including 1) a strongly nucleosome avoiding group of factors with binding events stereotyped at ~180 bases from the nearest nucleosome dyad (comprising C-FOS, NFYA and IRF3), 2) a class of factors that “nestle up” precisely to the expected end of nucleosome DNA contacts, which notably includes chromatin looping factors CTCF and cohesion complex subunits RAD21 and SMC3; 3) a large class of primarily TFs that have gradations of nucleosome avoiding or nucleosome-overlapping binding behavior, and 4) a class whose binding sites tend to overlap nucleosome-associated DNA. Interestingly, this final class includes chromatin remodeling factors such as CHD1 and SIN3A as well as RNA polymerase II, which appears to be enriched at the nucleosome boundary8. The interplay between precise nucleosome positioning and locations of DNA binding factor immediately suggests specific hypotheses for mechanistic studies, a potential advantage of ATAC-seq.
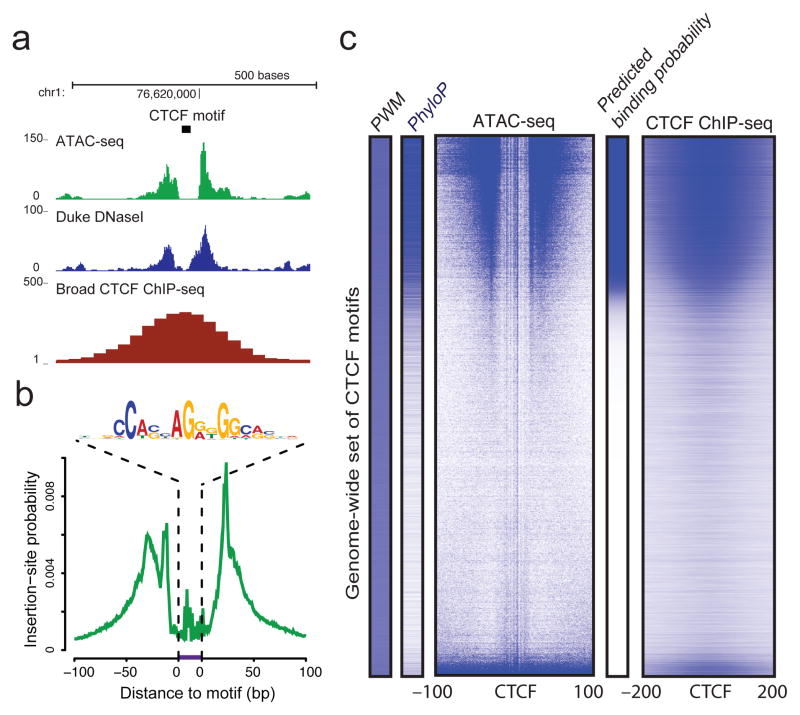
ATAC-seq footprints infer factor occupancy genome-wide
ATAC-seq enables accurate inference of DNA binding factor occupancy genome-wide. We reasoned that DNA sequences directly occupied by DNA-binding proteins are protected from transposition; the resulting sequence “footprint” reveals the presence of the DNA-binding protein at each site, analogous to DNase digestion footprints24. At a specific CTCF binding site on chromosome 1, we observed a clear footprint (a deep notch of ATAC-seq signal), similar to footprints seen by DNase-seq25,26, at the precise location of the CTCF motif that coincides with the summit of the CTCF ChIP-seq signal in GM12878 cells (Fig 4a). We averaged ATAC-seq signal over all expected locations of CTCF within the genome and observed a well-stereotyped “footprint” (Fig. 4b). Similar results were obtained for a variety of common TFs (for examples see Supplementary Fig. 8). We inferred the CTCF binding probability from motif consensus score, evolutionary conservation, and ATAC-seq insertion data to generate a posterior probability of CTCF binding at all loci (Fig. 4c)27. Results using ATAC-seq closely recapitulate ChIP-seq binding data in this cell line and compare favorably to DNase-based factor occupancy inference (see Supplementary Fig. 9), suggesting that factor occupancy data can be extracted from these ATAC-seq data allowing reconstruction of regulatory networks.
Figure 4. ATAC-seq assays genome-wide factor occupancy.
