Abstract
Mumps virus (MuV) causes acute infections in humans. In recent years, MuV has caused epidemics among highly vaccinated populations. The largest outbreak in the U.S. in the past 20 years occurred in 2005–2006 with over reported 5,000 cases which the majority of the cases was in vaccinated young adults. We sequenced the complete genome of a representative strain from the epidemic (MuV-IA). MuV-IA is a member of genotype G, the same genotype of MuV that was associated with the outbreak in the UK in 2004–2005. We constructed a reverse genetics system for MuV-IA (rMuV-IA), and rescued a virus lacking the open reading frame (ORF) of the SH gene (rMuVΔSH). rMuVΔSH infection in L929 cells induced increased NF-κB activation, TNF-α production and apoptosis compared to rMuV-IA. rMuVΔSH was attenuated in an animal model. These results indicated that the SH ORF of MuV plays a significant role in interfering with TNF-α signaling and viral pathogenesis during virus infection.
INTRODUCTION
Mumps virus (MuV) is a member of the family Paramyxoviridae. MuV causes acute parotitis in humans, characterized by lateral or bilateral swelling of the salivary glands (Carbone and Wolinsky, 2001). MuV is also notable as a neurotropic agent causing a number of central nervous system (CNS) manifestations ranging from mild meningitis to severe encephalitis. The incidence of mumps and its complications were dramatically reduced following the introduction of measles, mumps, rubella vaccine (MMR) in 1971 (Amexis et al., 2002; Carbone and Wolinsky, 2001). MMR vaccine containing the Jeryl Lynn (JL) strain, an attenuated strain of MuV, is highly efficacious and produces few adverse reactions.
In spite of high coverage with MMR, mumps outbreaks continue to occur in the US and other countries. The causes cited range from failure to vaccinate, vaccine failure, and emergence of strains of MuV capable of escaping vaccine-induced immunity (Crowley and Afzal, 2002; Lim et al., 2003; Otto et al., ; Strohle, Bernasconi, and Germann, 1996; Utz et al., 2004; Whelan et al.). In 2006, the U.S. experienced the largest mumps epidemic in nearly 20 years (Marin et al., 2008). The outbreak originated at a university in Iowa and spread to 11 other states. Over 5,000 mumps cases were reported in 2006 compared to an average of approximately 250 cases/year in the previous decade. In 2009–2010, a mumps outbreak occurred in the State of New York and the State of New Jersey in the US in which 88% of the patients had one-dose of mumps vaccine and 75% of the patients had two doses of vaccine (MMWR, 2010).
The SH gene of MuV is the most variable region in mumps genome. Based on the sequence variability of the SH protein, all current wild type MuVs have been classified into 12 genotypes (Jin et al., 2005). Sequence comparison of the SH proteins from the outbreaks indicated that both Iowa and New Jersey/New York outbreaks were caused by closely related mumps strains in genotype G. Interestingly, the genotype G viruses detected in the outbreaks in the U.S. were also closely related to mumps virus strains associated with the massive mumps epidemic in the UK in which over 56,000 cases were reported in the 2004–2005 (MMWR, 2006; Rota et al., 2009; Watson-Creed et al., 2006).
MuV, a paramyxovirus, is a negative stranded, non-segmented RNA virus with a genome of 15,384 nucleotides (Carbone and Wolinsky, 2001). The virus has seven genes but encodes nine known viral proteins. The nucleocapsid protein (NP), phosphoprotein (P) and large RNA polymerase (L) protein are important for transcription and replication of the viral RNA genome (Elango et al., 1988; Okazaki et al., 1992; Rima et al., 1980). The V/P gene encodes three proteins, I, V and P (Paterson and Lamb, 1990). Mutations in the P gene have been associated with increased virulence of mumps virus (Saito et al., 1996). The V protein plays important roles in inhibiting interferon signaling in infected cells (Kubota et al., 2002; Takeuchi et al., 1990; Ulane et al., 2003; Yokosawa et al., 2002). The fusion (F) protein, a glycoprotein, mediates both cell-to-cell and virus-to-cell fusion in a pH-independent manner that is essential for virus entry into cells (Waxham et al., 1987). The hemagglutinin-neuraminidase (HN), another viral glycoprotein, is also involved in virus entry (Tanabayashi et al., 1992) and mutations in the HN gene have been implicated in mumps virus virulence (Cusi et al., 1998). The matrix (M) protein plays an important role in virus assembly (Matsumoto, 1982). The small hydrophobic (SH) protein is a 57-residue type 1, hydrophobic integral membrane protein (Elango et al., 1988). It has been reported that MuV SH has a similar function to the SH protein of a closely related virus, parainfluenza virus 5 (PIV5, formerly known as simian virus 5, SV5) (Hiebert, Richardson, and Lamb, 1988) (Wilson et al., 2006). While the SH ORF of PIV5 is not essential for virus growth in tissue culture cells, a virus lacking SH induces higher levels of cell death and is attenuated in an animal model (He et al., 1998; He et al., 2001). SH of PIV5 inhibits TNF-α signaling (Lin et al., 2003; Wilson et al., 2006). However, the expression of the SH protein in cells infected with MuV has never been demonstrated and the role of SH has never been reported. In this work, we have generated a reverse genetics system based on a representative isolate from the outbreak in 2006 in the U.S. and studied the biologic properties of a rescued recombinant virus lacking the SH ORF. These results indicated that MuV SH ORF plays a role in interfering with TNF-α signaling during virus infection.
RESULTS
Sequence of the complete genome of MuV-IA
To better understand the genetic characteristics of viruses associated with recent outbreaks in the U.S., the complete genomic sequence of a representative isolate from the Iowa outbreak was determined. A set of primers was designed based on the consensus sequence derived from comparison of the genomic sequences of Jeryl Lynn, Urabe, 88–1961 and PetroNov. Viral RNA of MuV-IA was reverse-transcribed into cDNA using random hexamers, PCR reactions were then carried out using the set of primers and the products were sequenced using the corresponding primers. A second set of primers based on the sequencing results were then used to perform RT-PCR and the products overlapping with those of first round of sequencing fragments were sequenced using the primers. Leader and trailer sequences were determined by performing 5’/3’ RACE.
