Abstract
We have identified a new variant of human Stat5a, found at higher ratios to full-length Stat5a in invasive ductal carcinoma versus contiguous normal tissue. The variant, missing exon 5, inhibits p21 and Bax production and increases cell number. After prolactin stimulation, only full-length Stat5a interacts with the vitamin D and retinoid X receptors, whereas only Δ5 Stat5a interacts with activating protein 1–2 and specificity protein 1. Prolactin also oppositely regulates interaction of the two Stat5a forms with β-catenin. We propose that a change in splicing leading to upregulation of this new isoform is a pathogenic aspect of invasive ductal carcinoma.
1. Introduction
The signal transducer and activator of transcription (Stat) 5a is activated in response to a wide variety of cytokines. Those acting directly on mammary epithelium include growth hormone and prolactin (PRL) [1]. Once activated, Stat5a homo- or hetero-dimerizes and enters the nucleus where it functions as a transcription factor [2]. In the mammary gland, activation of Stat5a is important to both growth and differentiation [3,4]. Particularly because of the mediation of growth, the role of Stat5a in the development of breast cancer has been examined, and is complex. For example, mammary tumorigenesis is delayed when the Stat5a gene is deleted [5], suggesting a tumor-promoting role for Stat5a. At the same time, higher nuclear Stat5 activation is associated with a less invasive phenotype and a better prognosis [6,7]. Thus, a dual role for activated Stat5a, in which Stat5a promotes tumor initiation, but is also important for the maintenance of differentiation of more established tumors, has been proposed [8]. Having serendipitously identified a new N-terminal splice variant during cloning of the accepted full length variety, we asked whether the apparent duality of Stat5a in breast cancer might result from the presence of this previously unrecognized splice variant. The new variant of human Stat5a, missing thirty amino acids of the N-terminus corresponding to the whole of exon 5, was named Δ5 Stat5a. Compared to full length (FL) Stat5a, this alternatively-spliced form interacts differently with other transcription factors and Stat5a-responsive promoters. Increased expression of Δ5 Stat5a results in increased cell number. Furthermore, Δ5 Stat5a is expressed in invasive ductal carcinoma at greater ratios to FL Stat5a than in contiguous histologically-normal tissue. These results suggest that Δ5 Stat5a plays a role in tumor initiation, while FL Stat5a is likely more important to maintenance of tissue differentiation.
2. Materials and Methods
Additional experimental details can be found in the appropriate figure legends.
2.1 Cell Culture
HEK 293, MCF-7, MCF-10a, MCF-12a, T47D, MDA MB468, HUVEC, U2OS, SaOS, PC3 and DU145, were obtained from American Type Culture Collection (Manassus, VA), where they were authenticated by short tandem repeat DNA profiling. Cells were cultured in media and supplements from Invitrogen (Carlsbad, CA).
2.2 Cloning
Two pairs of primers were designed to amplify two fragments of the Stat5a coding sequence, an N-terminal fragment and a C-terminal fragment, and about 150 bp of overlay. For the 5'-terminal fragment, the forward and reverse primers were: 5'-GCT GCT CTC CGC TCC TTC CTG TAG TAA C-3' (S)/5'-GCT CAT TGC TGC CAA CAC TGA ACT G-3'(A) and for the 3'-terminal fragment, the forward and reverse primers were: 5'-AAT GAG AAC ACC CGC AAC GAG-3' (S) /CAA CAC GAC CGC TTC ACA TTG-3'(A). One more PCR reaction was designed to combine the two fragments using 5'-GAC ACG CGT ACC ATG GCG GGC TGG ATC CAG-3'(S)/ 5'-AAC GGT ACC A TGA GAG GGA GCC TCT GGC AGA-3'(A). The cleavage sites for Mlu I and KpnI (shown in boldface) were designed to ligate the STAT5a cDNA forms into desired plasmids.
2.3 Transient transfection and promoter constructs
For experiments examining promoter activation, dimerization, phosphorylation, and subcellular localization of Δ5 Stat5a versus FL Stat5a, transient transfections were performed using Lipofectamine™ 2000 (Invitrogen) a day after plating when the cells were 90%–95% confluent. The cells were examined 48 hours later. Controls included luciferase unlinked to either form of Stat5a such that this could be used as a measure of transfection efficiency. For the bioluminescence resonance energy transfer (BRET) experiments, the signal generated by unlinked luciferase together with unlinked green fluorescent protein (GFP), was used as a measure of background random molecular interactions. An artificial promoter containing 3 identical Stat5 response elements linked to a Renilla luciferase (Rluc) reporter [9] and a β-casein-luc plasmid, which contains 2.4 kb of the proximal β-casein promoter [10], were generous gifts from Drs. Linda Schuler (Madison, WI) and Jeffrey Rosen (Baylor, TX). To study p21 and Bax promotor activity, constructs containing the full-length p21 [11] or Bax [12,13] promoter regions linked to a luciferase reporter were used, as described previously [14]. To determine the potential differential ability of FL and Δ5Stat5a to tetramerize, promoters with two identical Stat5 response elements separated by 1–25 nucleotides linked to a luciferase reporter were constructed (see diagram in supplementary data figure 1). These were co-transfected with long form prolactin receptor (PRLR) and the FL Stat5a construct into HEK 293 cells and 24h later were exposed to PRL for a further 24h. After establishing the optimal spacer length to achieve synergy between the two Stat5 response elements in this system, the effect of Δ5 Stat5a and FL Stat5a on promoter activity with 1 and 2 GAS sites (the latter separated by the determined optimum) was determined under the same conditions.
2.4 Dicer substrate small interfering RNA (DsiRNA) for knockdown of Δ5 Stat5a
Cells were transfected with 1μg DsiRNA for Δ5Stat5a or the universal negative control (IDT Technology, San Diego, CA). The custom DsiRNA Δ5Stat5a duplex sequence was:
5'-CACGCAGCUCCAGUGCAGCUCUCCG-3' (S)
5'-CGGAGAGCUGCACUGGAGCUGCGUGGC-3`(A)
Cells were collected 24 hours after transfection and RNA/protein expression was analyzed by RT-PCR using the Δ5 Stat5a-specific primers and Western blot using the C-terminal antibody that recognizes both forms of the protein.
2.5 Bioluminescence Resonance Energy Transfer
BRET analyses were performed as previously described [15] using vectors carrying FL Stat5a or Δ5 Stat5a, the long form of the PRLR or Stat5b upstream of modified green fluorescent protein (GFP2) or Renilla luciferase (Rluc). Energy transfer was defined as the BRET ratio (Emission500nm–520nm-background500nm–520nm)/ (Emission385nm–420 nm-background 385nm–420 nm). Signals obtained from non-transfected cells and co-transfected plasmids expressing unlinked Rluc and GFP2 were considered background.
2.6 Viable Cell Number (MTS assay)
This assay, which measures reduction of MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazo lium, inner salt] by mitochondria in viable cells, was performed under strict conditions, as previously described [16].
2.7 Generation of FL and Δ5 Stat5a recombinant adenoviruses
Recombinant adenoviral vectors expressing either FL or Δ5 Stat5a were constructed according to the Stratagene (La Jolla, CA) protocol. Essentially, a replication-deficient E1- and E2-deleted Ad5 vector, AdEasy, was used as a backbone. This allowed for the addition of a hemagglutinin (HA) tag on the Stat5a molecules and regulation of expression by a human cytomegalovirus promoter/enhancer. The Stat5a cDNA sequences were amplified from a eukaryotic expression vector and subcloned into a shuttle vector, pShuttle-IRES-GFP-1. The cDNA sequences were then uploaded from the shuttle vector onto the adenovirus genome through homologous recombination. Once a recombinant was identified, bulk production used the recombination-deficient XL10-Gold® strain. Purified Ad plasmid DNA was digested with Pac I to expose its inverted terminal repeats, and then used to transfect AD293 cells (Stratagene) where deleted viral assembly genes are complemented. Titer in plaque-forming units (p.f.u) was determined following infection of AD293 cells. The multiplicity of infection (MOI) was defined as the ratio of the number of p.f.u used to the number of cells to be infected. To assess the efficiency of the adenovirus-mediated gene transfer, cells were co-infected with AdlacZ and stained for β-galactosidase.
