SUMMARY
Cytochrome c maturation (ccm) in many bacteria, archaea, and plant mitochondria requires eight membrane proteins, CcmABCDEFGH, called system I. This pathway delivers and attaches heme covalently to two cysteines (of Cys-Xxx-Xxx-Cys-His) in the cytochrome c. All models propose that CcmFH facilitates covalent attachment of heme to the apocytochrome; namely, that it is the synthetase. However, holocytochrome c synthetase activity has not been directly demonstrated for CcmFH. We report formation of holocytochromes c by CcmFH and CcmG, a periplasmic thioredoxin, independent of CcmABCDE (we term this activity CcmFGH-only). Cytochrome c produced in the absence of CcmABCDE is indistinguishable from cytochrome c produced by the full system I, with a cleaved signal sequence and two covalent bonds to heme. We engineered increased cytochrome c production by CcmFGH-only, with yields approaching those from the full system I. Three conserved histidines in CcmF (TM-His1, TM-His2, and P-His1) are required for activity, as are the conserved cysteine pairs in CcmG and CcmH. Our findings establish that CcmFH is the system I holocytochrome c synthetase. Although we discuss why this engineering would likely not replace the need for CcmABCDE in nature, these results provide unique mechanistic and evolutionary insights into cytochrome c biosynthesis.
Keywords: cytochrome c maturation, synthetase, heme trafficking, biosynthesis, pathway
INTRODUCTION
C-type cytochromes are heme proteins that carry out essential electron transfer reactions in organisms from every kingdom of life. These cytochromes function outside of the cytoplasmic membrane in prokaryotes, in the lumen of chloroplasts, and in the intermembrane space of mitochondria [for example, reviewed in (Allen, 2011; Hamel et al., 2009; Kranz et al., 2009; Mavridou et al., 2013; Sanders et al., 2010; Sawyer and Barker, 2012)]. Cytochromes c are characterized by covalent attachment between the heme and the apoprotein (via thioether linkages between the 2- and 4-vinyls of heme and two thiols of a conserved Cys-Xxx-Xxx-Cys-His motif in the apoprotein). Because c-type cytochromes are assembled at their site of action (separated from heme biosynthesis by a lipid bilayer), holocytochrome c formation poses unique challenges to heme trafficking and post-translational modification. Three major pathways (called systems I, II, and III) exist in nature to direct the covalent attachment of heme to cytochrome c (Kranz et al., 1998; Page et al., 1998). In many bacteria, plant and protozoal mitochondria, and archaea, holocytochrome c formation is carried out by the cytochrome c maturation (ccm) pathway, called system I, which comprises eight membrane proteins (in E. coli, CcmABCDEFGH) (Hamel et al., 2009; Kranz et al., 2009; Mavridou et al., 2013; Sanders et al., 2010; Sawyer and Barker, 2012).
At the center of heme trafficking in system I is the periplasmic heme chaperone protein, CcmE. CcmE forms a unique and well-studied covalent intermediate with heme, between a conserved histidine (His130, in E. coli CcmE) and the β carbon of the heme 2-vinyl (Lee et al., 2005; Stevens et al., 2003; Uchida et al., 2004). Heme in “holo” (heme bound) CcmE is transferred to the apocytochrome (putatively, by the action of CcmFH), but details of this aspect of system I are just emerging (San Francisco and Kranz, in press). CcmABCD (an ABC transporter complex) are involved in formation and release of holoCcmE, presumably now free to associate with CcmF (Christensen et al., 2007; Feissner et al., 2006a; Goldman et al., 1997; Schulz et al., 1999). CcmF forms an integral membrane complex with CcmH (Rayapuram et al., 2008; Ren et al., 2002; Richard-Fogal et al., 2009; Sanders et al., 2008), and is believed to be the site of thioether formation between the heme and the apocytochrome. CcmF contains a separate, stably bound non-covalent heme b (Richard-Fogal et al., 2009; San Francisco et al., 2011), which we have proposed is involved in reducing the incoming heme from holoCcmE (Kranz et al., 2009; Richard-Fogal et al., 2009; San Francisco et al., 2011). Reduced heme (Fe2+) is required for thioether formation with the apocytochrome (Barker et al., 1993; Nicholson and Neupert, 1989), and reduction of the heme in holoCcmE would also favor discharge of heme from the covalent His130 intermediate (Kranz et al., 2009; Richard-Fogal and Kranz, 2010; San Francisco et al., 2011). CcmG (Beckman and Kranz, 1993; Di Matteo et al., 2010; Ouyang et al., 2006) and CcmH (Di Matteo et al., 2007; Zheng et al., 2012) are membrane-tethered thioredox-active proteins that likely maintain the thiol groups of the apocytochrome (in the conserved Cys-Xxx-Xxx-Cys-His motif) in the reduced state (Meyer et al., 2005; Monika et al., 1997; Setterdahl et al., 2000; Turkarslan et al., 2008).
CcmF (in complex with CcmH) has been referred to extensively as the “cytochrome c heme lyase” or “cytochrome c synthetase” of system I, although the assignment of this function to CcmF is largely circumstantial. It is well-established that CcmF forms a complex with CcmH (Rayapuram et al., 2008; Ren et al., 2002; Richard-Fogal et al., 2009; Sanders et al., 2008), and there is evidence that CcmH interacts directly with the apocytochrome (Di Matteo et al., 2007; Di Silvio et al., 2013; Meyer et al., 2005; Verissimo et al., 2011). However, interaction(s) between holoCcmE (the protein assumed to deliver heme to site of thioether formation with the apocytochrome) and CcmFH have only recently been demonstrated (San Francisco and Kranz, in press). The absence of direct experimental evidence demonstrating that CcmFH is the cytochrome c synthetase thus constitutes a major gap in our understanding of the system I pathway.
Here, we report maturation of holocytochrome c by CcmFH and CcmG, in the absence of CcmABCDE. The cytochrome c produced by the CcmABCDE-independent pathway (here termed “CcmFGH-only”) is biochemically and spectroscopically indistinguishable from cytochrome c produced by the full system I. We engineer production of holocytochrome c by the CcmFGH-only pathway (by increasing cellular heme concentrations and expression of the apocytochrome and ccmFGH, by gene dosage) to levels approaching those of the full system I. Three of the conserved histidines in CcmF (TM-His1, TM-His2, and P-His1), as well as the conserved cysteine pairs in the thioredox proteins CcmG and CcmH, are required for holocytochrome formation by CcmFGH-only. These findings establish that CcmFH is the holocytochrome c synthetase for system I (with thioreduction mediated by CcmG). We discuss evolutionary and mechanistic implications, comparing CcmFGH-only with the system II cytochrome c synthetase, CcsBA.
RESULTS
CcmFH and CcmG can attach heme to cytochrome c in the absence of CcmABCDE
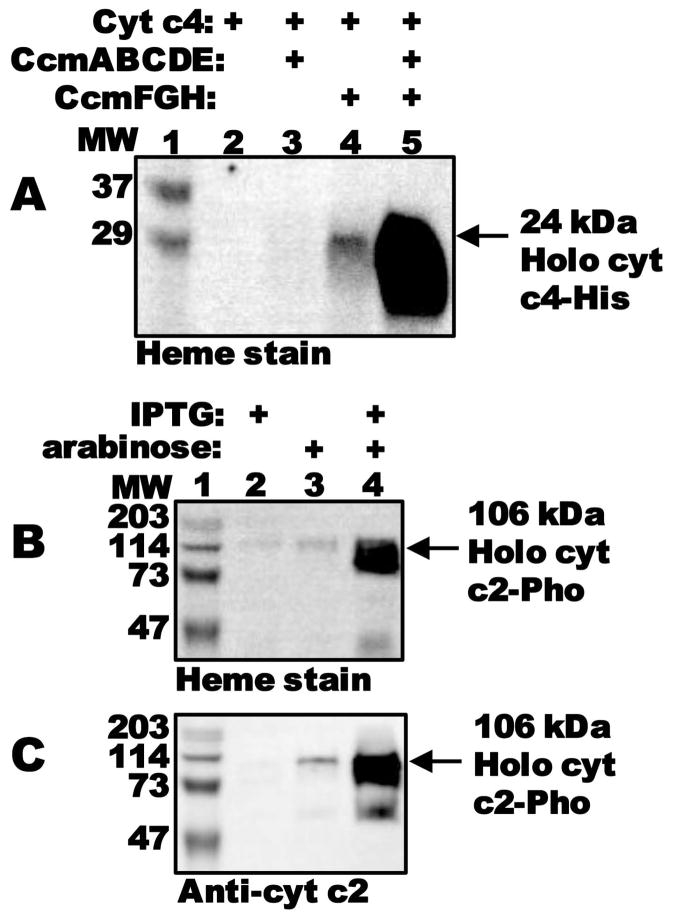
Conceptually, system I occurs in two steps: i) formation and release of holoCcmE (by CcmABCD), and ii) heme delivery to CcmFH (by holoCcmE), the putative site of holocytochrome c formation (hence, the “cytochrome c synthetase”). Studies in our lab (Feissner et al., 2006a; Goldman et al., 1997; Richard-Fogal and Kranz, 2010; Richard-Fogal et al., 2009) and others (Christensen et al., 2007; Ren and Thony-Meyer, 2001) have addressed step one, analyzing intermediates during formation of holoCcmE (such as the stable CcmCDE complex) and its release. However, less is known about the second step. To study the two steps independently, we engineered the IPTG-inducible pGEX with the genes encoding step one of the pathway, ccmABCDE, and an arabinose-inducible pBAD-based plasmid with the genes encoding step two, ccmFGH. Heme staining of BPER cell extracts revealed that expression of all Ccm proteins with the arabinose-inducible chromosmally-integrated cytochrome c4 (San Francisco et al., 2011), led to robust production of holocytochrome c4 (Fig 1A, lane 5). Note that, typically, only heme that is covalently bound remains with the cytochrome c after denaturing SDS-PAGE, and this heme is readily detectable by heme stain (Feissner et al., 2003). Surprisingly, we noticed that cells containing only CcmFGH (i.e., lacking ccmABCDE), also produced holocytochrome c4 (Fig 1A, lane 4). Thus, it appeared that holocytochrome formation could proceed in the absence of CcmABCDE, albeit, under these conditions, at levels less than one-tenth those when CcmABCDE were present. We confirmed chromosomal deletion of the ccmA-H operon in our E. coli Δccm strains (Feissner et al., 2006b; San Francisco et al., 2011) by genomic PCR (Fig S1).