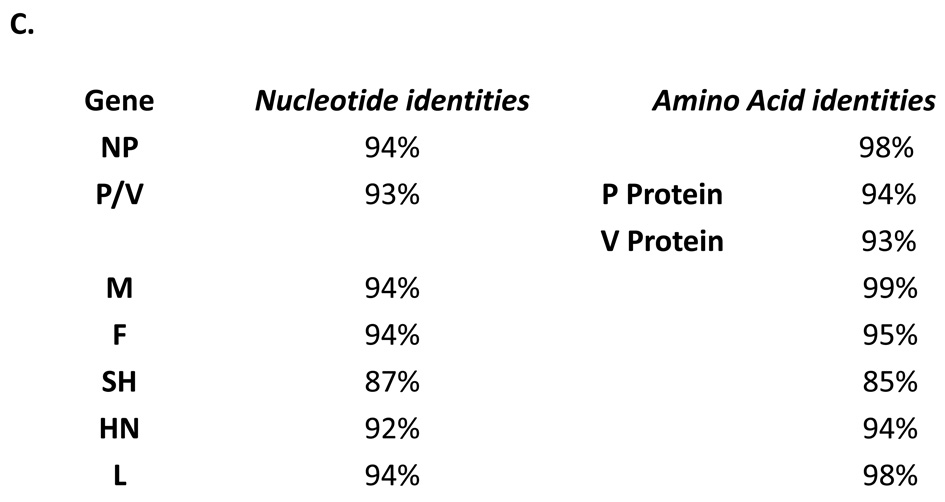
There is only one conserved change in the putative transmembrane domain of the SH protein when the SH protein sequence of MuV-IA was compared to other strains of mumps virus in genotype G (Figure 1A), confirming that MuV-IA belongs to genotype G (Rota et al., 2009). To further study the genomic divergence of MuV-IA, a phylogenetic tree was generated using the genomic sequence of MuV-IA and 32 full length genomic sequences from Genbank (Figure 1 B). Phylogenetic analysis indicated that MuV-IA is most closely related to the sequence of MuV Du/CRO05, a genotype G virus, which was isolated in Croatia in 2005 (Santak et al., 2006). A comparison of the predicted amino acid sequences between the protein coding regions of MuV-IA and Jeryl Lynn vaccine (major component) showed that while NP, M and L protein sequences are highly conserved with an identity of over 98%, there was more divergence among V, P, F, SH and HN proteins (Figure 1C). The predicted SH protein sequences had only 85% identity.
Figure 1. Analysis of MuV-IA genome.
(A). Alignment of the SH proteins. Sequences of three strains, Glouc1/UK96, UK01-22 and MuV-IA were shown. MuV-IA was different from Glouc1/UK96* and UK01-22 for only 5 nucleotides and 2 amino acids respectively. The transmembrane domain of mumps virus SH protein, outlined in rectangular box, was predicted using TMHMM server v.2.0 (CBS; Denmark). (B). Sequence comparison of different mumps viruses. The genome sequences of 32 strains of mumps viruses were obtained from NCBI Genbank and aligned with genome sequence of MuV-IA using MEGA 4 version 4.0.2. The 32 MuV Strains were (accession numbers are given in the brackets): 87–1004 (AF314560), SIPAR 02 (AF314558), Biken (AF314561), 87–1005 (AF314562), MuV(2001) (AF314559), Urabe 1004-10/2 (FJ375177), Urabe Gw7 (FJ375178), Hoshino (AB470486), Miyahara(1992)(NC_002200), MuV Miyahara (1992)(2) (AB040874), Y213(AB576764), Dg1062/Korea/98 (32172464), L3/Russia/Vector (AY508995), L-Zagreb master seed (AY685921), L-Zagreb vaccine strain (AY685920), 9218/Zg98(299766355), Novosbrisk genotype C (50404164), PetroNov genotype H (AY681495), 88–1961 (AF467767), Glouc1/UK96(AF280799), Du/CRO05 (EU370207), SP-A (FJ556896), SP (EU884413), SP(2006) (DQ649478), JL2 (AF3452901), Jeryl Lynn sub strain (FN31985), Enders (GU9800521), Jeryl Lynn major component (AF338106), MuV(2000) (AF201473), JL1 (FJ211586), RIT4385 (FJ211585), RIT4385(2) (FJ211584). (C). Sequence comparison between MuV-IA and Jeryl Lynn (JL) strain (major component). The gene and protein sequences of NP, P/V (encoding P protein and V protein), M, F, SH, HN and L of MuV-IA and Jeryl Lynn live vaccine major component were aligned using NCBI BLAST program. Nucleotide identities and amino acid identities were shown above. * The SH genes were aligned using MEGA 4.0.2.
Generation of an infectious cDNA clone for MuV-IA
To be able to study the pathogenesis of MuV-IA, a reverse genetics system was derived. Because RNA viruses exist as a quasi-species, the consensus sequence of the genome was used as the base for the recombinant MuV. A plasmid containing a mini-genome with luciferase reporter (Luc) reporter gene for mumps virus (pT7-MuV-Mini-Luc) similar to the PIV5 mini-genome expressing plasmid was constructed using rMuV-IA trailer and leader sequences (data not shown) (Lin et al., 2005). In addition, plasmids encoding NP, P and L in the pCAGGS vector have been obtained and confirmed by sequencing. To test the functionality of the plasmids, the plasmids were transfected into BSR-T7 cells. At 2 dpi, the cells were harvested and Luciferase (Luc) assays were performed. Luc activity was detected in the cell transfected with all plasmids, not ones missing P or L, indicating that the plasmids expressed functional P and L proteins (data not shown). RT-PCR was conducted to amplify DNA fragments representing the complete genome and inserted into individual plasmid vectors before being assembled into a full-length genome. The plasmid with the full length genome of MuV-IA expressed under the control of a T7 (pMuV-IA) promoter (pMuV-IA) was similar to the plasmid used to generate infectious PIV5 (He et al., 1997). pMuV-IA had changes in two nucleotides within the L ORF compared with consensus sequence of MuV-IA at positions of 11863 (T to C) and 12028 (C to T). However, neither of these nucleotide changes resulted in changes in the predicted L protein sequence. A recombinant MuV (rMuV-IA) was rescued using the plasmid containing the full-length genome of MuV-IA. BSRT-7 cells were co-transfected with pMuV-IA and plasmids expressing viral RNA polymerase components. Individual plaques were selected and amplified in Vero cells. The entire genome of the rescued virus was sequenced and found to match the input cDNA genome sequence (data not shown).
