Abstract
Genetic analyses of lung cancer have helped found new treatments in this disease. We conducted an integrative analysis of gene expression and copy number in 261 non-small cell lung cancers (NSCLC) relative to matched normal tissues to define novel candidate oncogenes, identifying 12q13-15 and more specifically the YEATS4 gene as amplified and overexpressed in ~20% of the NSCLC cases examined. Overexpression of YEATS4 abrogated senescence in human bronchial epithelial cells (HBECs). Conversely, RNAi-mediated attenuation of YEATS4 in human lung cancer cells reduced their proliferation and tumor growth, impairing colony formation and inducing cellular senescence. These effects were associated with increased levels of p21WAF1 and p53 and cleavage of PARP, implicating YEATS4 as a negative regulator of the p21-p53 pathway. We also found that YEATS4 expression affected cellular responses to cisplastin, with increased levels associated with resistance and decreased levels with sensitivity. Taken together, our findings reveal YEATS4 as a candidate oncogene amplified in NSCLC, and a novel mechanism contributing to NSCLC pathogenesis.
Keywords: YEATS4, NSCLC, oncogene, p53, integrative analysis
INTRODUCTION
Lung cancer is the leading cause of cancer death worldwide. The 5-year survival rate is a mere 15% and there exists a lack of therapies to effectively treat this deadly disease. However, within the last decade, characterization of lung cancer genomes has revealed a number of genes critical to tumorigenesis, resulting in significant changes to lung cancer treatment and a subsequent increase in progression free and overall survival for a subset of these patients. These successes have prompted a search for additional driver alterations, and have identified a number of recurrently mutated genes including TP53, CDKN2A, PTEN, NRAS, BRAF, PIK3CA, DDR2, KEAP1and NRF2 as well as gene fusions encompassing RET and ROS tyrosine kinases (1-5).
In addition to somatic mutations, copy number alterations such as recurrent amplifications and deletions occur in almost all lung cancers (6, 7). DNA amplification directly contributes to oncogene activation and the promotion of tumorigenesis, particularly for tumors driven by oncogene addiction. Oncogenes amplified at the DNA level therefore make ideal therapeutic targets as unlike loss of function tumor suppressor genes (TSG), they have the potential to be targeted directly. In NSCLC, recurrent amplifications of several regions activate known oncogenes. These include; 1q21.2 (ARNT), 3q26.3-q27 (PIK3CA & SOX2), 5p15.33 (TERT), 7p11.2 (EGFR), 7q31.1(MET), 8p12 (FGFR1) 8q24.21 (MYC), 12q14.1 (CDK4), 14q13.3 (NKX2-1) (7-13). Despite these discoveries, roughly 50% of lung cancers harbor no known targetable alterations, highlighting the need for a better understanding of the biology underlying lung tumorigenesis (2, 5).
To identify novel oncogenes in NSCLC, we performed a large scale integrative analysis of DNA copy number and gene expression on 261 lung tumors, spanning both major NSCLC subtypes; adenocarcinoma (AC) and squamous cell carcinoma (SqCC). Our approach was based on the rationale that oncogenes selectively amplified and biologically relevant to NSCLC tumor biology would: i) span regions of frequent high level amplification, ii) undergo frequent overexpression and iii) exert pro-tumorigenic functions in vitro and in vivo. Our analysis identified a recurrent amplicon at 12q15, within which we identified the candidate oncogene YEATS4/GAS41 (YEATS domain containing 4, glioma-amplified sequence 41). In vivo and in vitro functional assays were performed to characterize the biologic effects and investigate the oncogenic mechanism of YEATS4 in lung tumorigenesis. Based on the frequency of YEATS4 amplification and overexpression in NSCLC tumors and cell lines, its role in viability, anchorage independent growth, senescence and tumor formation, we propose that YEATS4 is novel candidate oncogene in lung cancer.
MATERIALS AND METHODS
NSCLC tumor samples and cell lines
261 formalin-fixed paraffin embedded and fresh-frozen lung tumors (169 AC and 92 SqCC) were obtained under informed, written consent with approval from the University of British Columbia-BC Cancer Research and University of Toronto Ethics Board from patients undergoing surgical resection at the Vancouver General Hospital and the Princess Margaret Hospital in Toronto(14). Tissue sections were micro-dissected with the guidance of lung pathologists and matched non-malignant lung tissue obtained for a subset of the primary tumors. DNA was extracted using standard phenol-chloroform procedures. RNA was extracted from tumor and matched non-malignant normal tissue using RNeasy Mini Kits (Qiagen) or Trizol reagent (Invitrogen). Quality and quantity of genomic material was assessed using a NanoDrop 1000 spectrophotometer and by gel electrophoresis and/or by Agilent 2100 Bioanalyzer. Demographic information for this cohort is summarized elsewhere (14). NSCLC cell lines H1993, H1355, H226, A549 were obtained from American Type Culture Collection and HCC4011 from Dr. Adi Gazdar and fingerprinted to confirm their identity (15). All lines were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum and 0.1% Penicillin-Streptomycin (Invitrogen). Immortalized normal human bronchial epithelial cells (HBEC) with (HBEC-KT53) and without p53 knockdown (HBEC-KT), courtesy of Dr. John Minna, were cultured in K-SFM media supplemented with 50ng/ul BPE and 5 ng/ul EGF (Invitrogen). Demographic data for the panel of cell lines used in this study can be found at http://edrn.jpl.nasa.gov/ecas/data/dataset/urn:edrn:UTSW_MutationData.
Array Comparative Genomic Hybridization and GISTIC analysis
Copy number profiles were generated for 261 NSCLC tumors using whole-genome tiling path array comparative genomic hybridization (aCGH), and were processed as previously described (16, 17). Probes were mapped to the March 2006 (Hg18) genomic coordinates and aCGH-Smooth was used to segment and smooth log2 ratio values(18). The corresponding segments and ratio values were analyzed using the GISTIC algorithm (19) and gene pattern software (http://www.broadinstitute.org/cancer/software/genepattern/) to identify regions of significant amplification across samples. Amplification threshold of 0.8, join segment size of 2, qv threshold 0.05 and removal of the X chromosome were the settings applied for analysis.
Gene expression profiling and data integration
Gene expression profiles were generated using custom Affymetrix microarrays for a subset (35 AC and 13SqCC) of the 261 tumors which had sufficient quantity and quality material for both tumor and matched non-malignant tissue. Data was normalized using the Robust Multichip Average algorithm in R(20). Genes were classified as over- or underexpressed if the mRNA fold change in tumors relative to matched non-malignant was greater or less than 2-fold. Mann-Whitney U tests with Benjamini Hochberg correction p<0.05 were used to compare expression of 12q15 genes between tumor and non-malignant tissue in 83 AC pairs (EDRN) and determine whether increased gene dosage resulted in increased gene expression. A Spearman’s correlation conducted using MATLAB software was used to determine the strength of the correlation between copy number and expression, with a coefficient >0.55 considered significant.
In vitro and in vivo assays were performed as previously described (21-23). Detailed information can be found in the supplemental methods.
