Abstract
Alternative pre-mRNA splicing determines the protein output of most neuronally expressed genes. Many examples have been described of protein function being modulated by coding changes in different mRNA isoforms. Several recent studies demonstrate that through the coupling of splicing to other processes of mRNA metabolism alternative splicing can also act as an on/off switch for gene expression. Other regulated splicing events may determine how an mRNA is utilized in its later cytoplasmic life by changing its localization or translation. These studies make clear that the multiple steps of post-transcriptional gene regulation are strongly linked. Together these regulatory process play key roles in all aspects of the cell biology of neurons, from their initial differentiation, to their choice of connections, and finally to their function with mature circuits.
Keywords: RNA binding proteins, nonsense-mediated mRNA decay, intron retention, RNA localization, post-transcriptional gene regulation
Splicing as a modulator of protein structure and function
Alternative mRNA processing in neurons was first observed soon after the first examples of alternative mRNAs arising from viral and immunoglobulin genes. The calcitonin and CGRP peptides were found to be encoded by the same locus, with neurons producing CGRP and thyroid cells producing calcitonin through changes in the splicing and polyadenylation of the primary transcript [1]. In addition to making a complete change in gene product, early analyses showed that alterations in splice site choice and hence in mRNA coding sequences can produce refined structural and functional modifications in proteins essential for neuronal development and activity (Figure 1). Well before the first genome sequences, many hundreds of new spliced isoforms were being described each year, often specific to the brain. It was clear that neurons (and also muscle) make extensive use of this mechanism to regulate protein function. Common alternative splicing patterns are exon cassettes that are either included or excluded from the final mRNA and alternative 5’ and 3’ splice sites that change the length of an exon [2]. Multiple changes in splicing can be combined to produce very complex patterns of expression and large numbers of mRNA and protein products from the same gene. Modern sequencing technologies have allowed the extent of mRNA and protein diversity generated at the RNA processing level to be quantified. It is now clear that nearly all multi-exon mammalian genes produce more than one mRNA product [3, 4]. Studies of tissue specific alternative exons found that they most commonly modify surface loops or unstructured binding motifs within proteins, and thus make specific changes in protein-protein interactions [5, 6]. This regulation is seen in all cell types, but is taken to particular extremes of complexity in the nervous system [3, 7-12].
Figure 1.
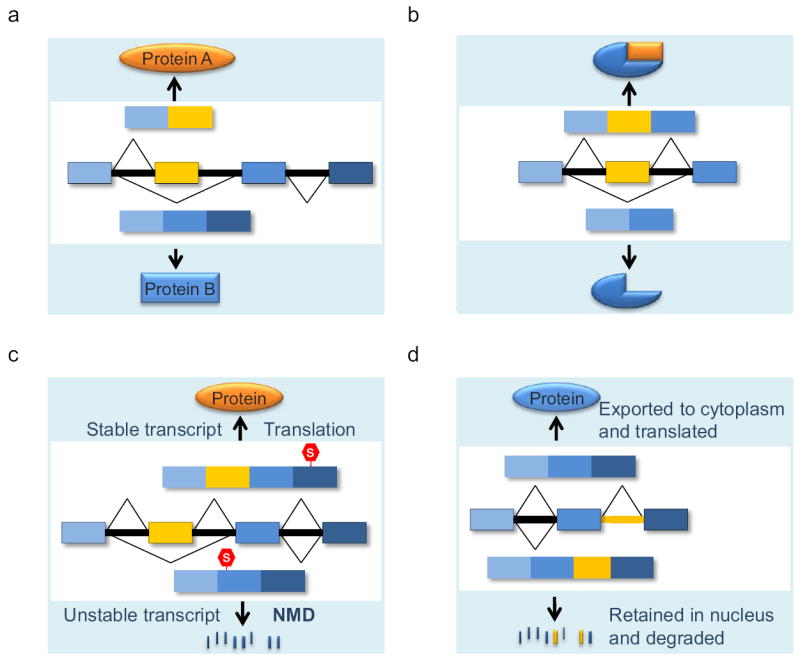
Patterns of alternative pre-mRNA splicing that affect gene expression in the mammalian nervous system. In the pre-mRNA, exons (boxes) are connected by introns (horizontal lines). Constitutive and alternative exons are blue and yellow boxes respectively. Exons undergo different patterns of joining to generate alternative mRNA isoforms. (a) Alternative pre-mRNA processing can produce transcripts encoding completely different proteins. (b) In many cases, alternative exons encode peptide segments that modify protein structure and activity. (c) Regulated alternative splicing coupled with the nonsense mediated mRNA decay (NMD) of one of the spliced forms can control overall expression of a gene. Stop codons are indicated by S in hexagons. (d) Regulated intron retention may inhibit nuclear export to control gene expression, or may affect subsequent cytoplasmic mRNA function. The retained intron is indicated by a yellow line.