2.8 Chromatin Immunoprecipitation Assay (ChIP Assay)
Cells were infected and treated as described in the figure legend. Human PRL was purified from Escherichia coli as described previously [17]. Cross-linking was performed with 1% formaldehyde in Dulbecco's phosphate-buffered saline (DPBS) at room temperature for 5–10 min. To arrest cross-linking, glycine was added directly to the medium to a final concentration of 125 mM for 10 min, and the cells were then rinsed twice with ice-cold DPBS. Cell extracts were harvested with lysis buffer (0.5 ml/35mm plate of 10 mM Tris-HCl, 1 nM EDTA, pH 8.0, 0.5% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride; 1 μg/ml aprotinin; 1 μg/ml leupeptin) and frozen in liquid nitrogen. Lysates were transferred to 15-ml tubes containing 25 μm glass beads and the lysate/bead mixture was sonicated (Sonic dismembranator, Fisher Scientific, Pittsburg, PA) in ice (9 pulses of 20 sec at a setting of 6 watts), yielding chromatin fragments of about 500bp in size. After centrifugation at 14000 × g for 4 min at 4°C, supernatants were collected. To provide a positive control (input), 200 μl of the supernatant was set aside. Each immunoprecipitation was performed with ~25μg chromatin in RIPA buffer (140 mM NaCl; 1 mM EDTA; 10 mM Tris-HCl; 1 mM phenylmethylsulfonylfluoride; 1% Triton X-100; 0.1% sodium dodecyl sulphate; 0.1% Na-deoxycholate, pH 8). To reduce non-specific background, each chromatin sample was pre-cleared with 60 μl of a suspension of protein A-sepharose, supplemented with 300 μg/ml sonicated salmon sperm DNA (Fermentas) and 1 mg/ml BSA, for 1 hour at 4°C on a rotating wheel. Chromatin complexes were immunoprecipitated for 16–18 hours at 4°C while rotating with 10 μg primary antibody (anti-HA, Santa Cruz, CA), or with a non-specific antibody to provide a negative control. Immune complexes were collected with 40 μl protein A-sepharose (10 mg/ml binding capacity), supplemented with 300 μg/ml sonicated salmon sperm DNA and 1 mg/ml BSA for 3 hours at 4°C on a rotating wheel, followed by centrifugation at 14000 × g for 30 seconds. Pellets were washed once with LiCl buffer (10 mM Tris-HCl, pH 8; 250 mM LiCl; 1 mM EDTA, pH 8; 0.5 % Nonidet P-40; 0.5% Na-Deoxycholate), and twice with TE buffer (10 mM Tris-HCl; 1 mM EDTA, pH 8). After standard ribonuclease and proteinase K treatment, overnight at 37°C, cross-linking was reversed by incubation for 6 hours at 45°C. DNA was purified by phenol/chloroform extraction and precipitated in the presence of 100 μg of glycogen per sample. PCR amplification was performed using primers specific for various regions of the β-casein promoter (see figure for regions). The primers used were as follows:
primer1:5'-ggaatgacaaaggcgacattgccaccaacc-3'(S)/5'-gcatctatgttcatcaggggtattggcata-3'(A)
primer 2: 5'-ctctaagctcatatttcttattccatgatt-3'(S)/5'-cagatatgaaggcaggataaagggtaaaat-3'(A)
primer 3: 5'-gattagtgtgactaaaaattgatgagcatt-3'(S)/5'-atatcatcctagatctatgaatgaggggga-3'(A)
primer 4: 5'-agctcaaacttggtcagtttcattttaata-3'(S)/5'-aagcagttaatcgaggaggatagagaattc-3'(A)
primer 5: 5-actgtcctccagtcattgtcttttgttatt-3'(S)/5'-atgacacactaattttgtgatttgaaataa-3'(A).
2.9 Processing of Tumor Tissues and Quantitative Real Time Polymerase Chain Reaction (qRT-PCR)
Breast tissues were homogenized in TRIzol (100mg/ml). RNA was extracted and the quality determined by OD 260/280nm (>1.8) by NanoDrop before conversion to cDNA. SYBR Green qPCR was performed with an ABI Prism 7700 sequence detector with normalization to β-actin. Primers, specific to FL or Δ5 Stat5a were: FL: 5'-TGG TCC GAG AAG CCA ACA-3'(S)/ 5'-AG GCT CTC CTG GTA CTG GAT-3'(A). Δ5:5'-GCA GCT CCA GTG CAG CTC TC-3'(S)/5'-CA AAC TGA GCT TGG ATC CTC-3'(A). Gel analysis of products demonstrated the specificity of these primers and qPCR demonstrated equivalent efficiencies (Average Ct for FL primers was 17.20 and for Δ5 primers was 17.27). The amplicon product is 1044 bp for FL and 1034 bp for Δ 5. The Δ 5 primers were also used for semi-quantitative PCR.
2.10 Gel electrophoresis and General Western Blotting
Cell lysates were prepared in the presence of protease and phosphatase inhibitors (Promega Madison, WI). In order to obtain well-separated bands, electrophoresis occurred for 4–5 hours in 7 cm 9% SDS minigels at 100V. Western blotting of multiple cell lines was with an antibody to the C-terminus of Stat5a (L20) that should recognize both forms equally. When applicable, anti-TATA binding protein (N12) was used to verify the purity of nuclear and cytoplasmic fractions. For phosphorylation, cells were transfected with plasmids to overexpress either FL Stat5a or the Δ5 Stat5a-GFP construct. Immunoprecipitation was with either anti-Stat5 (836) or anti-GFP (5384) and immunoblotting was with anti-phosphotyrosine (508) or anti-GFP. No immunoprecipitation occurred prior to blotting with anti-p21(397) or anti-BAX (625102). Localization was with an appropriate species-specific peroxidase-linked second antibody. All antibodies were from Santa Cruz Biotechnology except for anti-BAX (Biolegend, San Diego, CA). For each antibody, the catalogue number is provided in parentheses. Quantification of Western blots used photoshop image analysis software.
2.11 Transcription factor interactions
TF-TF Array (Panomics/Affymetrix, Santa Clara, CA): Cells were treated as described in the figure legend. Nuclear extracts were prepared (Active Motif, Carlsbad, CA) and incubated with biotin-labelled, double-stranded oligonucleotide probes at 15°C for 30 min. An antibody against the C-terminus of Stat5a that equally recognizes both forms was used to bind FL and Δ5 Stat5a and any associated transcription factor, along with the corresponding labelled probes. The oligo/protein/antibody complexes were precipitated using avidin-Dynabeads, eluted from the beads and hybridized with the TF-TF interaction array membrane (Array I, pre-spotted with 54 cis-elements) overnight. The hybridized spots on the membrane were incubated in peroxidase-labelled second antibody.
Co-immunoprecipitations: Cells were transfected with 3 μg plasmids (FL Stat5a-GFP, Δ5STAT5a-GFP or PCDNA 3.1 empty plasmid as control). Anti-GFP (2μg) was added to 200ug total cell lysate, and 20ug total cell lysate with control plasmid was used as control. Anti-Sp1(59G) and -RXR (D20) were from SantaCruz and anti-β-catenin was from BD Transduction (San Jose, CA).
2.12 Patient samples
Tumors and adjacent normal regions were obtained at surgery, identified by a pathologist as early invasive ductal carcinoma with no lymph node positivity, and forwarded as anonymous samples by institutions working as part of the National Cancer Institute's Cooperative Human Tissue Network (CHTN) http://www.chtn.nci.nih.gov/human-subjects/. Each branch of the network obtains written informed patient consent for the banking and distribution of samples for research. Most samples in this study were obtained from the Midwestern Division of CHTN and their statement about informed consent can be found at https://htrn.osu.edu/Services/TissueProcurement/Pages/default.aspx. Samples are distributed by CHTN branches after coding for anonymity. Use of anonymous patient samples in this study was approved by the University of California, Riverside, Institutional Review Board.
2.13 Statistical analyses
All statements are based on replicated experiments (a minimum of three times) except for the surveys of cell lines designed to determine the simple presence or absence of either Δ5 Stat5a mRNA or protein. For quantitative data, analysis was by ANOVA with post-tests and, where applicable, corrections for multiple comparisons.
