Abstract
Amyloid diseases, including Alzheimer's, Parkinson's, and the prion conditions, are each associated with a particular protein in fibrillar form. These amyloid fibrils were long suspected to be the disease agents, but evidence suggests that smaller, often transient and polymorphic oligomers are the toxic entities. Here we identify a segment of the amyloid-forming protein, alphaB crystallin, which forms an oligomeric complex exhibiting properties of other amyloid oligomers: beta-sheet-rich structure, cytotoxicity, and recognition by an anti-oligomer antibody. The X-ray-derived atomic structure of the oligomer reveals a cylindrical barrel, formed from six anti-parallel, protein strands, which we term a cylindrin. The cylindrin structure is compatible with a sequence segment from the Abeta protein of Alzheimer's disease. Cylindrins offer models for the hitherto elusive structures of amyloid oligomers.
Keywords: AlphaB crystallin, amyloid oligomer, cylindrin, steric zipper, amyloid
Studies from many laboratories have suggested that the molecular agents in amyloid-related conditions are not the associated protein fibrils that have long been taken as the defining feature of these disorders, but instead are lower molecular weight entities, often termed “small amyloid oligomers” (1–7). These oligomers are not generally stable aggregates; they appear as transient species during the conversion of their monomeric precursors to more massive, stable fibrils, and sometimes they appear as an ensemble of sizes and shapes. This polymorphic and time-dependent nature of small amyloid oligomers has made it difficult to pin down their assembly pathways, their stoichiometries, their atomic-level structures, their relationship to fibrils, and their pathological actions (1, 8–10). What has emerged is a consensus, minimal definition of small amyloid oligomers: they are non-covalent assemblies of several identical chains of proteins known also to form amyloid fibrils; the oligomers exhibit greater cytotoxicity than either the monomer or fibrils formed from the same protein; in many cases the oligomer is recognized by a “conformational” antibody (A11) that binds oligomers but not fibrils, regardless of the sequence of the constituent protein (5). This suggests that oligomers display common conformation features that differ from those of fibrils (11).
In seeking to better define small amyloid oligomers, we chose to work with alphaB crystallin (ABC). This protein is a chaperone (12–14) that forms amyloid fibrils (15), but the fibrils form more slowly than those of the Amyloid beta peptide (Abeta) or Islet Amyloid polypeptide (IAPP), so that the oligomeric state may be trapped prior to the onset of fibrillization. We have discovered a segment of ABC which forms a relatively stable small oligomer which satisfies the definition of a small amyloid oligomer given in the preceding paragraph.
We identified the oligomer-forming segment of ABC by inspection of its 3D structure (16) and by applying the Rosetta-Profile algorithm to its sequence. This algorithm identifies sequence segments that form the steric-zipper spines of amyloid fibrils (17, 18). We noted that two segments of high amyloidogenic propensity, with sequences KVKVLG and GDVIEV, share the same Gly residue 95 at the C-terminus of the first segment and the N-terminus of the second; moreover, the entire 11-residue segment KVKVLGDVIEV forms a hairpin loop in the 3D structure of ABC (Fig. 1A), with Gly at its center. As predicted, the second six-residue segment GDVIEV, termed G6V (see Table I which defines the structures described in this report), forms fibrils and microcrystals (Fig. S1). The microcrystals enabled us to determine the atomic structure of G6V (Fig. S2), which proved to be a standard Class 2 steric zipper (19), essentially an amyloid-like protofilament.
Fig. 1.