A number of mechanisms have been described that mediate the controlled use of particular splice sites [13-15]. The most common is through the action of specialized RNA binding proteins that recognize sequence elements in the pre-mRNA adjacent to regulated splice sites [13, 15] [16]. Dozens of such proteins exhibiting diverse domain structures are now known to affect splicing, and there are hundreds of potential regulatory RNA binding proteins encoded in mammalian genomes. These proteins can bind in either exons or introns and can either repress or activate the use of particular splice sites or exons. Genome-wide maps of RNA binding by these regulators are defining the relationship between their binding site positions and their regulatory effects. From these studies and earlier work, it is clear that the same protein can act positively on some exons and negatively on others depending on where it binds and what other factors are involved [17]. A number of splicing regulatory proteins show tissue specific expression and this can lead to the cell type specific activation or repression of particular splice sites. Regulators determining neuronal specific expression of alternative exons include members of the PTBP, NOVA, RBFOX, CELF, ELAV, MBNL, SRRM4 and other protein families [18, 19]. Many alternative exons have been recently acquired during evolution, whereas other exons show conserved tissue-specific regulation across vertebrate or mammalian species, or across insect species [8, 11] [20]. Exons within this highly conserved class are generally regulated by multiple factors and the RNA sequences surrounding them exhibit unusually high levels of conservation that maintain the presence of multiple short binding elements [21]. The assembly of complex combinations of splicing regulators onto these sequences gives rise to the unique tissue specificity of exon use [22]. Large scale analyses of splicing regulatory elements are being directed to understanding this splicing code and the prediction of where and when each exon is expressed in mRNA [7, 23, 24] [25].
mRNA variants are usually first described in tissue samples such as whole brain, or in cultured cell lines, and subsequently shown to be specific to particular cell types such as neurons or glia. Many of these alternative splicing choices are determined during development and different mRNA isoforms are characteristic of early or late neuronal differentiation [26] [19]. However, isoform expression can be much more specific than is sometimes appreciated. It was shown early on that individual neurons in a circuit or ganglion can express different spliced isoforms of important transcripts [27-29]. Dynamic regulation of splicing also occurs in mature neurons in response to stimuli or stressors [26] [30, 31] [32] [33]. Thus for variants described as neuronal, the choice of splicing pattern does not necessarily reflect a difference between neurons and other cells, but might have much more complex determinants within neurons. How this specificity at the level of neuronal subtypes or individual cells is directed is not clear.
Thousands of neuronally regulated mRNA isoforms have been described, but how particular splicing changes alter the activity of the encoded proteins is often not known, even when the splicing shows conserved regulation and conserved protein coding across species, and modifies key domains for protein activity (Figure 1b). For complex systems, it can be a major project to catalog all of the expressed isoforms [34] [12]. Understanding how protein activity varies between different isoforms must then be assessed case by case using assays appropriate to each gene product, a much slower process than simply identifying the isoforms. Nevertheless when examined closely, splicing changes often alter protein activity in significant ways. Key changes in cell adhesion and connectivity during development, in the efficacy of synaptic vesicle release, in the gating properties of ion channels, in the regulation of signaling pathways by neurotransmitter receptors, and in the coupling of kinase activity to cell excitation have all been attributed to changes in particular protein isoforms [10, 26, 35] [19]. However, even when a substantial change in protein activity is caused by a splicing choice, it is not always clear why this change is important to the cellular process affected or why it needs to be regulated with development or in mature cells. Again this requires focused genetic and cell biological studies of individual spliced isoforms (For examples see: [26, 36-40]).
Splicing as an on/off switch
Rather than modifying protein activity, regulated splicing choices can also control the overall expression of proteins. This was first described as a mechanism to enforce female specific expression of the Sex lethal (Sxl) and transformer (tra) genes in Drosophila, key regulators of somatic sexual development in the fly [41] [13]. Sxl and tra produce different mRNA isoforms in female and male flies, where the female but not the male or non-sex specific isoforms encode functional proteins. These studies focused on the dominantly acting female products, and it has not been entirely elucidated whether there are roles for the male transcripts or if they might be eliminated. Recent studies have found that the expression of important neuronal proteins is controlled by similar mechanisms, where the coupling of alternative splicing to downstream processes of mRNA maturation allows a splicing choice to act as an on/off switch for a final protein product. In these systems, because it is regulating overall expression, the cellular role of the splicing change is often clearer than when splicing modifies protein structure and function. As seen for modification of protein function by alternative splicing, the regulation of overall expression by splicing contributes both to neuronal development and to dynamic modulation of activity in mature cells.