3. Results
3.1 A new variant of human Stat5a
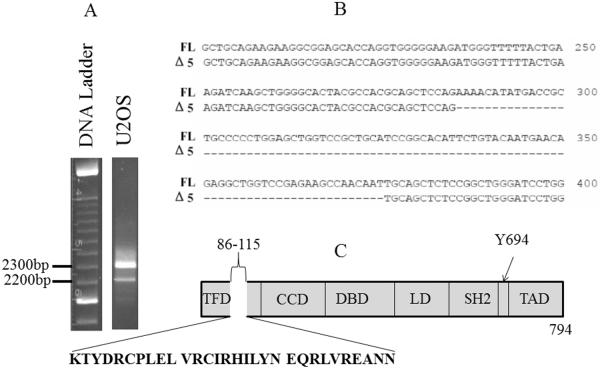
Stat5a cDNA was amplified by PCR. Unexpectedly, agarose electrophoresis showed two PCR products: one corresponded to FL Stat5a, but the other was shorter (result for U2OS osteosarcoma cells shown in figure 1A). Sequence analysis showed that the smaller fragment was shorter by 90 nucleotides and missing the region corresponding to exon 5 (figure 1B). Exon 5 codes for a small part of the N-terminal region (figure 1C). Modelling using the Geno3D program (http://geno3d-pbil.ibcp.fr) predicted an overall very similar structure in which the major difference was the absence of a small α helix, indicated by the white oval (figure 1D).
Figure 1. A new variant of Stat5a missing the sequence corresponding to exon 5.
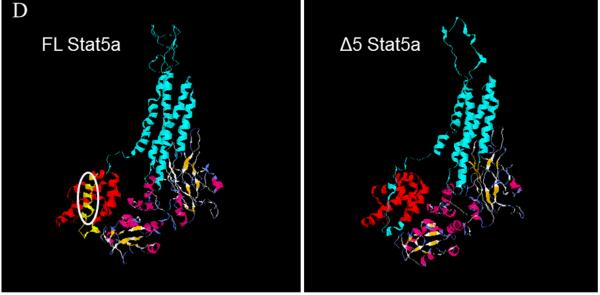
A: an ethidium bromide stained agarose gel illustrating two cDNA bands corresponding to full length (FL) and Δ5 Stat5a (Δ5), in this case derived from U2OS human osteosarcoma cells. B: Sequence alignment of the region of deletion between FL and Δ5 Stat5a. Broken line represents the missed nucleotides. C: Cartoon showing the missing portion of the Stat5a protein. TFD, tetramer forming domain; CCD, coiled coil domain; DBD, DNA binding domain; LD, linker domain; SH2, src homology domain 2; TAD, transactivation domain. The amino acid sequence of exon 5 is shown, as is the tyrosine phosphorylation site essential for dimerization. D: model of Δ5 Stat5a structure compared to FL Stat5a. Oval on FL model shows missing region, which is an α-helix shown in yellow at the N-terminus.
3.2 Delta 5 Stat5a is widely expressed
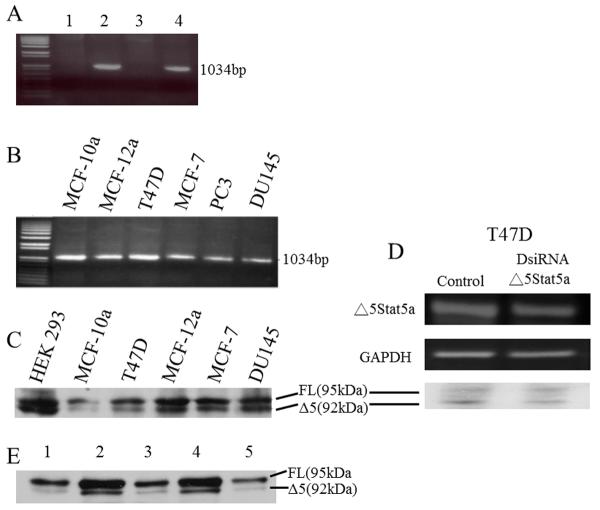
To rule out the possibility that the shorter product of Stat5a was due to an error during cloning or was specific to aberrant splicing in one cell line, multiple cell types were examined for potential expression of the variant using primers specific for the Δ5 form. The specificity of the primers is illustrated in figure 2A. The additional human cell lines examined were a second osteosarcoma line (SaOS), embryonic kidney (HEK 293), vascular endothelial (HUVEC), prostate cancer (PC3, DU145), and normal and cancerous breast epithelial (MCF-10a, MCF-12a, MCF-7, T47D) cells. Expression was evident in each of these cell types at the mRNA level (a selection is shown in figure 2B). It is apparent therefore that expression is not an oddity of a particular cell line, but occurs in cell lines considered relatively normal, as well as those that are cancerous. No equivalent form of Stat5b could be detected (negative data not shown).
Figure 2. Expression of Δ5 Stat5a at mRNA and Protein Level.
A: Ethidium bromide stained gel showing the specificity of the primers for FL and Δ5 Stat5a. Lane 1, primers specific to FL used with plasmid for Δ5; lane 2, primers specific for FL used with plasmid for FL(amplicon of 1044bp); lane 3, primers specific for Δ5 used with plasmid for FL; lane 4, primers specific for Δ5 used with plasmid for Δ5 (amplicon of 1034bp). B: ethidium bromide stained gel showing Δ5 Stat5a-specific amplicons produced from cDNA in each cell type. C: Western blot showing both FL and Δ5 Stat5a protein in mostly the same cell lines as shown in B. The antibody used was against the C-terminus of Stat5a and therefore recognized both FL and Δ5 Stat5a protein. D: Effect of DsiRNA knockdown of Δ5 Stat5a on mRNA (upper panel) and protein (lowest panel). 5×105 cells were seeded in 35mm wells. The next day, cells were transfected with 1μg DsiRNA or control. Cells were collected 24 hours after transfection and RNA/protein expression was analyzed by RT-PCR and Western blot. E: Western blot of FL and Δ5 Stat5a from five HUVEC cultures.
Because some mRNA splice variants are not expressed as proteins, the next question was whether Δ5 Stat5a was expressed at the protein level. Because the protein size is not very different from the FL Stat5a (only 30 amino acids in a 90kDa protein), several different electrophoresis conditions were tried to determine how to adequately separate the two forms. The Western blot in Figure 2C shows electrophoretic separation of the proteins and a sampling of the cell lines, all of which expressed a protein of the correct size, as well as the mRNA. To demonstrate that the lower protein band was indeed the Δ5 Stat5a variant, we used a DsiRNA. This was designed to target part of exon 4 and part of exon 6 and so was specific to the Δ5 Stat5a splice form. Transfection with the DsiRNA knocked down expression at the mRNA and protein levels (figure 2D). An analysis of five separate HUVEC cultures showed that Δ5 Stat5a constituted 38.4 ± 6.75% (SD) of total Stat5a. In other words, FL Stat5a was present at about a 2:1 ratio with Δ5 Stat5a in this low passage number, normal cell line (figure 2E). Similar analyses of T47D cells and DU145 cells showed more variable results. Testing of five independent cultures of T47D cells on one occasion gave a ratio of 1.15 ± 0.17 SD, while similar analysis of DU145 cells gave a ratio of 0.55 ± 0.23 SD. However greater variability than this in the ratio in cancerous cell lines is illustrated by the difference between the number for T47D and DU145 cells and the ratios shown in figure 2C. The cause of the variability from one occasion to another is unknown, but could be related to cell density at the time of sampling, growth curve kinetics, or passage number. Without a constant ratio, no conclusions can be drawn about ratios in relatively normal versus cancerous cell lines.