RESULTS:
Recurrently amplified regions in NSCLC
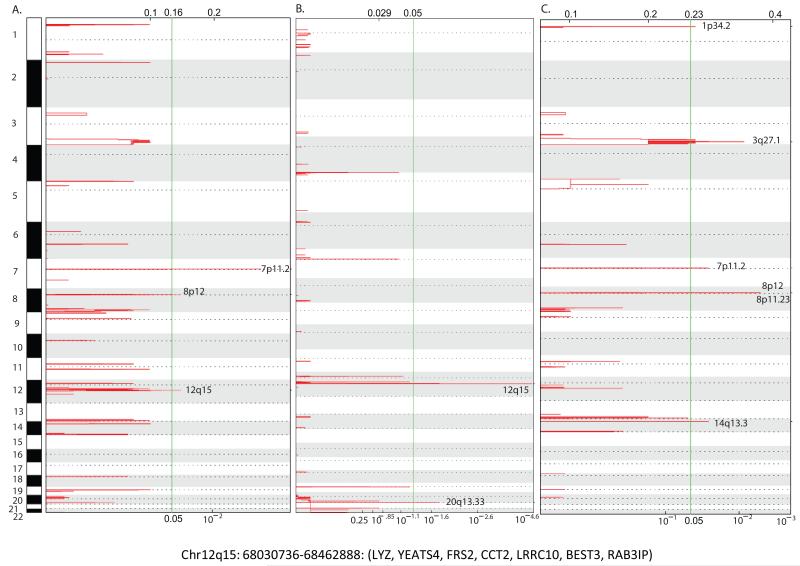
Copy number profiles for 169 AC and 92 SqCC were generated using aCGH. Significant regions of high level amplification (log2 ratio >0.8) were identified using the Genomic Identification of Significant Targets in Cancer (GISTIC) algorithm which calculates significance scores by considering both the amplitude and frequency of copy number alterations (19). GISTIC analysis of all 261 samples (NSCLC) identified 3 significant regions of focal amplification; 7p11.2 (q=0.00075), 8p12 (q= 0.036) and 12q15 (q=0.036). Subtype specific analysis revealed 2 regions of amplification across the 169 AC tumors; 12q15 (q= 4.5×10−5) and 20q13.33 (q=0.017) and 6 regions across the 92 SqCC tumors; 1p34.2 (q= 0.044), 3q27.1 (q=1.4 × 10−10), 7p11.2 (q=0.029), 8p11.23 (q=0.0042), 8p12 (q=0.0042) and 14q13.3 (q=0.03) (Fig. 1A-C). Amplification of these regions have been previously described in NSCLC indicating our tumors display patterns of alteration characteristic of lung cancer (2, 7, 26, 27).
Figure 1. Recurrent amplifications in NSCLC.
GISTIC plots for (A) 261 NSCLC, (B) 169 AC and (C) 92 SqCC. Chromosomes are depicted as rows and chromosome numbers are indicated. Red peaks indicate frequently amplified regions and the green vertical line indicates the false discovery rate threshold (q=0.05). Peaks extending beyond this line indicates a significant region. X-axis indicates the GISTIC score scale. Genomic coordinates and the genes located within the 12q15 amplicon are shown below.
While none of the regions identified were common between all three analyses, all of the regions identified in NSCLC were also significant in a subtype specific manner. Further examination of these amplicons revealed that known oncogenes EGFR and BRF2, both of which are known to be preferentially amplified in SqCC, (28) (21) were driving selection of the 7p11.2 and 8q12 amplicons, respectively. Intriguingly, the primary target of 12q15 amplification, which is believed to be MDM2- a ubiquitin ligase that targets TP53 for proteasomal degradation, and when overexpressed results in aberrant p53 inactivation, was excluded from both the focal and wide peak boundaries. The exclusion of MDM2 from this focal region suggested that a gene other than MDM2 may be driving selection of this amplicon. This combined with the fact that all other regions harbored known oncogenes 7p11.2 (EGFR), 8p11.23 (FGFR1), 8p12 (BRF2) 14q13.3 (NKX2-1), 20q13.3 (EEF1A) or are known to be subtype specific regions of amplification (1p34.2 and 3q in SqCC) (2, 7) prompted us to further explore the 12q15 amplicon.
Identification of YEATS4, the target of 12q15 amplification
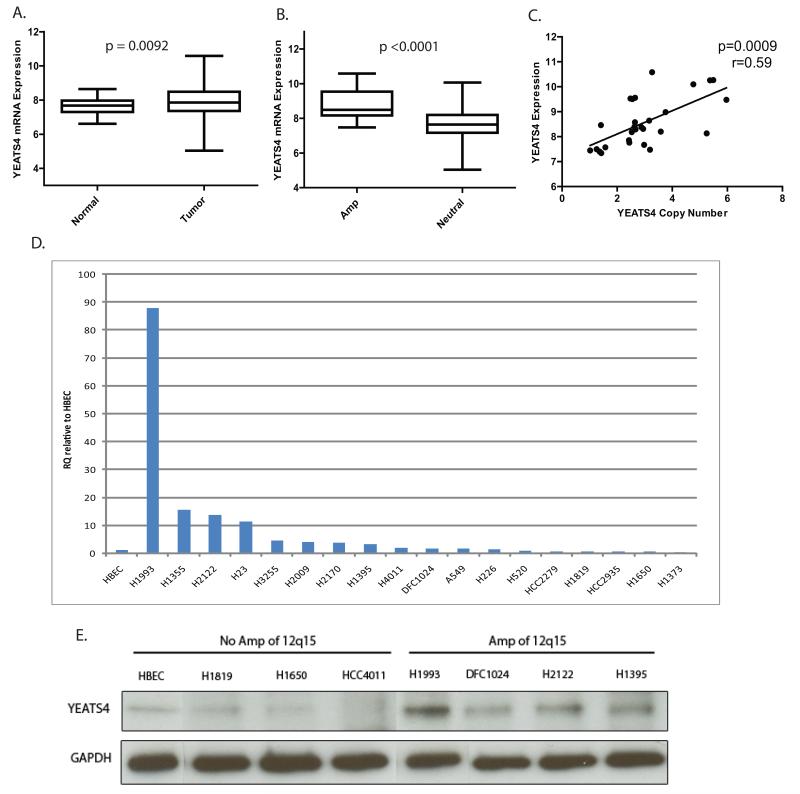
The peak amplified region of 12q15 spanned a 432 kb interval (68,030,736-68,462,888) and contained 7 genes; LYZ, YEATS4, FRS2, CCT2, LRRC10, BEST3, RAB3IP, none of which have been previously implicated in lung tumorigenesis (Fig. 1; Supplementary Table 1). Based on the notion that selectively amplified oncogenes would demonstrate elevated expression, we integrated copy number and gene expression data for adenocarcinoma tumors and matched non-malignant tissue. Due to the limited size of our dataset with both copy number and expression data, identification of the 12q15 driver gene was performed in the largest dataset available (EDRN, n=83). Of the 7 genes within the amplicon, only YEATS4 was both gained/amplified and concomitantly overexpressed in lung tumors relative to matched non-malignant tissues (Fig. 2A-C). While YEATS4 has not been previously described in lung cancer, it is a well-established oncogene in cancers of neural origin (29, 30) and frequently amplified in liposarcomas (31).
Figure 2. YEATS4 is recurrently amplified and overexpressed in NSCLC and is the target of 12q15 amplification.
