Abstract
Over a decade ago, the fMRI Data Center (fMRIDC) pioneered open-access data sharing in the task-based functional neuroimaging community. Well ahead of its time, the fMRIDC effort encountered logistical, sociocultural and funding barriers that impeded the field-wise instantiation of open-access data sharing. In 2009, ambitions for open-access data sharing were revived in the resting state functional MRI community in the form of two grassroots initiatives: the 1000 Functional Connectomes Project (FCP) and its successor, the International Neuroimaging Datasharing Initiative (INDI). Beyond providing open access to thousands of clinical and non-clinical imaging datasets, the FCP and INDI have also demonstrated the feasibility of large-scale data aggregation for hypothesis generation and testing. Yet, the success of the FCP and INDI should not be confused with widespread embracement of open-access data sharing. Reminiscent of the challenges faced by fMRIDC, key controversies persist and include participant privacy, the role of informatics, and the logistical and cultural challenges of establishing an open science ethos. We discuss the FCP and INDI in the context of these challenges, highlighting the promise of current initiatives and suggesting solutions for possible pitfalls.
Keywords: Open-access, open science, fMRI, R-fMRI, database, informatics, neuroinformatics
1. An Initial Call to Arms for Data Sharing
Over a decade ago, the fMRI Data Center (fMRIDC) pioneered open-access data sharing in the neuroimaging community. It’s objective was “speeding the progress and the understanding of cognitive processes and the neural substrates that underlie them” (Van Horn and Gazzaniga, 2002; Van Horn et al., 2004; Van Horn et al., 2001). The fMRIDC specifically aimed to establish a publicly accessible repository of peer-reviewed fMRI studies that contained all data necessary to interpret, analyze, and replicate the deposited studies. The creators of fMRIDC argued that such a resource not only enabled investigators to confirm and extend the findings of published studies, but it also facilitated meta-analytic and discovery science approaches, while providing broad training opportunities. To promote this type of open science culture, Michael Gazzaniga, then Editor-in-Chief of the Journal of Cognitive Neuroscience, made the decision to require all authors who were publishing in the Journal to deposit their data in fMRIDC and encouraged other journals to adopt the same policy. However, this ambitious policy was not uniformly embraced by the fMRI community and instead sparked enduring controversy (Nature Neuroscience Editorial, 2000; Nature Opinion, 2000).
Concerns regarding the fMRIDC highlighted two key obstacles to open-access data sharing that remain relevant to this day. First, critics questioned whether the field possessed the necessary technological or manpower resources to support the broad sharing of fMRI datasets. Depending on study design and sample size, storage of data from a single fMRI study can require 50MB to 30+GB, raising concerns about data storage, databasing needs, and how to distribute datasets of such scale (e.g., ftp-servers vs. shipping discs or hard-drives). The fMRIDC team was well equipped to handle large-scale datasets internally, having access to sufficient storage space (almost a petabyte combining spinning disks and tape backup) and ample processing power. However, in the year 2000, most internet technology was not capable of handling the transfer of large-scale datasets and the majority of end users possessed 30–40GB of storage space at most. The lack of standardization in imaging data storage formats, specification of experimental designs, and stimulus definitions for task-based fMRI further increased skepticism regarding the feasibility of the model proposed by the fMRIDC. The question of how to scale such efforts without marked innovation in informatics and standardization has remained open (Gardner et al., 2003; Gazzaniga et al., 2006; Koslow, 2000; Poline et al., 2012; The Governing Council of the Organization for Human Brain Mapping, 2001).
Besides logistical concerns, the prospect of open-access data sharing raised a host of sociocultural concerns (Gardner et al., 2003; Hirschfeld, 2012; Koslow, 2000; Pearce and Smith, 2011; Visscher and Weissman, 2011). These discomforts included: fears about loss of competitive advantage; concerns about how to recognize the value of data sharing in terms of promotion (e.g., tenure requirements) or grant impact factor scores; the possibility that the field would become mired in disputes regarding specific analyses in published papers; and “secret” fears that one’s data may be found to have contained embarrassing errors (Gardner et al., 2003; Milham, 2012; Poline et al., 2012; Visscher and Weissman, 2011). Beyond these concerns lay core questions: Should data sharing be optional or mandatory? When should sharing occur? Should data sharing be comprehensive or selective? Should data sharing efforts be required to use specific informatics platforms? Who should take the lead in advancing data sharing? The potential agents for resolving these issues include the agencies funding data collection, the researchers obtaining the data, the institutions employing researchers, and the journals publishing the findings. Gazzaniga and colleagues de facto proposed that journals should take the lead, but with the ensuing controversy and practical obstacles, other journals have not followed this example. As with the logistical concerns surrounding data sharing, these sociocultural issues remain unresolved (Poline et al., 2012; Visscher and Weissman, 2011).
Despite these controversies, the visionary and bold fMRIDC effort succeeded in gathering and sharing thousands of datasets from 2000 until its funding was discontinued in 2007. Even now the fMRIDC datasets remain available for download. Unable to achieve sustainable neuroimaging data sharing, the fMRIDC pioneered the idea and as the fMRIDC effort came to a close, a series of informatics-driven voluntary data-sharing initiatives began to emerge, powered by the Extensible Neuroimaging Archive Toolkit (XNAT) - an open source imaging informatics platform (e.g., Brainscape [http://www.brainscape.org/], OASIS [http://www.oasis-brains.org/], and XNAT Central [http://central.xnat.org/]; Marcus et al., 2010; Marcus et al., 2011; Marcus et al., 2007a; Marcus et al., 2007b). The XNAT-based initiatives sought to facilitate data sharing by providing researchers with tools for data management, quality assurance tasks, and data uploads. Their efforts avoided the controversies sparked by fMRIDC, but they did not evoke strong responses by investigators other than those directly involved in their development.
Against this background, two grassroots efforts towards open-access sharing of R-fMRI datasets emerged: the 1000 Functional Connectomes Project (FCP) and its successor the International Neuroimaging Data-sharing Initiative (INDI). We will briefly introduce the goals and design of each of these initiatives, and then review their progress, challenges and sustainability.
1.1. The 1000 Functional Connectomes Project
How reproducible is resting state fMRI across imaging centers? This question was initially posed by FCP co-founders Bharat Biswal and Michael P. Milham at the 1st Biennial Conference on Resting State Brain Connectivity, held in Magdeburg, Germany in 2008. They presented comparisons of results obtained across 5 imaging sites. Other investigators responded with enthusiasm by offering to contribute data as well as asking how they could access the aggregate data.
To guide the establishment of a consortium and set initial policies, an international steering committee was formed: Bharat Biswal, Randy Buckner, James Hyde, Rolf Kotter [deceased], Michael Milham (coordinating secretary), Marcus Raichle, Arno Villringer, and Yu-Feng Zang. The first major decision was the selection of a data sharing model. Committee members considered various options, ranging from “pay to play” (i.e., you must give data to receive data) and proposal curation (i.e., researchers must apply for permission to conduct an analysis, subject to coordination and approval of a publication committee), to unrestricted data sharing. Doubting the utility and practicality of more restrictive models, the committee unanimously decided in favor of unrestricted open-access sharing. Accordingly, on December 11th, 2009, the FCP publicly released data collected from over 1300 international participants at 30 international sites via the Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC; http://www.nitrc.org). Table 1 lists the available datasets in the FCP.