3.3 Delta 5 Stat5a homodimerizes and heterodimerizes with FL Stat5a/b and the PRLR
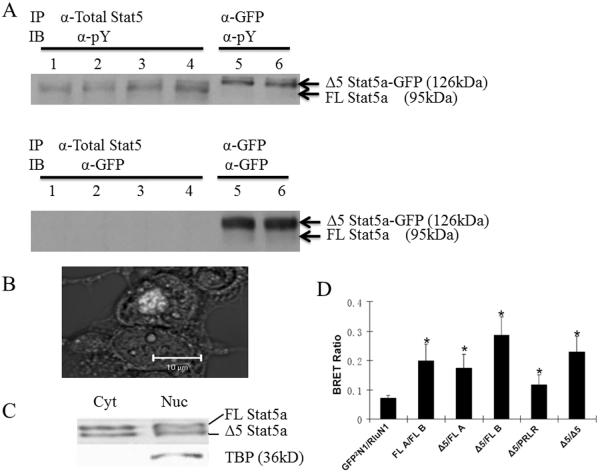
FL Stat5a binds to a tyrosine-phosphorylated transmembrane receptor and, after its own phosphorylation by a receptor-associated tyrosine kinase, then dimerizes with itself or Stat5b before translocation to the nucleus [8]. Because of the similarity in molecular weight between Stat5b and Δ5 Stat5a and the absence of any specific anti-phospho Stat5a antibody, we used a GFP2-tagged variety to determine whether Δ5 Stat5a was phosphorylated. The GFP2-tagged Δ5 Stat5a runs at a molecular weight of 126kDa and separates nicely from other forms of Stat5. Figure 3A shows phosphorylation of both FL and Δ5 Stat5a forms in response to serum in MCF7 cells. In these early studies of general function, serum stimulation was used in order to determine the likely general level of activation of this new form of Stat5a under most conditions. To determine whether nuclear translocation occurred, HEK293 cells were transiently transfected with a plasmid carrying Δ5 Stat5a-GFP2 cDNA. At the level of transfection employed, GFP-labeled Δ5 Stat5a was not very obvious when dispersed in the cytosol. However, when concentrated by nuclear translocation, the fluorescence becomes obvious. Figure 3B shows concentrated nuclear fluorescence in a successfully transfected cell. No nuclear translocation was observed in serum-starved cells (not shown). Nuclear translocation was further substantiated by producing nuclear and cytoplasmic fractions from T47D cells after PRL stimulation to demonstrate translocation of the endogenous rather than transfected forms. TATA binding protein was used as a marker for purity of the nuclear and cytoplasmic fractions (figure 3C). To determine the binding capabilities of Δ5 Stat5a, BRET analysis was performed. In this technique, the two molecules being examined for binding are tagged with either luciferase or GFP2. With close approximation (within 100 Å) and upon addition of cell permeant luciferase substrate, luminescence energy is transferred to GFP2 and then emitted at 510 nm. A BRET ratio greater than the control (GFP2N1/RluN1) shows binding. Thus, Δ5 Stat5a behaved like FL Stat5a in terms of its ability to interact with the long form of the PRLR, homodimerize, and heterodimerize with both FL Stat5a and Stat5b (figure 3D).
Figure 3. Phosphorylation, nuclear translocation and Initial Protein-Protein Interactions.
A: Western blot of immunoprecipitated protein extracts showing tyrosine phosphorylated (pY) FL and GFP-tagged Δ5 Stat5a from MCF7 cells. Lanes 1 and 2 show endogenous Stat5, lanes 3 and 4 endogenous plus additionally expressed transfected FL Stat5a, and lanes 5 and 6 the higher molecular weight transfected GFP-tagged Δ5 Stat5a. The lower panel shows the anti-GFP blot from the same lysate. B: confocal image (Zeiss 510) shows nuclear translocation of GFP2-tagged Δ5 Stat5a. Δ5 Stat5a-GFP2 was transiently expressed in HEK 293 Cells (in the presence of 10% FBS, which contains factors that can activate Stat5a). Fluorescence was examined 72 hours after transfection in a non-synchronized culture. The photograph is a merge of the fluorescence and phase contrast images. C: Nuclear translocation of endogenous FL and Δ5 Stat5a in T47D cells. Nuclear and cytoplasmic fractions were produced using the NE-PER kit (Pierce, ThermoFisher, Rockford, IL) after a 10 min incubation in PRL and then processed for Western blotting. TBP, TATA binding protein. D: Cultured HEK 293 cells were co-transfected with appropriate plasmids and a BRET2 assay was performed. The higher the BRET ratio, the greater the amount of energy transferred as a result of proximity. Δ5 Stat5a hetero-dimerized with FL Stat5a and Stat5b (compare Δ5/FLA and Δ5/FLB with FLA/FLB), and homo-dimerized with itself (Δ5/Δ5). It was also able to associate with the PRL receptor (Δ5/PRLR). * P<0.05 vs GFP2N1/RlucN1 (negative control testing random interactions between luciferase and GFP2).
3.4 Delta 5 Stat5a binds to Stat5 response elements (GAS sites)
To determine whether Δ5 Stat5a competed with FL for binding at response elements, we first used an artificial promoter consisting of multiple response elements linked to a luciferase reporter. This promoter construct was transiently transfected into HEK 293 cells along with expression vectors for FL and increasing amounts of Δ5 Stat5a. In addition to affording high efficiency transfection, HEK 293 cells were chosen since analyses showed a fairly stable ratio of approximately equal amounts of the two forms under normal circumstances. This allowed for a more straightforward analysis of response to increased expression of one form versus the other. Given that the plasmids only differed by 90 nucleotides, it is likely that expression levels were very similar. Certainly, under circumstances where we could distinguish the transfected from endogenous by using GFP-tagged constructs (see later figures), this was the case. Equal amounts of transfected DNA were achieved for each set of cells by using the GFP2N1 plasmid. When cells were incubated in serum and only the FL Stat5a was co-transfected, the luciferase construct was highly expressed. When tested for its ability to activate this artificial promoter, Δ5 Stat5a had only about 10% the activity of FL Stat5a (data not illustrated). However, when titrated against FL Stat5a, co-transfection with only one tenth the amount of Δ5 Stat5a construct reduced promoter activity in response to FL Stat5a 24 fold. Increasing amounts of Δ5 Stat5a showed a dose-related further decrease (figure 4A).
Figure 4. Competition at Stat5 Response Elements and Dominant Negative effect of Δ5 Stat5a on β-casein gene promoter and expression.
A: HEK 293 cells were transiently co-transfected with a multiple response element-luc reporter and constructs containing FL and/or Δ5 Stat5a cDNA, and incubated in serum. B: HEK 293 cells were transiently co-transfected with a β-casein promoter-luciferase construct and constructs containing FL and/or Δ5 Stat5a cDNA. In this case, all cells were simultaneously transfected with a long form PRLR construct. Relative light units (RLU) were measured 24 hours after incubation in 1μg/mL PRL. For each experiment, equal amounts of total DNA were transfected for each group. C: T47D cells were infected with an adenovirus (Adv) carrying FL or Δ5 Stat5a cDNA at MOI 30 for 2 hours. After 48 hours, regular and real time RT-PCR was performed to detect β-casein mRNA, normalized to β-actin mRNA. D: HEK 293 cells were transiently co-transfected with an artificial promoter containing 1 or 2 Stat 5 response elements (sequence in supplementary figure 1) linked to a luciferase reporter, FL or Δ5 Stat5a, and long form PRLR. After 24h, PRL (1μg/ml) was added for a further 24 hours. **,p<0.01, compared to Con; # p<0.05, ## p<0.01,compared to FL.
In order to use a more biologically-relevant promoter, a 2.4 kb β-casein promoter upstream of a luciferase reporter (β-casein-luc) was used. Also co-transfected was a plasmid coding for the long form of the PRLR so that we could examine the response to a specific stimulus over and above the mixture of stimuli present in serum. In these cells growing in serum-supplemented medium and stimulated with PRL, increased expression of FL Stat5a induced the β-casein-luc promoter compared to control, whereas increased expression of Δ5 Stat5a inhibited promoter activity (figure 4B). In cells co-transfected with a fixed amount of FL Stat5a plus increasing amounts of Δ5 Stat5a, Δ5 Stat5a acted as a dominant negative, although the degree of effect was not as great as for the artificial promoter.
In order to examine the effect of Δ5 Stat5a on endogenous expression of the β-casein gene, T47D breast cancer cells were infected with adenovirus carrying FL or Δ5 Stat5a cDNA. Infection with FL adenovirus tripled β-casein expression in response to PRL, whereas Δ5 Stat5a had no effect (figure 4C). Nevertheless, Δ5 Stat5a reduced FL-induced β-casein expression such that one fifth the amount of Δ5 Stat5a virus brought levels induced by FL Stat5a down to control values.