(A) Comparison of mRNA expression in 83 AC tumors and matched non-malignant tissue from the EDRN (p=0.0092). (B) YEATS4 expression between tumors with gain/amplification and tumors with neutral copy number (p<0.0001). (C) Spearman’s correlation of copy number and expression for tumors with copy number alterations of YEATS4 (r=0.59, p=0.009). Expression values for all plots are in log2 units. (D) RT-qPCR of YEATS4 expression in 18 NSCLC cell lines and non-malignant HBEC cells. (E) Immunoblot of YEATS4 in NSCLC lines with and without amplification of 12q15 with GAPDH as a loading control.
YEATS4 is frequently amplified and overexpressed in NSCLC
YEATS4 was amplified in 18% (47/261) and overexpressed in 31% (15/48) of cases from our cohort. While 12q15 was not significant in the GISTIC analysis of our 92 SqCC cases, to conclusively determine whether amplification of YEATS4 was specific to AC, we compared copy number and expression data for both subtypes. Although no statistical difference in YEATS4 copy number or expression was observed between subtypes (Supplemental Fig. 1B-D), on average AC tumors had a higher number of copies and greater fold change in expression compared to SqCC tumors. This suggests that while copy gain is a frequent event in both subtypes, it is likely a broader amplification event that occurs at a lower amplitude in SqCC relative to AC, which is why 12q15 failed to be identified by GISTIC in the SqCC tumors. Analysis of external datasets with both AC and SqCC data supported our findings, with gain/amplification and overexpression occurring at similar frequencies in both data sets (Table 1), indicating that amplification and overexpression of YEATS4 is not subtype specific.
Table 1.
Frequency of YEATS4 gain and amplification across multiple datasets
| Data Set | Histology | Samples with CN (n) |
Gain (n) (>2.3 copies, <5) |
Freq Gain (%) | Amplification (n) (>5 copies) |
Freq Amp (%) | Samples with Expression (n) |
Over- experssion |
Freq OE (%) |
|---|---|---|---|---|---|---|---|---|---|
| BCCRC | AC & SqCC |
169,92 | 28,19 | 16.5,20.1 | 5,2 | 3.0, 2.2 | 35,13 | 11, 4 | 31,30 |
| EDRN | AC | 83 | 18 | 21.7 | 4 | 4.8 | 83 | 15 | 18 |
| GSE25016 | AC & SqCC |
77,155 | 10,22 | 13, 14.2 | 0,1 | 0, 0.6 | N/A | N/A | N/A |
| dbGAP | AC | 354 | 18 | 5.1 | 4 | 1.1 | N/A | N/A | N/A |
| MSKCC | AC | 199 | 21 | 10.6 | N/A | N/A | N/A | N/A | N/A |
| Broad | NSCLC | 473 | 28 | 5.9 | 8 | 1.7 | N/A | N/A | N/A |
| TCGA | AC & SqCC |
277,201 | 19,22 | 6.9,10.9 | 15,6 | 5.4,3.0 | 35,17 | 11, 6 | 32, 35 |
| Sanger lung Lines |
NSCLC & SCLC |
128 | 42 | 32.8 | 1 | 0.78 | N/A | N/A | N/A |
| All Sanger cell lines |
All cancers |
508 | 109 | 21.5 | 8 | 1.5 | N/A | N/A | N/A |
To gain further insight into the prevalence of YEATS4 amplification, we investigated YEATS4 copy number and expression in publically available NSCLC tumor datasets. YEATS4 was gained (2.3-5 copies) or amplified (> 5 copies) at various frequencies across the five datasets, ranging from 5-22% and 0.4-5% respectively (Table 1). A broader analysis of 508 human cancer cell lines revealed YEATS4 copy gain/amp in 43/128 (33.6%) of lung cancer cell lines and in 117/508 (23%) of all cancer cell lines (Table 1). Expression analysis of the EDRN and TCGA data sets, revealed YEATS4 was overexpressed at comparable frequencies to our dataset; 18% (15/83) and 33% (14/42), respectively (Table 1). Taken together, these results show YEATS4 is frequently gained and overexpressed in NSCLC, irrespective of subtype, as well as gained in many other human cancers.
To validate array findings and verify YEATS4 is upregulated at the transcript level, we assessed YEATS4 expression by quantitative reverse transcriptase PCR (RT-qPCR) in a panel of 59 lung ACs relative to matched non-malignant tissue and in 18 NSCLC cell lines (2 SqCC and 16 AC) with reference to an immortalized normal human bronchial epithelial (HBEC) line. 15/59 (25.4%) tumors and 8/18 (44.4%) cell lines showed a two-fold or greater increase in YEATS4 expression relative to their matched control (Fig. 2D; Supplementary Fig. 1A). Moreover, analysis of the 35 AC samples with expression data revealed a strong correlation between array findings and PCR results (r=0.75, P<0.001, Pearson Correlation, data not shown), validating array findings and confirming frequent overexpression of YEATS4. Western blotting of cell lines with and without YEATS4 amplification revealed increased YEATS4 expression in lines with amplification, demonstrating that amplification drives overexpression at both the mRNA and protein level (Fig. 2E).
Association of YEATS4 and clinical features
Multivariate analysis of YEATS4 copy number and expression revealed no significant associations between any clinical features (age, sex, stage, smoking status, race). Survival analysis of the Director’s challenge expression datasets (32) using a cox-regression analysis revealed a trend towards poorer survival in patients with YEATS4 amplification, however this association failed to reach statistical significance in any of the datasets examined (data not shown).
YEATS4 displays oncogenic properties in vitro and in vivo
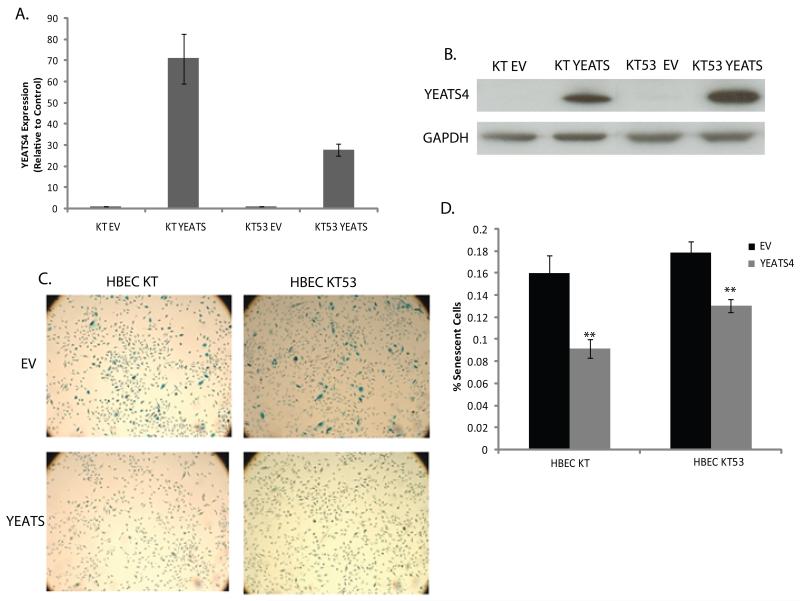
YEATS4 encodes a protein found in a number of multi-subunit protein complexes involved in chromatin modification and transcriptional regulation and has also been shown to be involved in the regulation of TP53. To assess its oncogenic potential, YEATS4 was stably transfected into two immortalized HBEC lines; HBEC-KT and HBEC-KT53 (KT-YEATS and KT53-YEATS). Empty vector transfected cells were used as controls (KT-EV and KT53-EV). YEATS4 gene and protein expression was confirmed by qPCR and western blot (Fig. 3A-B). Relative to controls, ectopic expression of YEATS4 had no effect on viability and failed to induce anchorage independent growth in HBECs (data not shown), indicating that in immortalized normal cells YEATS4 overexpression alone is incapable of inducing colony formation. However, a dramatic inhibition of senescence in overexpressing cells relative to controls was observed in both lines (Student’s t-test, p<0.05) (Fig. 3C-D), suggesting elevated YEATS4 expression is capable of inducing a phenotype associated with malignant transformation.