Table 1.
Available Open-Access Data Contributions in the FCP as of August 1 2012.
| Data Sample | Principal Investigators(s) |
|---|---|
| Ann Arbor, MI, USA | Christopher S. Monk, Rachael D. Seidler, Scott J. Peltier |
| Atlanta, GA, USA | Helen S. Mayberg |
| Baltimore, MD, USA | James J. Pekar, Stewart H. Mostofsky |
| Bangor, UK | Stan Colcombe |
| Beijing, China | Yu-Feng Zang |
| Berlin, Germany | Daniel Margulies |
| Cambridge, MA, USA | Randy L. Buckner |
| Cleveland, OH, USA | Mark J. Lowe |
| Dallas, TX, USA | Bart Rypma |
| Durham, NC, USA | David J. Madden |
| International Consortium for Brain Mapping (ICBM) | Allan C. Evans |
| Leiden, Netherlands | Serge A.R.B. Rombouts |
| Leipzig, Germany | Arno Villringer |
| Milwaukee, WI, USA | Shi-Jiang Li |
| Munich, Germany | Christian Sorg, Valentin Riedl |
| New Haven, CT, USA | Michelle Hampson |
| Newark, NJ, USA | Bharat B. Biswal |
| New York City, NY, USA | Michael Milham, F. Xavier Castellanos |
| Ontario, Canada | Peter Williamson |
| Oulu, Finland | Vesa J. Kiviniemi, Juha Veijola |
| Oxford, UK | Steve M. Smith, Clare Mackay |
| Palo Alto, CA, USA | Michael Greicius |
| Pittsburgh, PA, USA | Greg Siegle |
| Queensland, Australia | Katie McMahon |
| Saint Louis, MO, USA | Bradley L. Schlaggar, Steven E. Petersen |
| Taipei, Taiwan | Ching-Po Lin |
The response to the launch of the 1000 Functional Connectomes Project (http://www.fcon_1000.projects.nitrc.org) was gratifying, demonstrating the eagerness of investigators to access large-scale datasets and the feasibility of providing such data in an unrestricted manner. In the first two weeks, nearly 1000 website visits originated from 290 cities in 42 countries; by six months this had grown to 6600 visits from 1066 cities in 74 countries. Currently, the FCP and INDI repositories attract about 500 hits per week. Figures 1 and 2 illustrate the worldwide distribution and the number of weekly site visits. Almost three years later, some datasets have been downloaded more than 2000 times, with most exceeding 500 downloads. This broad interest was already translated into 38 publications using some or all of the FCP datasets (see Supplementary Information).
Figure 1.
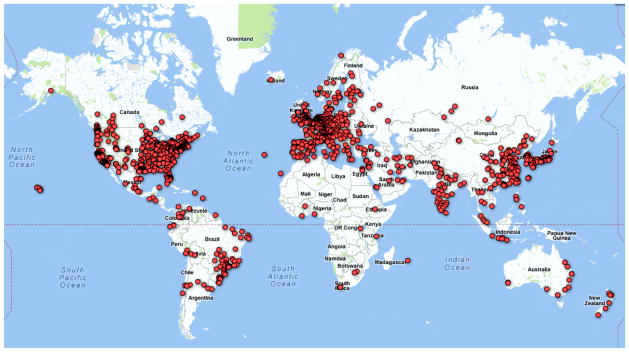
Overview of the origin of visitors (website hits) to the FCP and INDI repositories.
Figure 2.
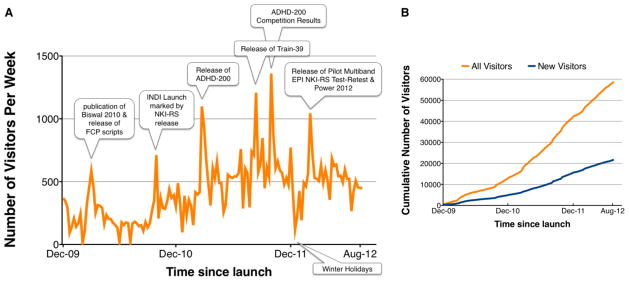
Number of visitors to the FCP and INDI repositories since their launch. A. Number of visitors (website hits) per week to the FCP and INDI repositories. Peaks in the number of website hits are relatable to specific FCP/INDI releases or events. B. Cumulative number of visitors (website hits) to the FCP and INDI repositories since their launch. While substantial numbers of new visitors continue to accrue, many users return repeatedly to the repository for more data or additional information.
Beyond demonstrating the feasibility of open data sharing, the inaugural FCP paper illustrated its scientific value (Biswal et al., 2010). Specifically, the aggregate FCP dataset revealed the striking universality of the brain’s intrinsic functional architecture, including stable loci of variation, detectable across participants and imaging sites. These findings were made all the more remarkable by the lack of prior coordination in data collection methods (scan parameters, etc.). Furthermore, although marked site-related variations were observed, statistically robust relationships with age and sex could be discerned, demonstrating that such site-related variation does not preclude data-exploration and effective discovery.
1.2. The International Neuroimaging Data-sharing Initiative
Although successful in providing a large-scale demonstration of the feasibility and scientific value of pooling and sharing R-fMRI data, the initial FCP effort was limited by the relative paucity of phenotypic information released with imaging datasets (only age, sex and handedness were provided). Most labs collect a wealth of information about their participants using a variety of measures, particularly those of psychological or clinical interests. The availability of such phenotypic information is necessary for the neuroscientific community’s efforts to rapidly identify links between inter-individual variations in the human connectome and behavioral phenotypes (Kelly et al., 2012). Equally important, the initial release was limited to presumed “healthy” individuals - thus precluding the generation of inferences about clinical populations from the FCP datasets. As such, it remains a challenge to make the aggregation and sharing of well-phenotyped imaging datasets a cultural norm.
In an effort to urge the R-fMRI community to tackle the challenge of sharing phenotypically rich datasets, including those from clinical populations, Michael P. Milham founded the International Neuroimaging Data-sharing Initiative (INDI), building on the FCP. An overview of datasets released through INDI can be found in Tables 2 and 3. INDI’s first goal was to test the feasibility of making more extensively phenotyped datasets available. Working towards this aim, INDI re-released the Cleveland and Beijing Normal datasets from the original FCP, however, this time the release included physiological data for the Cleveland dataset and both DTI and IQ scores for the Beijing sample. Similarly, the Nathan Kline Institute (NKI) Rockland Sample released a pilot “lifespan” sample that comprised R-fMRI and DTI scans from over 200 participants (ages: 6–85 years old) with accompanying data from more than 30 behavioral questionnaires, diagnostic psychiatric interviews and laboratory measures (e.g., hemogram, metabolic panel, lipid panels).
Table 2.