One mechanism for controlling expression through a splicing choice is for one isoform to be subject to mRNA decay rather than productive translation and thus be eliminated from cells when that splicing pattern is chosen [42] [43]. This can be accomplished through the coupling of alternative splicing to the nonsense mediated mRNA decay pathway (NMD, Figure 1c). NMD is a surveillance mechanism that prevents expression of potentially detrimental proteins from mutant or aberrantly spliced transcripts [44] [45]. During exon ligation, the exon junction complex (EJC) is deposited 20-24 nucleotides upstream of the spliced exon-exon junction [46] [47]. The EJC is composed of a tetrameric core of four proteins as well as auxiliary factors that accompany the final mRNA to the cytoplasm, and are thought to be stripped from the mRNA during translation. Most but not all mRNA reading frames terminate in the last exon downstream from all the EJCs. However, if translation should terminate more than 50 nucleotides upstream from an EJC, this acts as a signal to target the transcript to the NMD pathway [48]. A terminating ribosome recruits the NMD factor UPF1 that binds to the EJC auxiliary factors UPF2 and UPF3 to induce the decapping and endonucleolytic cleavage of the transcript and its degradation [49]. Aberrantly spliced mRNAs containing frameshifts or mRNAs containing frameshift or nonsense mutations that lead to premature translation termination before the last exon are usually degraded by NMD, thereby reducing the production of potentially dominant negative proteins. In the same manner, alternative splicing events that shift an mRNA reading frame, or which insert a new stop codon, can produce highly unstable mRNAs [42, 43]. In this manner the choice of splicing pattern switches expression from a stable mRNA to elimination of the gene product through mRNA decay (Figure 1c).
The coupling of alternative splicing to the NMD pathway (AS-NMD) is commonly seen in mRNAs for splicing regulators or other RNA binding proteins [42, 43] [50]. These transcripts often contain an alternative exon that is controlled by the encoded protein itself. Excess expression of the RNA binding protein activates an autoregulatory loop that produces the NMD targeted isoform and maintains homeostatic levels of the gene product. A similar negative feedback mechanism controls spermidine/spermine N1-acetyltransferase (SSAT) expression, where spermine-induced alternative splicing produces an NMD-targeted SSAT mRNA [51]. Although so far only seen in SSAT, AS-NMD provides a possible mechanism for end product inhibition of biosynthetic pathways. RNA binding proteins can also cross-regulate each other through this mechanism [52-55] [56, 57]. For example, the polypyrimidine tract binding protein PTBP1 represses a highly conserved exon in the transcript of its neuronal paralog PTBP2, leading to NMD of the Ptbp2 mRNA in undifferentiated cells. As neural progenitor cells differentiate, PTBP1 is repressed by the microRNA miR124 [56]. Through its effects on PTBP2 splicing, the loss of PTBP1 induces PTBP2 expression, resulting in a switch in these two similar proteins and an altered splicing program during early neuronal differentiation [52, 53, 58] [56].
Several new studies have now found that AS-NMD controls important functions in neuronal cell biology. DLG4 (also known as PSD-95) is a scaffold protein of the excitatory postsynaptic density where it clusters membrane receptors and channels with downstream signaling molecules [59]. DLG4 expression is enhanced late in neuronal differentiation when the protein plays essential roles in synapse maturation [60, 61]. It was recently found that developmental expression of Dlg4 is controlled at the RNA level by alternative splicing of its exon 18 [62]. Immature mammalian neurons and many non-neuronal cells express Dlg4 mRNA. However, this RNA is largely lacking exon 18, which shifts the reading frame, leading to translation termination at a conserved stop codon in exon 19 and hence to NMD of the Dlg4 mRNA transcript. Exon 18 splicing is controlled by the both PTBP1 and PTBP2 [58]. In non-neuronal cells and neural progenitors, exon 18 is repressed by PTBP1. During development, the depletion of PTBP1 induces expression of PTBP2, which maintains the repression of exon 18 and of DLG4 expression in the developing neuron. In late neuronal differentiation, PTBP2 is finally downregulated to allow exon 18 splicing, and this induces Dlg4 mRNA and protein expression to promote synapse maturation. The coupling of alternative splicing to nonsense mediated mRNA decay allows the PTB proteins to control the developmental expression of a key synaptic component. The PTB proteins are likely also controlling inhibitory synapse development through similar mechanisms; A PTB target exon, whose skipping should induce NMD, has also been described in a GABAB receptor transcript [56]. This mechanism of regulation thus provides a genetic on/off switch similar to the output of transcriptional or microRNA regulation.
That AS-NMD is a common mechanism for controlling synaptic protein expression is shown by another recent paper, examining synaptic components that are part of a post-transcriptional regulatory network controlled by the neuronal RNA binding proteins NOVA1 and NOVA2 [63] [64]. Correlating changes in overall gene expression with splicing changes in double Nova1/2 knockout mice, the authors identified AS-NMD targeted mRNAs for several synaptic proteins, including DLG3(SAP102) and SCN9A (NAV1.7 or NENA). Dlg3 exon 15, normally skipped in transcripts from wildtype brain, becomes included in the Nova double knockout brain. Inclusion of exon 15 results in a reading frameshift, leading to NMD of the transcript and significantly decreased DLG3 protein expression in the Nova1/2 null mice. Exon 17a of Scn9a also leads to frameshift-induced NMD when included in the mRNA. In contrast to Dlg3 exon 15, Scn9a exon 17a is normally included in wildtype brain - leading to constant degradation by the NMD pathway. Depletion of the NOVA proteins causes exon 17a skipping and increases SCN9A protein expression in the Nova1/2 null mice. Thus, by having opposite effects on NMD-inducing exons, the NOVA proteins stimulate Dlg3 expression and inhibit Scn9a expression. The physiological role of this coordinated AS-NMD regulation is not yet clear, but interestingly, these exons change in splicing after pilocarpine induced seizure indicating that they may be dynamically regulated in mature brain. The NOVA proteins may also contribute to the developmental specific expression of these synaptic proteins. For example, SCN9A is weakly expressed in the brain where the NOVA proteins are abundant, but is highly expressed in dorsal root ganglion, smooth muscle and thyroid tissue where NOVA1/2 are absent [65-67].