Figure 3. Overexpression of YEATS4 induces a malignant phenotype.
Ectopic expression of YEATS4 increases (A) mRNA expression (mean± SEM of triplicate replicates) and (B) protein levels relative to EV controls. GAPDH was used as a loading control. (C) β-Gal staining for cellular senescence in EV and YEATS4 expressing HBECs. Cells stained blue indicate senescence. Original magnification, 10×. (D) Quantification of cellular senescence in YEATS4 and control cells. The mean of the proportion of senescent cells (senescent cells/total cells) for YEATS4 and EV lines is shown for triplicate experiments ± SEM. ** p<0.01, Student’s t-test.
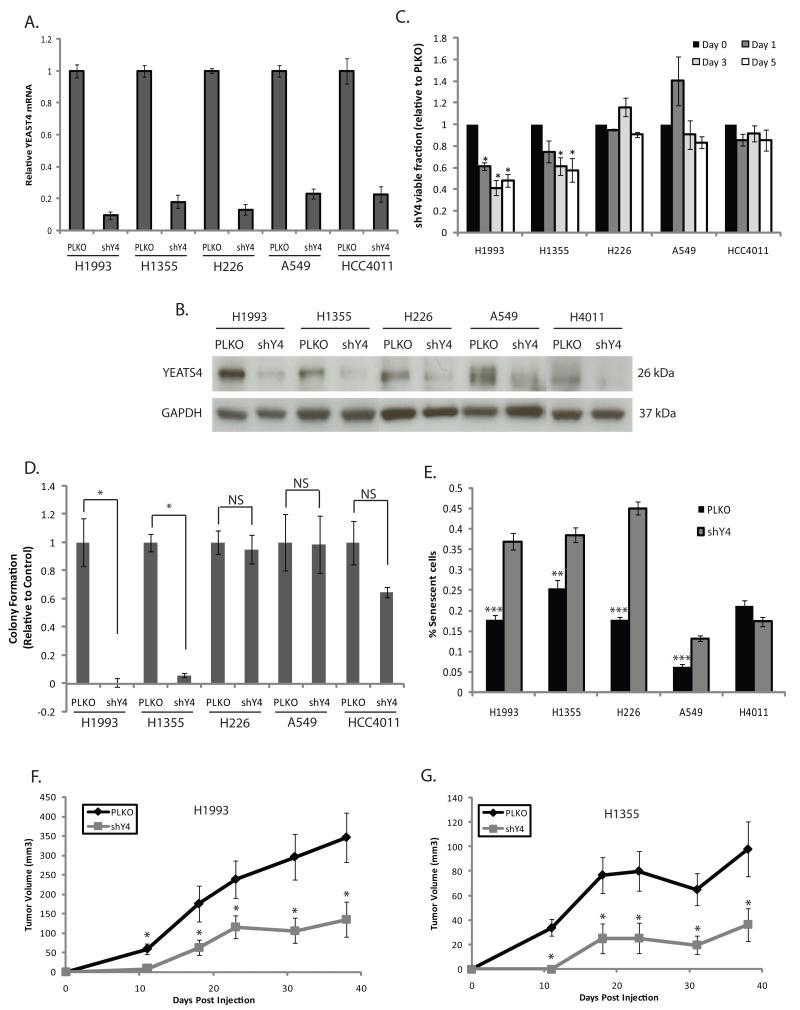
Complimentary knockdown experiments using lentiviral shRNAs were performed in lung cancer cell lines with (H1993, H1355, H226) and without (A549, HCC4011) YEATS4 amplification and various p53 backgrounds (Supplementary Table 2). Empty vector transfected cells were used as controls (PLKO) and knockdown was confirmed by qPCR and western blotting (Fig. 4A-B). Knockdown significantly decreased cell viability in H1993 and H1355 (p=0.0127 and p=0.0172, respectively), both of which harbour YEATS4 amplification and mutant p53(Fig. 4C), but had no effect on A549, HCC4011 or H266 lines (p = 0.428, p = 0.45 and p = 0.49, respectively) which do not harbor YEATS4 amplification (A549 &H4011), or have YEATS4 amplification with wild type (wt) p53 (H226) (Fig. 4C). Similarly, knockdown resulted in a significant decrease in anchorage-independent colony formation in H1993 (p = 7.26 ×10−6) and H1355 (p = 6.06 × 10−10) cells, but not in A549 (p = 0.97), H4011 (p = 0.21) or H226 (p = 0.74) cells, indicating wt p53 may abrogate the effect of YEATS4 knockdown on viability and colony formation in lines with amplification (Fig. 4D). A significant increase in senescence was observed in all three lines with amplification; H1993 (p = 5.71 × 10−6), H1355 (p = 0.0012) and H226 (p = 1.21 ×10−13) as well as moderate increase in A549 (p = 2.99× 10−7). No difference in senescence was observed in HCC4011 (p = 0.06) (Fig. 4E). The finding that A549 cells showed a modest increase in senescence is not surprising given the role of YEATS4 in the p53 pathway (discussed below) and the wt p53 background of this line, which enables pathway activation and cellular senescence.
Figure 4. YEATS4 knockdown impairs growth and induces senescence.
shRNA targeting YEATS4 significantly reduces (A) mRNA expression and (B) protein levels in all cell lines relative to controls (PLKO). GAPDH was used as a loading control. (C) Viability of cell lines with knockdown (shY4) relative to controls as measured by MTT. (D) Colony formation ability of shY4 cell lines relative to controls. (E) Quantification of cellular senescence based of β-Gal staining. Values reported as mean ± SEM of triplicate experiments.* p<0.05, ** p<0.01, *** p<0.001, Student’s t-test of shY4 cells relative to PLKO. (F,G) Effect of YEATS4 knockdown on tumor growth in mice injected with H1993 or H1355 PLKO and shY4 cells. Error bars indicate SEM of each group of 10 mice, * p<0.05
To explore the oncogenic potential of YEATS4 in vivo, tumor formation in NOD/SCID mice was examined by subcutaneous flank injections of H1993 and H1355 control and shY4 cells. Tumor formation was significantly reduced in shY4 cells of both cell lines at all time points (Fig. 4F,G). Our results demonstrate that knockdown of YEATS4 in cell lines with amplification effectively inhibits tumorigenesis, with a significant inhibition in viability, tumor and anchorage independent growth and increased cellular senescence, strongly supporting YEATS4 as an oncogene in NSCLC.