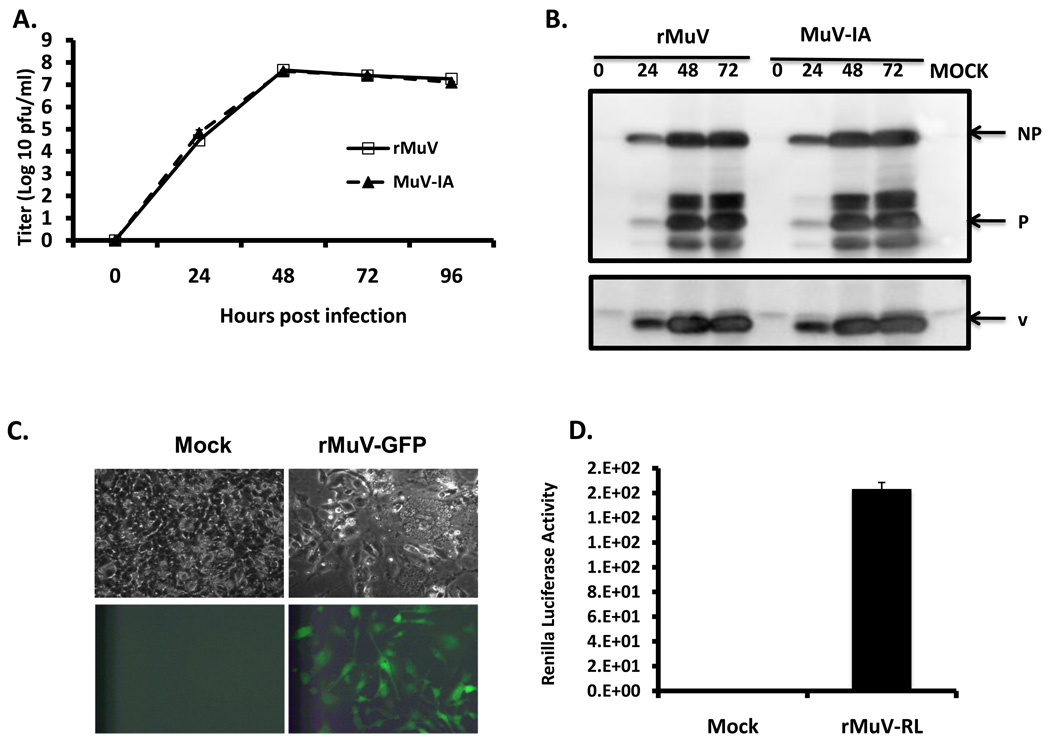
To compare time course of the growth of rMuV and MuV-IA, a multi-cycle growth assay was performed (Figure 2 A). Both viruses grew to similar peak titers in Vero cells. Viral titers in the supernatant of the infected cells increased exponentially during the first 2 days after infection, and reached a titer of 107 pfu/ml at 48 hpi. We also compared the growth of both viruses in HeLa cells (a human cell line), MDBK cells (a bovine cell line), and L929 cells (a murine cell line) and did not observe any obvious differences between these two viruses (data not shown). The viral protein expression levels in cells were also examined using Western blot (Figure 2 B) and the protein levels were similar at different time points after infection, indicating that the replication of rMuV resembles MuV-IA in tissue culture cells.
Figure 2. Analysis of rMuV, rMuV-EGFP and rMuV-RL.
(A). Growth rate of rMuV. rMuV was obtained by transfecting BSRT-7 cells with pMuV-IA, pCAGGS-NP, pCAGGS-P and pCAGGS-L as described in the Materials and Methods. Growth rates of rMuV (empty square) and MuV-IA (filled triangle) were compared as described in the Material and Methods. (B). Viral protein expression levels of rMuV and MuV-IA. 6 well plates of Vero cell were infected with mock, rMuV or MuV-IA at a MOI of 0.5. Cell lysates were subjected to immunoblotting with anti-NP, P or V. (C). Rescue of rMuV-EGFP. rMuV-EGFP was rescued in a similar manner as described for rMuV. Vero cell in 6 well plates were infected with mock or rMuV-EGFP at a MOI of 0.05 and photographed at 2 dpi. (D). Rescue of rMuV-RL. rMuV-RL was recovered from cloned DNA as described for rMuV. 24 well plates of Vero cell were infected with mock or rMuV-RL at MOI of 0.1. At 2 dpi, cells were assayed for renilla luciferase activity.
In addition, we rescued infectious recombinant viruses expressing either EGFP or Renila Luciferase (RL) protein as an extra gene. pMuV-EGFP was constructed by inserting an EGFP gene, flanked by gene start (GS) of SH and gene end (GE) of NP, between F gene and SH gene in pMuV-IA, pMuV-RL was constructed through substitution of coding sequence of EGEP with that of renilla luciferase (RL) in pMuV-EGFP. Expression of EGFP or RL in the infected Vero cells was detected (Figure 2 C, D).