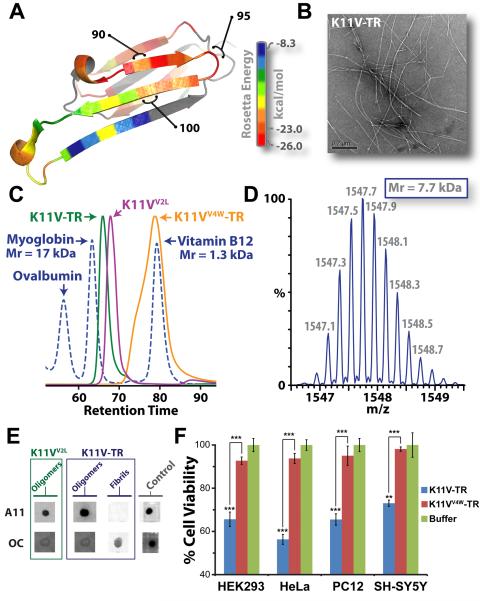
The cylindrins derived from alphaB crystallin (ABC), an amyloid-forming protein, exhibit the properties of oligomeric state, immunoreactivity, and cytotoxicity commonly ascribed to small amyloid oligomers. (A) Ribbon diagram of a single subunit of ABC (16), colored by propensity to form amyloid, with red being the highest and blue the lowest propensity. The segment from residue 90 to 100, termed K11V, forms the cylindrin. (B) Representative electron micrograph of amyloid fibrils formed by the tandem repeat V2L variant of K11V, K11V-TR. (C) Overlaid size exclusion chromatograms showing protein standards (blued dashed curve) and cylindrin segments. K11VV2L (purple curve; 1.2 kDa) and K11V-TR (green curve; 2.5 kDa) cylindrin segments migrate as oligomeric complexes. A mutant form of K11V-TR that disrupts oligomer formation of the cylindrin peptide, K11VV4W-TR (orange curve; 2.7 kDa), migrates as a dimeric/monomeric species. (D) Native nanoelectrospray mass spectrum of K11V-TR peak fractions from SEC-HPLC reveals trimeric tandem repeat cylindrin oligomers, confirming that the oligomeric complexes coincide in mass with the crystallized cylindrins. Expansion of the most abundanti on series of a +5 charge state correpsonding to a molecular mass of the three K11V-TR, coinciding with the crystallographic trimeric oligomer with a mass accuracy of 3.93 ppm is shown, with m/z labels. (E) Immuno-dot blot analysis of solutions of K11VV2L and K11V-TR, and K11V-TR fibrils with prefibrillar oligomer-specific, polyclonal antibody, A11 (5), and a mixture of fibril-specific monoclonal antibodies, OC (11). Solutions of cylindrin-forming segments are recognized by A11, whereas not by the OC antibody. In contrast, K11V-TR fibrils are recognized only by the OC antibody. Positive controls are shown to the right (5). (F) Cylindrin K11V-TR is toxic to four mammalian cells lines. Cell viability levels return to nearly 100% when we tested the control variant K11VV4W-TR. All samples were at a finalconcentration of 100 μM. Results represent mean ± SEM. Student's t-test (N = 4): **, P < 0.01; and ***,P < 0.001.
Table 1.
Structures and stoichiometries of amyloid-related oligomers discussed in this report, derived from alphaB crystallin.
| Protein/Peptide Segment (residue numbers)a | Structure | PDB ID | Oligomer Size by | Immunoreactivity | Toxicity | ||
|---|---|---|---|---|---|---|---|
| Crystallography | SEC | MS | |||||
| alphaB crystallin truncated (68–162) | Network of dimers linked by domain swapping | 3L1G | 2 to indefinitely large | 2–4b | 2–6 & 10–24b | - | Noc |
| K11V | Cylindrin | 3SGO | 6 | - | 6d | - | - |
| K11VV2L | Cylindrin V2L variant | 3SGP | 6 | 6 | 6e | A11 | Yes |
| K11V-TR | Cylindrin tandem repeat V2L variant | 3SGR | 3 | 3 | 3e | A11 | Yes |
| K11VV4W-TR | Cylindrin tandem repeat V2L/V4W variant | - | - | 1–2 | - | Nof | No |
| G6V | Fibril steric zipper | 3SGS | Indefinitely large | - | - | No | No |
Peptide segment amino acid sequences are provided in Table SI.
Data refer to truncated alphaB crystallin (16).
Data refer to full length alphaB crystallin (50).
Data not shown.
Data shown in Fig S3.
Very weak binding.
Data shown in Fig. S1E.
–, Sample was not tested or unknown.
SEC, Size exclusion chromatography. MS, Native mass spectrometry.
The hairpin, segment KVKVLGDVIEV (termed K11V) formed both amyloid fibrils and oligomers. Upon shaking at elevated temperature, K11V forms fibrils similar to those of the parent protein (ABC) from which the segment is derived (15) and similar to those of a tandem repeat of K11V (K11V-TR) (Fig. 1B, Fig. S1B–C, and Table S1). The fibrils range from 20 to 100 nm in diameter as viewed by electron microscopy (Fig. S1). X-ray diffraction of dried fibrils displayed rings at 4.8 and 12 Å resolution, consistent with the signature cross-beta pattern of amyloid fibrils (Fig. S1C). The amyloid fibrils of K11V-TR bind the specific amyloid dye congo-red, producing apple-green bifringerance under polarized light (Fig. S1D), and are immunoreactive with the fibril-specific, conformation-dependent antibody, OC (Fig. 1E and Fig. S1E) (20). Together these results prove that the segments G6V, K11V, and K11V-TR are all capable of converting to the amyloid state (21, 22), as is their parent protein, ABC.