Available Open-access Data Contributions in INDI Retrospective as of August 1 2012.
| Data Sample | Principal Investigator(s) | Sample Description |
|---|---|---|
| ADHD-200 | Jan Buitelaar F. Xavier Castellanos Daniel Dickstein Damien Fair David Kennedy Beatriz Luna Michael P. Milham Stewart Mostofsy Joel Nigg Julie B. Schweitzer Katerina Velanova Yu-Feng Wang Yu-Feng Zang |
776 resting-state fMRI and anatomical datasets aggregated across 8 independent imaging sites, 491 of which were obtained from typically developing individuals and 285 from children and adolescents with ADHD. Data includes ADHD symptom measures, demographic information, IQ scores, and lifetime medication status. |
| Beijing Enhanced | Yu-Feng Zang | 180 healthy controls from a community sample at Beijing Normal University in China. The data include IQ scores for a subset of participants (n=55) and a 64 directions DTI scan for all participants. |
| North Shore - LIJ | Ashesh Mehta Stephan Bickel Laszlo Entz |
6 patients with medically intractable epilepsy underwent implantation of intracranial electrodes for seizure onset localization prior to respective neurosurgery. Each participant received a 5-minute resting state scan 1 to 7 days before electrode implantation. |
| NYU Institute for Pediatric Neuroscience -Cocaine | Clare Kelly Michael Milham Adriana Di Martino Maarten Mennes F. Xavier Castellanos |
Data from 29 cocaine-dependent individuals and 24 healthy comparison participants to study structural and functional connectivity in cocaine addiction. |
| Cleveland CCF | Mark Lowe Erik Beall Michael Phillips |
31 adult controls. Data includes resting state scans and physiological measures (heart rate and breathing). |
| Train-39 | Art Kramer Michelle Voss Kirk Erickson Ruchika Prakash |
39 young adults trained on video game, Space Fortress. 13 functional scans and demographic information were acquired from each participant in order to assess how the brain acquires skills relevant to complex tasks. |
| Power 2012 | Jonathan Power Kelly Anne Barnes Avi Snyder Brad Schlaggar Steve Petersen |
Subjects used in the 2012 NeuroImage article by Power et al., entitled “Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion”. 77 children, adolescents, and adult controls. Data includes full scale IQ scores, MPRAGE and R-fMRI scans |
Table 3.
Available Open-Access Data Contributions in INDI Prospective as of August 1 2012.
| Data Sample | Principal Investigator(s) | Sample Description |
|---|---|---|
|
Beijing Eyes Open Eyes Closed Study |
Yu-Feng Zang | 48 healthy controls from a community sample from Beijing Normal University in China. Each participant has 3 resting state fMRI scans. During the first scan participants were instructed to rest with their eyes closed. The second and third resting state scan were randomized between resting with eyes open versus eyes closed. This dataset also contains a 64-direction DTI scan for every participant. Dataset Completed. |
| Beijing: Short TR Study | Yu-Feng Zang | 28 healthy, college-aged participants. Data includes long and short TR R-fMRI, MPRAGE, DTI, and demographic information. Dataset Completed. |
| NKI/Rockland Sample | Bharat Biswal F. Xavier Castellanos Barbara Coffey Stan Colcombe David Guilfoyle Matthew Hoptman Dan Javitt Harold S. Koplewicz Bennet Leventhal Larry Maayan Maarten Mennes Michael Milham Kate Nooner Nunzio Pomara |
207 psychiatrically evaluated individuals from a community sample. Phenotypic measures include intelligence testing, psychiatric diagnostic interview, executive function performance testing, and a battery of psychiatric, cognitive, and behavioral assessments. Neuroimaging data include resting state fMRI, DTI, and MPRAGE. Dataset Completed. |
| NYU Institute for Pediatric Neuroscience Sample | Michael Milham Adriana Di Martino Clare Kelly Maarten Mennes F. Xavier Castellanos |
49 psychiatrically screened individuals (ages 6 to 55 years old). Data includes R-FMRI, MPRAGE, DTI, demographic information, and IQ measures. Anticipated release frequency: 25 – 50 individuals quarterly. |
| Virginia Tech Carilion Research Institute | Cameron Craddock Stephen LaConte The Neuro Bureau |
25 psychiatrically screened individuals (ages 18 to 65 years old) from community sample. Data includes T1 weighted anatomical scan and two R-fMRI scans. Anticipated release frequency: 25+ participants quarterly. |
| Berlin School of Mind and Brain | Daniel Margulies Arno Villinger The Neuro Bureau |
50 individuals (ages 18 to 60 years old) from a community sample. Data includes two resting state scans, MPRAGE scan, and demographic information for all participants. A subset of participants completed the ICS and PANAS affective behavior scales. Anticipated release of data from 25+ participants quarterly. |
| The Quiron-Valencia Sample | Luis Marti-Bonmati Maria de la Iglesia Vaya The Spanish Resting State Network |
45 participants from a community sample. Data includes MPRAGE and R-fMRI scans and demographic information. Anticipated release frequency: 25+ participants quarterly. |
| The NKI-RS Multiband Imaging Test-Retest Pilot Dataset | Bharat Biswal F. Xavier Castellanos Barbara Coffey Stan Colcombe David Guilfoyle Matthew Hoptman Dan Javitt Harold S. Koplewicz Bennet Leventhal Larry Maayan Maarten Mennes Michael Milham Kate Nooner Nunzio Pomara |
24 participants (scanned twice, one week apart) from a community sample. Data includes MPRAGE, R-fMRI, multiband DTI, breath hold, eye movement, visual stimulation scans and demographic information. Anticipated release frequency: 25+ participants quarterly. |
Additionally, INDI aims to promote the sharing of datasets from a broad array of clinical populations. The collection of large well-powered clinical samples will be particularly labor intensive and costly due to the challenges of recruiting and properly assessing clinical populations, of which prevalence can range between < 0.1% and 28.8% (Kessler et al., 2005). This is especially true for fields such as child and adolescent psychiatry, where stigma and misconceptions surrounding research participation must be overcome and challenges to data collection and integrity are often encountered in the scanning environment (e.g., anxiety, inability to follow instructions, hyperkinesis). Rather than being dependent on a slow and imperfect process of synthesizing results via meta-analyses of small studies, INDI makes it feasible to aggregate the raw imaging data, if investigators are willing to make them available for such community-based efforts.
INDI chose to promote not only the sharing of single-study clinical samples, but also the sharing of large-scale clinical datasets using the aggregation model of the FCP. The first of these datasets was released in 2011 by the ADHD-200 Consortium (http://fcon_1000.projects.nitrc.org/indi/adhd200). Comprised of 8 independent imaging sites, the Consortium aggregated and released a sample consisting of previously collected R-fMRI scans, anatomical scans and accompanying phenotypic data for 571 typically developing children and 351 children with Attention Deficit Hyperactivity Disorder (ADHD). Again, the research protocols at the contributing sites were developed independently, without prior coordination. As such, differences in clinical assessment and imaging protocols, as well as the application and interpretation of diagnostic criteria, undoubtedly exist. Despite these challenges, scientific results have already begun to emerge from the composite clinical sample (e.g., Tomasi and Volkow, 2012). In August 2012, the Autism Brain Imaging Data Exchange (ABIDE; http://fcon_1000.projects.nitrc.org/indi/abide/), will release data via INDI that includes about 500 individuals with autism, and a comparable number of matched typically developing controls. ABIDE will take advantage of greater standardization of research diagnoses in the autism community to provide a wealth of phenotypic measures with considerable overlap among contributing sites (Gotham et al., 2009; Gotham et al., 2008; Lord et al., 2000).