With the novel variant described here, the alteration in structure is the absence of a small alpha helix. We therefore considered the possibility that this would affect the ability of Δ5 Stat5a to form tetrameric complexes [18]. To be sure we had a system in which we could demonstrate the advantage of tetramerization with the FL Stat5a, we titrated the number of residues between two Stat5 response elements linked to a luciferase reporter to be sure that we had the optimal distance for maximal activity (supplementary figure 1). The optimal number of residues was seven (supplementary figure 2). We then compared responses with one versus two optimally-spaced (7 residues apart) Stat5 response elements. With no enhancement due to tetramerization, one would expect double the activity with two versus one Stat5 response element. With enhancement, one would expect a result greater than double. Figure 4D shows the result for both FL and Δ5 Stat5a. With FL Stat5a, two Stat5 response elements gave a result that was 3.5 fold the result with 1. However, even though the response was much reduced with Δ5 Stat5a, two Stat5 response elements still gave a result that was 3.5 fold the result with 1. There was therefore no demonstrable effect on the ability of Δ5 Stat5a to tetramerize.
3.5 Delta 5 Stat5a and FL Stat5a show differences in binding to regions of the β-casein promoter
The differences in degree of effect when titrating Δ5 Stat5a against FL between the artificial promoter and the 2.4 kb β-casein promoter or endogenous β-casein gene could have been the result of differences in stimulus or possibly that FL and Δ5 Stat5a had different relative affinities for different Stat5 response elements in the β-casein promoter. To test the latter possibility, T47D cells were infected with adenovirus carrying FL or Δ5 Stat5a cDNA tagged with an HA sequence and subjected to ChIP analysis. The relative amount of β-casein promoter DNA associated with the anti-HA immunoprecipitated complexes was determined by PCR. Five pairs of primers (figure 5A) were designed to amplify regions containing putative/proven Stat5 response element sequences in the promoter. In figure 5B, the input columns show strong signals for each primer set. Neither FL nor Δ5 Stat5a bound to the region covered by primer set 5. However, FL Stat5a bound to the sequences amplified by each of the other sets of primers. By contrast, no signal was obtained for primer sets 1 and 2 when Δ5 Stat5a was used. This result demonstrates that the two forms of Stat5a differentially distinguish among Stat5a binding sites, suggesting differential effects on gene expression.
Figure. 5. DNA and other transcription factor binding properties of Δ5 and FL Stat5a.
Cultured T47D cells were infected with adenovirus carrying FL or Δ 5 Stat 5a cDNA tagged with HA (at the C-terminus) at MOI 30 for 2 hours. Forty eight hours post-infection, cells were exposed to PRL for 10 min. Cross-linking was performed and 9.4Kb of the β-casein promoter was used as the target. A: Five pairs of primers (primers 1, 2, 3, 4 and 5) were used to amplify regions containing different predicted Stat5 binding sites indicated by vertical bars. B: The samples were immunoprecipitated by an antibody against HA. Positive and negative controls are indicated as Input and IgG, respectively. C: Co-immunoprecipitation using GFP-tagged versions of Δ5 Stat5a and FL Stat5a as a function of time of incubation in 100 ng/ml PRL. Immunoprecipitation was with anti-GFP, followed by blotting with anti-GFP, β-catenin, RXR and Sp1. Grouped Westerns are from the same cell lysate for which the associated GFP staining is serving as loading control. D: Quantification of unstimulated interaction with Sp1 and RXR with FL and Δ5 Stat5a from multiple lysates. C, control. Results were normalized to the maximal interaction in each case, which was given the value 100.*, p<0.001.
3.6 Delta 5 Stat5a differs from FL Stat5a in its binding to other transcription factors
The three-dimensional model would predict very little if any effect of the deletion of exon 5 on the ability to bind DNA. Selectivity for different Stat5 binding sites might therefore result from interaction with other transcription factors. A blast search using the sequence of the helix missing in the Δ5 form, showed the greatest identity (53%) to an A blade region of the WD40 domain of β-transducin (β-TrCP1) [19]. β-TrCP1 is a highly conserved protein that controls the stability of proteins involved in transcription [19]. We therefore used an array to probe the interaction between FL or Δ5 Stat5a and 54 other transciption factors in T47D breast cancer cells. Even though both forms of Stat5a are present in T47D cells, infection was used to enhance expression of one form over the other so that potential differences in their interactions could be observed. Data presented in Table 1 show that Δ5 Stat5a interacts with mostly different transcription factors than FL Stat5a. This method cannot be considered quantitative since the oligonucleotide probes cannot be assumed to have equal efficiency, and on the array some spots saturate before others have developed; therefore interactions can only be recorded as positive. Both FL and Δ5 Stat5a interacted with activating protein-2 (AP-2) and the early growth response protein (EGR). For the other 10 positive spots, each only bound either FL or Δ5 Stat5a.
Table I.
Transcription factors interacting with FL and/or Δ5 Stat5a
| AP2 | EGR | CBF | NF1 | VDR | RXR | Stat3 | NFκB | MRE | AP1-2 | Sp1 | TR | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FL | + | + | + | + | + | + | + | |||||
|
| ||||||||||||
| Δ5 | + | + | + | + | + | + | + | |||||
T47D cells were infected with adenovirus carrying FL or Δ5 Stat5a cDNA. Ten minutes prior to nuclear extraction, the cells were incubated in the presence of PRL (500 ng/ml) to stimulate a Stat5-mediated response. Nuclear extracts were processed as described in methods and FL or Δ5 Stat5a-binding partners were identified using a TF-TF membrane array pre-spotted with 54 cis-elements in duplicate overnight. Results were replicated on three separate occasions. AP-1= Fos, FosB, Fra1, Fra2, Jun, JunB; AP-2, Acting enhancer binding protein 2; CBF, CAAT box general; EGR, early growth response; NF-1, NF-1 nuclear factor 1; RXR, retinoid X receptor; VDR, vitamin D receptor; Stat3, Signal Transducer and activator of transcription-3; SP1, Sp1 transcription factor; NFkB, nuclear factor of kappa light polypeptide gene enhancer in B-cells 1; TR, thyroid hormone receptor; MRE, which means that a metal response factor interacted.
Three transcription factors/interacting proteins were further explored by co-immunoprecipitation (figure 5C). In T47D cells, it is clear that an interaction with RXR is specific to FL Stat5a and an interaction with Sp1 is specific to Δ5 Stat5a (quantified in figure 5D). PRL stimulation of the cells dissociated the complex between RXR and FL Stat5a since the intensity of the band decreased as a function of time of incubation. By contrast, there was no clear dependence of Sp1 binding to Δ5 Stat5a on the presence of PRL.
Since the missing portion in Δ5 Stat5a resembled part of a protein that binds β-catenin (β-TrCP1), we also examined the possibility that β-catenin bound differentially to the Stat5a forms. However, both forms bound β-catenin. Interestingly though, PRL undid the interaction of β-catenin with Δ5 Stat5a and promoted the interaction of β-catenin with FL Stat5a. In MCF7 cells, less distinct (perhaps due to a lower expression of PRL receptors), but generally similar patterns were observed for β-catenin.
Thus, absence of the thirty amino acids corresponding to exon 5 has a major effect on complex formation with other transcription factors, although nuances are present in different cell lines.
3.7 Cell number-related effects
Given the differential interaction with other transcription factors, we asked whether Δ5 Stat5a had a differential effect on two Stat5a-responsive promoters, one related to cell cycle inhibition and the other to apoptosis. Overexpression of both forms of Stat5a in MCF7 cells inhibited promoter activity, but Δ5 Stat5a was far more efficacious, essentially eliminating activity at both the p21 and Bax promoters (figure 6A). Corresponding effects on protein were also observed (insets), and similar results were observed in T47D cells (data not shown). To test whether this translated to an effect on cell number, a variety of cells was infected at less than 50% confluency and incubated for 7 days in medium containing serum, and in one case in the absence or presence of PRL. The outcome varied in degree with cell line; in some instances, FL Stat5a had no effect and in others, it increased cell number. However, in all cases Δ5 Stat5a increased cell number and always more than FL Stat5a. Furthermore, PRL amplified the effect of increased expression of Δ5 Stat5a (figure 6B). Because each form of Stat5a associated with β-catenin, we also examined the effect of increased expression of each form on levels of β-catenin and its membrane-binding partner, E-cadherin. As shown in figure 6C, increased expression of both forms of Stat5a increased protein levels of E-cadherin, but only Δ5 Stat5a also increased β-catenin.
Figure 6. Effect of increased expression of Stat5a and Δ5 Stat5a on p21, Bax, β-catenin, E-cadherin and cell number.