YEATS4 suppresses p53 and p21
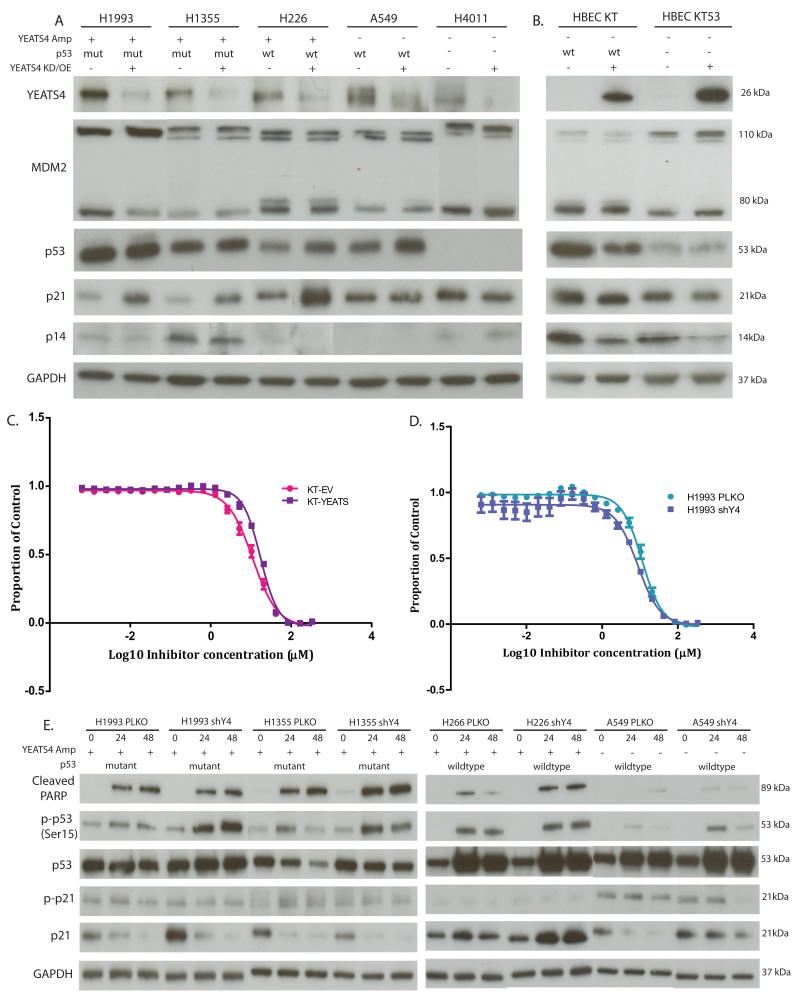
Inactivation of the p53 pathway is one of the most frequent alterations in lung cancer, with somatic mutations occurring in approximately 50% of all cases (28, 33). p53 is a key tumor suppressor that regulates cell cycle, DNA repair, apoptosis, and senescence and inhibits aberrant proliferation and the propagation of damaged cells. A study by Park et al, showed that under normal, unstressed conditions, YEATS4 binds to and inhibits the promoters of p14 and p21, subsequently repressing the p53 tumor suppressor pathway (34). To assess whether this interaction occurs in NSCLC, we assessed these proteins in cell lines with YEATS4 manipulation. Upon YEATS4 knockdown, p21 and p53 protein levels were increased, with the greatest increases in expression of p21 and p53 observed in cell lines harbouring YEATS4 amplification or wt p53 respectively (Fig. 5A). No change in p14 levels was observed upon knockdown. Overexpressing lines showed a modest reduction of p21 and p14 as well as a reduction of p53 levels in HBEC-KT (Fig. 5B). MDM2 levels remained unchanged following knockdown or overexpression of YEATS4, indicating that the observed changes in p21, p14 and p53 were a direct result of YEATS4 manipulation.
Figure 5. YEATS4 alters p21 and p53 protein levels.
(A) Knockdown of YEATS4 increases expression of p21 in cell lines with YEATS4 amplification and increases p53 in all lines that express p53. (B) Overexpression of YEATS4 reduces p14 and p21 levels in both HBEC lines, and p53 only in the HBEC KT line. Dose-response curves of HBEC KT (C) and H1993 (D) cells treated with 2-fold dilutions of cisplatin for 72 hours. Viability is shown as a proportion of treated cells against untreated controls (mean ± SEM of triplicate experiments). (E) Immunoblot of PLKO and shY4 cell lines treated with 40μm of Cisplatin for 0, 24 or 48 hours. Cisplatin treatment induces apoptosis as measured by the increase in cleaved PARP, p53 and phosphorylated p53 (Ser15). GAPDH was used as a loading control for all blots.
YEATS4 alters the sensitivity of cell lines to cisplatin and nutlin
To determine whether the downstream effects of YEATS4 manipulation alters cellular sensitivity to chemotherapy, cell lines were treated with serial dilutions of cisplatin, a commonly prescribed first line chemotherapy for lung cancer patients that crosslinks DNA triggering apoptosis, or nutlin, a cis-imidazoline analog that inhibits the interaction of p53 and MDM2, stabilizing p53. Based on the observed effects on p53 and p21 protein levels following manipulation of YEATS4 expression and the notion that cells with YEATS4 amplification may be dependent on YEATS4 for growth and survival, we hypothesized that HBEC-KT/KT53-Y cells would be more resistant to treatment, while shY4 cells harbouring YEATS4 amplification would be more sensitive.
As expected, HBEC-KT-YEATS and HBEC-KT53-YEATS lines were significantly more resistant to both cisplatin and nutlin than their control counterparts (Fig. 5C; Table 3). Differences in sensitivity were less consistent in the lung cancer cell lines, likely due to the fact these cell lines harbour numerous genomic alterations which could influence drug sensitivities. While H1993 shY4 cells were significantly more sensitive to Cisplatin (IC50 PLKO:11.45 vs. shY4:8.65) (Fig. 5D) supporting our hypothesis, knockdown in both H1355 and H226, showed the opposite trend resulting in greater resistance relative to controls (Table 2). As anticipated, A549 and HCC4011 shY4 cells showed no difference in sensitivity (Table 2). As specimens with mutant p53 are resistant to nutlin, only A549 and H226 were treated. Similar to the cisplatin results, A549 shY4 cells showed no significant difference in sensitivity to nutlin (PLKO: 7.58 vs. shY4:6.91, p=0.84), while H226 shY4 cells were unexpectedly significantly more resistant (PLKO:3.27, shY4:4.33, p=0.033) (Table 2). Analysis of lung cancer cell line IC50 data from the Sanger drug sensitivity project failed to reveal a significant association between YEATS4 amplifcation and response to cisplatin or nutlin. However, based on the fact that transformed bronchial epithelial cells which harbour minimal genetic alterations were significantly more resistant to cisplatin and nutlin following overexpression of YEATS4, and H1993 shY4 cells (which harbor the greatest amplification of YEATS4) were more sensitive to cisplatin compared to controls, we feel this data supports the notion that YEATS4 alters the in vitro sensitivity of lung cells to cisplatin and nutlin.
Table 2.