Under physiological conditions, the segment K11V, K11V-TR, and a sequence variant with Leu replacing Val at position 2 (K11VV2L), all form stable small oligomers, intermediate in size between monomer and fiber. For each sequence, we determined the number of molecules in the oligomers by size exclusion chromatography (SEC-HPLC) and native mass spectrometry experiments. Purified recombinant K11VV2L, and K11V-TR, a tandem repeat of K11VV2L eluted as oligomeric complexes by SEC (Fig. 1C). For example, the K11V-TR complex was estimated to be ~8 kDa in mass, corresponding roughly to three tandem segment chains. As an additional check on the stoichiometry of the tandem repeat K11V-TR oligomer, we subjected peak fractions to native nanoelectrospray mass spectrometry. Mass spectra clearly showed abundant ions of K11V-TR oligomers with masses corresponding to three peptide chains (Fig. 1D and S3). Furthermore, we were able to isolate ions of the K11V-TR oligomer and perform collision induced dissociation (CID) of this trimeric peptide complex into monomeric units of mass equal to the K11V-TR peptide (Fig. S4). Similar experiments show that K11V and K11VV2L form hexameric oligomers (Table 1 and Fig. S3). Thus native mass spectrometry is consistent with SEC-HPLC in suggesting a stoichiometry of a K11V oligomer of six chains and a K11V-TR oligomer of three tandem chains. These results are consistent with crystallography and energetic considerations (see below).
These ABC K11V oligomers exhibit molecular properties in common with amyloid oligomers from other disease-related proteins. We probed blots of the recombinant segments with the polyclonal A11, amyloid-oligomer-specific conformational antibody (5). Both single and tandem repeat segments are recognized by the A11 antibody (Fig. 1E and Fig. S1E). Using a cell viability assay on mammalian cells, we observed oligomers to be toxic, displaying dose-response effects similar to those of Abeta involved in Alzheimer's disease (2, 23, 24) (Fig. 1F and Fig. S5). To test if membrane disruption is responsible for this toxicity, as suggested for human Islet Amyloid Polypeptide (hIAPP) (25, 26), we performed liposome dye-release experiments. The hIAPP peptide clearly diminished liposome integrity leading to dye release, but the K11V-TR did not exhibit this trend (Fig. S6). In contrast to oligomeric solutions, no toxicity was observed for the fibrils of G6V. Thus ABC segments in oligomeric form are cytotoxic, but suggest a more complicated mechanism of toxicity than membrane disruption.
We next determined the crystal structures of various ABC K11V oligomers. A screen produced X-ray grade crystals of K11V, but structure determination by molecular replacement with fiber-like probes failed, suggesting that the ABC segment oligomers possess a previously unobserved type of amyloid structure. Turning to the SAD method for phase determination, we synthesized K11V derivatives with Br substitutions at positions 2 or 8 of the K11V sequence, K11V-Br2 and K11V-Br8, with the leucine-resembling non-natural amino acid (2-bromoallyl)-glycine. Both derivatives crystallized and led to structure determinations (Table S2) at 1.4Å resolution. Molecular replacement based on these structures led to the closely related structures of K11V itself, as well as K11V-TR and K11VV2L.