A: MCF-7 cells were transiently transfected with expression and promoter-reporter constructs. Results (luciferase activity) are normalized to the value in the control (C) transfections. Insets are Western blots that were all run on the same gel, were processed for Western blotting and presentation together before rearrangment to display in the same order as the rest of the figure. B: MCF7, MDA-MB 468, T47D, and PC3 cells were infected with adenovirus carrying Δ5 or FL Stat5a cDNA at MOI 30 for 2 hours and relative viable cell number was assayed 7 days later. For PC3 cells, some cells were also stimulated with PRL. For T47D cells, actual cell numbers are presented as an extra control for possible non-specific effects of infection on the MTS assay. n=16 for each replicate. *, p<0.01,** p<0.02, *** p< 0.005, **** p< 0.0001 versus control. C: Western blot from the T47D cells.
3.8 Higher expression of Δ5 Stat5a was detected in breast cancer tissue
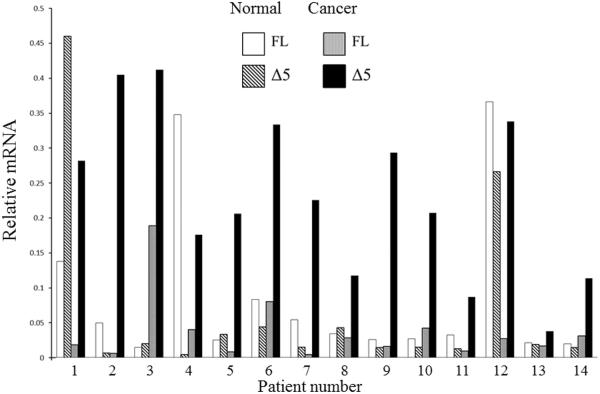
Overexpression can produce anomalous results and effects in cell lines, even if they had constant ratios of the two forms, may not be representative of primary tumors. We therefore sought to determine the expression of Δ5 Stat5a and FL Stat5a in patient samples. Samples were obtained from regions of invasive ductal carcinoma and from adjacent histologically-normal tissue. Results of qRT-PCR are shown in figure 7. The relative level of mRNA for each form in the two regions is presented for each patient. This is converted to a ratio in table 2; the ratio of Δ5 Stat5a to FL Stat5a was always higher in the cancerous region, although the magnitude of difference varied considerably. The difference between the ratios in normal versus cancerous tissue was significant with a p value = 0.007. With one exception, the ratio was either equal or showed greater amounts of FL Stat5a in the histologically-normal tissue.
Figure 7. Expression of Δ5 and FL Stat5a mRNA in Invasive Ductal Carcinoma and Histologically-Normal Adjacent Breast Tissue.
Results are shown as the relative amount of mRNA (determined by qRT-PCR with normalization to β-actin) for each form in the normal and cancerous region. Paired samples from 14 patients were analysed. The resultant ratios of the forms can be found in table 2.
Table II.
: Δ5 Stat5a to FL Stat5a mRNA expression ratio in 14 patients.
| Patient | Normal Δ5:FL | Cancer Δ5:FL | Fold Cancer v normal |
|---|---|---|---|
| 1 | 3.3 | 15.3 | 4.64 |
| 2 | 0.14 | 67.5 | 4.82 |
| 3 | 1.3 | 2.2 | 1.7 |
| 4 | 0.01 | 4.35 | 435 |
| 5 | 1.31 | 24.03 | 18.34 |
| 6 | 0.53 | 4.16 | 7.85 |
| 7 | 0.28 | 49.35 | 176 |
| 8 | 1.25 | 4.12 | 3.3 |
| 9 | 0.57 | 18.16 | 31.86 |
| 10 | 0.55 | 4.89 | 8.9 |
| 11 | 0.4 | 9.06 | 22.6 |
| 12 | 0.73 | 12.35 | 16.9 |
| 13 | 0.88 | 2.27 | 2.58 |
| 14 | 0.74 | 3.61 | 4.88 |
| Mean | 0.86 | 15.8 | 52.8 |
Pre-ratio values for 10 of these patients can be found in figure 7. The ratio in the cancer tissue is different from the ratio in the normal tissue with a p value of 0.007.
4. Discussion
4.1 Why has Δ5 Stat5a not been previously observed?
Cloning of Stat5a from a human cell line has uncovered a novel splice variant widely expressed in both normal and cancerous human epithelial cell lines, including those of the endothelium, embryonic kidney, breast and prostate; this variant is also present in osteosarcoma cell lines. Delta 5 Stat5a is therefore not an aberration particular to cancerous cells, even though its production is clearly increased in one or more cell type in primary human breast cancer tissue. Delta5 only differs from FL Stat5a by 30 amino acids, or 3.8% of the total mass. This likely explains its lack of previous discovery. For example, since FL Stat5a and Δ5 Stat5a are so close in size, it is likely that they were not resolved on previous Western blots. Similarly, since 96% of the sequence is identical, RTPCR primers have likely amplified both forms together. If two bands have been observed in protocols using anti-pan Stat5 antibodies, it is likely that the slightly smaller band was assumed to be Stat5b. Moving forward, with the correct choice of primers and electrophoresis conditions, each form can be separately analyzed by qRT-PCR, and Western blot. Unfortunately, since the alternate splicing does not cause a frame shift and there is no major change in predicted structure, specific antibodies to Δ5 Stat5a cannot be produced. This precludes an immunohistochemical analysis comparing the level of expression and nuclear localization of both forms in breast cancer tissue.
4.2 Differential promoter activities
To determine the cellular consequence of increased expression of Δ5 Stat5a, we examined its ability to behave like FL Stat5a. This included the ability to bind to a suitable receptor, become phosphorylated, homo-dimerize, hetero-dimerize with FL Stat5a or Stat5b, and translocate into the nucleus. In all of these aspects, Δ5 Stat5a behaved like FL Stat5a. However, when tested for its ability to activate an artificial promoter, it had 10% the activity of FL. Moreover, when titrated against FL, Δ5 Stat5a acted as a dominant negative. Dominant negative activity has been described for other variants [20–22]. However, these variants have not been found in mammary gland [23], despite specific searching. All other variants of Stat5a are significantly smaller and are missing part of the C-terminus. In our Western analysis of Δ5 Stat5a, we used an antibody against the C-terminus and hence would not have identified these forms. Moreover, DsiRNA specific to Δ5 Stat5a knocked down the Δ5 Stat5a band on a Western blot.
4.3 Interaction with different Stat5 response elements
Dominant negative activity was also observed with 2.4 kb of the β-casein promoter in a transfected system, as well as with the endogenous β-casein promoter in breast cancer cells, although the relative potency as a dominant negative was reduced with the more complex and natural promoter compared to the artificial promoter. One possible explanation for this was that Δ5 was not able to compete with FL Stat5a at all of the response elements in the natural promoter. Analysis by ChIP assay showed that this was indeed the case for the complete endogenous promoter since only FL Stat5a bound to the regions covered by primer sets 1 and 2. However, this did not explain the difference between the artificial promoter and the 2.4 kb promoter since the 2.4 kb promoter only contained the region covered by primer sets 4 and 5, where both forms of Stat5a behaved similarly. This could have been related to different cell systems, different stimuli, different sequences of Stat5 response elements, or different interactions with other transcription factors. Analysis of the Stat5 response element sequences also showed that one of the two elements covered by primer set 2 was identical to the element covered by primer set 4, and yet Δ5 Stat5a bound to region 4 and not to region 2. Differential binding could therefore not be explained by differences in the response element sequences. The lack of binding by either form to region 5 in the endogenous promoter was unexpected since this contained a sequence identical to that used in the artificial promoter. The reason for this is unknown, but could conceivably be related to the HA tag, even though this was C-terminal and away from the DNA binding site, or to the specifics of the cell line used in which this site could be masked by binding of other proteins. These proteins could include the B cell lymphoma 6 protein, which binds similar sequences [24], or proteins that bind adjacent sites, such as brachyury, a transcription factor expressed during the epithelial to mesenchymal transition [25,26].