Cisplatin and Nutlin IC50s
| Cell Line | YEATS4 | Cisplatin | Nutlin | Trend | ||||
|---|---|---|---|---|---|---|---|---|
| IC50 | SEM | t Test | IC50 | SEM | t Test | |||
| H1993 | PLKO | 11.45 | 1.11 | |||||
| H1993 | shY4 | 8.649 | 0.488 | 0.004 | Sensitive | |||
| H1355 | PLKO | 9.111 | 0.491 | |||||
| H1355 | shY4 | 15.91 | 0.905 | 1.6E-06 | Resistant | |||
| H226 | PLKO | 5.788 | 0.276 | 3.266 | 0.130 | |||
| H226 | shY4 | 9.965 | 0.716 | 0.0003 | 4.325 | 0.376 | 0.033 | Resistant in both |
| A549 | PLKO | 10.88 | 0.452 | 7.58 | 0.316 | |||
| A549 | shY4 | 11.4 | 0.705 | 0.204 | 6.908 | 0.123 | 0.084 | Not significant in both |
| H4011 | PLKO | 8.952 | 0.326 | |||||
| H4011 | shY4 | 10.32 | 0.566 | 0.055 | Not significant | |||
| HBEC KT | EV | 11.09 | 1.472 | 16.55 | 1.405 | |||
| HBEC KT | YEATS | 17.32 | 0.696 | 0.007 | 26.01 | 2.696 | 0.023 | Resistant in both |
| HBEC KT53 | EV | 15.41 | 1.612 | 19.63 | 1.367 | |||
| HBEC KT53 | YEATS | 20.38 | 0.718 | 0.029 | 24.85 | 1.385 | 0.034 | Resistant in both |
Sensitivity to cisplatin is not mediated solely through the p53-p21 pathway
To gain further insight into the potential mechanisms of altered sensitivity to cisplatin, we treated cell lines with 40μM cisplatin for 48 hours, and collected protein lysates at 0, 24 and 48 hours post treatment. As expected, cisplatin treatment of HBECs resulted in an increase in p53, p53 Ser15 phosphorylation (a marker of stabilization), p21 and induced apoptosis as measured by cleaved PARP. However, no differences between HBEC-EV and HBEC-YEATS cells were observed for any of the proteins examined (Supplementary Fig. 2). In shY4 cells with amplification, treatment with cisplatin led to a greater induction of p53 and phospho-p53 (Ser15), and in H226 also led to a significant increase in p21 levels relative to control cells (Fig. 5E). As no significant differences in protein levels between HBEC-EV and HBEC-YEATS were observed, despite a significant increase in resistance following overexpression, our results suggests that while the p53-p21 signaling pathway may be involved, resistance is likely mediated through the interaction of YEATS4 with other signaling pathways.
YEATS4 knockdown phenotypes are independent of p21 signaling in mutant p53 cells
To explore the effect of increased p21 expression on the observed phenotypes following knockdown, siRNA knockdown of CDKN1A was performed on shY4 and PLKO cells for cell lines with YEATS4 amplification (H1993, H1355 & H226). Knockdown of CDKN1A showed no effect on viability or colony formation in any of the lines (data not shown), but significantly altered senescence levels in the presence of wt p53 (Supplemental Fig. 2A). CDKN1A siRNA reduced senescence in both H226 shY4 and PLKO cells relative to non-targeting control siRNA treated cells, such that the percent of senescent H226 shY4-p21 cells was similar to H226 PLKO-NTC (Supplemental Fig. 2B). The findings from these experiments suggest that in a wildtype p53 background, the increase in senescence following YEATS4 knockdown occurs in a p53 dependent manner and is the direct result of increased p21 expression. As CDKN1A knockdown failed to rescue viability, colony formation and senescence in cell lines with mutant p53, these findings further support the notion that the phenotypes observed following knockdown of YEATS4 are not solely due to changes in p53-p21 signaling. Based on these findings, and the prominent role of Rb in senescence, we investigated whether the increased senescence following YEATS knockdown could be due to altered Rb signaling. We observed modest reductions in Rb Ser807/811 phosphorylation following YEATS knockdown which in mutant p53 cell lines appears to be due in part to reduced levels of p27 (Supplemental Fig. 2C).
Identification of additional cellular networks regulated by YEATS4
In an attempt to gain a better understanding of other pathways YEATS4 is involved in, we performed expression profiling on shY4 and PLKO cells for the three cell lines with YEATS4 amplification. To identify significantly enriched pathways/networks and gene sets affected by YEATS4 knockdown, Ingenuity Pathway Analysis (IPA) and Gene Set Enrichment Analysis (GSEA) were performed. A total of 32 genes (27 overexpressed and 5 underexpressed) were differentially expressed between knockdown and control cells across all three cell lines. Due to the small number of input genes, none of the significantly enriched canonical pathways passed multiple testing correction. However, network analysis, which assesses regulatory relationships existing between genes and proteins, identified two networks associated with pro-tumorigenic functions; (1) cancer and (2) cell death, survival, cell cycle and cell morphology. These networks were centered around known targets or binding partners of YEATS4 including p53, CDKN1A and MYC (Supplementary Fig. 3), further supporting our in vitro findings. Pre-ranked GSEA revealed significant enrichment of a number of transcription factor gene sets including MYCN, which has been shown to be a binding partner of YEATS4 and all 6 serum response factor (SRF) gene sets. SRF is a ubiquitously expressed transcription factor implicated in cell proliferation, differentiation, metastasis and clinically associated with castration-resistant prostate cancer (35, 36). Interestingly, PDLIM7 which contains a serum response element and is transcribed upon induction of SRF, was shown to inhibit p53 and p21 through the inhibition of MDM2 self ubiquitination. While neither MYCN, SRF or PDLIM7 were differentially disrupted at the mRNA level following knockdown, our downstream analysis suggests the target genes of these two transcription factors could be involved in YEATS4 mediated tumorigenesis and warrant investigation in future studies to elucidate additional mechanisms through which YEATS4 promotes tumorigenesis.
DISCUSSION
While single dimensional genomic analyses have been instrumental in cancer gene discovery, this type of analysis often overlooks genes disrupted at low frequencies, and is unlikely to distinguish causal from passenger events. The integration of multiple parallel genomic dimensions enables the identification of genes with concurrent DNA and expression alterations, which are likely selected for due to their roles in driving cancer phenotypes (37). Towards this end, we integrated copy number and gene expression data in an attempt to identify novel oncogenes important in lung tumorigenesis. While our analysis revealed gains/amplifications in a number of regions previously reported in NSCLC, the amplicon at 12q15 was the only one without a candidate driver gene located within the amplicon boundaries and was therefore the only regions we pursued further. Integration of expression and copy number data for the 7 genes located within 12q15 identified YEATS4 as the candidate target gene of this amplicon.
First identified and isolated in the glioblastoma multiforme cell line TX3868, YEATS4 is a highly conserved nuclear protein essential for cell viability that is frequently amplified in gliomas, astrocytomas and liposarcomas (29, 31, 38). A member of a protein family characterized by the presence of an N-terminal YEATS domain, YEATS4 shares high homology with transcription factor family members AF-9 and ENL(39). Like other family members, YEATS4 is involved in chromatin modification and transcriptional regulation through its incorporation into multi-subunit complexes; specifically the human TIP60/TRRAP and SRCAP complexes (40, 41), which mediate the incorporation of an H2A variant histone protein into nucleosomes, altering chromatin structure and controlling transcriptional regulation.