These studies and others indicate that AS-NMD is more widely used to modulate overall expression from a gene than was previously appreciated [68-70]. The regulation of two of the four DLG family proteins (DLG3 and DLG4) by this mechanism indicates that the other Dlg genes, Dlg1 (Sap97) and Dlg2 (Psd-93), as well as other members of the membrane-associated guanylate kinase (MAGUK) family might bear closer examination. There are also several earlier examples of important transcripts in neurons subject to possible NMD regulation. The axon guidance receptor gene Robo3 produces two spliced variants, Robo3.1 and Robo3.2 that differ by the presence of a retained intron in Robo3.2 [71]. The Robo3.2 reading frame terminates within the retained intron and upstream from a splice junction making it a potential NMD target. Although a role for NMD has not been shown in Robo3.2 expression, the expression of ROBO3.2 protein appears to be translationally regulated upon midline crossing by the axon. How this might result from its unusual mRNA structure is not yet clear [72]. Another neuronal mRNA targeted by the NMD pathway is the immediate early gene Arc, whose encoded protein regulates the surface expression of AMPA-type glutamate receptors. The Arc mRNA is unusual in containing two exon-exon junctions in its 3’ UTR, making it a “constitutive” NMD target. Depletion of EIF4AIII, a core component of the EJC, increases Arc mRNA and protein abundance in neurons [73]. For both Robo3.2 and Arc, the splicing of the NMD-inducing isoforms does not appear to change. Instead, the presence of the EJC on these transcripts appears to limit protein expression from these mRNAs in a controllable manner. These transcripts provide additional examples of the important interplay of splicing and NMD in controlling neuronal gene expression. One poorly understood feature of NMD regulation is that the unstable isoform is often not completely eliminated and yet its protein product is usually not observed. Why these mRNAs do not produce product is not known and indicates interesting additional regulatory connections between the different steps of gene expression.
In addition to cassette exons, another pattern of alternative splicing is the retention of a complete intron in the final mRNA. However, pre-mRNAs are generally held in the nucleus until introns are fully spliced, as the interaction of the U1snRNP with 5’ splice sites or of U2AF with 3’ splice sites can maintain the nuclear localization of incompletely spliced mRNAs [74, 75]. Studies have also shown that splicing and EJCs stimulate mRNA nuclear export [76]. These aspects of mRNA quality control make intron retention different from other patterns of alternative splicing, such as changes in exon use that do not generally leave partial or complete introns in the final mRNA. For partially spliced transcripts to act as mRNAs, mechanisms must be in place to allow their nuclear export. This is best understood for HIV and other retroviruses where incompletely spliced transcripts required for the viral lifecycle are targeted for export by special viral proteins such as Rev [77] [78]. Alternative splicing to retain introns can have similar consequences to other patterns of alternative splicing such as altering the coding of the mRNA. However, regulated intron retention can also act as an on/off switch for gene expression by inhibiting nuclear export of the intron-retained transcript. The splicing factor, SRSF1 negatively regulates its own expression through this mechanism, as well as by the AS-NMD mechanism [79]. In neuronal cells, a similar mechanism was found in apolipoprotein E4 (Apoe), whose intron 3 is normally retained to restrict APOE expression. The splicing of intron 3 is induced in response to excitotoxic stimuli to increase APOE expression [80].
A recent paper demonstrates that multiple genes encoding presynaptic proteins (Stx1b, Vamp2 and Sv2a) employ regulated intron retention to control their developmental expression [81]. The splicing of 3’-terminal introns within these gene transcripts is repressed by PTBP1 in nonneuronal cells and neural progenitor cells. Retention of these introns inhibits export of the mRNA products to the cytoplasm and leads to their nuclear degradation. As neurons differentiate and PTBP1 expression decreases, the 3’-terminal introns are excised to allow normal nuclear export and cytoplasmic accumulation of the fully spliced mRNA. The authors show that knockdown of Ptbp1 leads to increases in these transcripts, and for the Stx1b transcript this response requires pyrimidine rich elements in the retained intron. This study adds new features to the PTB regulatory network and raises interesting mechanistic questions regarding how PTBP1 can cause intron retention instead of other changes in the splicing pattern. Mutation of the splice sites within the Stx1b intron allows export of the Stx1b transcripts, consistent with previous studies on the role of splice sites in keeping intron-containing transcripts in the nucleus [74, 75]. The elimination of the intron-retained Stx1b mRNA requires the nuclear exosome and the nuclear pore associated protein Tpr/Mlp1 [82]. It will be interesting to examine how this decay pathway is initiated and the roles of the U1 snRNP and PTBP1, which presumably remain bound to the Stx1b intron until it is degraded. This study demonstrates a new mechanism for a splicing regulator to control the overall expression of a gene. In this case, the splicing choice is coupled to nuclear RNA decay processes rather than cytoplasmic NMD (Figure 1d).