Rescue of a recombinant mumps virus lacking the SH ORF
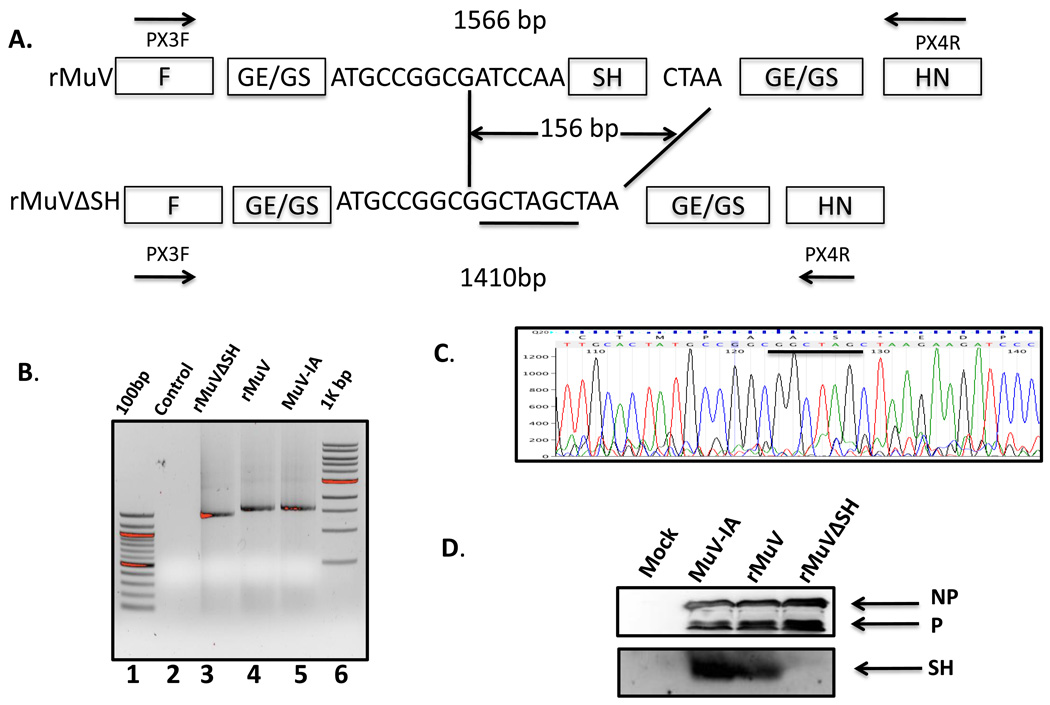
To study the function of the SH protein of MuV, 156 nucleotides in the SH gene open reading frame (ORF) of the SH gene were deleted from pMuV-IA. The truncated SH ORF contained a short ORF encoding 5 amino acid residues flanked by the original SH ORF start and gene end (pMuV-IAΔSH, Figure 3 A). An infectious MuV lacking the SH ORF was rescued (rMuVΔSH) (Figure 3B and C) and the genome was sequenced, which matched the input cDNA sequence (data not shown). The rMuVΔSH genome was of 15,228 nt in size, complying with “the rule of six” (Kolakofsky et al., 1998). To confirm that wtMuV and rMuV did express a SH protein and that rMuVΔSH did not, cell lysates of infected Vero cells were examined by immunoblotting with anti-SH as well as anti- NP and anti-P (Figure 3 D). SH was detected in MuV-IA and rMuV-infected cells, but not in rMuVΔSH-infected cells, confirming the lack of the SH protein expression in rMuVΔSH-infected cells.
Figure 3. Generation of a MuV lacking SH (rMuVΔSH).
(A). A schematics of rMuVΔSH. The SH ORF was replaced with a 5 amino acid coding sequence containing an Nhe I site (underlined). (B). Confirmation of rMuVΔSH by RT-PCR. After plaque purification, viral RNA was extracted and was subjected to RT-PCR, in which two primers flanking the SH gene were used to perform a PCR to confirm the deletion. Lane 1 and Lane 6 are 100bp and 1kb DNA ladder respectively; Lane 2 is the negative control—PCR without polymerase. Lane 3, 4 and 5 are PCR products from rMuVΔSH, rMuV and wtMuV-infected cells respectively. (C). Confirmation of rMuVΔSH by sequencing. PCR products were sequenced. The inserted sequence is underlined. (D). Expression of SH in MuV-IA, rMuV and rMuVΔSH-infected cells. Vero cells were mock infected or infected with MuV-IA, rMuV or rMuVΔSH. Cell lysates were subjected to immunoblotting with anti-NP, P or SH.
Analysis of rMuV and rMuVΔSH
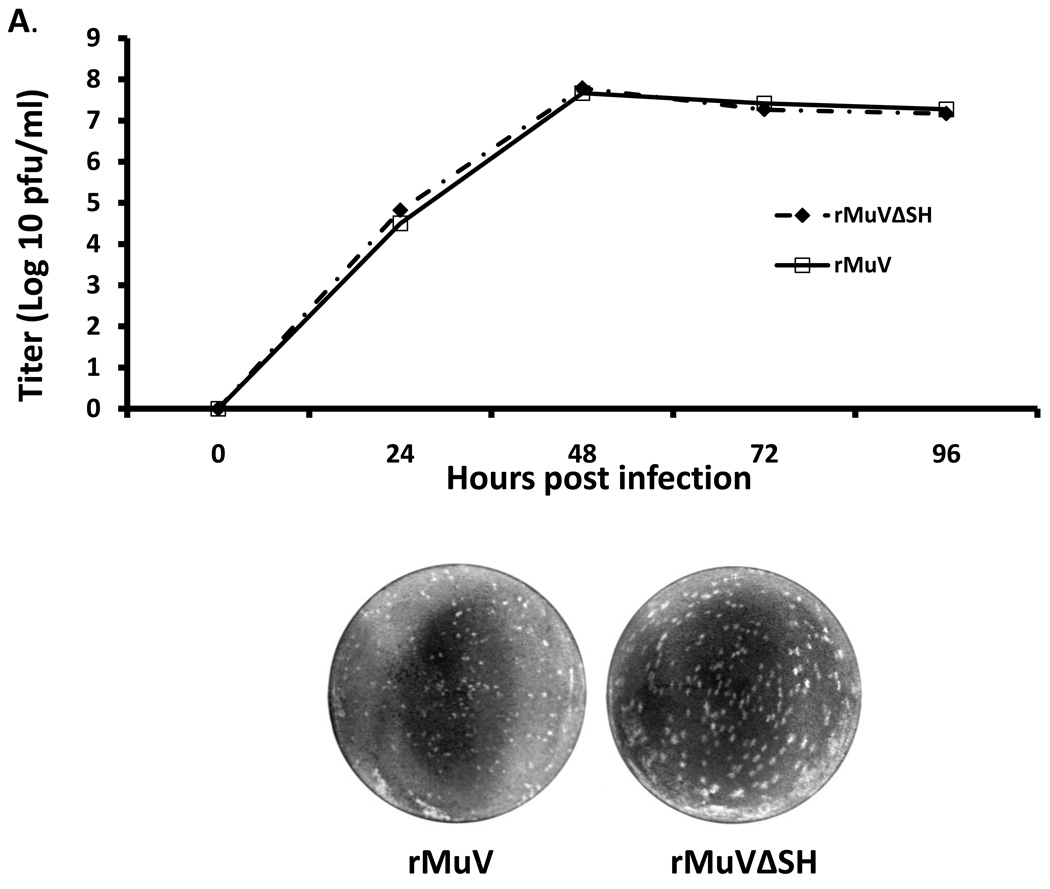
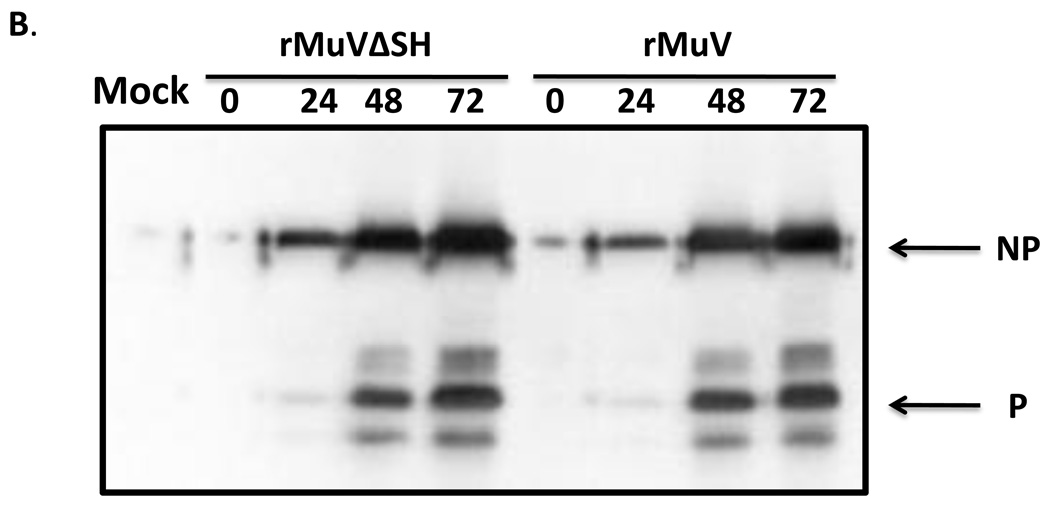
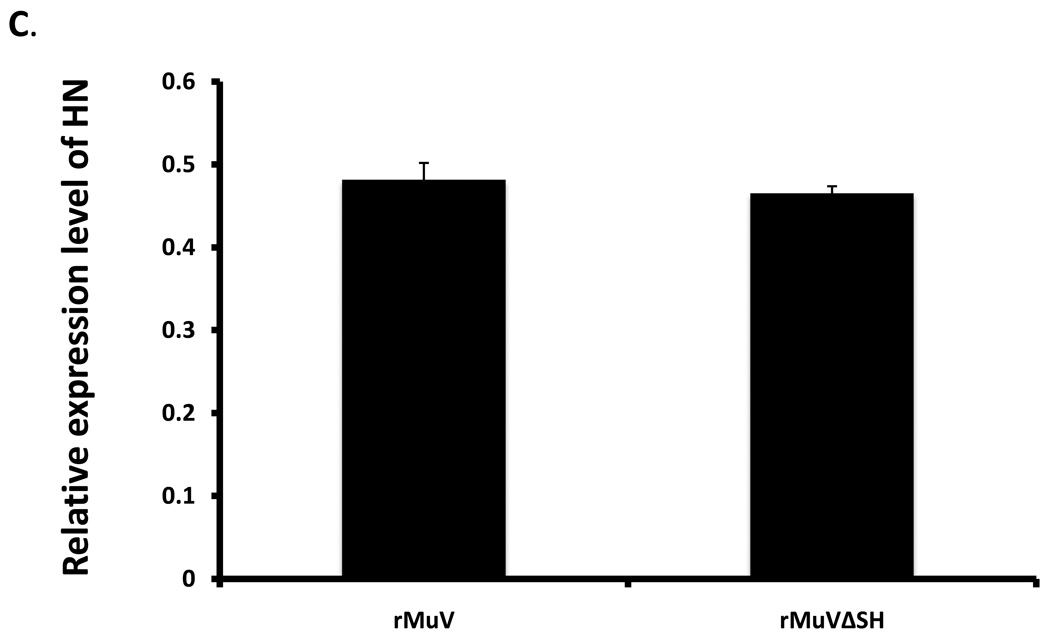