Fig. 1.
CcmFGH attaches heme to cytochrome c in the absence of CcmABCDE. (A) Heme staining of BPER cell extracts showing synthesis of 24-kDa holocytochrome c4:His6 as a function of expression of separate Ccm components, as indicated. (B) Heme staining and (C) anti-cytochrome c2 immunoblot of BPER cell extracts showing synthesis of 106-kDa holocytochrome c2:Pho by CcmFGH as a function of induction with IPTG and/or arabinose. One hundred μg total protein was loaded in each lane. MW, molecular weight standards (lanes 1).
The holocytochrome c synthetase activity of CcmFGH-only is not limited to the di-heme cytochrome c4, but also extends to the mono-heme cytochrome c2 (Fig 1B and C, lane 4). The cytochrome c2 plasmid used here has an in-frame C-terminal fusion to E. coli alkaline phosphatase (Pho), expressed from an IPTG-inducible promoter (Beckman et al., 1992). We conclude that CcmFGH, in the absence of other Ccm components, can attach heme to two unrelated (other than the Cys-Xxx-Xxx-Cys-His motif) cytochrome c substrates. Furthermore, since alkaline phosphatase is secreted to the periplasm, covalent heme attachment to the cytochrome c2:Pho fusion protein must occur in the periplasm (i.e., where CcmFGH function).
Cytochrome c produced by CcmFGH-only is identical to cytochrome c produced by full system I
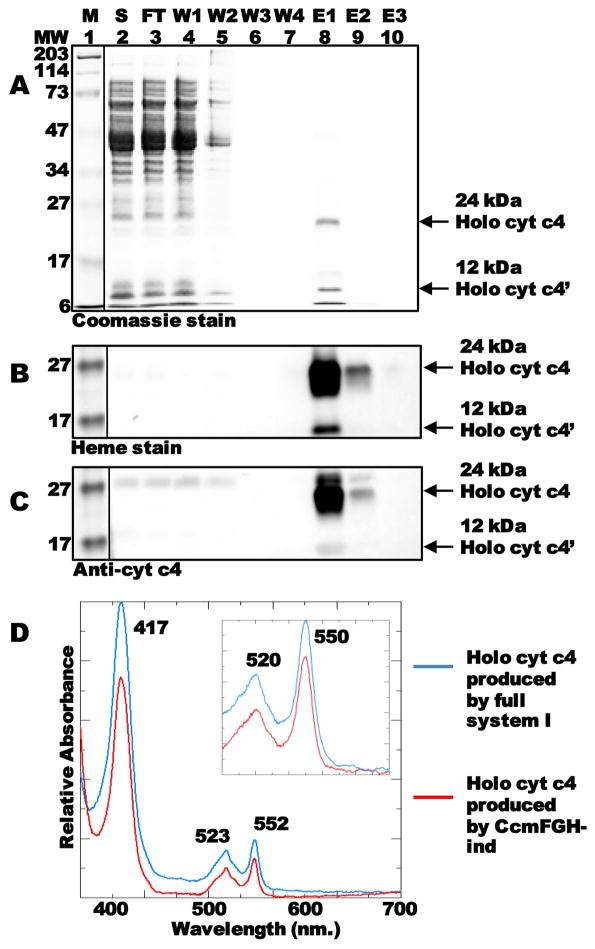
We wanted to further characterize the di-heme holocytochrome c4 produced by CcmFGH-only. To confirm proper periplasmic secretion (and cleavage of the periplasmic signal sequence), and to analyze the spectral properties of the cytochrome c produced by CcmFGH-only, we grew 1L cultures and purified hexahistidine-tagged holocytochrome c4 from the soluble fraction (Fig 2A–C). The full-length 24 kDa holocytochrome c4 and the proteolyzed 12 kDa holocytochrome c4′ were detectable by Coomassie blue staining of SDS-PAGE (Fig 2A, lane 8 “E1”). Note that it is common for endogeneous proteolysis of the 24 kDa cytochrome to occur, yielding two 12 kDa mono-heme forms (Feissner et al., 2006b); however, only the C-terminal of these products is purified (along with the full-length 24 kDa protein) since the hexahistidine tag is at the C-terminus of the protein. The purified 24 and 12 kDa forms contained covalent heme (Fig 2B, lane 8), and reacted with cytochrome c4 antisera (Fig 2C, lane 8). The UV-Vis absorption spectrum (Fig 2D, red line) and reduced pyridine hemochrome (Fig 2D, inset, red line) of the purified cytochrome c4 are consistent with two covalent attachments between the thiols of cytochrome c (in Cys-Xxx-Xxx-Cys-His) and the vinyls of heme. The spectral features of cytochrome c4 produced by CcmFGH-only (e.g., 552-nm absorption maximum in the spectrum of the reduced sample, and 550-nm absorption maximum in the reduced pyridine hemochrome spectrum) are indistinguishable from those of the cytochrome c4 produced when all system I proteins are present (CcmABCDEFGH; Fig 2D, blue line; Fig 2D, inset, blue line). Mass spectral analysis of cytochrome c4 purified from cells expressing CcmFGH-only or the full system I identified species of the expected molecular weights in each preparation (i.e., within 2 Daltons of the published molecular weights for the full-length holocytochrome c4 and proteolyzed holocytochrome c4′ (Feissner et al., 2006b); Fig S2). This further confirmed covalent attachment of heme and proper cleavage of the periplasmic secretion signal (Fig S2B). In these large-scale cultures (under the growth and induction conditions described in experimental procedures), yields of holocytochrome c via CcmFGH-only were approximately one-sixth those of the full system I.
Fig. 2.
Purification and characterization of cytochrome c produced by CcmFGH in the absence of CcmABCDE. (A) Coomassie blue staining of purified cytochrome c4:His6 showing full length 24-kDa holocytochrome c4 and proteolyzed 12-kDa holocytochrome c4′. (B) Heme staining of purified cytochrome c4 showing 24-kDa and 12-kDa forms. (C) Anti-cytochrome c4 immunoblot showing 24-kDa and 12-kDa holocytochrome c4. For (A)–(C), abbreviations are M, molecular weight standards; S, soluble fraction; FT, flow through; W1, wash 1; W2, wash 2; W3, wash 3; W4, wash 4; E1, elution 1; E2, elution 2; E3, elution 3. (D) UV-Vis absorption spectra of purified, sodium dithionite-reduced holocytochrome c4 produced by the full system I (blue line) or in the absence of CcmABCDE (red line). (Inset) Sodium dithionite-reduced pyridine hemochrome spectrum of purified holocytochrome c4 produced by the full system I (blue line) or in the absence of CcmABCDE (red line) from 500–600 nm. Absorption maxima are indicated.