Beyond the gathering and sharing of published or archived datasets, as promoted by the fMRIDC and FCP initiatives, INDI has advanced a model for prospective, pre-publication data sharing (Birney et al., 2009). Investigators are invited to contribute pre-publication datasets as collection occurs using a variety of distribution schedules (e.g., weekly, monthly, quarterly; see Table 3). Undoubtedly the most radical of the INDI initiatives, it allows researchers to prospectively contribute their data in part or in whole. This allows for the retention of some degree of appropriate exclusivity, even as they “work in the open.” Speeding the availability of datasets is expected to minimize potentially wasteful redundancies, optimize opportunities for harmonization across labs, and ensure that data are shared while they retain their greatest value (i.e., while the imaging methods are still considered state-of-the-art). Additionally, the phenotypically rich imaging data enables testing of emerging hypotheses via replication – an essential process that is too often neglected due to the increasing emphasis on novelty in grant evaluations.
The prototype for the prospective data sharing effort launched by INDI was the pilot Nathan S. Kline Institute for Psychiatric Research-Rockland Sample (NKI-RS). During 2011, the NKI-RS distributed over 200 deeply phenotyped R-fMRI and DTI datasets via weekly uploads. Designed to reflect the interests of the many NKI investigators, the NKI-RS includes a variety of phenotypic measures. By sharing the data as well as the data collection protocols, this effort makes it possible to design other initiatives that include shared measures, thus accelerating the potential for the field to achieve large, aggregated datasets in a cost effective manner. As discussed below, funding agencies will play an important role in determining whether such efforts will continue to emerge by valorizing efforts that generate data used by the broader community to address questions beyond those initially posed by the original principal investigators (e.g., Biomedical Informatics Research Network [BIRN], Functional BIRN, Alzheimer’s Disease Neuroimaging Initiative [ADNI], National Institutes of Health [NIH] MRI Study of Normal Brain Development, the Enhanced NKI-Rockland Sample, and the Human Connectome Project [HCP]).
2. Towards Sustainable Data Sharing
From our perspective, the three key, outstanding issues with regard to open-access data sharing are: (1) Respect for participant privacy and confidentiality; (2) Logistics of data accessibility; and (3) Long-term sustainability. In the following sections we discuss initial solutions employed by the FCP and INDI to address these issues with the goal of facilitating the evolution of sustainable data sharing.
2.1. Protecting Privacy in An Open Science Community
Individuals around the world voluntarily participate in research protocols to advance scientific understanding and its practical applications. At the core of this contract is the promise of privacy and respect of confidentiality –with respect to both an individual’s enrollment in a specific study and any information obtained in the course of participation. The most salient risk of open-access data sharing is the very real potential of breaching participant privacy (Navarro, 2008). Image headers can contain a wealth of information about participants (e.g., name or identification code, date of birth, scan date, weight, etc.) some of which can be used to identify specific individuals. Additionally, high-resolution structural images can enable identification via 3D facial reconstruction.
Realizing the need to minimize the potential for breach of privacy as the sine qua non of open-access data sharing, the FCP steering committee agreed to full anonymization of all datasets in accordance with the U. S. Health Insurance Portability and Accountability Act (HIPAA). Specifically, the 18 types of protected health information (PHI) identified by HIPAA (Gunn et al., 2004) are removed from all datasets prior to upload to the FCP site for distribution. The general consensus is that once fully de-identified in compliance with HIPAA, a dataset is no longer considered to be subject to the same rules governing human research (Freymann et al., 2012). With that said, a few local ethics boards have required investigators to re-consent participants to obtain explicit agreement that their data may be released in any form, even if the data are fully de-identified and anonymized (the coding algorithm is destroyed so no links can be traced between the released data and personal identifiers). This inconsistency reflects a need for more explicit guidance by oversight and funding agencies including the US National Institutes of Health (NIH), the National Science Foundation (NSF), and their international counterparts. Additionally, it highlights the need for researchers around the world to adjust their consent process immediately to inform participants that their brain imaging and phenotypic data may be shared, whether in the short run, or one or more years after study completion and publication of initial findings.
In some cases full HIPAA de-identification may not be possible. The Enhanced NKI-Rockland Sample in INDI, for example, uses a community ascertained recruitment strategy in which all participants must be residents of Rockland County, NY. This specification of location beyond resolution at the level of a state is not consistent with HIPAA. Also, there is concern that releasing highly dimensional datasets increases the possibility of identification, especially as the number of included phenotypic variables rises. The Netflix competition (http://www.netflixprize.com and http://www.wikipedia.org/wiki/Netflix_Prize) provides an example of such possibility, as some competitors were able to identify individuals using anonymous datasets. Although more cumbersome, data usage agreements, such as those specified by the National Database For Autism Research (NDAR) or the Alzheimer’s Disease Neuroimaging Initiative (ADNI), permit sharing of datasets which are not fully de-identified by imposing restrictions on prospective users. Data usage agreements are intended to facilitate research and must be enhanced to protect participant privacy, while not impeding specific research efforts.
2.2. Improving Mechanisms for Data Contribution and Distribution
An optimal sharing platform provides a powerful, easy-to-use database that is readily searchable, while automatically maintaining quality control and data integrity (Poline et al., 2012). At the same time, the platform should make it easy for researchers to share data, ideally through a simple one-click upload, with automatic data verification thereafter (Poline et al., 2012). In 2009 when the 1000 Functional Connectomes Project was formed, several informatics and databasing resources were in their early phases of development (e.g., XNAT, HID). However, none were suitable for the task at hand. Like the fMRIDC, the FCP elected to compensate for the lack of informatics infrastructure by using manpower to facilitate data contribution, preparation and distribution. Hard-drives and compact discs were received from contributing sites around the world. Their contents were extracted and organized into an easy to use directory structure employing a common NIFTI-based image format. Accompanying data were simply stored as .csv files. The organized data were then combined by imaging site into large archive files (.tar) that could be easily downloaded by users. For data distribution, the FCP steering committee considered a variety of options. The option ultimately selected was to use the Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC; http://www.nitrc.org), which was an open science resource supported by the NIH Blueprint for Neuroscience Research program. Although NITRC had not been primarily intended for data sharing, its file upload/download interface for software made setup as a data repository relatively straightforward. Importantly, rapid turn-around user support and news forums facilitated dialogues between FCP organizers and users.