To investigate the growth rate of rMuVΔSH, a multiple-cycle growth curve and protein expression levels were examined in Vero cells, and the titers of the viruses released from rMuVΔSH-infected Vero cells remained similar to rMuV-infected Vero cells at all time points (Figure 4A). When the infected cells were lysed and viral protein expression levels were compared using Western blot, the protein levels of NP and P in rMuVΔSH and rMuV-infected cells were similar (Figure 4B), indicating that the SH ORF was not essential for viral gene expression, or virus release in Vero cells, consistent with the previous findings (5). The HN gene is downstream of the SH gene. To examine whether there is any significant impact of the deletion of the SH ORF on the expression level of HN, expression levels of HN and NP of infected cells were examined using flowcytometry. As shown in Figure 4C, relative expression level of HN in rMuV-infected cells and in rMuVΔSH-infected cells were similar, suggesting that the deletion of the SH ORF sequence did not affect expression of the HN protein. Furthermore, mRNA expression levels of HN were examined using real-time RT-PCR. No significant difference was observed between rMuV and rMuVΔSH (Figure 4D). Interestingly, rMuVΔSH formed larger plaques in Vero cells compared to rMuV (Figure 4A).
Figure 4. Growth rates and viral protein expression of rMuVΔSH and rMuV.
(A). Growth rates of rMuVΔSH and rMuV. Vero cell were infected with rMuVΔSH (filled diamond) and rMuV (empty square) at MOI of 0.01. Supernatants were used for plaque assay. (B). Viral protein expression levels in rMuVΔSH and rMuV infected cells. Vero cells were mock infected or infected with rMuVΔSH or rMuV at a MOI of 0.5. (C). HN expression level in rMuVΔSH and rMuV infected cells. Vero cells were mock infected or infected with rMuVΔSH or rMuV at a MOI of 0.5. Cells were collected at 24hpi and expression levels of total HN and NP were examined using flow cytometry. Mean fluorescence intensity (MFI) of HN and NP were calculated. Y-axis represents the relative ratio of MFI of HN normalized by MFI of NP. (D). HN mRNA level in rMuVΔSH or rMuV infected Vero cells. Viral RNA was extracted from rMuVΔSH or rMuV-infected Vero cells, reverse transcribed with oligodT and used for real time PCR. HN and F mRNA levels were calculated and HN mRNA level was normalized by F mRNA level. Y-axis represents ratio of HN mRNA verses F mRNA.