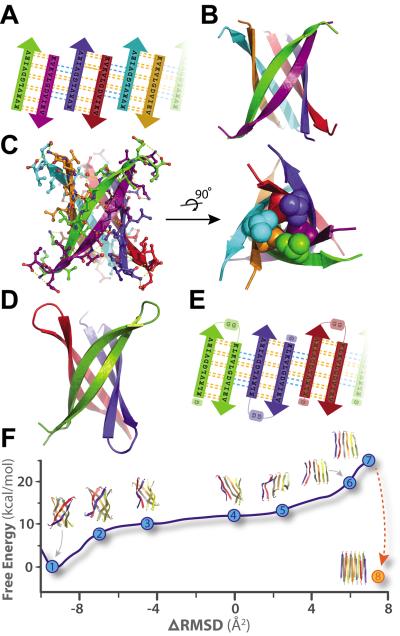
The structure of K11V, the amyloid-related oligomer, is a six-stranded anti-parallel barrel of cylindrical shape, consistent in mass with our solution studies, which we term a cylindrin. The cylindrin (Fig. 2) is distinctly different in structure from either the native structure of ABC (Fig. 1A) or from its G6V segment (Fig. S2), a dual beta-sheet steric zipper. It is also distinct from other structures currently in the Protein Data Bank (see SOM Text, Proposed and Similar Cylindrin Models), but resembles several previously proposed beta-barrel models (27–31). Each strand of the cylindrin is bonded to one neighboring strand by a strong interface and to a second by a weak interface. The strong interface (between purple and green chains, Fig. 2B–C) is formed by twelve hydrogen bonds, and splays outward at the ends. The weak interface is formed by eight hydrogen bonds, four from the main-chain, two mediated through sidechain interactions, and two through a water bridge (Fig. 2C). The axial channel of the cylindrin is closed by the hydrophobic interactions of two inward pointing sets of three valine sidechains, and is devoid of water (Fig. 2C). The surface area buried per residue (Ab) in the strand packing interface of the cylindrin is 87 Å2, smaller than the 131 Å2 value for the strand-to-strand interface of the steric zipper of GNNQQNY (32). Similarly the cylindrin packing interface has a shape complementarity (Sc) value of 0.75, somewhat smaller than the value of 0.80 for the GNNQQNY interface (Table S3). Thus the cylindrin structure has features in common with a steric zipper in being formed from hydrogen bonded beta-strands and having a dry interior, but it is cylindrical rather than nearly flat, and is probably less stable, as suggested by the lower Ab and Sc values.
Fig. 2.
Crystal structures of cylindrins and computed free energy change of the simulated structural transition from cylindrin to a fibril. Each colored beta-strand (arrow) is composed of eleven amino acid residues from ABC of sequence KVKVLGDVIEV (K11V). (A) Schematic of unrolled cylindrin (outside view), illustrating strand-to-strand registration. Hydrogen bonds between the main chains of neighboring strands are shown by yellow dashed lines; hydrogen bonds mediated by water bridges or side chains are shown by blue dashed lines. (B) Ribbon representation of the cylindrin crystal structure. Pairs of strands form anti-parallel dimers, which assemble around a three-fold axis down the barrel axis of the cylindrin. The height of the cylindrin is 22 Å. The inner dimension of the cylindrin, around the waist from Cα to Cα, is 12 Å, and at the splayed ends is 22 Å. (C) The cylindrin with sidechains shown as atoms, and hydrogenbonds in yellow. Twelve backbone hydrogen bonds stabilize the strong interface between tightlytwisted anti-parallel strands (e.g. between green and purple chains). The weaker interface between the pairs of tightly twisted strands is formed by four main-chain hydrogen bonds, with an additional two hydrogen bonds coming from a water bridge and two hydrogen bonds from side chain interactions (e.g. between purple and blue chains). The dry interior of the cylinder is closed by triplets of Val residues, shown as spheres, at the top and bottom. (D) Crystal structure of K11V-TR formed by three chains of 25residues each. (E) Schematic of unrolled K11V-TR cylindrin (outside view). Similar hydrogen bondingpatterns are formed as in (A). (F) The computed Gibbs free energy at 300K for a cylindrin forced to afibril. The reaction coordinate (ΔRMSD) measures the difference in root-mean-squared deviation fromthe two end points: the cylindrin and the in-register anti-parallel beta-sheet (IAB). The cylindrin set thefree energy minimum (point 1). The transition was initiated by disrupting the weak interface (points 2–3). As the cylindrin unrolls, the weak interface requires complete dissociation of backbone hydrogen bonds (points 4–5), whereas the strong interfaces maintains hydrogen bonding (point 6). The IAB has a higher free energy than the cylindrin (point 7), and when two associate and interdigitate to form a steric-zipper (point 8) the free energy drops to 5.2 kcal/mol/peptide lower than the cylindrin (Table S4).