Like the fMRIDC, the FCP/INDI model has strived for simplicity while incorporating technological advances. However, it remains to be seen whether such a simple model is sustainable. The FCP/INDI represents a largely unfunded effort. The workload is directly proportional to the number of datasets contributed. If the entire imaging community actually starts sharing data, the current data preparation and organization system will be overwhelmed. Widespread imaging data sharing will be of little use, if it results in a massive warehouse of spreadsheets and imaging datasets for which there is no efficient means of orderly aggregation. Moreover, the challenges of data organization will increase as the phenotypical richness of the datasets expands (e.g., Enhanced NKI-RS and Brain Genomic Superstruct). The mixed success of equivalent efforts in genetics (e.g., dbGaP [http://www.ncbi.nlm.nih.gov/gap/]; Mailman et al., 2007) underscores this point (Walker et al., 2011; Wooten and Huggins, 2011). Finally, data hosting is not without cost. NIH funding for NITRC absorbed the cost of data hosting for the FCP and INDI, but this funding has very definite limits.
Much like a decade ago, most users are not ready to handle phenotypically rich datasets in an orderly and efficient matter. For instance, the soon to be released Enhanced NKI-RS will contain more than 1000 phenotypic variables. Such growth in complexity poses new challenges. Fortunately, solutions are already underway. NITRC has recently established NITRC-IR, an XNAT based platform that is currently hosting copies of both the 1000 Functional Connectomes Project and the ADHD-200 datasets in a database format (http://www.nitrc.org/ir/). Perhaps most exciting is the upcoming INDI release of the Autism Brain Imaging Data Exchange (ABIDE; http://fcon_1000.projects.nitrc.org/indi/abide/; Di Martino et al., in preparation). Faced with the challenge of sharing over 500 datasets from children with autism spectrum disorders and a matching number of typically developing children, with 30–50 phenotypic variables per imaging site, the INDI team contacted the leading informatics platforms in the field (COINS: http://coins.mrn.org/, Scott et al., 2011; LORIS: http://cbrain.mcgill.ca/loris, Das et al. 2012; LONI IDA; http://pipeline.loni.ucla.edu/; NITRC-IR/XNAT: http://www.nitrc.org/ir/). They each agreed to simultaneously host the ABIDE datasets prepared by the INDI team. Such coordination is intended to give users an opportunity to sample emerging technologies and encourage investigators to explore the value of these platforms not only for sharing, but also for their own internal infrastructure. Again, the NKI-Rockland Sample provides an informative model: The initial NKI-RS effort was carried out using paper and pencil, with data stored in the usual mix of Excel and SPSS style .csv files. In contrast, the Enhanced NKI-RS effort relies primarily on web-based data collection for phenotyping, using the COINS system for data capture and integration.
When laboratories adopt such informatics platforms, the process of data sharing will be substantially improved. Ideally, these platforms will include one-click upload buttons that send data to a central facility for organization, quality control and open-access hosting (Poline et al., 2012). However, the ability to completely automate data sharing may not be possible at the moment, as visual inspection remains a key step in quality control (i.e., a quarantine and inspection process prior to release). Nonetheless, the expected reduction in errors and increases in local and overall efficiency make such a development an important goal.
2.3. Prioritizing a Culture of Open Science: Mandates, Funding and Credits1
To the extent that the FCP/INDI can be characterized as having succeeded, our efforts challenge the 2009 dismissal of the fMRIDC and open data sharing as a “failed experiment” (Friston, 2009). Beyond the various data releases that now constitute the FCP and INDI repositories, a number of ancillary initiatives by INDI and its partners have served to further promote an open science agenda in the imaging community (see Table 4). More than ever, discussions about open science and data sharing are taking place in the pages of premier scientific journals, with increasing enthusiasm and insistence (Akil et al., 2011; Boulton, 2012; Buckner, 2010; Ghosh et al., 2012; Khamsi, 2012; Milham, 2012; Pastrana, 2010; Piwowar, 2011).
Table 4.
FCP and INDI ancillary community events to promote open science.
| Date | Community Event | More information |
|---|---|---|
| March 2010 | Release of the FCP scripts | Release of the processing scripts that were used for the analysis presented in Biswal et al. 2010. The scripts were optimized for the data structure of the FCP, but can be easily applied to a user’s specific data (http://fcon_1000.projects.nitrc.org). |
| March 2011 | Launch of the ADHD-200 competition | Global Competition launched by the ADHD-200 Consortium to accompany release of the ADHD-200 datasets. The competition required participants to train diagnostic classifiers using the available ADHD-200 datasets. Three months later, in June 2011, new, unlabeled data were released and participants had to use their classifiers to predict the ADHD status of the new datasets. Twenty-one international teams submitted their diagnostic predictions for scoring. Results were announced in October 2011 and the winners were recognized by NIDA at the 2011 annual Society for Neuroscience pre-conference symposium. (http://fcon_1000.projects.nitrc.org/indi/adhd200/results.html) |
| April 2011 | Release of the ADHD-200 datasets preprocessed by the NeuroBureau | The Neuro Bureau (http://www.neurobureau.org) is a forum and collaborative initiative that supports open neuroscience by promoting the sharing of ideas, data, and methods across disciplines. They preprocessed the complete ADHD-200 dataset and publicly re-released the preprocessed data. That way, users (and participants in the ADHD-200 competition) with restricted computing power are still able to use the data. |
| May 2011 | Launch of the Child Mind Institute Librarian Initiative | Comprehensive hand-vetted and sorted reference libraries for various literatures, including Resting State fMRI and Diffusion Tensor Imaging are distributed via Mendeley, with monthly updates. (http://www.mendeley.com/profiles/cmi-librarian/) |
| June 2011 | The NeuroBureau Brain Art Exhibition at the annual meeting of the Organization for Human Brain Mapping in Quebec, Canada | To expand the horizon of brain scientists, the NeuroBureau, supported by the Child Mind Institute and Stavros Niarchos Foundation, organized an exhibition including art pieces related to the brain and the mind. The exhibition was hosted at the annual meeting of the OHBM in Quebec, Canada, a meeting attended by over 2000 neuroimagers from across the world. |
| June 2011 | The NeuroBureau Brain Art Competition | Related to the brain art exhibition, the NeuroBureau invited neuroimagers to submit their most beautiful figures or graphical representations for a ‘Brain Art’ competition. More than 30 neuroimagers participated in the competition. The Brain Art competition, and the Cirque Du Cerveau Gala and award ceremony were supported by the Child Mind Institute and Stavros Niarchos Foundation. (http://neurobureau.projects.nitrc.org/BrainArt/Competition.html; http://neurobureau.projects.nitrc.org/BrainArt/Awards_Venue.html) |
| June 2012 | 2nd NeuroBureau Brain Art Exhibition at the annual meeting of the Organization for Human Brain Mapping in Beijing, China | Following the successful 2011 Brain Art Exhibition, the Neurobureau, again supported by the Child Mind Institute and the Stavros Niarchos Foundation, organized the second edition of the Brain Art Exhibition at the 2012 annual meeting of the OHBM in Beijing, China, |
| June 2012 | 2nd NeuroBureau Brain Art Competition | In the second edition of the Brain Art competition, over 30 neuroimagers participated in the categories: best abstract, best human connectome, best educational, most humorous, and best video representation of the brain. The Brain Art competition was supported by the Child Mind Institute and Stavros Niarchos Foundation. |
| July-August 2012 | Launch of the INDI Summer of Sharing | The 2012 INDI Summer of Sharing is an effort dedicated to the sustenance and acceleration of data and analytic resource sharing among imaging community members. It bundles a series of efforts including data releases, processing pipeline releases, as well as a links to other sharing initiatives (http://fcon_1000.projects.nitrc.org/indi/summerofsharing2012.html). |
| August 2012 | Release of the C-PAC processing pipeline | The Configurable Pipeline for the Analysis of Connectomes (C-PAC) is a configurable plug-and-play Nipype-based pipeline package developed by the INDI team to accomplish a broad array of resting-state fMRI analyses. |
| September 2012 | BrainHack Unconference | The 2012 BrainHack Unconference was organized by the NeuroBureau and is aimed at seeding open, collaborative projects in neuroimaging (http://www.brainhack.org). The unconference took the form of a 3-day hackaton and was centered on data from the FCP and INDI. Over 50 international neuroimagers participated. |
At the same time, successful demonstrations of open-access data sharing should not be confused with the fact that the imaging community is still far from having truly implemented open-access data sharing, or the broader agenda of open science (e.g., open analytics). Considering what will be necessary to achieve a fully open science culture in the imaging community returns us to the key question raised by the fMRIDC: How do we adopt open science as a priority for our community? At present, open-access data sharing is appreciated by many, but supported by a “Gang of the Few” (Milham, 2012). The efforts of these few researchers are laudable. However, it is important to understand that unless the larger community adopts the practice of open-access data sharing, the breadth of available datasets will remain limited, and due to a lack of representativeness potential biases can be introduced into findings obtained with the shared datasets.