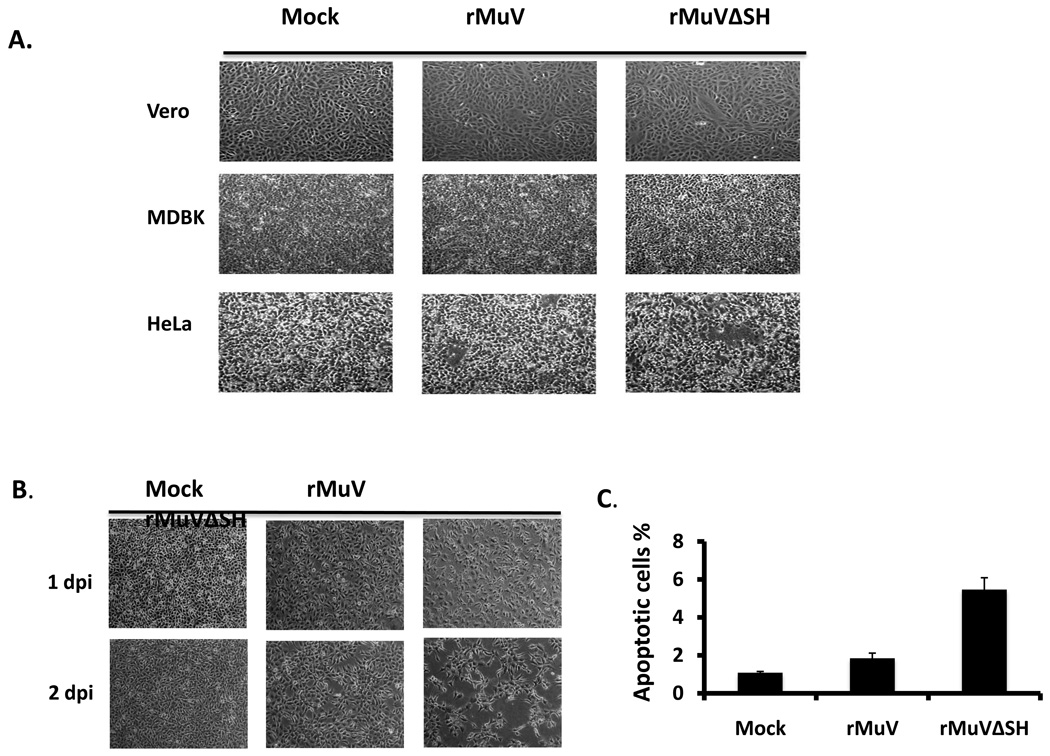
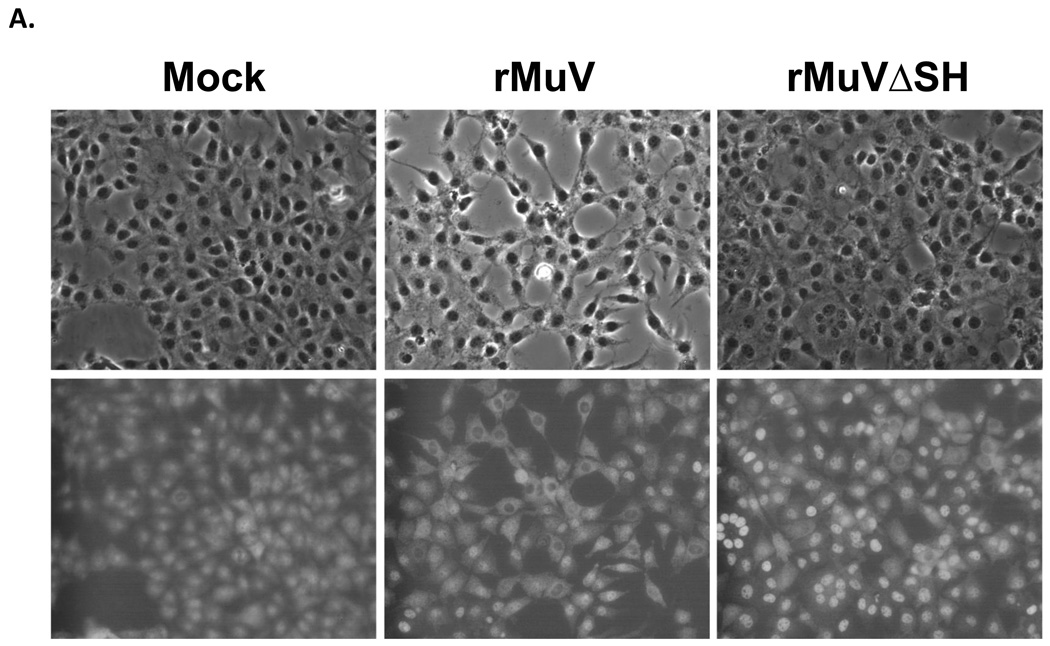
rMuVΔSH induced cytopathic effect in L929 cells
We compared infection of Vero, MDBK and HeLa cells with rMuVΔSH and rMuV. At 1day post infection, there were no observable differences in rMuVΔSH- or rMuV-infected Vero and MDBK cells. Previous studies in our lab showed that the SH ORFs of PIV5 and RSV played a role in blocking TNF-α signaling. To test the hypothesis that mumps virus SH ORF has a role in regulating the TNF-α signaling pathway, we investigated the phenotype of rMuVΔSH in L929 cells, which undergo apoptosis after TNF-α treatment. rMuVΔSH infection led to significantly more cell death than infections with rMuV or wtMuV. The phenotype was evident at 2-day post infection (Figure 5B). To investigate whether the cytopathic effects (CPE) observed in rMuVΔSH infected L929 cells was caused by apoptosis, TUNEL assay was performed. At 1dpi, infection with rMuVΔSH resulted in a higher percentage of infected cells with apoptosis than rMuV (Figure 5C), indicating that the lack of SH led to increased apoptosis in infected cells.
Figure 5. Induction of cell death by rMuVΔSH.
(A). Cytopathic effects of rMuVΔSH or rMuV in tissue culture cell lines. Vero cells, MDBK cells or HeLa cells were mock infected or infected with rMuVΔSH or rMuV at MOI of 0.01 and photographed at 1 dpi. (B). Cytopathic effects of rMuVΔSH infection in L929 cells. L929 cells were mock infected, or infected with rMuVΔSH, or rMuV at a MOI of 3 and were photographed at 1dpi and 2dpi. (C). rMuVΔSH infection induced apoptosis in L929 cells. L929 cells were infected as in B. At 1dpi, both floating cells and attached cells were collected, fixed, permeabilized and used for TUNEL assay.