Splicing as a determinant of cytoplasmic mRNA utilization
Alterations in 5’ and 3’ UTRs allow changes in how the mRNA from a gene is subsequently utilized by the translation machinery [83-86]. Alternative 5’ and 3’ UTRs are often produced through changes in transcription initiation site, or the choice of polyadenylation site, but can also involve changes in spliceosome assembly. Moreover, as seen in viral transcripts, some mRNAs containing introns can be exported to the cytoplasm. Besides altering coding of the mRNA, these introns may serve other roles in neurons. Neurons maintain a specialized system for transporting mRNAs along dendritic processes to synapses. These localized mRNAs contain special sequences, or zipcodes, that mediate their dendritic targeting and once transported are under translational control in response to synaptic transmission [87] [88]. MRNA localization elements are frequently found in 5’ or 3’ UTRs. However, unspliced introns have been observed in dendritic RNA. A recent report found that the dendritic localization of Gria3, Gria4 and Grik1 transcripts may rely on retained intron sequences [89]. The ID element class of SINE retrotransposons is enriched in these retained-introns and may serve as the cis-acting localization element. However, another study found that unlike the more characterized RNA localization elements, an ID element was not sufficient to mediate transport of a fused EGFP reporter mRNA to dendrites in brain, suggesting the presence of additional regulatory elements in these RNAs [90]. How these intron-containing cytoplasmic transcripts escape the nucleus is not clear. Factors serving an equivalent function to HIV Rev may act on these RNAs to bypass normal controls on maturation. However, a recent study demonstrated a new mRNA export pathway in Drosophila that involves budding through the nuclear envelope of assembled RNA granules destined for dendritic localization [91]. There is precedent in mammalian cells for similar nuclear budding in the maturation of herpes viruses [92]. Thus, intron containing mRNAs may circumvent normal controls on export using this pathway. Also interesting is what role the intron-retained RNAs play once they are at their final destination. Spliceosomal components have not been convincingly found in the dendrite. Thus, once out of the nucleus, mRNAs are unlikely to undergo further splicing, at least as mediated by the spliceosome. However, fully spliced mRNAs are also present in the dendrite, along with those containing introns. If the introns contain the localization elements, these transcripts may serve to target fully spliced mRNAs to dendrites when they are packaged into the same mRNA granule. Such hitchhiking of one RNA with another capable of targeting is well documented for localized Oskar mRNAs in the Drosophila embryo [93-95]. The partially spliced mRNAs may encode modified protein products or may be subject to mRNA decay through NMD or other mechanisms. The assembly of different mRNAs into a transport granule implied by hitchhiking opens up many additional possibilities for changes in mRNA structure to determine cytoplasmic mRNA fate.
Besides intron retention, it is likely that other patterns of alternative splicing also affect the sorting of transcripts to distinct subcellular compartments or to different translational fates. Alternative polyA sites that determine different 3’ UTR lengths are commonly seen to alter regulation by miRNAs [85, 86]. These sites have been observed to change during neuronal differentiation and with neuronal activity [96-98]. Changes in splicing that alter 5’ and 3’ UTRs are also common. These examples highlight that choices made during the nuclear maturation of an mRNA strongly influence how all subsequent processes act upon it.
Concluding remarks
These recent studies have illuminated new means for alternative splicing choices to determine gene function. They underscore what was clear from earlier work – that splicing regulation affects nearly all aspects of neuronal development and physiology. This work adds new questions to many long-standing ones regarding neuronal mRNA metabolism. The precise role of individual splice variants is still only known for a few transcripts. Understanding how splicing changes protein and cellular function will require extensive physiological and genetic studies of individual gene products. The role of splicing regulatory networks in the overall biology of excitable cells is also not known. It is clear that there are coordinated changes in splicing driven by individual regulators, and that these changes occur at precise developmental transitions or in response to particular stimuli. But why individual transcripts need to be coregulated and how they contribute to a coordinated change in cellular function is not understood. On a more precise level of development, how do changes in splicing contribute to neuronal subtype identity? The diversity of protein structures created by splicing is enormous. Complex changes in isoform expression have the potential to tune individual cells to match their precise functions, but it is not understood how this might be important in the development of different kinds of cells or in defining their later function within circuits. Add these questions about the biology of splicing to the longstanding ones regarding the integrated mechanisms controlling mRNA maturation, export, localization, translation and decay, and it is clear that we will be finding fascinating new connections between RNA biology and neuronal cell biology for quite some time.