Engineering optimal biosynthesis of holocytochrome c produced by CcmFGH-only
To engineer increased cytochrome c maturation in the absence of CcmABCDE, we piloted the following strategies: i) increasing the concentration of inducer (arabinose; “↑ arab” condition), ii) raising intracellular heme levels by addition of the heme biosynthetic precursor δ-aminolevulinic acid (ALA; “ALA” condition), iii) decreasing oxygen tension by reduced shaking during growth (“↓ rpm” condition), and iv) engineering the cytochrome c4 gene for increased expression (by gene dosage) from a single pBAD-based vector, downstream of ccmF:His6GH (“p-c4” condition). Addition of ALA and increasing cytochrome c4 gene dosage led to the largest increases in yields of holocytochrome c (Fig 3A, lanes 4 and 6, respectively; Fig 3B, red and orange lines; quantified in Fig 3C). Additively, with each of the four modifications together, levels of holocytochrome c4 produced by CcmFGH-only increased approximately 6-fold (up to 0.3 mg per L culture), similar to yields of holocytochrome c4 when CcmABCDE are also present with the chromosomally integrated cytochrome c4 gene (Fig 3A, compare lanes 7 and 9; Fig 3B, compare light blue and brown lines; quantified in Fig 3C). In cells carrying CcmABCDE in the absence of CcmFGH with the chromosomally integrated cytochrome c4 gene, no holocytochrome c formation was detected (Fig 3A, lane 8). In the presence of CcmABCDE and CcmFGH (i.e., full system I) and each of the four modifications described above, yields of cytochrome c4 were approximately 1 mg per L culture (Fig 3A, lane 10; Fig 3B, top blue line; quantified in Fig 3C). Analysis of membrane fractions by immunoblot confirmed the presence of CcmF and CcmH, respectively (except in the strain carrying only pGEX CcmABCDE; Fig 3D and 3E, lane 8), and the absence of CcmE (except for those cells carrying pGEX CcmABCDE; Fig 3F). These results demonstrate that, under optimized conditions, CcmFGH-only is a robust synthetase capable of producing substantial levels of periplasmic holocytochrome c.
Fig. 3.
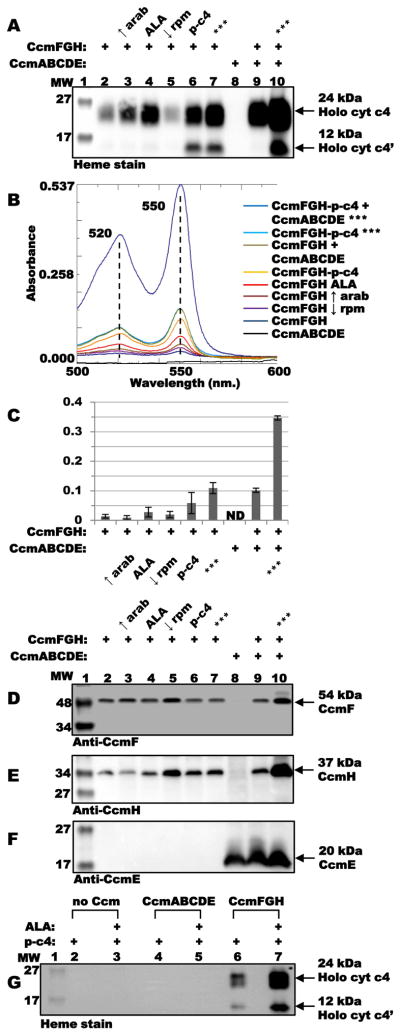
Optimization of holocytochrome c4 produced by CcmFGH in the absence of CcmABCDE. (A) Heme staining of TALON-purified proteins showing relative levels of holocytochrome c4 synthesized by CcmFGH-only (lanes 2–7), CcmABCDE (lane 8), and CcmFGH + CcmABCDE (lanes 9 and 10) for each indicated condition/additive. “↑ arab,” increase in final arabinose inducer concentration from 0.2 % to 0.4 %; “ALA,” addition of ALA to 50 μg mL−1; “↓ rpm,” decrease in shaking during growth from 230 rpm to 120 rpm; “p-c4,” plasmid-borne cytochrome c4 engineered into pBAD CcmFGH; “***,” each of the above four additives/conditions together. Arrows denote full-length 24-kDa and proteolyzed 12-kDa holocytochrome c4. (B) Sodium dithionite-reduced pyridine hemochrome spectra of TALON-purified proteins for each condition, from 500–600 nm. Absorption maxima are indicated. (C) Quantification of yields of holocytochrome c4 (based on 550 nm. absorption in reduced pyridine hemochrome) for each additive/condition from three independent experiments. Error bars denote standard deviation. ND, none detected. (D) Anti-CcmF immunoblot of DDM-solubilized membrane fractions showing 54-kDa CcmF. (E) Anti-CcmH immunoblot of DDM-solubilized membrane fractions showing 37-kDa CcmH. (F) Anti-CcmE immunoblot of DDM-solubilized membrane fractions showing 20-kDa CcmE. Labels for (D) – (F) are as in (A). (G) Heme staining of TALON-purified proteins showing relative levels of plasmid-borne holocytochrome c4 (“p-c4”) synthesized in the absence of Ccm proteins (lanes 2 and 3), by CcmABCDE (lanes 4 and 5), and by CcmFGH-only (lanes 6 and 7) in the presence or absence of ALA (50 μg mL−1). Arrows denote full-length 24-kDa and proteolyzed 12-kDa holocytochrome c4.
To establish that the cytochrome c4 was not self-assembling (i.e., non-enzymatically binding and attaching heme) under the optimized conditions, E. coli Δccm containing only the cytochrome c4 gene (in the pBAD plasmid) was induced with arabinose in the presence or absence of ALA, and the hexahistidine-tagged cytochrome c4 was purified and analyzed (Fig 3G and Fig S3). No covalently bound heme was detected by heme stain (Fig 3G, lanes 2 and 3) or by absorption spectroscopy (Fig S3). To test whether CcmABCDE could act as a synthetase under the optimized conditions, we co-expressed the cytochrome c4 (in pBAD) and CcmABCDE (in pGEX) in the presence or absence of ALA. Again, holocytochrome c4 was not produced at detectable levels, as determined by heme stain (Fig 3G, lanes 4 and 5) and absorption spectroscopy (Fig S3), while the CcmFGH-only yielded readily-detectable holocytochrome c4 (Fig 3G, lanes 6 and 7; Fig S3).
The role of conserved P-His and TM-His residues in CcmF for holocytochrome c assembly via CcmFGH-only
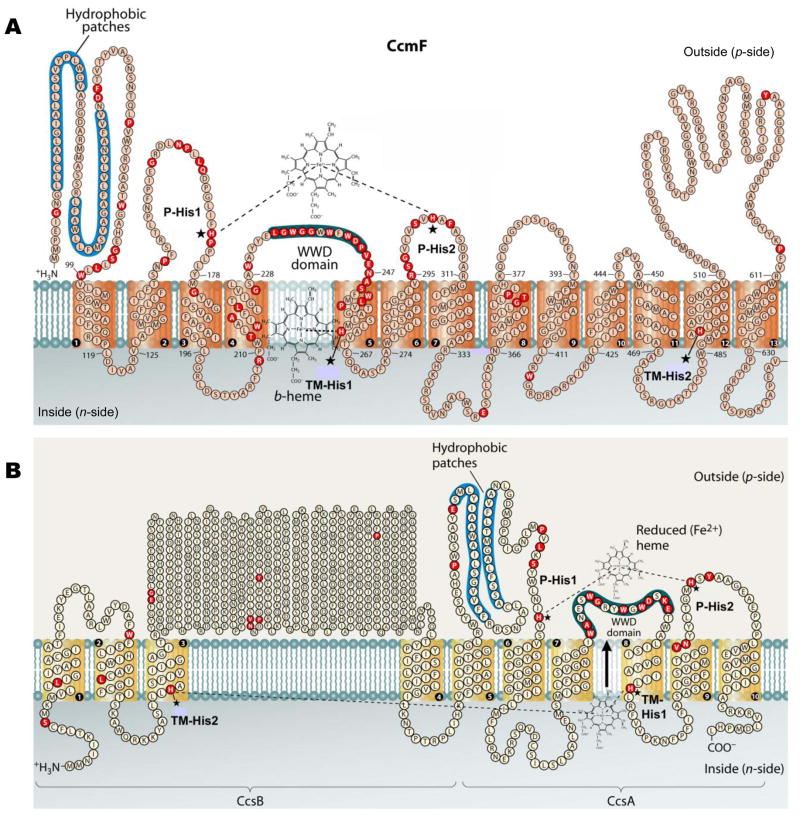
CcmF contains four conserved histidine residues: TM-His1 and TM-His2 in transmembrane domains 5 and 12, respectively, and P-His1 and P-His2, located in periplasmic loops flanking the WWD domain (see Fig 4A). P-His1 and P-His2 are proposed to ligate heme from holoCcmE when it is bound in the WWD domain, en route to covalent attachment to apocytochrome c; TM-His1 and TM-His2 are ligands to the stable heme b (San Francisco et al., 2011), which may play a role in reducing the holoCcmE heme (bound in the WWD domain) prior to covalent attachment. In the context of the full system I, each of the four histidines is essential for holocytochrome c formation (Richard-Fogal et al., 2009; San Francisco et al., 2011). To test whether holocytochrome c formation via CcmFGH-only required these four histidines, we engineered substitutions at each residue (in pRGK388) and assayed for heme attachment to cytochrome c4. Substitutions at TM-His1, TM-His2, and P-His1 abolished holocytochrome formation to undetectable levels, indicating that these residues are absolutely required for heme attachment to cytochrome c via the CcmABCDE-independent pathway (Fig 5A, lanes 3, 4, and 6; quantified in Fig 5B). Substitution of P-His2 with glycine supported holocytochrome formation at approximately 20 % levels of WT (Fig 5A, lane 5; quantified in Fig 5B). This contrasts with the absolute requirement for P-His2 in the context of the full system I (i.e., in the presence of CcmABCDE), and may be suggestive of a holoCcmE-specific role for P-His2 in cytochrome c formation (see Discussion).