To provide adequate cylindrin material for biochemical and toxicological studies, we generated a synthetic gene to express in bacteria a tandem repeat, K11V-TR, of the well diffracting K11VV2L segment, covalently linked through a double glycine linker and containing an additional N-terminal glycine (Fig. S7 and Table S1). This K11V-TR peptide reduces the complexity of the cylindrin assembly process from six to three chains (Fig. 2D–E). We were able to determine the K11V-TR crystal structure, even though the glycine linkers produce some disorder in the crystals (Table S2). Other than the glycine linkers and the Val to Leu replacement, the cylindrical bodies of the six-stranded K11V and the three double-stranded K11V-TR oligomers are essentially identical. Energetic considerations suggest that the cylindrin should be stable in solution: the surface area buried per interchain interface of K11V-TR is 841 Å2, nearly as much as for the foldon trimerization domain, 1092 Å2 (PDB 1RFO), and cylindrin forms twice as many hydrogen bonds between neighboring chains as does the foldon domain.
For a negative control of cylindrin structure and properties, we generated a variant form of the tandem segment, K11VV4W-TR in which the V4W substitution occurs in both repeats (Table S1). This substitution was predicted on the basis of the K11V crystal structure to disrupt oligomer formation through steric clash of core, buried residues. This variant peptide eluted in the mass range of a dimeric/monomeric species by SEC-HPLC and displayed dramatically reduced cell toxicity (Fig. 1F and S5).
To compare cylindrins to fibers, we define a cylindrin as a toxic, amyloid-related, oligomeric, cylindrically shaped beta-barrel formed from anti-parallel, extended protein strands and having the cylinder filled with packed sidechains. A cylindrin resembles a steric zipper in having a packed interior, but differs from a steric zipper in an important respect which may illuminate the reaction pathway from oligomers to fibrils. When unrolled into a beta-sheet, each anti-parallel pair of strands in the cylindrin sheet (Fig. 2A) is out of register with neighboring pairs by 6 residues (shear number is 6) (Fig. S8) (33). In contrast, the beta-strands in full amyloid fibers (22, 34, 35) and short steric-zippers (19) are in-register. This means that a cylindrin unrolled into a sheet, would not be an in-register structure, ready to bond with an identical sheet to form the steric zipper spine of an amyloid fiber. The transition from cylindrin to steric zipper involves breaking of hydrogen bonds, and re-registration of the strands into an in-register structure, as we have simulated by targeted molecular dynamics, followed by free energy perturbation in explicit solvent (Fig. 2F and SOM Text, Molecular Dynamic Simulations and Calculations). We chose the end target as an antiparallel sheet, based on FTIR experiments (SOM Text, Fibril Model of the Cylindrin Sequence). These calculations suggest that the transition from cylindrin to an anti-parallel fiber-like structure involves crossing a high free energy implying that fibers may nucleate from monomers without passing through cylindrin-like oligomeric states (36–38); that is, cylindrin is likely to be off-pathway to fiber formation.
An important question is whether the ABC cylindrin is a possible model for amyloid oligomers formed by well-studied toxic proteins, such as Abeta and hIAPP. There is evidence that amyloid oligomers share common structural features. For example, studies have suggested oligomers are beta-sheet rich (38–40), and several toxic oligomers are recognized by the A11 conformational antibody (41), which also recognizes the cylindrin. A11 also recognizes alpha-hemolysin, a soluble beta barrel protein (42). Thus the cylindrin structure may represent the common structural core of amyloid oligomers (SOM Text, Potential Cylindrin A11 Epitopes). To investigate this possibility, we used the Rosetta-Profile method (18) to ask if other toxic sequences, or segments of them, are compatible with the cylindrin structure. We found that the C-terminal segment of Abeta is reasonably compatible with the cylindrin structure, and with a two-residue registration shift between pairs of anti-parallel strands, a very good fit with the cylindrin structure is obtained (Fig. S11). This finding in itself does not imply that this is the structure of the Abeta toxic species, but it is in agreement with the observation of hexamers of Abeta oligomers by native mass spectrometry analysis (43).
The ABC cylindrin may represent one of many possible assemblies of cylindrin-like structures. The number of strands or shear number may vary. For example, Abeta oligomers have been described ranging in size from dimers and tetramers to hexamers, and dodecamers (43–45). Those with larger numbers of strands could have open central channels as modeled by others (46, 47), whereas cylindrins having smaller numbers of strands would have dry interfaces similar to crystallographically (48, 49) and computationally (44) derived models. Parallel assemblies could also form cylindrin-like structures, such as those previously modeled (27, 46). In fact, strong evidence for the extreme polymorphism of amyloid oligomers suggests that cylindrin-like assemblies could exist in a variety of structures with a variety of properties, including varying stabilities and toxicities (39, 44).