TNF-α played a critical role in rMuVΔSH-induced apoptosis
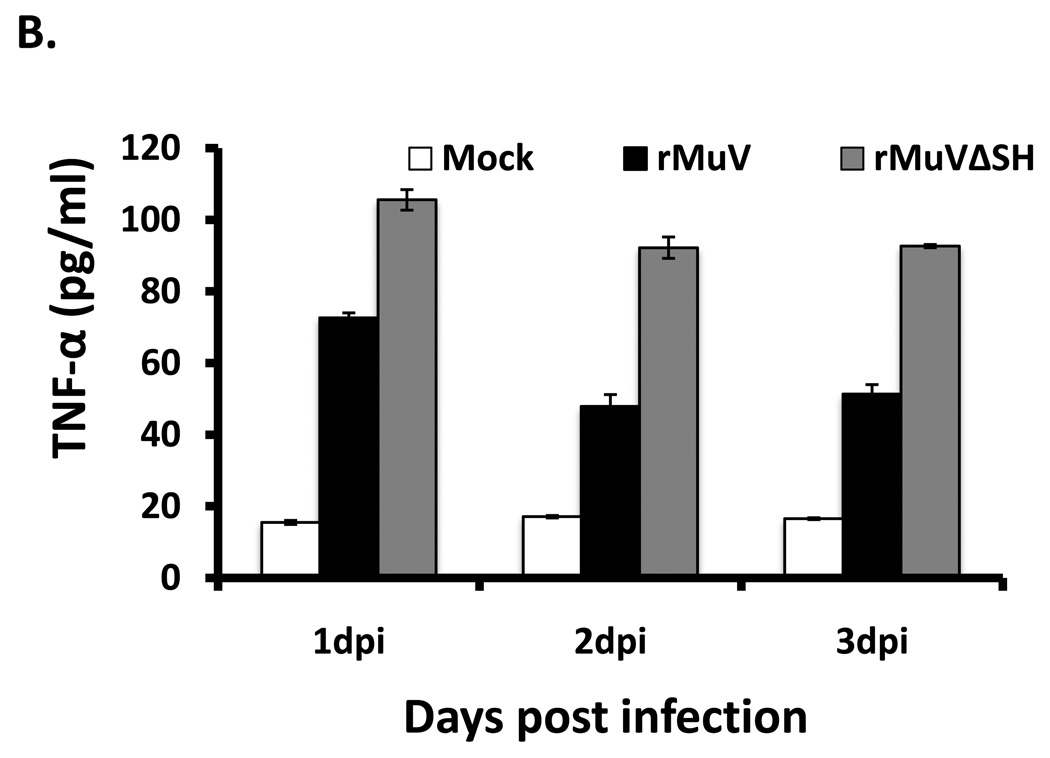
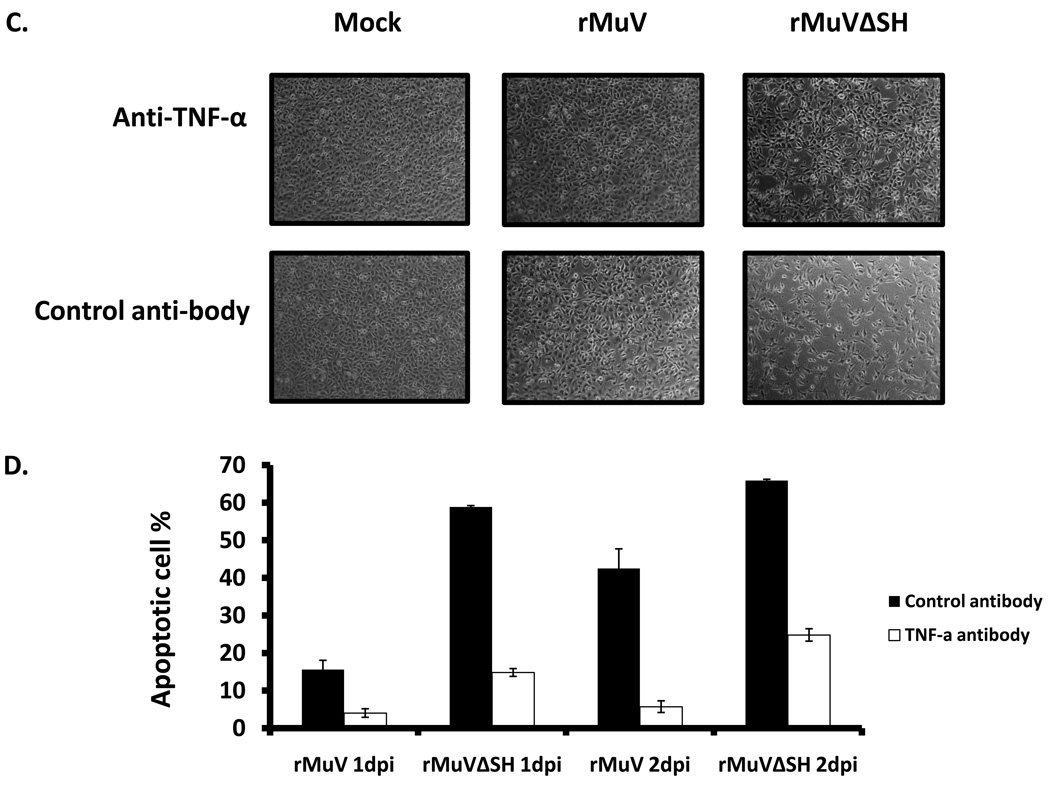
To test whether apoptosis in rMuVΔSH infected L929 cells resulted from an elevated TNF-α, we examined activation of NF-κB in rMuVΔSH-infected L929 cells by examining nuclear translocation of p65, a key subunit of NF-κB. NF-κB factors are localized in the cytoplasm. On activation, for example by TNF-α stimulation, p65 is translocated into the nucleus (Baud and Karin, 2001). A higher level of p65 nuclear localization was observed in rMuVΔSH-infected L929 cells (Figure 6 A), indicating activation of NF-κB. To investigate whether the production of TNF-α was increased in rMuVΔSH-infected cells, supernatants of infected were collected and levels of TNF-α were measured using ELISA. TNF-α production level was up regulated in rMuVΔSH-infected cells (Figure 6B). To determine whether the increased TNF-α played a role in increased apoptosis in rMuVΔSH-infected cells, the infected cells were treated with neutralizing antibody against TNF-α. Anti-TNF-α reduced CPE in rMuVΔSH-infected cells, while the control antibody had no effect (Figure 6C), indicating that TNF-α played a critical role in rMuVΔSH induced cell death. This was confirmed with TUNEL assay (Figure 6D). At 1 dpi, with control antibody treatment, rMuVΔSH induced almost 4-fold higher apoptotic rate than rMuV. Treatment of anti-TNF-α antibody effectively blocked cell death in infected cells (Figure 6C, D).