Fig. 4.
Topology of the system I CcmF protein from Escherichia coli (A) and the system II CcsBA fusion protein from Helicobacter hepaticus (B). Possible histidine axial ligands to heme are starred, and are designated P-His1, P-His2, TM-His1, and TM-His2. The highly conserved WWD domain and the hydrophobic patches are shaded. Completely conserved amino acids (red) were identified by individual protein alignments using CcmF or the CcsB and CcsA ORFs from selected organisms, as described in (Kranz et al., 2009). Diagram is modified from (Kranz et al., 2009).
Fig. 5.
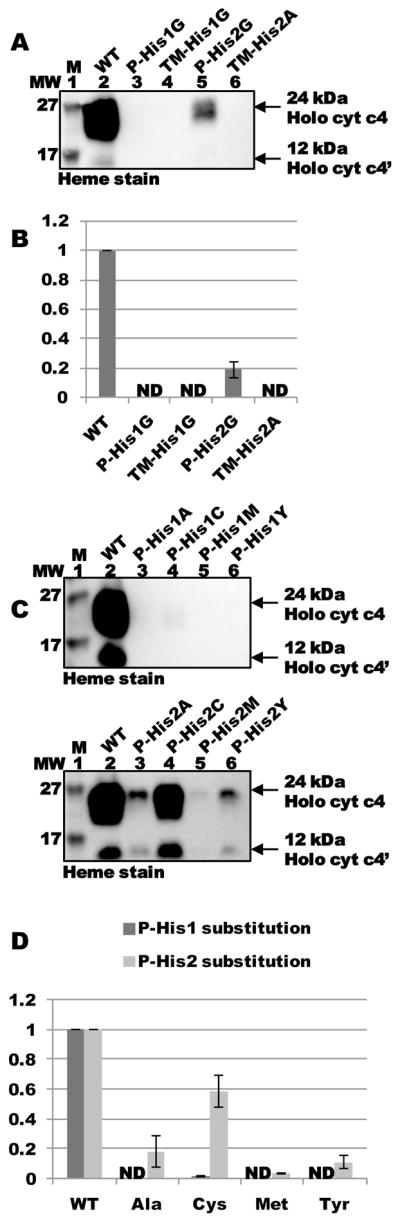
Effect of substitutions at conserved His residues in CcmF on holocytochrome c formation in the absence of CcmABCDE. (A) Heme staining of purified holocytochrome c4 assembled by CcmFGH-only (“WT”) and site-directed variants of CcmF. (B) Quantification of the results of heme staining (24 kDa holocytochrome c4) from purified fractions from three independent experiments. (C) Heme staining of purified holocytochrome c4 assembled by CcmFGH-only (“WT”) and the indicated site-directed variants at CcmF P-His1 (upper panel) or P-His2 (lower panel). (D) Quantification of the results of heme staining from purified fractions from three independent experiments. For (B) and (D), synthesis of holocytochrome c4 is relative to CcmFGH-only (“WT”), which has been set at 100 %. Error bars denote standard deviation. ND, none detected.
To test whether other substitutions could support holocytochrome formation at P-His1 or P-His2, we engineered alanine, cysteine, methionine, or tyrosine substitutions at P-His1 or P-His2. While no P-His1 substitution supported holocytochrome c formation (Fig 5C, upper panel), we discovered that P-His2Cys supported cytochrome c assembly at approximately 60 % WT levels (Fig 5C, lower panel, lane 4; quantified in Fig 5D). P-His2Ala was similar to substitution with glycine (20 % function relative to WT; Fig 5C, lower panel, lane 3; quantified in Fig 5D) while methionine and tyrosine substitutions were 3 % and 10 % WT levels, respectively (Fig 5C, lower panel, lanes 5 and 6; quantified in Fig 5D). The inability for any engineered substitution at P-His1 to support holocytochrome formation indicates that this residue cannot vary from the natural histidine, similar to results with full system I.
Conserved cysteines in CcmG and CcmH are required for holocytochrome formation via CcmFGH-only
CcmG and CcmH each contain a conserved pair of thioredox-active cysteines that are required for cytochrome c synthesis in the context of the full system I (Fabianek et al., 1998; Fabianek et al., 1999; Robertson et al., 2008). To test whether holocytochrome c formation by CcmFGH-only required these thioredoxin functions, we engineered serine substitutions at the conserved cysteine pairs in CcmG or CcmH and assayed for cytochrome c formation. Similar to the full system I, in the absence of the conserved cysteine pair in either CcmG or CcmH, no holocytochrome formation was observed (Fig 6A, lanes 3 and 4; quantified in Fig 6B). Thus, the redox-active cysteine pairs of CcmG and CcmH are required for holocytochrome c formation via CcmFGH-only.
Fig. 6.
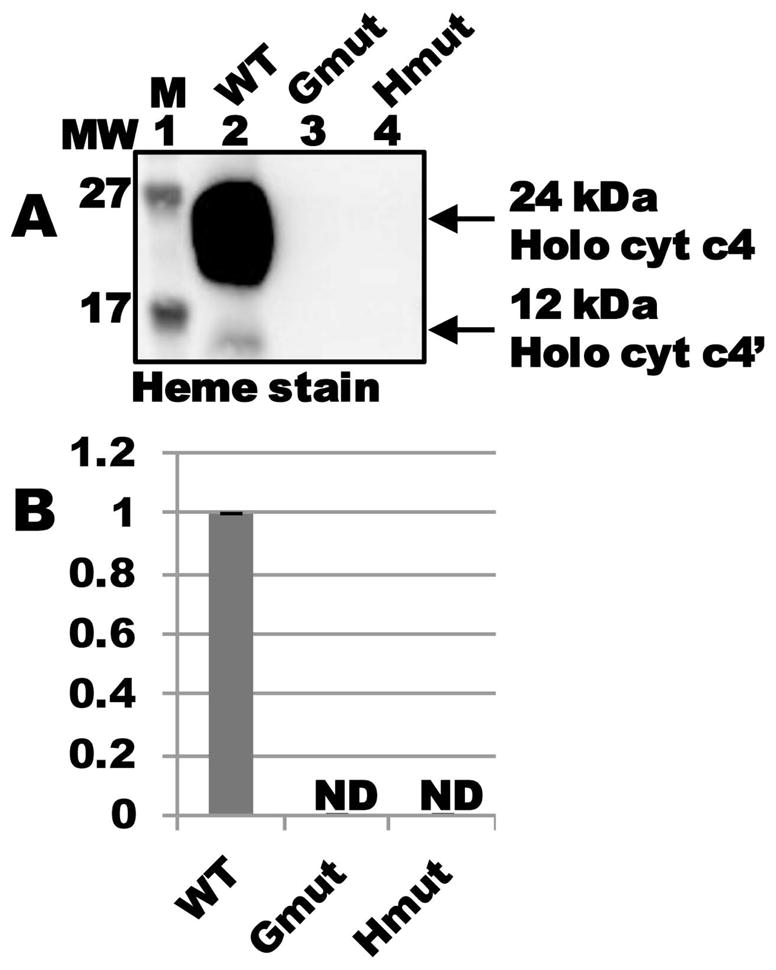
Conserved cysteines in CcmG and CcmH are required for holocytochrome c formation by CcmFGH in the absence of CcmABCDE. (A) Heme staining of purified holocytochrome c4 assembled by CcmFGH-only (“WT”) and the indicated site-directed variants at CcmG and CcmH. “Gmut” is a Cys80Ser/Cys83Ser double mutant, and “Hmut” is a Cys43Ser/Cys46Ser double mutant. (B) Quantification of the results of heme staining (24 kDa holocytochrome c4) from purified fractions from three independent experiments. Synthesis of holocytochrome c4 is relative to CcmFGH-only (“WT”), which has been set at 100 %. ND, none detected.