4.4 Interaction with other transcription factors
Since the details of the Stat5 response element sequences were not the key, we considered differences resulting from association with other transcription factors. With an array allowing measurement of an interaction between either FL or Δ5 Stat5a and 54 other transcription factors, we observed specific interaction of one or other form with 12 transcription factors/complexes in PRL-stimulated T47D breast cancer cells. Of these 12, only activating protein 2 (AP-2) and the early growth response (EGR) protein were common to both forms of Stat5a. Of the remaining 10 transcription factors, FL bound to CAAT box general (CBF), nuclear factor-1 (NF-1), the vitamin D receptor (VDR-DR3), retinoid X receptor (RXR-DR1) and signal transducer and activator of transcription 3 (Stat3). By contrast, Δ5 Stat5a bound to activating protein-1–2 (AP-1–2) but not AP-1-1, nuclear factor kappa light chain enhancer of activated B-cells (NFκB), metal response factor (MRF), thyroid hormone receptor (TR) and specific protein-1 (SP-1).
Further analysis of RXR and Sp1 by co-immunoprecipitation confirmed the array results and, in the case of RXR, demonstrated a dependence on PRL. It will take considerable work to determine the functional consequences of the multiple and differentially-regulated interactions. However, with that caveat stated, and knowing that the Δ5 form has increased expression in invasive breast cancer, it is tempting to speculate that a heterodimer between FL Stat5a and the VDR [27] or RXR [28] may be important for maintaining differentiation, whereas an interaction between Δ5 Stat5a and some component of the AP-1 complex may be important in tumor formation [29–33].
What is clear from these results is that the two forms of Stat5a interact differently with other transcription factors. The region of Stat5 traditionally considered to be important for protein-protein interactions is the coiled coil domain [34], whereas more recent data [e.g. 35] and the data herein show an important role for the N-terminal domain. Further, our data show a specific role for a small N-terminal α helix.
4.5 Effects on cell number
Inhibition of p21 and Bax expression would be expected to reduce cell cycle control at the G0/G1 checkpoint [36] and to decrease apoptosis [37], respectively, both effects likely contributing to the increased cell number when Δ5 Stat5a expression was increased in cancer cells. Also, without this checkpoint control, damaged DNA could be replicated [36], thereby increasing the likelihood of oncogenic change with increased Δ5 Stat5a. The increase in cell number from increased expression of Δ5 Stat5a was variable among cell lines and not absolutely specific to the Δ5 form, although the Δ5 form always increased cell number, whereas the FL Stat5a did not. The increase in cell number with Δ5 Stat5a ranged from a very modest effect in MDA MB468 cells to 2-fold control with T47D cells when unstimulated in a 7 day period. Factors that might limit the degree of response include 1) the amount and ratio of FL to Δ5 Stat5a already present in the cell, and 2) the degree of growth and survival dependence of each cell line on autocrine loops and serum factors that utilize Stat5a signaling. However, a constant feature in the four lines tested was greater cell number with increased Δ5 Stat5a. Additional stimulation with PRL further amplified this effect in the cell line tested.
4.6 Interactions with β-catenin
Despite the absence of a sequence in the Δ5 form that is known to form part of a β-catenin binding site in another protein, both Δ5 and FL Stat5a bound β-catenin. B-catenin is in large part sequestered by E-cadherin at the membrane as part of adherens junctions. Adherens junctions contribute to contact-inhibition of epithelial proliferation. While increased expression of both forms of Stat5a increased levels of E-cadherin, only Δ5 also increased levels of β-catenin, thereby increasing the likelihood of free β-catenin in cells with increased expression of Δ5 Stat5a. When free from association with E-cadherin, β-catenin can bind to transcription factors that promote oncogenesis [38]. In the case of FL Stat5a, PRL promoted β-catenin binding, whereas for the Δ5 form, PRL decreased β-catenin binding. Thus, assuming that the β-catenin not bound to the Stat5a forms was in fact free, PRL stimulation in cells expressing more Δ5 Stat5a would be expected to promote cell proliferation, whereas PRL stimulation in cells expressing more FL Stat5a may have the opposite effect. However, this prediction remains to be rigorously tested.
4.7 Primary tumors
The effects on cell number, p21 and Bax expression, induction of β-catenin, and differential association with multiple transcription factors are all consistent with a role for Δ5 Stat5a in tumor progression, as is the increased expression of Δ5 Stat5a in invasive ductal carcinoma of the breast. For the primary tumors, the actual level of expression varied over a large range. This could be the result of 1) early splicing changes in regions that were still histologically normal in some patient samples, and/or 2) inherent epithelial to stromal variability in composition of samples from the breast. Nevertheless, a constant feature of cancerous versus adjacent histologically-normal tissue was an increase in the ratio of Δ5 to FL Stat5a.
5. Conclusion
While under particular experimental circumstances increased expression of each form of Stat5a could stimulate β-casein expression and an increase in cell number, results support the idea that FL Stat5a is primarily used to promote β-casein expression, whereas the Δ5 form inhibits this activity and promotes an increase in cell number. Given these different activity emphases and different associations with other transcription factors, it is crucial that the two forms be separately analyzed in future if we are to understand the role of Stat5a in cancer progression. We propose that inadvertent analysis of FL and Δ5 Stat5a together, and varying ratios of the two forms in different cell lines used by individual investigators, may explain some apparently contradictory roles for PRL and Stat5a activation in breast cancer.
Supplementary Material
Acknowledgements
We thank Drs Linda Schuler (University of Wisconsin, Madison, WI) and Jeffrey Rosen (Baylor College of Medicine, Houston, TX) for provision of the artificial 3XGAS-luc and 2.4kb β-casein promoter-luc, and Drs. Leonard Freedman and Robert Vogel (Merck and Co. Inc., West Point, PA) and Drs Xuan Liu (Department of Biochemistry, University of California, Riverside), and Melanie Cobb (University of Texas Southwestern Medical Center) for the p21-luc and Bax-luc, respectively. This work was supported by United States Public Health Service grant DK61005 and California Breast Cancer Research Program grant 171B-0053 to AMW, National Natural Science Foundation of China Grant, 81172497, and Jishou University Grant, jsdxkyzz101010 to DT. The funding sources had no role in study design, execution, or interpretation, nor any role in the writing of the manuscript or decision to submit for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement The authors declare no conflicts of interest.