In addition to its role in transcriptional regulation, yeast two hybrid screens have revealed a number of YEATS4 binding partners. These include MYC, MYCN, TACC1, TACC2, NuMa, AF10, PFDN1 and KIAA1009 (42-46). Analysis of expression data before and after YEATS4 knockdown showed no effect on expression of any binding partners, suggesting that YEATS4 does not control the expression of its binding partners at the mRNA level. To date, the majority of work surrounding YEATS4 has focused primarily on the identification of YEATS4 binding partners with only a few studies having explored the phenotypic effects of YEATS4 amplification, none of which have been performed in lung (34, 46).
Our study is the first to show gain/amplification and overexpression of YEATS4 in NSCLC and the first to implicate amplification of YEATS4 in lung cancer tumorigenesis. We observed frequent gain/amplification of YEATS4 in multiple independent tumor cohorts in addition to our own, as well as a strong correlation between gain and overexpression in both tumors and cell lines (Fig. 2). Analysis of the catalogue of somatic mutations in cancer (COSMIC) revealed YEATS4 is rarely mutated in lung (0.23%) or any cancer type (0.17%) suggesting that DNA amplification is the predominant mechanism of activation. In addition to the genomic evidence supporting selection of YEATS4 in NSCLC, we demonstrate the oncogenic potential of YEATS4 both in vitro and in vivo (Fig. 3&4). Ectopic expression resulted in a significant reduction in senescence, suggesting overexpression of YEATS4 is sufficient to induce phenotypic changes characteristic of malignant transformation, (Fig. 3) while knockdown in cell lines with amplification and mutant p53 showed reduced viability and colony formation along with an increase in senescence, consistent with oncogenic function. While wt p53 abrogates the effects on viability and colony formation on YEATS4 knockdown lines with amplification, a significant increase in senescence was still observed. In addition to these phenotypic effects, we also demonstrated that YEATS4 inhibits p21 thereby repressing p53 activity, consistent with the findings of Park and Roeder who demonstrated this interaction in unstressed conditions (34). siRNA-mediated knockdown of CDKN1A failed to rescue viability, colony formation and senescence in mutant p53 backgrounds suggesting the phenotypic effects of YEATS4 amplification occur through a mechanism other than p21.
MDM2, an E3 ubiquitin ligase, is the major negative regulator of p53, mediating its ubiquitination and subsequent degradation (47, 48). Overexpression results in inactivation of p53 and is a common mechanism of p53 inactivation in cancer. MDM2 is frequently amplified and overexpressed in human cancers including lung cancer, and is largely considered to be the driver gene of the 12q15 amplicon (7, 49). We were therefore intrigued to discover that despite being frequently gained in our dataset, MDM2 did not fall within the boundaries of the 12q15 amplicon identified in our cohort. This led us to suppose that an alternative oncogene was being selected for in this region. When looking at high resolution copy number profiles, while the majority of cases showed identical copy number for both YEATS4 and MDM2, a small number of cases (3/83) had more copies of YEATS4 than MDM2, suggesting YEATS4 is selected as the target of amplification in these samples and that amplification of YEATS4 is not merely a passenger event of MDM2 amplification. Of note, 4/83 cases had higher level gain/amplification of MDM2 relative to YEATS4. For cancers with amplification of 12q15 spanning both YEATS4 and MDM2, these genes may work synergistically to suppress p53, however further experimentation is required to investigate this hypothesis. Along with the many tumor promoting effects of YEATS4, of immediate clinical interest is our discovery of a YEATS4 dependent mechanism of reduced cisplatin and nutlin sensitivity, which appears to occur at least in part through inhibition of p21 and subsequent suppression of the p53 pathway.
In summary, we have shown YEATS4 is frequently amplified and overexpressed in NSCLC and together with its multiple oncogenic functions suggests it is a novel oncogene in lung cancer. Given the effect of YEATS4 on the p53 pathway, we propose that MDM2 is not the sole driver gene targeted by amplification of 12q15 and that suppression of the p53 pathway can be achieved through amplification of YEATS4. Additional investigation into the signaling pathways altered as a result of YEATS4 amplification will provide further insight into the mechanism underlying YEATS4-mediated tumorigenesis.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank May Zhang and Miwa Suzuki for their assistance.
GRANT SUPPORT
Funding provided by the Canadian Institutes for Health Research (MOP 86731, 94867), Canadian Cancer Society (CCS20485), NCI Early Detection Research Network and the Canary Foundation. LAP & KLT are supported by Vanier Canada Graduate Scholarship, WWL by CIHR Jean-Francois Saint Denis Fellowship in Cancer Research, EAV by CIHR Frederick Banting & Charles Best Canada Graduate Scholarship and RC by Banting Postdoctoral Fellowship.
Funding: Grant supported from the Canadian Institutes for Health Research (MOP 86731, 94867), Canadian Cancer Society (CCS20485), NCI Early Detection Research Network and the Canary Foundation.
Footnotes
Disclosure: The authors have no conflicts of interest to disclose
REFERENCES
- 1.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammerman PS, Hayes DN, Wilkerson MD, Schultz N, Bose R, Chu A, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohno T, Ichikawa H, Totoki Y, Yasuda K, Hiramoto M, Nammo T, et al. KIF5B-RET fusions in lung adenocarcinoma. Nature medicine. 2012;18:375–7. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S, et al. RET, ROS1 and ALK fusions in lung cancer. Nature medicine. 2012;18:378–81. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 5.Pao W, Hutchinson KE. Chipping away at the lung cancer genome. Nature medicine. 2012;18:349–51. doi: 10.1038/nm.2697. [DOI] [PubMed] [Google Scholar]
- 6.Lockwood WW, Chari R, Coe BP, Girard L, Macaulay C, Lam S, et al. DNA amplification is a ubiquitous mechanism of oncogene activation in lung and other cancers. Oncogene. 2008;27:4615–24. doi: 10.1038/onc.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–8. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–42. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirsch FR, Varella-Garcia M, Franklin WA, Veve R, Chen L, Helfrich B, et al. Evaluation of HER-2/neu gene amplification and protein expression in non-small cell lung carcinomas. Br J Cancer. 2002;86:1449–56. doi: 10.1038/sj.bjc.6600286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okudela K, Suzuki M, Kageyama S, Bunai T, Nagura K, Igarashi H, et al. PIK3CA mutation and amplification in human lung cancer. Pathol Int. 2007;57:664–71. doi: 10.1111/j.1440-1827.2007.02155.x. [DOI] [PubMed] [Google Scholar]
- 11.Onozato R, Kosaka T, Kuwano H, Sekido Y, Yatabe Y, Mitsudomi T. Activation of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers. J Thorac Oncol. 2009;4:5–11. doi: 10.1097/JTO.0b013e3181913e0e. [DOI] [PubMed] [Google Scholar]