DISCUSSION
CcmF has been referred to as the “cytochrome c heme lyase” or the “cytochrome c synthetase” in nearly every review on system I in the last decade [for recent reviews, see (Hamel et al., 2009; Kranz et al., 2009; Mavridou et al., 2013; Sanders et al., 2010; Sawyer and Barker, 2012)]. However, direct experimental evidence demonstrating this activity for CcmF has been lacking. Here, we demonstrate that holocytochrome c formation can occur completely (and robustly) in the absence of the CcmABCDE components of the system I pathway, thus establishing that the cytochrome c synthetase activity of system I inheres in CcmFH (and CcmG). We suspect that the synthetase activity of CcmFH (with CcmG) has gone unnoticed thus far due to challenges in expressing these membrane proteins and engineering conditions for optimal biogenesis. Only of late have we been able to express and purify CcmF in sufficient quantities to begin biochemical characterization of this protein (Richard-Fogal et al., 2009; San Francisco et al., 2011). Indeed, the discovery that CcmF contained a stable heme b (coordinated by TM-His1 and TM-His2) was made only recently (Richard-Fogal et al., 2009). Thus, we attribute our observation of this “new” CcmFGH-only synthetase activity to our recent ability to successfully modulate expression of stable, functional CcmFH and CcmG in E. coli, and control the expression of substrate cytochromes c (e.g., the di-heme cytochrome c4 and mono-heme cytochrome c2 used here).
It is important to state that the CcmFGH-only synthetase activity we describe here would not substitute for the natural complete system I that is present in a wide range of organisms. Indeed, many mutations in CcmABCDE in different organisms have been shown to result in deficiencies in cytochrome c maturation [for example, (Beckman et al., 1992; Kranz, 1989; Page et al., 1997; Ramseier et al., 1991; Schulz et al., 1998)]. The CcmFGH-only activity is a ramification of engineering and optimization to establish the minimal components of system I that possess synthetase activity. This metabolic engineering, which included i) increasing expression by gene dosage (for both ccm and apocytochrome genes), ii) promoter and ribosome binding site changes (i.e., inducible promoters in pBAD and pGEX-based vectors), and iii) increasing endogenous synthesis of heme (by ALA addition), indicated that CcmFH, along with CcmG (but not CcmABCDE—i.e., holoCcmE) functions as the synthetase; that is, CcmFH attaches heme to the CXXCH motif of the apocytochrome. Even for organisms that depend on a single cytochrome c for respiration (which are rare), the above engineering approaches would likely be much less “beneficial” for holocytochrome c assembly than delivery of heme by holoCcmE (i.e., the natural CcmABCDE step of system I). Consider also that the full system I is able to use heme at low levels and can rely on holoCcmE as a heme reservoir, which are certainly important in the natural “lifestyles” of many organisms. Nevertheless, the results described here are the most compelling thus far to designate CcmF/H as the cytochrome c synthetase of system I.
Prior in vitro studies by Ferguson and colleagues showed that holoCcmE* (lacking its TMD) could transfer its heme directly to apocytochrome c (Daltrop et al., 2002). The conditions for the in vitro transfer involved 18 hours of incubation and employed the stable apocytochrome c552 from a thermophile as an acceptor. The authors indicate that their results “mimic the molecular pathway in vivo”. To test whether we could engineer this transfer in vivo, we over-expressed CcmABCDE (thus, holoCcmE) and the cytochrome c4 in the presence of increased heme (i.e., added ALA). No detectable holocytochrome c4 was formed in the absence of CcmFGH (Fig 3G, lane 8 and Fig S3). We conclude that in vivo, holoCcmE does not act as a “synthetase” itself. Although there is little doubt that in system I the heme from holoCcmE is transferred to apocytochrome c, and that the heme in holoCcmE binds to the CcmF WWD domain (San Francisco and Kranz, in press), the synthetase activity is ultimately provided by the CcmF/H complex.
Requirements for holocytochrome c formation by CcmFGH-only versus the full system I
Many of the requirements for cytochrome c assembly by the full system I extend to CcmFGH-only. In system I, the thio-active proteins CcmG and CcmH each contain a conserved pair of cysteines that are required for holocytochrome formation (Fabianek et al., 1998; Fabianek et al., 1999; Robertson et al., 2008). CcmG and CcmH are involved in maintaining the thiols of the apocytochrome (at Cys-Xxx-Xxx-Cys-His) in the reduced state, which is a requirement for thioether formation with the heme vinyls. The conserved cysteines in CcmG (which are reduced by the membrane protein DsbD) likely function to reduce the thiols of CcmH (Meyer et al., 2005; Monika et al., 1997; Setterdahl et al., 2000; Turkarslan et al., 2008). CcmH also has a direct apocytochrome binding function (Di Matteo et al., 2007; Di Silvio et al., 2013; Meyer et al., 2005; Verissimo et al., 2011), and it is proposed that, via interaction with CcmF, CcmH physically positions the apocytochrome (with reduced cysteine thiols) for covalent heme attachment (Meyer et al., 2005; Verissimo et al., 2011). We discovered that mutation of either of the cysteine pairs in CcmG or CcmH abolished holocytochrome formation by CcmFGH-only (Fig 6). The requirement for the conserved redox-active cysteine pairs in CcmG and CcmH demonstrates that the CcmFGH-only pathway is “intact” with regard to handling of the apocytochrome. Although other explanations are possible, we favor the hypothesis that the lower cytochrome c maturation activity of CcmFGH-only (relative to full system I) is likely due to less efficient heme delivery (i.e., the absence of the holoCcmE “heme reservoir” (Feissner et al., 2006b); see below). This finding also highlights the significant evolutionary advantage of the full system I.
Holocytochrome c formation by the full system I requires conserved histidines in CcmF: mutation of any of the four (Fig 4A) renders the system non-functional (Ren et al., 2002; Richard-Fogal et al., 2009; San Francisco et al., 2011). We have shown experimentally that TM-His1 and TM-His2 are ligands to the b-heme (San Francisco et al., 2011), while P-His1 and P-His2 are proposed to ligate the incoming heme from holoCcmE when it is bound in the WWD domain (Kranz et al., 2009; Richard-Fogal et al., 2009; San Francisco et al., 2011). For CcmFGH-only, we found that TM-His1, TM-His2, and P-His1 were each indispensable for holocytochrome formation. However, CcmFGH-only was only partially dependent on P-His2 (Ala or Gly substitutions at this position retained approximately 20 % activity, and Cys, 60 %, relative to WT). Since these same substitutions show no activity in the context of the full system I, we propose that P-His2 may have a function that is specific to the system I heme chaperone, holoCcmE. The lack of a requirement for P-His2 reflects, possibly, a different mechanism of heme trafficking to the WWD domain in CcmFGH-only. Below, we suggest two possible trafficking routes for the endogeneous heme that is attached to cytochrome c by CcmFGH-only.
Similarities between CcmFGH-only and CcsBA: evolutionary insights
The assignment of the synthetase (heme lyase) activity of system I to CcmF has been based in part on its topological similarity to the cytochrome c synthetase of system II, CcsBA (see Fig 4). System II is present in many gram positive (and other) bacteria, cyanobacteria, and the chloroplasts of plants and algae (Ahuja et al., 2009; Simon and Hederstedt, 2011; Xie and Merchant, 1998). CcsBA, when expressed heterologously in E. coli Δccm is sufficient for assembly of cytochrome c (Feissner et al., 2006b; Frawley and Kranz, 2009; Kern et al., 2010; Richard-Fogal et al., 2012); thus, it is the system II holocytochrome c synthetase. CcsBA is proposed to traffick heme across the cytoplasmic membrane, in addition to facilitating thioether formation between the heme and apocytochrome (Frawley and Kranz, 2009; Hamel et al., 2003; Kranz et al., 2009; Merchant, 2009). Thus, it is a heme transporter and a holocytochrome c synthetase. Comparison of the membrane topologies of CcmF and CcsBA reveals several similar features (see Fig 4): each contains four completely conserved His residues (two in transmembrane domains, here called TM-His1 and TM-His2; and two in extra-cytoplasmic loops, here called P-His1 and P-His2) and the conserved WWD domain, a hydrophobic, extra-cytoplasmic feature that has been shown (in the system I protein CcmC) to interact directly with heme (Richard-Fogal and Kranz, 2010). Additionally, both CcsBA (Frawley and Kranz, 2009) and CcmF (Richard-Fogal et al., 2009) bind a non-covalent heme b (see Fig 4). However, in CcsBA, this heme b is ultimately attached to cytochrome c, whereas the stable heme b in CcmF (in the context of the full system I) may play a role in reducing heme bound in the WWD domain (from holoCcmE).
The topological similarities between CcmF and CcsBA, coupled with the finding here that CcmF (together with CcmH and CcmG) is a cytochrome c synthetase, raise several interesting questions. First, how is heme trafficked to the site of thioether formation with the apocytochrome (presumably, at the CcmF WWD domain with axial ligation by P-His1) in CcmFGH-only synthesis? While we cannot rule out other hypotheses, we envision two possible scenarios: i) heme enters the WWD domain of CcmF directly from the cytoplasmic membrane outer leaflet, possibly, (where it is subsequently reduced by the transmembrane heme b), or ii) similar to the proposed mechanism for CcsBA, the transmembrane heme b may be channeled to the external WWD domain for attachment to the apocytochrome. While experiments addressing these hypotheses will be challenging, the possibility that the CcmFGH-only synthetase may be CcsBA-like (with respect to the channeling of heme from the transmembrane binding site to the WWD domain) is intriguing from an evolutionary perspective.