(a) CTCF footprints observed in ATAC-seq and DNase-seq data, at a specific locus on chr1. (b) Aggregate ATAC-seq footprint for CTCF (motif shown) generated over binding sites within the genome (c) CTCF predicted binding probability inferred from ATAC-seq data, position weight matrix (PWM) scores for the CTCF motif, and evolutionary conservation (PhyloP). Right-most column is the CTCF ChIP-seq data (ENCODE) for this GM12878 cell line, demonstrating high concordance with predicted binding probability.
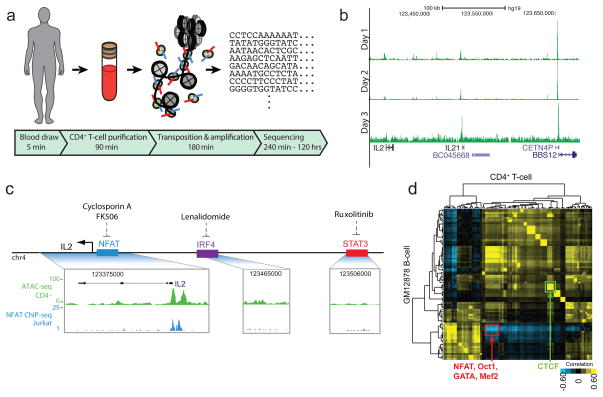
ATAC-seq enables epigenomic analysis on clinical timescales
Because ATAC-seq is rapid, information rich, and compatible with small numbers of cells, we reasoned it may serve as a powerful tool for personalized epigenomics in the clinic. Specifically, we envision “personal epigenomics” as genome-scale information about chromatin generated from an individual from a standard clinical sample in a clinical timescale. We applied ATAC-seq to assay the personal T-cell epigenome of a healthy volunteer via standard serial blood draws, to demonstrate a workflow capable of generating ATAC-seq libraries in clinical timescales. Using rapid T-cell enrichment and sample handling protocols (see Methods), the total required time from blood draw to sequencing was approximately 275 minutes (Fig. 5a). When coupled with ongoing improvements to sequencing and analysis turn-around times, we envision ATAC-seq will offer the possibility of a daily turn-around time for a personal epigenomic map. To explore this possibility, we performed ATAC-seq on three consecutive days via standard blood draws from a single individual (Fig. 5b). As an exercise to consider how personal epigenomic maps may contain personalized regulatory information, we investigated ATAC-seq profile at the IL2 locus. IL-2 is a key cytokine that drives T-cell growth and functions in inflammatory and autoimmune diseases28. Furthermore, distinct drugs29–31 inhibit the activities of different transcription factors that bind putative IL2 enhancers in a context-dependent manner. In principle, one might wish to identify the causal transcription factor pathway in order to rationally target inhibition without exposing the patient to drugs unlikely to serve the therapeutic goal of IL-2 blockade. ATAC-seq shows that in the proband’s T-cells, only NFAT, but not two other drug targets, is engaging IL2 (Fig. 5c), providing clinically relevant information on the regulatory state of this individual.
Figure 5. ATAC-seq enables real-time personal epigenomics.
(a) Work flow from standard blood draws. (b) Serial ATAC-seq data from proband T-cells over three days. (c) Example of application of ATAC-seq data (green track) to prioritize candidate TF drug targets. Among identified TF binding sites proximal to cytokine gene IL2 that can be targeted by FDA-approved drugs, only NFAT is engaged in proband T-cells. ATAC-seq footprint prediction is confirmed by alignment with published NFAT ChIP-seq data (blue track, data from ref 35). (d) Cell type-specific regulatory network from proband T cells compared with GM12878 B-cell line. Each row or column is the footprint profile of a TF versus that of all other TFs in the same cell type. Color indicates relative similarity (yellow) or distinctiveness (blue) in T versus B cells. NFAT is one of the most highly differentially regulated TFs (red box) whereas canonical CTCF binding is essentially similar in T and B cells.