It is our belief that funding agencies and scientific journals must work together to advance the implementation of an open science agenda in the imaging community. The maintenance and sharing of data is costly. Funding agencies must approach this responsibility seriously by providing the additional costs associated with open science, with the long-term perspective of excellent return on investment. This may be a hard proposition at a time of economic stringency but, in the long term, this will save money that can be put to use in generating much needed data that can be readily used by many in the open science model. Still, the mere distribution of funds to support open science is not enough. Funding agencies must track scientific impact beyond simple citation numbers. This can be done by monitoring how the data and analytical methods are made available by individual investigators and then used by the broader community to actually advance our understanding of brain function and disease states. This can be reinforced by using such metrics for evaluation when competing continuations are refunded (i.e., favoring refunding grants that successfully shared their prior data). Similarly, for new grants, review criteria should be expanded to consider the potential value of the data generated for the broader community – beyond the hypotheses or specific aims of the proposing investigator(s).
To move this effort forward, funding agencies will have to decide whether to promote the implementation of data sharing by incentivization or mandates (Tenopir et al., 2011). The NIMH and NIDA have issued administrative grant supplements to encourage investigators to share data (Gardner et al., 2003; Kennedy, 2003), although the metrics and where data should be deposited remain unclear. Likewise, the NIH especially mandates principal investigators with grants over $500.000 to share their data, yet release frequency, dates or formats are not specified. Mandated sharing already exists in molecular genetics and for certain grants relating to autism spectrum disorders (e.g., NDAR: http://ndar.nih.gov/). On the other hand, mandates raise complex issues. For example, does all phenotypic data need to be shared along with the imaging data, or only a common set of data elements? Should there be a time-limited embargo before open data sharing must occur? Or, should data sharing be coincident with publication? And most importantly, is a mandate to openly share imaging datasets sufficient to implement open sharing? Arguably, mandates for investigators to employ agency approved informatics systems and obtain proper consent for sharing (present or future) are necessary and less invasive steps towards the successful implementation of sharing in the imaging field.
Beyond the facilitative role of funding agencies journal play a key role in advancing an open science culture in the imaging community (Nature Opinion, 2002). As shown by Gazzaniga and colleagues, journals can require the sharing of any datasets used in a given publication. Once a seemingly radical idea (Committee on Responsibilities of Authorship in the Biological Sciences National Research Council, 2003), researchers are increasingly realizing the potential for such measures to increase the quality and impact of a publication by facilitating the replication of findings between groups in a cost effective manner, and by making truly precise meta-analyses possible. Digital object identifiers (DOI’s) are potentially attractive means of tracking shared data and facilitating the crediting of data sharing for researchers. Importantly, measures to credit contributions to open science initiatives (e.g., data, analytic tools) will only be of value if academic institutions reform themselves to actively encourage and reward participation in open science (e.g., in determination of faculty requirements, departmental funding allocations, tenure decisions). In sum, journals are well positioned to encourage or enforce data sharing in the community, though only if they take on this mission in concert with funding agencies and academic institutions – otherwise the field will re-experience fMRIDC’s fate.
3. Conclusions
The fMRIDC provided an initial model for open-access data sharing within the neuroimaging community. Arguably ahead of its time, the community did not embrace the fMRIDC efforts due to a variety of logistical and sociocultural concerns. A decade after the launch of the fMRIDC, the FCP and INDI have provided definitive demonstrations of the utility of data sharing, beyond the purposes of replication. They highlight the feasibility of forming large-scale datasets capable of accelerating the pace of neuroscientific and psychiatric discovery while also facilitating methodological innovation. Increasingly well-received by the imaging community, open-access data sharing still faces many practical challenges. Informatics platforms need to reach the level of functionality necessary to support large-scale data sharing. Likewise, researchers must be incentivized to use informatics platforms rather than more commonplace and rudimentary data management approaches. Simultaneously, privacy policies must be formally agreed upon by leading funding agencies in order to balance privacy protection against the immense value of open-access data sharing. Researchers need to incorporate explicit consent for future sharing in their informed consent process. Most importantly, funding agencies, journals and research institutions will have to more clearly prioritize open science, through a combination of incentives, mandates and credits. Despite these enduring challenges, the FPC/INDI efforts have surpassed our expectations in terms of user response, suggesting the community is amenable to adopting open-access data sharing as a model for conducting competitive science. In the meantime, we conclude with our favorite exhortation: Share that brain!
Supplementary Material
Highlights.
The FCP and INDI reinvigorated open-access data sharing in the neuroimaging community
Open-access data sharing is far from universally accepted in the fMRI community
Researchers must be incentivized to use informatics platforms
Researchers need to obtain explicit participant consent for future data sharing
Funding agencies, journals and research institutions need to prioritize open science
Acknowledgments
We would like to thank the FCP steering committee and all the contributors to the 1000 Functional Connectomes Project and INDI (including the ADHD-200 and ABIDE consortiums). Their vision and contributions have made these efforts successful. The FCP and INDI are eternally grateful to the NITRC team for their hosting and web support, particularly Christian Haselgrove, David Kennedy, and Nina Preuss. We would like to give special thanks to: Randy Buckner for his helpful discussions with Michael P. Milham during the conceptualization of INDI; Randy Buckner for suggesting the “1000 Connectomes Project” as a name for the initiative, and a helpful reviewer of Biswal et. al. (2010) for motivating us to add the word ‘Functional’; Yu-Feng Zang for leading the FCP and INDI in terms of dataset contributions; Xi-Nian Zuo for his many efforts in the analyses for the 1000 Functional Connectome Project Consortium paper (Biswal et al., 2010); and David Van Essen and Avi Snyder for ad-hoc contributions to the FCP Steering Committee. Finally, we would like to thank Ayesha Anwar, Cameron Craddock, Caitlin Hinz, Adriana Di Martino, Clare Kelly, Bennet Leventhal, Daniel Lurie, Jack Van Horn, Chao-Gan Yan, and Zhen Yang for helpful comments on earlier versions of this manuscript.