The complex phylogenetic distribution of systems I and II suggest significant lateral transfer (Goldman and Kranz, 1998), making it difficult to discern clear evolutionary patterns. We have shown previously that system I can mature cytochromes c at 5-fold lower heme concentrations than can system II (Richard-Fogal et al., 2007), and that holoCcmE can function as a “heme reservoir” (i.e., when heme synthesis is completely inhibited) (Feissner et al., 2006a). On the basis of these results, we have speculated that the holoCcmE reservoir aspect of system I may have evolved for cytochrome c synthesis under conditions of iron (or heme) scarcity. The ability to use heme at very low levels, and as a reservoir when no iron is present, might represent a possible selection advantage. Since the CcmFGH-only synthetase lacks the holoCcmE reservoir, it could be considered a “prototype” of an intermediate, or a “transition,” on the evolutionary path from system II to the full system I. CcmFGH-only synthesis thus provides a unique opportunity to directly compare holocytochrome c formation (under low heme conditions) by system I with and without the holoCcmE reservoir.
CcmFGH-only: a viable candidate for in vitro reconstitution of holocytochrome c assembly
One of the remaining challenges in the field of cytochrome c biogenesis is in vitro reconstitution of each of the systems for holocytochrome formation. Reconstitution of the synthetase reactions would be especially informative, as they are common to all three major pathways. To date, system II has been a more attractive candidate for in vitro reconstitution than system I, since it consists of a single protein complex. Our findings here demonstrate that the system I synthetase (CcmFGH-only) is now a viable candidate for in vitro reconstitution as well. The CcmFGH-only synthetase activity reported here thus represents a significant advance for the field, in terms of the proof of its function, the unique evolutionary and mechanistic insights it provides, and the potential for in vitro reconstitution.
EXPERIMENTAL PROCEDURES
Bacterial Growth Conditions
Unless otherwise noted, Escherichia coli strains (Table S1) were grown at 37°C by shaking at 230 rpm in Luria-Bertani broth (LB; Difco) supplemented with the appropriate antibiotics (Sigma-Aldrich) and other media additives at the following concentrations: carbenicillin, 50 μg mL−1; chloramphenicol, 20 μg mL−1; gentamicin, 10 μg mL−1; kanamycin, 20 μg mL−1; Isopropyl β-D-1-thiogalactopyranoside (IPTG; Gold Biotechnology), 1 mM; arabinose (Gold Biotechnology), 0.2 % (wt/vol); δ-aminolevulinic acid (ALA; Sigma-Aldrich), 50 μg ml−1.
Protein Expression and Purification
E. coli Δccm strains RK103 (Feissner et al., 2006b) or RK111 (Δccm carrying the arabinose-inducible chromosomal integrate of the cyt c4:His6 gene) (San Francisco et al., 2011) (Table S1) were used for expression. Starter cultures were inoculated from a single colony and grown overnight in 10 mL of LB with the appropriate antibiotics. 1 L of LB was inoculated with the 10 mL starter culture and grown to an OD600 of 1.8, then induced with IPTG and/or arabinose for 14–16 hr. Cells were harvested at 5,000 × g and frozen at −80°C. Cell pellets were thawed and resuspended in phosphate-buffered saline (PBS; 100 mM NaCl, 7.5 mM Na2HPO4, 2 mM NaH2PO4, pH 7) and treated with 1 mM phenylmethylsulfonyl fluoride (PMSF; Sigma-Aldrich) and 100 μg ml−1 egg white lysozyme (Sigma-Aldrich) for 30 min while shaking on ice. Cells were disrupted by repeated sonication on ice for 30 sec bursts (5 × 30 sec on, 30 sec off) on a Branson 250 sonicator (50% duty, 8 output). Crude sonicate was centrifuged at 24,000 × g for 20 min to clear cell debris, and the soluble and membrane fractions were separated by centrifugation at 100,000 × g for 45 min. Soluble fractions (L; load) were passed over TALON resin per the manufacturer’s recommendations and washed in 1 x modified TALON buffer with increasing concentrations of imidazole (wash 1 (W1), 0 mM imidazole; wash 2 (W2), 2 mM imidazole; wash 3 (W3), 5 mM imidazole). Bound hexahistidine-tagged protein was eluted in 1 x modified TALON buffer containing 150 mM imidazole (E; elution). Total protein concentration was determined using the Nanodrop 1000 spectrophotometer (Thermo Scientific). For growth by shaking at 120 rpm (“↓ rpm” condition), starter cultures were initiated from a single colony and grown overnight in 100 mL of LB with the appropriate antibiotics. 1 L of LB was inoculated with the 100 mL starter culture and grown to an OD600 of 1.8, then induced with IPTG and/or arabinose for 14–16 hr. For the “↑ arabinose” condition, expression was induced with 0.4 % arabinose rather than 0.2 %. For the “ALA” condition, 50 μg mL−1 ALA was added to culture at the time of induction. For analysis of crude membrane fractions, membrane pellets (isolated by centrifugation at 100,000 × g) were solubilized in a modified 1x TALON (Clontech) buffer (50 mM Tris-HCL, pH 7; 300 mM NaCl) with 1 % (wt/vol) dodecyl maltoside (DDM, Anatrace) by agitation on ice for 1 hr, and then centrifuged at 24,000 × g for 20 min to remove unsolubilized material.
Heme stains and other methods
Heme stains and immunoblots were performed as described previously (Feissner et al., 2003; Feissner et al., 2006b). Proteins were separated by 12.5 % SDS-PAGE and transferred to Hybond C nitrocellulose membranes (GE Healthcare). For immunoblots, anti-CcmF antibodies were used at a dilution of 1:10000, anti-CcmE antibodies at 1:10000, anti-cytochrome c4 antibodies at 1:10000, and anti-CcmH antibodies at 1:5000. Protein A peroxidase (Sigma-Aldrich) was used as the secondary label. The chemiluminescent signal for heme stains was developed using the SuperSignal Femto kit (Thermo Scientific) or, for immunoblots, the Immobilon Western kit (Millipore), and detected with an LAS-1000 Plus detection system (Fujifilm-GE Healthcare). The abundance of holocytochrome c4 was determined by pyridine extraction as described in (Berry and Trumpower, 1987), or by densitometry analysis of the chemiluminescent signal from heme staining using the Science Lab 99-Image Gauge version 3.4 software (Fujifilm-GE Healthcare) as described in (Richard-Fogal et al., 2007). Protein purity was assessed by Coomassie Blue staining of SDS-PAGE.
UV/Vis absorption spectroscopy
UV-visible absorption spectra were recorded with a Shimadzu UV-2101 PC UV-Vis scanning spectrophotometer at room temperature as described previously (Frawley and Kranz, 2009). All spectra were recorded in the same buffer (modified 1x TALON buffer) in which the proteins were purified. Chemically reduced spectra were generated by addition of a few grains of sodium dithionite (sodium hydrosulfite).
Construction of plasmids
All oligonucleotide primer sequences, plasmids, and strains are given in Table S1. The single pBAD-based plasmid containing ccmF:His6GH and cyt c4:His6 was constructed by polymerase chain reaction (PCR) amplification of the cyt c4:His6 gene (and the upstream ribosome binding site; RBS) from pRGK332 (Feissner et al., 2006b) and insertion of the resulting fragment into the single PstI site downstream of ccmF:His6GH in pRGK388 (Richard-Fogal et al., 2009) to create pRGK448. The resulting plasmids were screened by restriction digest to confirm correct orientation of the insert, and then sequenced. All substitutions in pRGK388 (i.e., CcmG (Cys80Ser/Cys83Ser), CcmH (Cys43Ser/Cys46Ser), CcmF(His261Gly), and CcmF(His491Ala)) were engineered using the QuikChange I Site-Directed Mutagenesis Kit (Agilent Technologies) per the manufacturer’s recommendations. All oligonucleotides were synthesized by Sigma-Aldrich. Each of the final constructs was sequenced to verify the mutation(s).
Proteomics analysis
Proteomics analyses were carried out by the Washington University Resource for Biomedical and Bio-organic Mass Spectrometry Facility. Briefly, TALON-purified holocytochrome c4:His6 samples (produced by the full system I or by CcmFGH-only) were loaded onto an Agilent 2.1 X 15 mm C8 Zorbax Cartridge-Column and eluted using a 9.5 min gradient with a flow rate of 200 μL/min. A positive ion mass spectrum was acquired using a Bruker Maxis Q-ToF mass spectrometer equipped with an electrospray ionization source. Protein molecular ions were identified with multiple charge states and deconvoluted to identify the molecular weight(s) of the main species.