Using ATAC-seq footprints we generated the occupancy profiles of 89 transcription factors in proband T-cells, enabling systematic reconstruction of regulatory networks. With this personalized regulatory map, we compared the genomic distribution of the same 89 transcription factors between GM12878 and proband CD4+ T-cells. Transcription factors that exhibit large variation in distribution between T-cells and B-cells are enriched for T-cell specific factors (Fig. 5d). This analysis shows NFAT is differentially regulating, while canonical CTCF occupancy is highly correlated within these two cell types (Fig. 5d). Supporting this interpretation, we note specific loci where NFAT is localized nearby to known T-cell specific genes such as CD28 and a novel lincRNA RP11-229C3.2 (Supplementary Fig. 10). Additionally, ATAC-seq of CD4+ and CD8+ T-cells, and monocytes isolated by fluorescence-activated cell sorting (FACS) from a single blood draw created an interpretative framework for the personal epigenomes, and demonstrated that ATAC-seq is compatible with cellular enrichment using surface markers (Supplementary Fig. 11). Separately, allele-specific chromatin accessibility has been shown to be particularly relevant to our understanding of human disease32. As a proof of principle we also used ATAC-seq to identify candidate allele-specific open chromatin regions within the GM12878 cell line (Supplementary Fig. 12). These results demonstrate the feasibility of generating detailed personalized gene regulatory networks from clinical samples, opening the door for future diagnostic applications.
Discussion
Epigenomic studies of chromatin accessibility have yielded tremendous biological insights, but are currently limited in application by their complex workflows and large cell number requirements. While, improvements of existing methods may enable them to reach the similar performance, we believe ATAC-seq offers substantial advantages over existing technologies due to its speed, simplicity, and low input cell number requirement. ATAC-seq is an information rich assay, allowing simultaneous interrogation of factor occupancy, nucleosome positions in regulatory sites, and chromatin accessibility genome-wide. These insights are derived from both the position of insertion and the distribution of insert lengths captured during the transposition reaction. While extant methods such as DNase- and MNase-seq can provide some subsets of the information in ATAC-seq, they each require separate assays with large cell numbers, which increases the time, cost, and limits applicability to many systems. ATAC-seq also provides insert size “fingerprints” of biologically relevant genomic regions, suggesting that it capture information on chromatin compaction. We expect ATAC-seq to have broad applicability, significantly add to the genomics toolkit, and improve our understanding of gene regulation, particularly when integrated with other powerful rare cell techniques, such as FACS, laser capture microdissection (LCM) and recent advancements in RNA-seq33,34.
One potentially exciting application of ATAC-seq is to generate “personal epigenomic” profiles on a timescale compatible with clinical decision-making. We have optimized procedures to transform a clinical blood sample to completed sequencing library in 275 minutes. The reduced input requirements and rapid workflows, when coupled with the recent introduction of rapid-turnaround high-throughput sequencing instruments, such as the MiSeq and HiSeq2500, will enable investigation of personalized epigenetic landscapes of selected tissues both in the lab and the clinic. Of course, deeper analyses to generate global regulatory networks and other inferences will take additional time, and we anticipate that bioinformatic analyses—not the molecular biology or sequencing—will become the bottleneck of epigenomic studies in the future. We show that ATAC-seq is compatible with FACS, enabling studies on carefully sorted and rare subpopulations from primary tissues. We expect cellular subpopulations selected at different points in development and aging, and human diseases, including cancer, autoimmunity, and neuropsychiatric disorders are viable applications. In summary, we believe that the attractive combination of speed, simplicity and low input requirements of ATAC-seq will enable new gene regulatory insights into biology and medicine.
Online Methods
The ATAC-seq protocol has three major steps:
Prepare nuclei: To prepare nuclei, we spun 50,000 cells at 500 × g for 5 minutes, followed by a wash using 50 μL of cold 1x PBS and centrifugation at 500 × g for 5 minutes. Cells were lysed using cold lysis buffer (10 mM Tris-Cl, pH 7.4, 10 mM NaCl, 3 mM MgCl2 and 0.1% IGEPAL CA-630). Immediately after lysis, nuclei were spun at 500 × g for 10 minutes using a refrigerated centrifuge. To avoid losing cells during the nuclei prep, we used a fixed angle centrifuge and carefully pipetted away from the pellet after centrifugations.
Transpose and purify: Immediately following the nuclei prep, the pellet was resuspended in the transposase reaction mix (25 μL 2x TD buffer, 2.5 μL Transposase (Illumina) and 22.5 μL of nuclease free water). The transposition reaction was carried out for 30 minutes at 37 °C. Directly following transposition the sample was purified using a Qiagen Minelute kit.