Supplementary Material
Acknowledgments
We thank L. Goldschmidt for the 3D profile scan of alphaB crystallin, Jacques-Philippe Colletier, Daniel Anderson, Gary Fujii, James Stroud, Howard Chang, Stuart Sievers, Jonathan Weissman, Justin Benesch, Georg Hochberg, and Carol V. Robinson for useful discussion, Arnie Berk and Dawei Guo for help with tissue culture experiments, Cory Ralston at the Advanced Light Source (ALS) 8.2.2 and Kanagalaghatta Rajashankar and beam-line staff at Argonne Photon Source (APS), Northeastern Collaborative Access Team beamlines 24-ID-E/C with data collection. The last is supported by award RR-15301 from the National Center for Research Resources of the National Institutes of Health. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357. We thank the NIH Chemistry Biology Interface Training program (award 5T32GM008496) sponsorship for A. Laganowsky, and UCLA Dissertation Year fellowships awarded to A. Laganowsky and M. Zhao. We thank the award MCB-0445429 from the National Science Foundation, award 1R01-AG029430 from the National Institutes of Health, award NIH-016570 from Alzheimer's Disease Research (ADRC) at UCLA, and HHMI for support. Atomic coordinates and structure factors have been deposited in the Protein Data Bank with the following accession codes: K11V (3SGO); K11V-Br2 (3SGM); K11V-Br8 (3SGN); K11VV2L (3SGP); K11V-TR (3SGR); and GDVIEV (3SGS).
References
- 1.Caughey B, Lansbury PT. Annu Rev Neurosci. 2003;26:267. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 2.Xue WF, et al. J Biol Chem. 2009 Dec 4;284:34272. doi: 10.1074/jbc.M109.049809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kodali R, Wetzel R. Curr Opin Struct Biol. 2007 Feb;17:48. doi: 10.1016/j.sbi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Kirkitadze MD, Bitan G, Teplow DB. J Neurosci Res. 2002 Sep 1;69:567. doi: 10.1002/jnr.10328. [DOI] [PubMed] [Google Scholar]
- 5.Kayed R, et al. Science. 2003 Apr 18;300:486. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 6.Bitan G, Fradinger EA, Spring SM, Teplow DB. Amyloid. 2005 Jun;12:88. doi: 10.1080/13506120500106958. [DOI] [PubMed] [Google Scholar]
- 7.Glabe CG. J Biol Chem. 2008 Oct 31;283:29639. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiti F, Dobson CM. Annu Rev Biochem. 2006;75:333. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 9.Chiti F, Dobson CM. Nat Chem Biol. 2009 Jan;5:15. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- 10.Rochet JC, Lansbury PT., Jr. Curr Opin Struct Biol. 2000 Feb;10:60. doi: 10.1016/s0959-440x(99)00049-4. [DOI] [PubMed] [Google Scholar]
- 11.Kayed R, et al. Mol Neurodegener. 2007;2:18. doi: 10.1186/1750-1326-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horwitz J. Proc Natl Acad Sci U S A. 1992 Nov 1;89:10449. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jehle S, et al. Proceedings of the National Academy of Sciences of the United States of America. 2011 Apr 19;108:6409. doi: 10.1073/pnas.1014656108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ecroyd H, Carver JA. Cell Mol Life Sci. 2009 Jan;66:62. doi: 10.1007/s00018-008-8327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meehan S, et al. J Mol Biol. 2007 Sep 14;372:470. doi: 10.1016/j.jmb.2007.06.060. [DOI] [PubMed] [Google Scholar]
- 16.Laganowsky A, et al. Protein Sci. 2010 May;19:1031. doi: 10.1002/pro.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson MJ, et al. Proc Natl Acad Sci U S A. 2006 Mar 14;103:4074. doi: 10.1073/pnas.0511295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldschmidt L, Teng PK, Riek R, Eisenberg D. Proc Natl Acad Sci U S A. 2010 Feb 23;107:3487. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawaya MR, et al. Nature. 2007 May 24;447:453. [Google Scholar]