Current financial support for the INDI team is provided by gifts from Joseph P. Healy and the Stavros Niarchos Foundation to the Child Mind Institute, and an endowment provided to the NYU Child Study Center by Phyllis Green and Randolph Cowen. Additional current support includes NIMH awards to MPM (R03 MH096321, R01MH094639) and FXC (R01MH083246). Past support to MPM was provided by the Leon Levy Foundation and to FXC by the Stavros Niarchos Foundation. NITRC is funded by an NIH Blueprint for Neurosciences Research (neuroscienceblueprint.nih.gov) contract to TCG, Inc.
Footnotes
Michael P. Milham (MPM) was a participant in the NIH Blueprint Workgroup for Neuro-Image Data Sharing, where many of the issues raised in this section were discussed in detail. The viewpoints expressed here were undoubtedly shaped by MPM’s participation in the event. As such, the authors would like to express their appreciation of the event and properly acknowledge it.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akil H, Martone ME, Van Essen DC. Challenges and Opportunities in Mining Neuroscience Data. Science. 2011;331:708–712. doi: 10.1126/science.1199305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Hudson TJ, Green ED, Gunter C, Eddy S, Rogers J, Harris JR, Ehrlich SD, Apweiler R, Austin CP, Berglund L, Bobrow M, Bountra C, Brookes AJ, Cambon-Thomsen A, Carter NP, Chisholm RL, Contreras JL, Cooke RM, Crosby WL, Dewar K, Durbin R, Dyke SOM, Ecker JR, El Emam K, Feuk L, Gabriel SB, Gallacher J, Gelbart WM, Granell A, Guarner F, Hubbard T, Jackson Sa, Jennings JL, Joly Y, Jones SM, Kaye J, Kennedy KL, Knoppers BM, Kyrpides NC, Lowrance WW, Luo J, MacKay JJ, Martín-Rivera L, McCombie WR, McPherson JD, Miller L, Miller W, Moerman D, Mooser V, Morton CC, Ostell JM, Ouellette BFF, Parkhill J, Raina PS, Rawlings C, Scherer SE, Scherer SW, Schofield PN, Sensen CW, Stodden VC, Sussman MR, Tanaka T, Thornton J, Tsunoda T, Valle D, Vuorio EI, Walker NM, Wallace S, Weinstock G, Whitman WB, Worley KC, Wu C, Wu J, Yu J. Prepublication data sharing. Nature. 2009;461:168–170. doi: 10.1038/461168a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal BB, Mennes M, Zuo XN, Gohel S, Kelly C, Smith SM, Beckmann CF, Adelstein JS, Buckner RL, Colcombe S, Dogonowski AM, Ernst M, Fair D, Hampson M, Hoptman MJ, Hyde JS, Kiviniemi VJ, Kotter R, Li SJ, Lin CP, Lowe MJ, Mackay C, Madden DJ, Madsen KH, Margulies DS, Mayberg HS, McMahon K, Monk CS, Mostofsky SH, Nagel BJ, Pekar JJ, Peltier SJ, Petersen SE, Riedl V, Rombouts SA, Rypma B, Schlaggar BL, Schmidt S, Seidler RD, Siegle GJ, Sorg C, Teng GJ, Veijola J, Villringer A, Walter M, Wang L, Weng XC, Whitfield-Gabrieli S, Williamson P, Windischberger C, Zang YF, Zhang HY, Castellanos FX, Milham MP. Toward discovery science of human brain function. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton G. Open your minds and share your results. Nature. 2012;486:441. doi: 10.1038/486441a. [DOI] [PubMed] [Google Scholar]
- Buckner RL. Human functional connectivity: New tools, unresolved questions. Proceedings of the National Academy of Sciences. 2010;107:10769–10770. doi: 10.1073/pnas.1005987107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Committee on Responsibilities of Authorship in the Biological Sciences National Research Council. Sharing Publication-Related Data and Materials: Responsibilities of Authorship in the Life Sciences. The National Academies Press; 2003. [PubMed] [Google Scholar]
- Das S, Zijdenbos AP, Harlap J, Vins D, Evans AC. LORIS: a web-based data management system for multi-center studies. Frontiers in neuroinformatics. 2012;5:37. doi: 10.3389/fninf.2011.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freymann JB, Kirby JS, Perry JH, Clunie DA, Jaffe CC. Image data sharing for biomedical research--meeting HIPAA requirements for De-identification. Journal of digital imaging: the official journal of the Society for Computer Applications in Radiology. 2012;25:14–24. doi: 10.1007/s10278-011-9422-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ. Modalities, modes, and models in functional neuroimaging. Science. 2009;326:399–403. doi: 10.1126/science.1174521. [DOI] [PubMed] [Google Scholar]
- Gardner D, Toga AW, Ascoli GA, Beatty JT, Brinkley JF, Dale AM, Fox PT, Gardner EP, George JS, Goddard N, Harris KM, Herskovits EH, Hines ML, Jacobs GA, Jacobs RE, Jones EG, Kennedy DN, Kimberg DY, Mazziotta JC, Miller PL, Mori S, Mountain DC, Reiss AL, Rosen GD, Rottenberg DA, Shepherd GM, Smalheiser NR, Smith KP, Strachan T, Van Essen DC, Williams RW, Wong ST. Towards effective and rewarding data sharing. Neuroinformatics. 2003;1:289–295. doi: 10.1385/NI:1:3:289. [DOI] [PubMed] [Google Scholar]
- Gazzaniga MS, Van Horn JD, Bloom F, Shepherd GM, Raichle M, Jones E. Continuing progress in neuroinformatics. Science. 2006;311:176. doi: 10.1126/science.311.5758.176a. [DOI] [PubMed] [Google Scholar]
- Ghosh SS, Klein A, Avants B, Millman KJ. Learning from open source software projects to improve scientific review. Frontiers in computational neuroscience. 2012;6:18. doi: 10.3389/fncom.2012.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotham K, Pickles A, Lord C. Standardizing ADOS scores for a measure of severity in autism spectrum disorders. Journal of autism and developmental disorders. 2009;39:693–705. doi: 10.1007/s10803-008-0674-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotham K, Risi S, Dawson G, Tager-Flusberg H, Joseph R, Carter A, Hepburn S, McMahon W, Rodier P, Hyman SL, Sigman M, Rogers S, Landa R, Spence MA, Osann K, Flodman P, Volkmar F, Hollander E, Buxbaum J, Pickles A, Lord C. A replication of the Autism Diagnostic Observation Schedule (ADOS) revised algorithms. Journal of the American Academy of Child and Adolescent Psychiatry. 2008;47:642–651. doi: 10.1097/CHI.0b013e31816bffb7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn PP, Fremont AM, Bottrell M, Shugarman LR, Galegher J, Bikson T. The Health Insurance Portability and Accountability Act Privacy Rule: a practical guide for researchers. Medical care. 2004;42:321–327. doi: 10.1097/01.mlr.0000119578.94846.f2. [DOI] [PubMed] [Google Scholar]
- Hirschfeld G. Open science: Data sharing is harder to reward. Nature. 2012;487:302. doi: 10.1038/487302c. [DOI] [PubMed] [Google Scholar]