Genomic PCR
E. coli strains (WT E. coli K-12 MG1655; Δccm (RK103); Δccm cyt c4:His6 chromosomal integrate (RK111); and RK111 carrying pBAD ccmF:His6GH) were streaked onto LB plates containing the appropriate antibiotics and incubated at 37°C overnight. Single colonies were resuspended in 100 μL H2O by vortexing, placed at 90°C for 10 min to lyse cells, and centrifuged at 16 000 × g for 5 min to remove cell debris. 1 μL of the supernatant (containing cellular DNA) was used as the template in each of four PCRs with the following primer sets: A) delCcmF-right + delCcmH-left, B) delCcmC-right + delCcmF-left, C) delCcmB-right + delCcmE-left, D) menBflank-right + menBflank-left. Oligonucleotide primer sequences are given in Table S1. PCRs were performed using GoTaq Green Master Mix (Promega) for 25 cycles, per the manufacturer’s recommendations. PCR products were visualized by ethidium bromide staining of agarose gels (0.8 %).
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health Grant R01 GM47909 to R.G.K. The mass spectral analysis (shown in Fig S2) was supported in part by National Institute of General Medical Sciences Grant 8 P41 GM103422-35 from the National Institutes of Health.
References
- Ahuja U, Kjelgaard P, Schulz BL, Thony-Meyer L, Hederstedt L. Haem-delivery proteins in cytochrome c maturation System II. Mol Microbiol. 2009;73:1058–1071. doi: 10.1111/j.1365-2958.2009.06833.x. [DOI] [PubMed] [Google Scholar]
- Allen JW. Cytochrome c biogenesis in mitochondria--Systems III and V. Febs J. 2011;278:4198–4216. doi: 10.1111/j.1742-4658.2011.08231.x. [DOI] [PubMed] [Google Scholar]
- Barker PD, Ferrer JC, Mylrajan M, Loehr TM, Feng R, Konishi Y, Funk WD, MacGillivray RT, Mauk AG. Transmutation of a heme protein. Proc Natl Acad Sci U S A. 1993;90:6542–6546. doi: 10.1073/pnas.90.14.6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman DL, Trawick DR, Kranz RG. Bacterial cytochromes c biogenesis. Genes Dev. 1992;6:268–283. doi: 10.1101/gad.6.2.268. [DOI] [PubMed] [Google Scholar]
- Beckman DL, Kranz RG. Cytochromes c biogenesis in a photosynthetic bacterium requires a periplasmic thioredoxin-like protein. Proc Natl Acad Sci U S A. 1993;90:2179–2183. doi: 10.1073/pnas.90.6.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry EA, Trumpower BL. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal Biochem. 1987;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- Christensen O, Harvat EM, Thony-Meyer L, Ferguson SJ, Stevens JM. Loss of ATP hydrolysis activity by CcmAB results in loss of c-type cytochrome synthesis and incomplete processing of CcmE. Febs J. 2007;274:2322–2332. doi: 10.1111/j.1742-4658.2007.05769.x. [DOI] [PubMed] [Google Scholar]
- Daltrop O, Stevens JM, Higham CW, Ferguson SJ. The CcmE protein of the c-type cytochrome biogenesis system: unusual in vitro heme incorporation into apo-CcmE and transfer from holo-CcmE to apocytochrome. Proc Natl Acad Sci U S A. 2002;99:9703–9708. doi: 10.1073/pnas.152120699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Matteo A, Gianni S, Schinina ME, Giorgi A, Altieri F, Calosci N, Brunori M, Travaglini-Allocatelli C. A strategic protein in cytochrome c maturation: three-dimensional structure of CcmH and binding to apocytochrome c. J Biol Chem. 2007;282:27012–27019. doi: 10.1074/jbc.M702702200. [DOI] [PubMed] [Google Scholar]
- Di Matteo A, Calosci N, Gianni S, Jemth P, Brunori M, Travaglini-Allocatelli C. Structural and functional characterization of CcmG from Pseudomonas aeruginosa, a key component of the bacterial cytochrome c maturation apparatus. Proteins. 2010;78:2213–2221. doi: 10.1002/prot.22733. [DOI] [PubMed] [Google Scholar]
- Di Silvio E, Di Matteo A, Malatesta F, Travaglini-Allocatelli C. Recognition and binding of apocytochrome c to P. aeruginosa CcmI, a component of cytochrome c maturation machinery. Biochim Biophys Acta. 2013;1834:1554–1561. doi: 10.1016/j.bbapap.2013.04.027. [DOI] [PubMed] [Google Scholar]
- Fabianek RA, Hennecke H, Thony-Meyer L. The active-site cysteines of the periplasmic thioredoxin-like protein CcmG of Escherichia coli are important but not essential for cytochrome c maturation in vivo. J Bacteriol. 1998;180:1947–1950. doi: 10.1128/jb.180.7.1947-1950.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabianek RA, Hofer T, Thony-Meyer L. Characterization of the Escherichia coli CcmH protein reveals new insights into the redox pathway required for cytochrome c maturation. Arch Microbiol. 1999;171:92–100. doi: 10.1007/s002030050683. [DOI] [PubMed] [Google Scholar]
- Feissner R, Xiang Y, Kranz RG. Chemiluminescent-based methods to detect subpicomole levels of c-type cytochromes. Anal Biochem. 2003;315:90–94. doi: 10.1016/s0003-2697(02)00658-9. [DOI] [PubMed] [Google Scholar]
- Feissner RE, Richard-Fogal CL, Frawley ER, Kranz RG. ABC transporter-mediated release of a haem chaperone allows cytochrome c biogenesis. Mol Microbiol. 2006a;61:219–231. doi: 10.1111/j.1365-2958.2006.05221.x. [DOI] [PubMed] [Google Scholar]
- Feissner RE, Richard-Fogal CL, Frawley ER, Loughman JA, Earley KW, Kranz RG. Recombinant cytochromes c biogenesis systems I and II and analysis of haem delivery pathways in Escherichia coli. Mol Microbiol. 2006b;60:563–577. doi: 10.1111/j.1365-2958.2006.05132.x. [DOI] [PubMed] [Google Scholar]
- Frawley ER, Kranz RG. CcsBA is a cytochrome c synthetase that also functions in heme transport. Proc Natl Acad Sci U S A. 2009;106:10201–10206. doi: 10.1073/pnas.0903132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman BS, Beckman DL, Bali A, Monika EM, Gabbert KK, Kranz RG. Molecular and immunological analysis of an ABC transporter complex required for cytochrome c biogenesis. J Mol Biol. 1997;268:724–738. doi: 10.1006/jmbi.1997.0992. [DOI] [PubMed] [Google Scholar]
- Goldman BS, Kranz RG. Evolution and horizontal transfer of an entire biosynthetic pathway for cytochrome c biogenesis: Helicobacter, Deinococcus, Archae and more. Mol Microbiol. 1998;27:871–873. doi: 10.1046/j.1365-2958.1998.00708.x. [DOI] [PubMed] [Google Scholar]
- Hamel P, Corvest V, Giege P, Bonnard G. Biochemical requirements for the maturation of mitochondrial c-type cytochromes. Biochim Biophys Acta. 2009;1793:125–138. doi: 10.1016/j.bbamcr.2008.06.017. [DOI] [PubMed] [Google Scholar]
- Hamel PP, Dreyfuss BW, Xie Z, Gabilly ST, Merchant S. Essential histidine and tryptophan residues in CcsA, a system II polytopic cytochrome c biogenesis protein. J Biol Chem. 2003;278:2593–2603. doi: 10.1074/jbc.M208651200. [DOI] [PubMed] [Google Scholar]
- Kern M, Scheithauer J, Kranz RG, Simon J. Essential histidine pairs indicate conserved haem binding in epsilonproteobacterial cytochrome c haem lyases. Microbiology. 2010;156:3773–3781. doi: 10.1099/mic.0.042838-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz R, Lill R, Goldman B, Bonnard G, Merchant S. Molecular mechanisms of cytochrome c biogenesis: three distinct systems. Mol Microbiol. 1998;29:383–396. doi: 10.1046/j.1365-2958.1998.00869.x. [DOI] [PubMed] [Google Scholar]
- Kranz RG. Isolation of mutants and genes involved in cytochromes c biosynthesis in Rhodobacter capsulatus. J Bacteriol. 1989;171:456–464. doi: 10.1128/jb.171.1.456-464.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz RG, Richard-Fogal C, Taylor JS, Frawley ER. Cytochrome c biogenesis: mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol Mol Biol Rev. 2009;73:510–528. doi: 10.1128/MMBR.00001-09. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Pervushin K, Bischof D, Braun M, Thony-Meyer L. Unusual heme-histidine bond in the active site of a chaperone. J Am Chem Soc. 2005;127:3716–3717. doi: 10.1021/ja044658e. [DOI] [PubMed] [Google Scholar]
- Mavridou DA, Ferguson SJ, Stevens JM. Cytochrome c assembly. IUBMB Life. 2013;65:209–216. doi: 10.1002/iub.1123. [DOI] [PubMed] [Google Scholar]