- 12.Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto H, Shigematsu H, Nomura M, Lockwood WW, Sato M, Okumura N, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008;68:6913–21. doi: 10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lockwood WW, Wilson IM, Coe BP, Chari R, Pikor LA, Thu KL, et al. Divergent genomic and epigenomic landscapes of lung cancer subtypes underscore the selection of different oncogenic pathways during tumor development. PloS one. 2012;7:e37775. doi: 10.1371/journal.pone.0037775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gazdar AF, Girard L, Lockwood WW, Lam WL, Minna JD. Lung cancer cell lines as tools for biomedical discovery and research. Journal of the National Cancer Institute. 2010;102:1310–21. doi: 10.1093/jnci/djq279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishkanian AS, Malloff CA, Watson SK, DeLeeuw RJ, Chi B, Coe BP, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 17.Khojasteh M, Lam WL, Ward RK, MacAulay C. A stepwise framework for the normalization of array CGH data. BMC bioinformatics. 2005;6:274. doi: 10.1186/1471-2105-6-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jong K, Marchiori E, Meijer G, Vaart AV, Ylstra B. Breakpoint identification and smoothing of array comparative genomic hybridization data. Bioinformatics. 2004;20:3636–7. doi: 10.1093/bioinformatics/bth355. [DOI] [PubMed] [Google Scholar]
- 19.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20007–12. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 21.Lockwood WW, Chari R, Coe BP, Thu KL, Garnis C, Malloff CA, et al. Integrative genomic analyses identify BRF2 as a novel lineage-specific oncogene in lung squamous cell carcinoma. PLoS medicine. 2010;7:e1000315. doi: 10.1371/journal.pmed.1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lockwood WW, Thu KL, Lin L, Pikor LA, Chari R, Lam WL, et al. Integrative genomics identified RFC3 as an amplified candidate oncogene in esophageal adenocarcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:1936–46. doi: 10.1158/1078-0432.CCR-11-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thu KL, Pikor LA, Chari R, Wilson IM, Macaulay CE, English JC, et al. Genetic disruption of KEAP1/CUL3 E3 ubiquitin ligase complex components is a key mechanism of NF-kappaB pathway activation in lung cancer. J Thorac Oncol. 2011;6:1521–9. doi: 10.1097/JTO.0b013e3182289479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coe BP, Chari R, MacAulay C, Lam WL. FACADE: a fast and sensitive algorithm for the segmentation and calling of high resolution array CGH data. Nucleic acids research. 2010;38:e157. doi: 10.1093/nar/gkq548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chari R, Lonergan KM, Pikor LA, Coe BP, Zhu CQ, Chan TH, et al. A sequence-based approach to identify reference genes for gene expression analysis. BMC medical genomics. 2010;3:32. doi: 10.1186/1755-8794-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao X, Weir BA, LaFramboise T, Lin M, Beroukhim R, Garraway L, et al. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res. 2005;65:5561–70. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]
- 27.Job B, Bernheim A, Beau-Faller M, Camilleri-Broet S, Girard P, Hofman P, et al. Genomic aberrations in lung adenocarcinoma in never smokers. PloS one. 2010;5:e15145. doi: 10.1371/journal.pone.0015145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herbst RS, Heymach JV, Lippman SM. Lung cancer. The New England journal of medicine. 2008;359:1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer U, Heckel D, Michel A, Janka M, Hulsebos T, Meese E. Cloning of a novel transcription factor-like gene amplified in human glioma including astrocytoma grade I. Human molecular genetics. 1997;6:1817–22. doi: 10.1093/hmg/6.11.1817. [DOI] [PubMed] [Google Scholar]
- 30.Munnia A, Schutz N, Romeike BF, Maldener E, Glass B, Maas R, et al. Expression, cellular distribution and protein binding of the glioma amplified sequence (GAS41), a highly conserved putative transcription factor. Oncogene. 2001;20:4853–63. doi: 10.1038/sj.onc.1204650. [DOI] [PubMed] [Google Scholar]
- 31.Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL, et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet. 2010;42:715–21. doi: 10.1038/ng.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nature medicine. 2008;14:822–7. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Human mutation. 2007;28:622–9. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 34.Park JH, Roeder RG. GAS41 is required for repression of the p53 tumor suppressor pathway during normal cellular proliferation. Molecular and cellular biology. 2006;26:4006–16. doi: 10.1128/MCB.02185-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nature cell biology. 2009;11:257–68. doi: 10.1038/ncb1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prencipe M, Madden SF, O’Neill A, O’Hurley G, Culhane A, O’Connor D, et al. Identification of transcription factors associated with castration-resistance: is the serum responsive factor a potential therapeutic target? The Prostate. 2013;73:743–53. doi: 10.1002/pros.22618. [DOI] [PubMed] [Google Scholar]
- 37.Chari R, Thu KL, Wilson IM, Lockwood WW, Lonergan KM, Coe BP, et al. Integrating the multiple dimensions of genomic and epigenomic landscapes of cancer. Cancer metastasis reviews. 2010;29:73–93. doi: 10.1007/s10555-010-9199-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zimmermann K, Ahrens K, Matthes S, Buerstedde JM, Stratling WH, Phi-van L. Targeted disruption of the GAS41 gene encoding a putative transcription factor indicates that GAS41 is essential for cell viability. The Journal of biological chemistry. 2002;277:18626–31. doi: 10.1074/jbc.M200572200. [DOI] [PubMed] [Google Scholar]
- 39.Schulze JM, Wang AY, Kobor MS. YEATS domain proteins: a diverse family with many links to chromatin modification and transcription. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2009;87:65–75. doi: 10.1139/O08-111. [DOI] [PubMed] [Google Scholar]
- 40.Cai Y, Jin J, Florens L, Swanson SK, Kusch T, Li B, et al. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. The Journal of biological chemistry. 2005;280:13665–70. doi: 10.1074/jbc.M500001200. [DOI] [PubMed] [Google Scholar]
- 41.Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Molecular and cellular biology. 2004;24:1884–96. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piccinni E, Chelstowska A, Hanus J, Widlak P, Loreti S, Tata AM, et al. Direct interaction of Gas41 and Myc encoded by amplified genes in nervous system tumours. Acta biochimica Polonica. 2011;58:529–34. [PubMed] [Google Scholar]
- 43.Lauffart B, Howell SJ, Tasch JE, Cowell JK, Still IH. Interaction of the transforming acidic coiled-coil 1 (TACC1) protein with ch-TOG and GAS41/NuBI1 suggests multiple TACC1-containing protein complexes in human cells. The Biochemical journal. 2002;363:195–200. doi: 10.1042/0264-6021:3630195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harborth J, Weber K, Osborn M. GAS41, a highly conserved protein in eukaryotic nuclei, binds to NuMA. The Journal of biological chemistry. 2000;275:31979–85. doi: 10.1074/jbc.M000994200. [DOI] [PubMed] [Google Scholar]
- 45.Debernardi S, Bassini A, Jones LK, Chaplin T, Linder B, de Bruijn DR, et al. The MLL fusion partner AF10 binds GAS41, a protein that interacts with the human SWI/SNF complex. Blood. 2002;99:275–81. doi: 10.1182/blood.v99.1.275. [DOI] [PubMed] [Google Scholar]
- 46.Schmitt J, Fischer U, Heisel S, Strickfaden H, Backes C, Ruggieri A, et al. GAS41 amplification results in overexpression of a new spindle pole protein. Genes, chromosomes & cancer. 2012;51:868–80. doi: 10.1002/gcc.21971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marine JC, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell death and differentiation. 2010;17:93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- 48.Manfredi JJ. The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes & development. 2010;24:1580–9. doi: 10.1101/gad.1941710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic acids research. 1998;26:3453–9. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.