PCR: Following purification, we amplified library fragments using 1x NEBnext PCR master mix and 1.25 μM of custom Nextera PCR primers 1 and 2 (see table below), using the following PCR conditions: 72°C for 5 minutes, 98°C for 30 seconds, followed by thermocycling at 98°C for 10 seconds, 63°C for 30 seconds and 72°C for 1 minute. To reduce GC and size bias in our PCR we monitored the PCR reaction using qPCR in order to stop amplification prior to saturation. To do this, we amplified the full libraries for 5 cycles, after 5 cycles we took an aliquot of the PCR reaction and added 10 μl of the PCR cocktail with Sybr Green at a final concentration of 0.6x. We ran this reaction for 20 cycles, to determine the additional number of cycles needed for the remaining 45 μL reaction. The libraries were purified using a Qiagen PCR cleanup kit yielding a final library concentration of ~30 nM in 20 μL. Libraries were amplified for a total of 10–12 cycles.
Low cell number protocol
To prepare the 500 and 5,000 cell reactions we used the same protocol with some notable exceptions: The transposition reaction was done in a 5 μL instead of 50 μL reaction. Also, we eliminated the Qiagen Minelute purification prior to PCR and instead took the 5 μL reaction immediately after transposition directly into the 50 μL PCR.
Library QC and quantitation
During the ATAC-seq protocol, we chose to avoid a size selection step to maximize the library complexity. The sequenced insert size is a distribution between 40 to 1 kb with a mean of ~120 bps. From bioanalyzer and gels we observed fragments >2 kb, which would make Qubit and other mass-based quantitation methods hard to interpret. For this reason we quantified our libraries using qPCR based methods.
CD4+ enrichment from peripheral blood
One green-top tube of whole blood was obtained from 1 normal volunteer three times over a 72-hour period, under a Stanford University IRB-approved protocol. Informed consent was obtained. 5mL of blood at each timepoint was negatively selected for CD4+ cells, using RosetteSep Human CD4+ T Cell Enrichment Cocktail (StemCell Technology). RosetteSep cocktail was incubated with the blood at 50 μL/mL for 20 min, diluted in an equal volume of PBS with 2% FBS, and underlayed with 15 mL Ficol-Paque Plus (GE). Blood was centrifuged for 20 minutes at 1200 × g without break, negatively selected cells were removed from the density medium: plasma interface, and cells were washed X2 in PBS with 2% FBS.
FACS sorting peripheral blood leukocytes and GM cells
GM 12878 cells were stained with DAPI NucBlue Fixed Cell Stain (molecular probes) and live cells were sorted using a FACSAria (BD Biosciences) using a 100 μm nozzle. One peripheral blood sample (buffy coat) was stained with BD Bioscience antibodies CD14-A-488 (M5E2, 1:20), CD3-PE-Cy7 (SK7, 1:20), CD4-APC-Cy7 (RPA-T4, 1:20), and CD8 (RPA-T8, 1:20) for 20 minutes in the dark at RT. Cells were lysed using BDpharmLyse 1:10 dil in diH20 (BD) for 15 min, centrifuged for 5 minutes, washed with PBS 2% FBS X 2, and resuspended in PBS with 2% FBS. 50,000 CD3+CD8+, CD3+CD4+, and CD14+ cell populations were sorted into PBS with 10%FBS.
Analysis
Primary data processing
Data was collected using either 34 × 8 × 34 reads from a MiSeq or 50 × 8 × 50 reads on a HiSeq. Reads were aligned to hg19 using BOWTIE36 using the parameters –X2000 and –m1. These parameters ensured that fragments up to 2 kb were allowed to align (-X2000) and that only unique aligning reads were collected (−m1). For all data files duplicates were removed using Picard.
For peak calling and footprinting, we adjusted the read start sites to represent the center of the transposon binding event. Previous descriptions of the Tn5 transposase show that the transposon binds as a dimer and inserts two adapters separated by 9 bps (main text ref. 11). Therefore, all reads aligning to the + strand were offset by +4 bps, and all reads aligning to the – strand were offset −5 bps.
ATAC-seq peak calling
We used ZINBA to call all reported ATAC-seq peaks in this manuscript. ZINBA was run using a window size of 300 bp and an offset 75 bp. Alignability was used to model the zero-inflated component and the ATAC-seq read count for the background and enriched components. Enriched regions were identified as those with a posterior probability >0.8.