8. References
- 1].Tan SH, Nevalainen MT. Signal transducer and activator of transcription 5A/B in prostate and breast cancers. Endocr Relat Cancer. 2008;15:367–390. doi: 10.1677/ERC-08-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2].Watson CJ, Neoh K. The Stat family of transcription factors have diverse roles in mammary gland development. Semin Cell Dev Biol. 2008;19:401–406. doi: 10.1016/j.semcdb.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 3].Liu X, Robinson GW, Wagner K-U, Garrett L, Wyhshaw-Boris A, Henninghausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 4].Furth PA, Nakles RE, Millman S, Diaz-Cruz ES, Cabrera MC. Signal transducer and activator of transcription 5 as a key signaling pathway in normal mammary gland developmental biology and breast cancer. Breast Cancer Res. 2011;13:220. doi: 10.1186/bcr2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5].Ren S, Cai HR, Li M, Furth PA. Loss of Stat5a delays mammary cancer progression in a mouse model. Oncogene. 2002;21:4335–4339. doi: 10.1038/sj.onc.1205484. [DOI] [PubMed] [Google Scholar]
- 6].Nevalainen MT, Xie J, Torhorst J, Bubendorf L, Haas P, Kononen J, Sauter G, Rui H. Signal Transducer and Activator of Transcription-5 Activation and Breast Cancer Prognosis. J Clin Oncol. 2004;22:2053–2060. doi: 10.1200/JCO.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 7].Sultan AS, Xie J, LeBaron MJ, Ealley EL, Nevalainen MT, Rui H. Stat5 promotes homotypic adhesion and inhibits invasive characteristics of human breast cancer cells. Oncogene. 2005;24:746–760. doi: 10.1038/sj.onc.1208203. [DOI] [PubMed] [Google Scholar]
- 8].Wagner K-U, Schmidt JW. The two faces of Janus kinases and their respective STATs in mammary gland development and cancer. J Carcinog. 2011;10:32. doi: 10.4103/1477-3163.90677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9].Brockman JL, Schroeder MD, Schuler LA. PRL activates the cyclin D1 promoter via the Jak2/Stat pathway. Mol Endocrinol. 2002;16:774–784. doi: 10.1210/mend.16.4.0817. [DOI] [PubMed] [Google Scholar]
- 10].Lee KF, Atiee SH, Rosen JM. Differential regulation of rat β-casein-chloramphenicol acetyltransferase fusion gene expression in transgenic mice. Mol Cell Biol. 1989;9:560–565. doi: 10.1128/mcb.9.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11].Liu M, Lee MH, Cohen M, Bommakanti M, Freedman LP. Transcriptional activation of the cdk inhibitor p21 by vitamin D3 leads to the induced differentiation of the myelomonocytic cell line U937. Genes Dev. 1996;10:142–153. doi: 10.1101/gad.10.2.142. [DOI] [PubMed] [Google Scholar]
- 12].Samuels-Lev Y, O'Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S, Campargue I, Naumovski L, Crook T, Lu X. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell. 2001;8:781–794. doi: 10.1016/s1097-2765(01)00367-7. [DOI] [PubMed] [Google Scholar]
- 13].Senoo M, Matsumura Y, Habu S. Identification of a novel retrovirus long terminal repeat (LTR) that is targeted by p51A (TAp63g) and selective dominant-negative activity of p73L (DNp63a) toward p53-responsive promoter activities. Biochem Biophys Res Commun. 2001;286:628–634. doi: 10.1006/bbrc.2001.5429. [DOI] [PubMed] [Google Scholar]
- 14].Ueda EK, Lo HL, Bartolini P, Walker AM. S179D Prolactin Primarily Uses the Extrinsic Pathway and Mitogen-Activated Protein Kinase Signaling to Induce Apoptosis in Human Endothelial Cells. Endocrinology. 2006;147:4627–4637. doi: 10.1210/en.2006-0348. [DOI] [PubMed] [Google Scholar]
- 15].Tan DY, Johnson DA, Wu W, Zeng LF, Chen YH, Chen WY, Vonderhaar BK, Walker AM. Unmodified Prolactin (PRL) and S179D PRL-initiated Bioluminescence Resonance Energy Transfer between Homo- and Hetero-pairs of Long and Short Human Prolactin Receptors in Living Human Cells. Mol Endocrinology. 2005;19:1291–1303. doi: 10.1210/me.2004-0304. [DOI] [PubMed] [Google Scholar]
- 16].Huang KT, Chen YH, Walker AM. Inaccuracies in MTS assays: major distorting effects of medium, serum albumin, and fatty acids. Biotechniques. 2004;37:406–412. doi: 10.2144/04373ST05. [DOI] [PubMed] [Google Scholar]
- 17].Chen TJ, Kuo CB, Tsai KF, Liu JW, Chen DY, Walker AM. Development of recombinant human prolactin receptor antagonists by molecular mimicry of the phosphorylated hormone. Endocrinology. 1998;139:609–616. doi: 10.1210/endo.139.2.5758. [DOI] [PubMed] [Google Scholar]
- 18].Moriggl R, Sexl V, Kenner L, Duntsch C, Stangl K, Gingras S, Hoffmeyer A, Bauer A, Piekorz R, Wang D, Bunting KD, Wagner EF, Sonneck K, Valent P, Ihle JN, Beug H. Stat5 tetramer formation is associated with leukemogenesis. Cancer Cell. 2005;7:87–99. doi: 10.1016/j.ccr.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 19].Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a β-TrCP1-Skp1-β-Catenin complex:destruction motif binding and lysine specificity of the SCFβ-TrCp1 ubiquitin ligase. Mol Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 20].Hendry L, John S. Regulation of Stat signaling by proteolytic processing. Eur J Biochem. 2004;271:4613–4620. doi: 10.1111/j.1432-1033.2004.04424.x. [DOI] [PubMed] [Google Scholar]
- 21].Azam M, Erdjument-Bromage H, Kreider BL, Xia M, Quelle F, Basu R, Saris C, Tempst P, Ihle JN, Schindler C. Interleukin 3 signals through multiple isoforms of Stat 5. Embo J. 1995;14:1402–1411. doi: 10.1002/j.1460-2075.1995.tb07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22].Meyer J, Jucker M, Ostertag W, Stocking C. Carboxyl-truncated STAT5beta is generated by a nucleus-associated serine protease in early hematopoietic progenitors. Blood. 1998;91:1901–1908. [PubMed] [Google Scholar]
- 23].Iavnilovitch E, Eilon T, Groner B, Barash I. Expression of a carboxy terminally truncated Stat5 with no transactivtion domain in the mammary glands of transgenic mice inhibits cell proliferation during pregnancy, delays onset of milk secretion, and induces apoptosis upon involution. Mol Reprod Develop. 2006;73:841–849. doi: 10.1002/mrd.20479. [DOI] [PubMed] [Google Scholar]
- 24].Tran TH, Utama FE, Lin J, Yang N, Sjolund AB, Ryder A, Johnson KJ, Neilson LM, Liu C, Brill KL, Rosenberg AL, Witkiewicz AK, Rui H. Prolactin inhibits BCL6 expression in breast cancer through a Stat5a-dependent mechanism. Cancer Res. 2010;70:1711–1721. doi: 10.1158/0008-5472.CAN-09-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25].Krukovskaia LL, Polev DE, Nosovalu K, Baranova AV, Koliubaeva SN, Kozlov AP. Investigation of transcription factor brachyury (T) expression in human normal and tumor tissues. Vopr Onkol. 2008;54:739–743. [PubMed] [Google Scholar]
- 26].Fernando RI, Castillo MD, Litzinger M, Hamilton DH, Palena C. Il-8 signaling plays a critical role in the epithelial-mesenchymal transition of human carcinoma cells. Cancer Res. 2011;71:5296–5306. doi: 10.1158/0008-5472.CAN-11-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27].Lopes N, Paredes J, Costa JL, Ylstra B, Schmitt F. Vitamin D and the mammary gland: a review on its role in normal development and breast cancer. Breast Cancer Res. 2012;14:211. doi: 10.1186/bcr3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28].Tang XH, Gudas LJ. Retinoids, retinoic acid receptors, and cancer. Annu Rev Pathol. 2011;6:345–364. doi: 10.1146/annurev-pathol-011110-130303. [DOI] [PubMed] [Google Scholar]
- 29].Ozanne BW, Spence HJ, McGarry LC, Hennigan RF. Transcription factors control invasion: AP-1 the first among equals. Oncogene. 2007;26:1–10. doi: 10.1038/sj.onc.1209759. [DOI] [PubMed] [Google Scholar]
- 30].Gutzman JH, Rugowski DE, Nikolai SE, Schuler LA. Stat5 activation inhibits prolactin-induced AP-1 activity: distinct prolactin-initiated signals in tumorigenesis dependent on cell context. Oncogene. 2007;26:6341–6348. doi: 10.1038/sj.onc.1210454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31].Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 32].Wisdom R. AP-1: one switch for many signals. Exp Cell Res. 1999;253:180–185. doi: 10.1006/excr.1999.4685. [DOI] [PubMed] [Google Scholar]
- 33].Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 34].John S, Vinkemeier U, Soldaini E, Darnell JE, Jr, Leonard WJ. The significance of tetramerization in promoter recruitment by Stat5. Mol Cell Biol. 1999;19:1910–1918. doi: 10.1128/mcb.19.3.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35].Engblom D, Kornfeld JW, Schwake L, Tronche F, Reimann A, Beug H, Hennighausen L, Moriggl R, Schütz G. Direct glucocorticoid receptor-Stat5 interaction in hepatocytes controls body size and maturation-related gene expression. Genes Dev. 2007;21:1157–1162. doi: 10.1101/gad.426007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36].Cazzalini O, Scovassi AI, Savio M, Stivala LA, Prosperi E. Multiple roles of the cell cycle inhibitor p21(CDKN1A) in the DNA damage response. Mutat Res. 2010;704:12–20. doi: 10.1016/j.mrrev.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 37].Maddika S, Ande SR, Panigrahi S, Paranjothy T, Weglarczyk K, Zuse A, Eshraghi M, Manda KD, Wiechec E, Los M. Cell survival, cell death and cell cycle pathways are interconnected: implications for cancer therapy. Drug Resist Updat. 2007;10:13–29. doi: 10.1016/j.drup.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 38].Thakur R, Mishra DP. Pharmacological modulation of beta-catenin and its applications in cancer therapy. J Cell Mol Med. 2013;4:449–456. doi: 10.1111/jcmm.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.