- 20.Sarsoza F, et al. Acta Neuropathol. 2009 Oct;118:505. doi: 10.1007/s00401-009-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kajava AV, Baxa U, Steven AC. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2010 May;24:1311. doi: 10.1096/fj.09-145979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenwald J, Riek R. Structure. 2010 Oct 13;18:1244. doi: 10.1016/j.str.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 23.Austen BM, et al. Biochemistry. 2008 Feb 19;47:1984. doi: 10.1021/bi701415b. [DOI] [PubMed] [Google Scholar]
- 24.Picone P, et al. Biophys J. 2009 May 20;96:4200. doi: 10.1016/j.bpj.2008.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engel MF, et al. Proc Natl Acad Sci U S A. 2008 Apr 22;105:6033. doi: 10.1073/pnas.0708354105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khemtemourian L, Engel MF, Liskamp RM, Hoppener JW, Killian JA. Biochim Biophys Acta. 2010 Sep;1798:1805. doi: 10.1016/j.bbamem.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 27.Pryciak PM, Conway JD, Eiserling FA, Eisenberg D. In: Protein Structure, Folding, and Design. Oxender DL, editor. vol. 39. Alan R. Liss; New York: 1986. pp. 243–246. [Google Scholar]
- 28.Salemme FR, Weatherford DW. J Mol Biol. 1981 Feb 15;146:119. doi: 10.1016/0022-2836(81)90369-7. [DOI] [PubMed] [Google Scholar]
- 29.Salemme FR, Weatherford DW. J Mol Biol. 1981 Feb 15;146:101. doi: 10.1016/0022-2836(81)90368-5. [DOI] [PubMed] [Google Scholar]
- 30.Shafrir Y, Durell S, Arispe N, Guy HR. Proteins. 2010 Dec;78:3473. doi: 10.1002/prot.22853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conway JD. UCLA. 1986. [Google Scholar]
- 32.Nelson R, et al. Nature. 2005 Jun 9;435:773. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murzin AG, Lesk AM, Chothia C. J Mol Biol. 1994 Mar 11;236:1369. doi: 10.1016/0022-2836(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 34.Benzinger TL, et al. Proc Natl Acad Sci U S A. 1998 Nov 10;95:13407. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tycko R. Annu Rev Phys Chem. 2011 May;62:279. doi: 10.1146/annurev-physchem-032210-103539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Last NB, Rhoades E, Miranker AD. Proc Natl Acad Sci U S A. 2011 Jun 7;108:9460. doi: 10.1073/pnas.1102356108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu JW, et al. J Biol Chem. 2010 Feb 26;285:6071. doi: 10.1074/jbc.M109.069542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kar K, Jayaraman M, Sahoo B, Kodali R, Wetzel R. Nat Struct Mol Biol. 2011 Mar;18:328. doi: 10.1038/nsmb.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chimon S, et al. Nat Struct Mol Biol. 2007 Dec 2; doi: 10.1038/nsmb1345. [DOI] [PubMed] [Google Scholar]
- 40.Ono K, Condron MM, Teplow DB. Proc Natl Acad Sci U S A. 2009 Sep 1;106:14745. doi: 10.1073/pnas.0905127106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomic JL, Pensalfini A, Head E, Glabe CG. Neurobiol Dis. 2009 Sep;35:352. doi: 10.1016/j.nbd.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshiike Y, Kayed R, Milton SC, Takashima A, Glabe CG. Neuromolecular Med. 2007;9:270. doi: 10.1007/s12017-007-0003-6. [DOI] [PubMed] [Google Scholar]
- 43.Bernstein SL, et al. Nat Chem. 2009 Jul;1:326. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmed M, et al. Nat Struct Mol Biol. 2010 May;17:561. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bitan G, Vollers SS, Teplow DB. J Biol Chem. 2003 Sep 12;278:34882. doi: 10.1074/jbc.M300825200. [DOI] [PubMed] [Google Scholar]
- 46.Jang H, et al. J Mol Biol. 2010 Dec 17;404:917. doi: 10.1016/j.jmb.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT., Jr. Nature. 2002 Jul 18;418:291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 48.Liu C, et al. J Am Chem Soc. 2011 May 4;133:6736. doi: 10.1021/ja200222n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Streltsov VA, Varghese JN, Masters CL, Nuttall SD. J Neurosci. 2011 Jan 26;31:1419. doi: 10.1523/JNEUROSCI.4259-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dehle FC, Ecroyd H, Musgrave IF, Carver JA. Cell Stress Chaperones. 2010 Nov;15:1013. doi: 10.1007/s12192-010-0212-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.