ATAC-seq insertion size enrichment analysis within chromatin annotations
First, the distribution of paired-end sequencing fragment sizes overlapping each chromatin state (http://www.ensembl.org/info/docs/funcgen/regulatory_segmentation.html) were computed. The distributions were then normalized to the percent maximal within each state and enrichment was computed relative to the genome-wide set of fragment sizes.
Nucleosome positioning
To generate the nucleosome position data track, we chose to split reads into various bins. Reads below 100 bps were considered nucleosome free, reads between 180 and 247 bps were considered to be mononucleosomes, reads between 315 and 473 bps were considered to be dinucleosomes and reads between 558 and 615 were considered to be trinucleosomes (for determining cutoffs see Supplementary Figure 7). Dinucleosome reads were split into two reads and Trinucleosome reads were split into three reads. Reads were analyzed using Danpos and Dantools22 using the parameters -p 1, -a 1, -d 20, -clonalcut 0. The background used was nucleosome free reads (reads less than 100 bps), allowing an effective negative weighting of these reads. This analysis allows calling multiple overlapping nucleosomes. Although generating nucleosome tracks using simple insert size cutoffs may yield false positives due to other nucleosome sized features, i.e. enhanaceosomes, we observed that we faithfully recapitulated global features on nucleosome position genome-wide (Fig 2c,d main text).
ChIP-seq peak calling and clustering
ChIP-seq data was downloaded form the UCSC ENCODE repository, for a complete list of data use see supplementary table 2. Peaks where called using GEM37, the parameters used where -k_min 6 -k_max 20. Inputs where used as a control for peak calling. Binding events were annotated by distance to the nearest dyad in bins of 10 bps. Factors were then hierarchically clustered using Euclidean distance and normalized by gene and centered by mean38.
Footprinting using CENTIPEDE
The genome-wide set of motifs were obtained from the ENCODE motif repository (http://www.broadinstitute.org/~pouyak/encode-motif-disc/). The input for CENTIPEDE (main text ref. 27) included the PWM score, conservation (PhyloP) and ATAC-seq counts within +/−100bp of each genomic region matching a motif. ChIP-seq data was obtained from the UCSC ENCODE repository.
Comparison of transcription factor regulatory networks
Transcription factor regulatory networks were constructed by comparing the GENCODE v14 genes with the genome-wide set of posterior probabilities estimated by CENTIPEDE for the respective cell-types. The extent of a transcription factor regulating each gene was determined by taking the sum of the weighed posterior probabilities for a given transcription factor mapping to the same chromosome. For each mapped motif the posterior probability was weighted based on the distance to the transcription start site for each gene. Comparison of transcription factor regulatory networks was computed as the correlation of each transcription factor in a given cell type with all transcription factors in the other cell type. The resulting correlation matrix was hierarchically clustered using the Pearson correlation coefficient and complete linkage38.
Candidate IL2 enhancer analysis
We inspected ENCODE data on UCSC genome browser to identify putative IL2 enhancers in one or more cell types that may be responsive to FDA approved immomodulatory drugs. We scanned the intergenic region upstream of IL2 in hg19 for (i) enhancer-associated histone marks (H3K4me1 and H3K27ac), (ii) binding by one or more TFs as confirmed by ChIP-seq, and (iii) the TF pathway can be targeted by a human therapeutic. This analysis identified IRF4 and STAT3 binding sites in addition to the known NFAT-responsive elements (main text ref 28).
Supplementary Material
Acknowledgments
We thank members of Greenleaf and Chang labs for discussion, A. Burnet and S. Kim lab and the Stanford flow-cytometry core facility for assistance with FACS sorting, A. Schep for modeling Tn5 insertion preference, and V. Risca for graphics. Supported by NIH (H.Y.C., W.J.G., J.D.B.), including RC4NS073015, U01DK089532, and U19AI057229, Scleroderma Research Foundation (H.Y.C.), and California Institute for Regenerative Medicine (H.Y.C.). H.Y.C. acknowledges support as an Early Career Scientist of the Howard Hughes Medical Institute. GM12878 cells were a gift from the Snyder laboratory.
Footnotes
Accession codes: Raw data have been uploaded to GEO with the accession number: GSE47753.