- Merchant SS. His protects heme as it crosses the membrane. Proc Natl Acad Sci U S A. 2009;106:10069–10070. doi: 10.1073/pnas.0905189106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer EH, Giege P, Gelhaye E, Rayapuram N, Ahuja U, Thony-Meyer L, Grienenberger JM, Bonnard G. AtCCMH, an essential component of the c-type cytochrome maturation pathway in Arabidopsis mitochondria, interacts with apocytochrome c. Proc Natl Acad Sci U S A. 2005;102:16113–16118. doi: 10.1073/pnas.0503473102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monika EM, Goldman BS, Beckman DL, Kranz RG. A thioreduction pathway tethered to the membrane for periplasmic cytochromes c biogenesis; in vitro and in vivo studies. J Mol Biol. 1997;271:679–692. doi: 10.1006/jmbi.1997.1227. [DOI] [PubMed] [Google Scholar]
- Nicholson DW, Neupert W. Import of cytochrome c into mitochondria: reduction of heme, mediated by NADH and flavin nucleotides, is obligatory for its covalent linkage to apocytochrome c. Proc Natl Acad Sci U S A. 1989;86:4340–4344. doi: 10.1073/pnas.86.12.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang N, Gao YG, Hu HY, Xia ZX. Crystal structures of E. coli CcmG and its mutants reveal key roles of the N-terminal beta-sheet and the fingerprint region. Proteins. 2006;65:1021–1031. doi: 10.1002/prot.21184. [DOI] [PubMed] [Google Scholar]
- Page MD, Pearce DA, Norris HA, Ferguson SJ. The Paracoccus denitrificans ccmA, B and C genes: cloning and sequencing, and analysis of the potential of their products to form a haem or apo- c-type cytochrome transporter. Microbiology. 1997;143 ( Pt 2):563–576. doi: 10.1099/00221287-143-2-563. [DOI] [PubMed] [Google Scholar]
- Page MD, Sambongi Y, Ferguson SJ. Contrasting routes of c-type cytochrome assembly in mitochondria, chloroplasts and bacteria. Trends Biochem Sci. 1998;23:103–108. doi: 10.1016/s0968-0004(98)01173-6. [DOI] [PubMed] [Google Scholar]
- Ramseier TM, Winteler HV, Hennecke H. Discovery and sequence analysis of bacterial genes involved in the biogenesis of c-type cytochromes. J Biol Chem. 1991;266:7793–7803. [PubMed] [Google Scholar]
- Rayapuram N, Hagenmuller J, Grienenberger JM, Bonnard G, Giege P. The three mitochondrial encoded CcmF proteins form a complex that interacts with CCMH and c-type apocytochromes in Arabidopsis. J Biol Chem. 2008;283:25200–25208. doi: 10.1074/jbc.M802621200. [DOI] [PubMed] [Google Scholar]
- Ren Q, Thony-Meyer L. Physical interaction of CcmC with heme and the heme chaperone CcmE during cytochrome c maturation. J Biol Chem. 2001;276:32591–32596. doi: 10.1074/jbc.M103058200. [DOI] [PubMed] [Google Scholar]
- Ren Q, Ahuja U, Thony-Meyer L. A bacterial cytochrome c heme lyase. CcmF forms a complex with the heme chaperone CcmE and CcmH but not with apocytochrome c. J Biol Chem. 2002;277:7657–7663. doi: 10.1074/jbc.M110979200. [DOI] [PubMed] [Google Scholar]
- Richard-Fogal C, Kranz RG. The CcmC:heme:CcmE complex in heme trafficking and cytochrome c biosynthesis. J Mol Biol. 2010;401:350–362. doi: 10.1016/j.jmb.2010.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard-Fogal CL, Frawley ER, Feissner RE, Kranz RG. Heme concentration dependence and metalloporphyrin inhibition of the system I and II cytochrome c assembly pathways. J Bacteriol. 2007;189:455–463. doi: 10.1128/JB.01388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard-Fogal CL, Frawley ER, Bonner ER, Zhu H, San Francisco B, Kranz RG. A conserved haem redox and trafficking pathway for cofactor attachment. Embo J. 2009;28:2349–2359. doi: 10.1038/emboj.2009.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard-Fogal CL, San Francisco B, Frawley ER, Kranz RG. Thiol redox requirements and substrate specificities of recombinant cytochrome c assembly systems II and III. Biochim Biophys Acta. 2012;1817:911–919. doi: 10.1016/j.bbabio.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson IB, Stevens JM, Ferguson SJ. Dispensable residues in the active site of the cytochrome c biogenesis protein CcmH. FEBS Lett. 2008;582:3067–3072. doi: 10.1016/j.febslet.2008.07.052. [DOI] [PubMed] [Google Scholar]
- San Francisco B, Bretsnyder EC, Rodgers KR, Kranz RG. Heme ligand identification and redox properties of the cytochrome c synthetase, CcmF. Biochemistry. 2011;50:10974–10985. doi: 10.1021/bi201508t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Francisco B, Kranz RG. Interaction of HoloCcmE with CcmF in Heme Trafficking and Cytochrome c Biosynthesis. Journal of Molecular Biology. doi: 10.1016/j.jmb.2013.10.025. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders C, Turkarslan S, Lee DW, Onder O, Kranz RG, Daldal F. The cytochrome c maturation components CcmF, CcmH, and CcmI form a membrane-integral multisubunit heme ligation complex. J Biol Chem. 2008;283:29715–29722. doi: 10.1074/jbc.M805413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders C, Turkarslan S, Lee DW, Daldal F. Cytochrome c biogenesis: the Ccm system. Trends Microbiol. 2010;18:266–274. doi: 10.1016/j.tim.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer EB, Barker PD. Continued surprises in the cytochrome c biogenesis story. Protein Cell. 2012;3:405–409. doi: 10.1007/s13238-012-2912-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz H, Hennecke H, Thony-Meyer L. Prototype of a heme chaperone essential for cytochrome c maturation. Science. 1998;281:1197–1200. doi: 10.1126/science.281.5380.1197. [DOI] [PubMed] [Google Scholar]
- Schulz H, Fabianek RA, Pellicioli EC, Hennecke H, Thony-Meyer L. Heme transfer to the heme chaperone CcmE during cytochrome c maturation requires the CcmC protein, which may function independently of the ABC-transporter CcmAB. Proc Natl Acad Sci U S A. 1999;96:6462–6467. doi: 10.1073/pnas.96.11.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setterdahl AT, Goldman BS, Hirasawa M, Jacquot P, Smith AJ, Kranz RG, Knaff DB. Oxidation-reduction properties of disulfide-containing proteins of the Rhodobacter capsulatus cytochrome c biogenesis system. Biochemistry. 2000;39:10172–10176. doi: 10.1021/bi000663t. [DOI] [PubMed] [Google Scholar]
- Simon J, Hederstedt L. Composition and function of cytochrome c biogenesis System II. Febs J. 2011;278:4179–4188. doi: 10.1111/j.1742-4658.2011.08374.x. [DOI] [PubMed] [Google Scholar]
- Stevens JM, Daltrop O, Higham CW, Ferguson SJ. Interaction of heme with variants of the heme chaperone CcmE carrying active site mutations and a cleavable N-terminal His tag. J Biol Chem. 2003;278:20500–20506. doi: 10.1074/jbc.M212925200. [DOI] [PubMed] [Google Scholar]
- Turkarslan S, Sanders C, Ekici S, Daldal F. Compensatory thio-redox interactions between DsbA, CcdA and CcmG unveil the apocytochrome c holdase role of CcmG during cytochrome c maturation. Mol Microbiol. 2008;70:652–666. doi: 10.1111/j.1365-2958.2008.06441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida T, Stevens JM, Daltrop O, Harvat EM, Hong L, Ferguson SJ, Kitagawa T. The interaction of covalently bound heme with the cytochrome c maturation protein CcmE. J Biol Chem. 2004;279:51981–51988. doi: 10.1074/jbc.M408963200. [DOI] [PubMed] [Google Scholar]
- Verissimo AF, Yang H, Wu X, Sanders C, Daldal F. CcmI subunit of CcmFHI heme ligation complex functions as an apocytochrome c chaperone during c-type cytochrome maturation. J Biol Chem. 2011;286:40452–40463. doi: 10.1074/jbc.M111.277764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Merchant S. A novel pathway for cytochromes c biogenesis in chloroplasts. Biochim Biophys Acta. 1998;1365:309–318. doi: 10.1016/s0005-2728(98)00085-1. [DOI] [PubMed] [Google Scholar]
- Zheng XM, Hong J, Li HY, Lin DH, Hu HY. Biochemical properties and catalytic domain structure of the CcmH protein from Escherichia coli. Biochim Biophys Acta. 2012;1824:1394–1400. doi: 10.1016/j.bbapap.2012.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.