Highlights.
Alternative splicing coupled to mRNA decay can control gene expression.
Choice of splicing pattern can also determine later mRNA utilization.
These aspects of splicing regulation profoundly affect neuronal cell biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Amara SG, et al. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982;298:240–244. doi: 10.1038/298240a0. [DOI] [PubMed] [Google Scholar]
- 2.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan Q, et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature genetics. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 5.Buljan M, et al. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Molecular cell. 2012;46:871–883. doi: 10.1016/j.molcel.2012.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis JD, et al. Tissue-specific alternative splicing remodels protein-protein interaction networks. Molecular cell. 2012;46:884–892. doi: 10.1016/j.molcel.2012.05.037. [DOI] [PubMed] [Google Scholar]
- 7.Barash Y, et al. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 8.Barbosa-Morais NL, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science New York, N Y. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- 9.Castle JC, et al. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nature genetics. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grabowski PJ, Black DL. Alternative RNA splicing in the nervous system. Progress in neurobiology. 2001;65:289–308. doi: 10.1016/s0301-0082(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 11.Merkin J, et al. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science New York, N Y. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park JW, Graveley BR. Complex alternative splicing. Advances in experimental medicine and biology. 2007;623:50–63. doi: 10.1007/978-0-387-77374-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annual review of biochemistry. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 14.Braunschweig U, et al. Dynamic Integration of Splicing in the Cell Nucleus. Cell. 2013 doi: 10.1016/j.cell.2013.02.034. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nature reviews Molecular cell biology. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabut M, et al. SnapShot: The splicing regulatory machinery. Cell. 2008;133:192, e191. doi: 10.1016/j.cell.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 17.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nature reviews Genetics. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nature reviews Genetics. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Norris AD, Calarco JA. Emerging Roles of Alternative Pre-mRNA Splicing Regulation in Neuronal Development and Function. Front Neurosci. 2012;6:122. doi: 10.3389/fnins.2012.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keren H, et al. Alternative splicing and evolution: diversification, exon definition and function. Nature reviews Genetics. 2010;11:345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 21.Matlin AJ, et al. Understanding alternative splicing: towards a cellular code. Nature reviews Molecular cell biology. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 22.Modafferi EF, Black DL. Combinatorial control of a neuron-specific exon. Rna. 1999;5:687–706. doi: 10.1017/s1355838299990155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, et al. VERSE: a varying effect regression for splicing elements discovery. Journal of computational biology : a journal of computational molecular cell biology. 2012;19:855–865. doi: 10.1089/cmb.2012.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. Rna. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang C, et al. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science New York, N Y. 2010;329:439–443. doi: 10.1126/science.1191150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, et al. Neuronal regulation of alternative pre-mRNA splicing. Nature reviews. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- 27.Benjamin PR, Burke JF. Alternative mRNA splicing of the FMRFamide gene and its role in neuropeptidergic signalling in a defined neural network. BioEssays : news and reviews in molecular, cellular and developmental biology. 1994;16:335–342. doi: 10.1002/bies.950160508. [DOI] [PubMed] [Google Scholar]
- 28.Sommer B, et al. Flip and flop: a cell-specific functional switch in glutamateoperated channels of the CNS. Science New York, N Y. 1990;249:1580–1585. doi: 10.1126/science.1699275. [DOI] [PubMed] [Google Scholar]
- 29.Buck LB, et al. Alternative splicing in individual Aplysia neurons generates neuropeptide diversity. Cell. 1987;51:127–133. doi: 10.1016/0092-8674(87)90017-1. [DOI] [PubMed] [Google Scholar]
- 30.Lynch KW. Regulation of alternative splicing by signal transduction pathways. Advances in experimental medicine and biology. 2007;623:161–174. doi: 10.1007/978-0-387-77374-2_10. [DOI] [PubMed] [Google Scholar]
- 31.Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nature reviews Molecular cell biology. 2004;5:727–738. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- 32.Liu G, et al. A conserved serine of heterogeneous nuclear ribonucleoprotein L (hnRNP L) mediates depolarization-regulated alternative splicing of potassium channels. The Journal of biological chemistry. 2012;287:22709–22716. doi: 10.1074/jbc.M112.357343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iijima T, et al. SAM68 regulates neuronal activity-dependent alternative splicing of neurexin-1. Cell. 2011;147:1601–1614. doi: 10.1016/j.cell.2011.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emerick MC, et al. Profiling the array of Ca(v)3.1 variants from the human T-type calcium channel gene CACNA1G: alternative structures, developmental expression, and biophysical variations. Proteins. 2006;64:320–342. doi: 10.1002/prot.20877. [DOI] [PubMed] [Google Scholar]