- Kelly C, Biswal BB, Craddock RC, Castellanos FX, Milham MP. Characterizing variation in the functional connectome: promise and pitfalls. Trends in cognitive sciences. 2012;16:181–188. doi: 10.1016/j.tics.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy DN. Share and share alike. Neuroinformatics. 2003;1:211–213. doi: 10.1385/NI:1:3:211. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Archives of general psychiatry. 2005;62:593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- Khamsi R. Diagnosis by default. Nature medicine. 2012;18:338–340. doi: 10.1038/nm0312-338. [DOI] [PubMed] [Google Scholar]
- Koslow SH. Should the neuroscience community make a paradigm shift to sharing primary data? Nature neuroscience. 2000;3:863–865. doi: 10.1038/78760. [DOI] [PubMed] [Google Scholar]
- Lord C, Risi S, Lambrecht L, Cook EH, Jr, Leventhal BL, DiLavore PC, Pickles A, Rutter M. The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. Journal of autism and developmental disorders. 2000;30:205–223. [PubMed] [Google Scholar]
- Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, Popova N, Pretel S, Ziyabari L, Lee M, Shao Y, Wang ZY, Sirotkin K, Ward M, Kholodov M, Zbicz K, Beck J, Kimelman M, Shevelev S, Preuss D, Yaschenko E, Graeff A, Ostell J, Sherry ST. The NCBI dbGaP database of genotypes and phenotypes. Nature genetics. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Fotenos AF, Csernansky JG, Morris JC, Buckner RL. Open access series of imaging studies: longitudinal MRI data in nondemented and demented older adults. Journal of cognitive neuroscience. 2010;22:2677–2684. doi: 10.1162/jocn.2009.21407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Harwell J, Olsen T, Hodge M, Glasser MF, Prior F, Jenkinson M, Laumann T, Curtiss SW, Van Essen DC. Informatics and data mining tools and strategies for the human connectome project. Frontiers in neuroinformatics. 2011;5:4. doi: 10.3389/fninf.2011.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Olsen TR, Ramaratnam M, Buckner RL. The Extensible Neuroimaging Archive Toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007a;5:11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- Marcus DS, Wang TH, Parker J, Csernansky JG, Morris JC, Buckner RL. Open Access Series of Imaging Studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. Journal of cognitive neuroscience. 2007b;19:1498–1507. doi: 10.1162/jocn.2007.19.9.1498. [DOI] [PubMed] [Google Scholar]
- Milham MP. Open neuroscience solutions for the connectome-wide association era. Neuron. 2012;73:214–218. doi: 10.1016/j.neuron.2011.11.004. [DOI] [PubMed] [Google Scholar]
- Nature Neuroscience Editorial. A debate over fMRI data sharing. Nature Neuroscience. 2000;3:845–846. doi: 10.1038/78728. [DOI] [PubMed] [Google Scholar]
- Nature Opinion. Whose scans are they, anyway? Nature. 2000;406:443–443. doi: 10.1038/35020214. [DOI] [PubMed] [Google Scholar]
- Nature Opinion. How to encourage the right behaviour. Nature. 2002;416:1–1. doi: 10.1038/416001b. [DOI] [PubMed] [Google Scholar]
- Navarro R. An ethical framework for sharing patient data without consent. Informatics in primary care. 2008;16:257–262. doi: 10.14236/jhi.v16i4.701. [DOI] [PubMed] [Google Scholar]
- Pastrana E. Collective brain maps. Nature Methods. 2010;7:253–253. [Google Scholar]
- Pearce N, Smith A. Data sharing: not as simple as it seems. Environmental Health. 2011;10 doi: 10.1186/1476-069X-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwowar HA. Who shares? Who doesn’t? Factors associated with openly archiving raw research data. PloS one. 2011;6:e18657. doi: 10.1371/journal.pone.0018657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poline JB, Breeze JL, Ghosh S, Gorgolewski K, Halchenko YO, Hanke M, Haselgrove C, Helmer KG, Keator DB, Marcus DS, Poldrack RA, Schwartz Y, Ashburner J, Kennedy DN. Data sharing in neuroimaging research. Frontiers in neuroinformatics. 2012;6:9. doi: 10.3389/fninf.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A, Courtney W, Wood D, de la Garza R, Lane S, King M, Wang R, Roberts J, Turner JA, Calhoun VD. COINS: An Innovative Informatics and Neuroimaging Tool Suite Built for Large Heterogeneous Datasets. Frontiers in neuroinformatics. 2011;5:33. doi: 10.3389/fninf.2011.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenopir C, Allard S, Douglass K, Aydinoglu AU, Wu L, Read E, Manoff M, Frame M. Data sharing by scientists: practices and perceptions. PloS one. 2011;6:e21101. doi: 10.1371/journal.pone.0021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Governing Council of the Organization for Human Brain Mapping. Neuroimaging databases. Science. 2001;292:1673–1676. doi: 10.1126/science.1061041. [DOI] [PubMed] [Google Scholar]
- Tomasi D, Volkow ND. Abnormal functional connectivity in children with attention-deficit/hyperactivity disorder. Biol Psychiatry. 2012;71:443–450. doi: 10.1016/j.biopsych.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horn JD, Gazzaniga MS. Opinion: Databasing fMRI studies towards a ‘discovery science’ of brain function. Nature reviews Neuroscience. 2002;3:314–318. doi: 10.1038/nrn788. [DOI] [PubMed] [Google Scholar]
- Van Horn JD, Grafton ST, Rockmore D, Gazzaniga MS. Sharing neuroimaging studies of human cognition. Nature neuroscience. 2004;7:473–481. doi: 10.1038/nn1231. [DOI] [PubMed] [Google Scholar]
- Van Horn JD, Grethe JS, Kostelec P, Woodward JB, Aslam JA, Rus D, Rockmore D, Gazzaniga MS. The Functional Magnetic Resonance Imaging Data Center (fMRIDC): the challenges and rewards of large-scale databasing of neuroimaging studies. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2001;356:1323–1339. doi: 10.1098/rstb.2001.0916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher KM, Weissman DH. Would the field of cognitive neuroscience be advanced by sharing functional MRI data? BMC Med. 2011;9:34. doi: 10.1186/1741-7015-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L, Starks H, West KM, Fullerton SM. dbGaP data access requests: a call for greater transparency. Science translational medicine. 2011;3:113cm134. doi: 10.1126/scitranslmed.3002788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooten EC, Huggins GS. Mind the dbGAP: the application of data mining to identify biological mechanisms. Molecular interventions. 2011;11:95–102. doi: 10.1124/mi.11.2.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