Author contributions: J.B., P.G., L.Z. performed the research. All authors designed experiments and interpreted the data. H.Y.C. and W.J.G. wrote the paper.
References
- 1.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD, Lorch Y. Chromatin structure and transcription. Annu Rev Cell Biol. 1992;8:563–587. doi: 10.1146/annurev.cb.08.110192.003023. [DOI] [PubMed] [Google Scholar]
- 3.Mellor J. The dynamics of chromatin remodeling at promoters. Molecular Cell. 2005;19:147–157. doi: 10.1016/j.molcel.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 4.Boyle AP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valouev AA, et al. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goryshin IY. Tn5 in vitro transposition. 1998;273:7367–7374. doi: 10.1074/jbc.273.13.7367. [DOI] [PubMed] [Google Scholar]
- 11.Adey A, et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gangadharan S, Mularoni L, Fain-Thornton J, Wheelan SJ, Craig NL. DNA transposon Hermes inserts into DNA in nucleosome-free regions in vivo. Proceedings of the National Academy of Sciences. 2010;107:21966–21972. doi: 10.1073/pnas.1016382107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc. 2010 doi: 10.1101/pdb.prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simon JM, Giresi PG, Davis IJ, Lieb JD. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nature Protocols. 2012;7:256–267. doi: 10.1038/nprot.2011.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Consortium TEP. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements) Methods. 2009;48:233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman MM, et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013;41:827–841. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prioleau MN, Nony P, Simpson M, Felsenfeld G. An insulator element and condensed chromatin region separate the chicken β-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J. 1999;18:4035–4048. doi: 10.1093/emboj/18.14.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghirlando R, Litt MD, Prioleau MN, Recillas-Targa F, Felsenfeld G. Physical properties of a genomic condensed chromatin fragment. Journal of Molecular Biology. 2004;336:597–605. doi: 10.1016/j.jmb.2003.12.049. [DOI] [PubMed] [Google Scholar]
- 20.Kornberg RD, Lorch Y. Chromatin and transcription: where do we go from here. Current Opinion in Genetics & Development. 2002;12:249–251. doi: 10.1016/s0959-437x(02)00293-9. [DOI] [PubMed] [Google Scholar]
- 21.Zhou J, Fan JY, Rangasamy D, Tremethick DJ. The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat Struct Mol Biol. 2007;14:1070–1076. doi: 10.1038/nsmb1323. [DOI] [PubMed] [Google Scholar]
- 22.Chen K, et al. DANPOS: Dynamic analysis of nucleosome position and occupancy by sequencing. Genome Research. 2013;23:341–351. doi: 10.1101/gr.142067.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kundaje A, et al. Ubiquitous heterogeneity and asymmetry of the chromatin environment at regulatory elements. Genome Research. 2012;22:1735. doi: 10.1101/gr.136366.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hesselberth JR, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Meth. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyle AP, et al. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Research. 2011;21:456–464. doi: 10.1101/gr.112656.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neph S, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pique-Regi R, et al. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Research. 2011;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fraser J, Irving B, Crabtree G, Weiss A. Regulation of interleukin-2 gene enhancer activity by the T cell accessory molecule CD28. Science. 1991;251:313–316. doi: 10.1126/science.1846244. [DOI] [PubMed] [Google Scholar]
- 29.Flanagan WM, Corthésy B, Bram RJ, Crabtree GR. Nuclear association of a T-cell transcription factor blocked by FK-506 and cyclosporin A. Nat Rev Mol Cell Biol. 1991;352:803–807. doi: 10.1038/352803a0. [DOI] [PubMed] [Google Scholar]
- 30.Lopez-Girona A, et al. Lenalidomide downregulates the cell survival factor, interferon regulatory factor-4, providing a potential mechanistic link for predicting response. British Journal of Haematology. 2011;154:325–336. doi: 10.1111/j.1365-2141.2011.08689.x. [DOI] [PubMed] [Google Scholar]
- 31.Verstovsek S, et al. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. N Engl J Med. 2010;363:1117–1127. doi: 10.1056/NEJMoa1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Meth. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 34.Shalek AK, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jolma A, et al. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Research. 2010;20:861–873. doi: 10.1101/gr.100552.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Y, Mahony S, Gifford DK. High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Comput Biol. 2012;8:e1002638. doi: 10.1371/journal.pcbi.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.