- 35.Black DL, Grabowski PJ. Alternative pre-mRNA splicing and neuronal function. Progress in molecular and subcellular biology. 2003;31:187–216. doi: 10.1007/978-3-662-09728-1_7. [DOI] [PubMed] [Google Scholar]
- 36.Bark C. SNAP-25 and gene-targeted mouse mutants. Ann N Y Acad Sci. 2009;1152:145–153. doi: 10.1111/j.1749-6632.2008.04009.x. [DOI] [PubMed] [Google Scholar]
- 37.Beffert U, et al. Modulation of synaptic plasticity and memory by Reelin involves differential splicing of the lipoprotein receptor Apoer2. Neuron. 2005;47:567–579. doi: 10.1016/j.neuron.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Hattori D, et al. Robust discrimination between self and non-self neurites requires thousands of Dscam1 isoforms. Nature. 2009;461:644–648. doi: 10.1038/nature08431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raj B, et al. Cross-regulation between an alternative splicing activator and a transcription repressor controls neurogenesis. Molecular cell. 2011;43:843–850. doi: 10.1016/j.molcel.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 40.Yano M, et al. Nova2 regulates neuronal migration through an RNA switch in disabled-1 signaling. Neuron. 2010;66:848–858. doi: 10.1016/j.neuron.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker BS. Sex in flies: the splice of life. Nature. 1989;340:521–524. doi: 10.1038/340521a0. [DOI] [PubMed] [Google Scholar]
- 42.McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends in biochemical sciences. 2008;33:385–393. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 43.Lareau LF, et al. The coupling of alternative splicing and nonsense-mediated mRNA decay. Advances in experimental medicine and biology. 2007;623:190–211. doi: 10.1007/978-0-387-77374-2_12. [DOI] [PubMed] [Google Scholar]
- 44.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nature reviews Genetics. 2012;13:246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang L, Wilkinson MF. Regulation of nonsense-mediated mRNA decay. Wiley interdisciplinary reviews RNA. 2012;3:807–828. doi: 10.1002/wrna.1137. [DOI] [PubMed] [Google Scholar]
- 46.Tange TO, et al. The ever-increasing complexities of the exon junction complex. Current opinion in cell biology. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 47.Le Hir H, Seraphin B. EJCs at the heart of translational control. Cell. 2008;133:213–216. doi: 10.1016/j.cell.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 48.Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Current opinion in cell biology. 2005;17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 49.Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Current opinion in cell biology. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 50.Ni JZ, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes & development. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hyvonen MT, et al. Polyamine-regulated unproductive splicing and translation of spermidine/spermine N1-acetyltransferase. Rna. 2006;12:1569–1582. doi: 10.1261/rna.39806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boutz PL, et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes & development. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spellman R, et al. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Molecular cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valacca C, et al. Sam68 regulates EMT through alternative splicing-activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. The Journal of cell biology. 2010;191:87–99. doi: 10.1083/jcb.201001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dredge BK, Jensen KB. NeuN/Rbfox3 nuclear and cytoplasmic isoforms differentially regulate alternative splicing and nonsense-mediated decay of Rbfox2. PloS one. 2011;6:e21585. doi: 10.1371/journal.pone.0021585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Makeyev EV, et al. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Molecular cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rossbach O, et al. Auto- and cross-regulation of the hnRNP L proteins by alternative splicing. Molecular and cellular biology. 2009;29:1442–1451. doi: 10.1128/MCB.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keppetipola N, et al. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit Rev Biochem Mol Biol. 2012;47:360–378. doi: 10.3109/10409238.2012.691456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sheng M, Hoogenraad CC. The postsynaptic architecture of excitatory synapses: a more quantitative view. Annual review of biochemistry. 2007;76:823–847. doi: 10.1146/annurev.biochem.76.060805.160029. [DOI] [PubMed] [Google Scholar]
- 60.El-Husseini AE, et al. PSD-95 involvement in maturation of excitatory synapses. Science New York, N Y. 2000;290:1364–1368. [PubMed] [Google Scholar]
- 61.Elias GM, et al. Differential trafficking of AMPA and NMDA receptors by SAP102 and PSD-95 underlies synapse development. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20953–20958. doi: 10.1073/pnas.0811025106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng S, et al. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nature neuroscience. 2012;15:381–388. doi: 10.1038/nn.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eom T, et al. NOVA-dependent regulation of cryptic NMD exons controls synaptic protein levels after seizure. eLife Sciences. 2013;2 doi: 10.7554/eLife.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ule J, Darnell RB. Functional and mechanistic insights from genome-wide studies of splicing regulation in the brain. Advances in experimental medicine and biology. 2007;623:148–160. doi: 10.1007/978-0-387-77374-2_9. [DOI] [PubMed] [Google Scholar]
- 65.Jo T, et al. Voltage-gated sodium channel expressed in cultured human smooth muscle cells: involvement of SCN9A. FEBS letters. 2004;567:339–343. doi: 10.1016/j.febslet.2004.04.092. [DOI] [PubMed] [Google Scholar]
- 66.Sangameswaran L, et al. A novel tetrodotoxin-sensitive, voltage-gated sodium channel expressed in rat and human dorsal root ganglia. The Journal of biological chemistry. 1997;272:14805–14809. doi: 10.1074/jbc.272.23.14805. [DOI] [PubMed] [Google Scholar]
- 67.Buckanovich RJ, et al. Nova, the paraneoplastic Ri antigen, is homologous to an RNA-binding protein and is specifically expressed in the developing motor system. Neuron. 1993;11:657–672. doi: 10.1016/0896-6273(93)90077-5. [DOI] [PubMed] [Google Scholar]
- 68.Gehman LT, et al. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nature genetics. 2011;43:706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O’Brien JE, et al. Rbfox proteins regulate alternative splicing of neuronal sodium channel SCN8A. Molecular and cellular neurosciences. 2012;49:120–126. doi: 10.1016/j.mcn.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zubovic L, et al. Mutually exclusive splicing regulates the Nav 1.6 sodium channel function through a combinatorial mechanism that involves three distinct splicing regulatory elements and their ligands. Nucleic acids research. 2012;40:6255–6269. doi: 10.1093/nar/gks249. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Weischenfeldt J, et al. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes & development. 2008;22:1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Black DL, Zipursky SL. To cross or not to cross: alternatively spliced forms of the Robo3 receptor regulate discrete steps in axonal midline crossing. Neuron. 2008;58:297–298. doi: 10.1016/j.neuron.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Giorgi C, et al. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 74.Legrain P, Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 75.Takemura R, et al. Multiple factors in the early splicing complex are involved in the nuclear retention of pre-mRNAs in mammalian cells. Genes to cells : devoted to molecular & cellular mechanisms. 2011;16:1035–1049. doi: 10.1111/j.1365-2443.2011.01548.x. [DOI] [PubMed] [Google Scholar]
- 76.Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nature reviews Molecular cell biology. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 77.Pollard VW, Malim MH. The HIV-1 Rev protein. Annual review of microbiology. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 78.Cullen BR. Nuclear mRNA export: insights from virology. Trends in biochemical sciences. 2003;28:419–424. doi: 10.1016/S0968-0004(03)00142-7. [DOI] [PubMed] [Google Scholar]
- 79.Sun S, et al. SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nature structural & molecular biology. 2010;17:306–312. doi: 10.1038/nsmb.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu Q, et al. Intron-3 retention/splicing controls neuronal expression of apolipoprotein E in the CNS. J Neurosci. 2008;28:1452–1459. doi: 10.1523/JNEUROSCI.3253-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yap K, et al. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes & development. 2012;26:1209–1223. doi: 10.1101/gad.188037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schmid M, Jensen TH. Nuclear quality control of RNA polymerase II transcripts. Wiley interdisciplinary reviews RNA. 2010;1:474–485. doi: 10.1002/wrna.24. [DOI] [PubMed] [Google Scholar]
- 83.Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rojas-Duran MF, Gilbert WV. Alternative transcription start site selection leads to large differences in translation activity in yeast. Rna. 2012;18:2299–2305. doi: 10.1261/rna.035865.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Di Giammartino DC, et al. Mechanisms and consequences of alternative polyadenylation. Molecular cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lutz CS, Moreira A. Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. Wiley interdisciplinary reviews RNA. 2011;2:23–31. doi: 10.1002/wrna.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xing L, Bassell GJ. mRNA localization: an orchestration of assembly, traffic and synthesis. Traffic. 2013;14:2–14. doi: 10.1111/tra.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Buckley PT, et al. Cytoplasmic intron sequence-retaining transcripts can be dendritically targeted via ID element retrotransposons. Neuron. 2011;69:877–884. doi: 10.1016/j.neuron.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Khanam T, et al. Can ID repetitive elements serve as cis-acting dendritic targeting elements? An in vivo study. PloS one. 2007;2:e961. doi: 10.1371/journal.pone.0000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Speese SD, et al. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell. 2012;149:832–846. doi: 10.1016/j.cell.2012.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Johnson DC, Baines JD. Herpesviruses remodel host membranes for virus egress. Nature reviews Microbiology. 2011;9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 93.Jambor H, et al. Dimerization of oskar 3’ UTRs promotes hitchhiking for RNA localization in the Drosophila oocyte. Rna. 2011;17:2049–2057. doi: 10.1261/rna.2686411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marchand V, et al. An intracellular transmission control protocol: assembly and transport of ribonucleoprotein complexes. Current opinion in cell biology. 2012;24:202–210. doi: 10.1016/j.ceb.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 95.Hachet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428:959–963. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- 96.Flavell SW, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hilgers V, et al. ELAV mediates 3’ UTR extension in the Drosophila nervous system. Genes & development. 2012;26:2259–2264. doi: 10.1101/gad.199653.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mansfield KD, Keene JD. Neuron-specific ELAV/Hu proteins suppress HuR mRNA during neuronal differentiation by alternative polyadenylation. Nucleic acids research. 2012;40:2734–2746. doi: 10.1093/nar/gkr1114. [DOI] [PMC free article] [PubMed] [Google Scholar]