Figure 6. The role of TNF-α in rMuVΔSH-mediated cell death.
(A). rMuVΔSH infection activated P65 in L929 cells. L929 cells on glass cover slips were infected with mock, rMuVΔSH or rMuV at a MOI of 10. At 1dpi, L929 cells on the cover slips were stained with anti-P65. (B). rMuVΔSH infection in L929 cells induced TNF-α production. L929 cells were mock infected or infected with rMuVΔSH, or rMuV at a MOI of 5. TNF-α in the media was measured using ELISA. (C). Treatment with anti-TNF-α antibody reduced CPE in rMuVΔSH infected L929 cells. L929 cells were mock infected or infected with rMuVΔSH or rMuV at a MOI of 5. Cells were cultured in media containing TNF-α neutralizing antibody or control antibody and were photographed at 1dpi. (D). TNF-α antibody treatment inhibited apoptosis in rMuVΔSH infected L929 cells. L929 cells in 6 well plates were infected and treated as in C. At 1 day and 2 days post infection, cells were collected and used for TUNEL assay.
SH of MuV-IA blocked TNF-α signaling in vitro
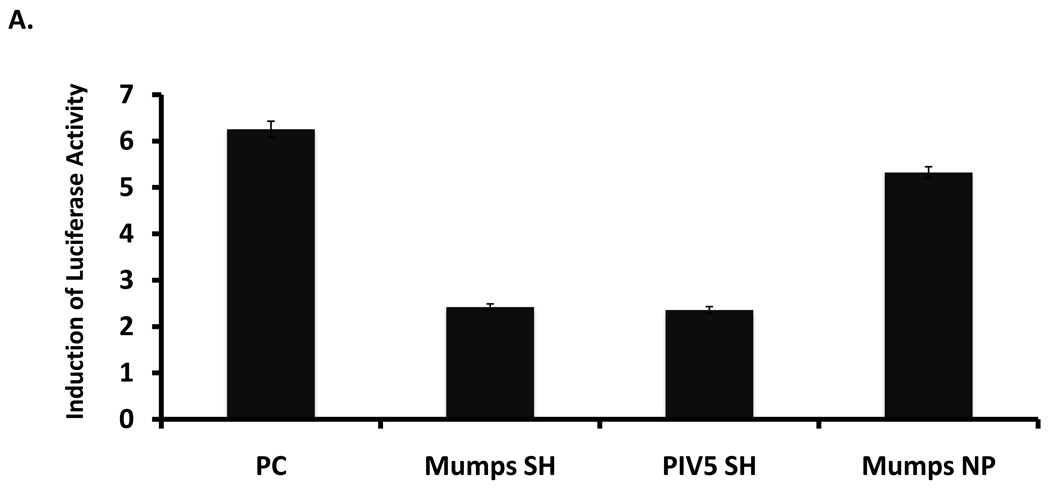
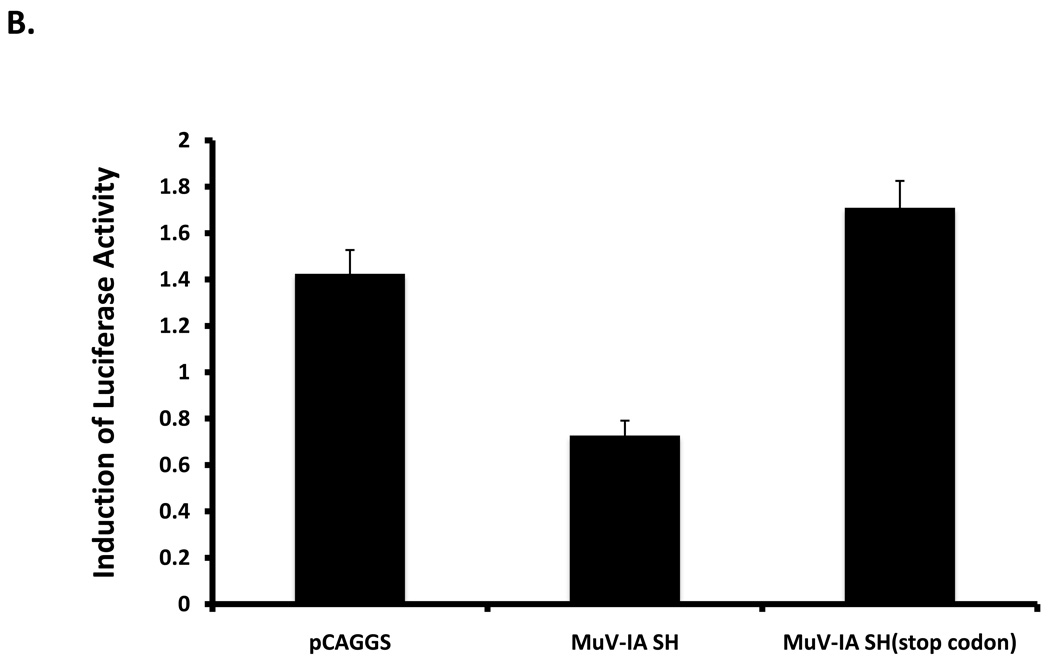
To investigate whether MuV-IA SH expressed alone can block TNF-α signaling, a plasmid encoding SH of MuV-IA was co-transfected with a NF-κB promoter-luciferase reporter system into L929 cells. At 1-day post transfection, cells were treated with TNF-α. TNF-α signaling was blocked by SH of MuV-IA as well as SH of PIV5, but not by NP of MuV-IA (Figure 7A) or the sequence of the SH ORF (Figure 7B), indicating that the SH protein can block TNF-α-mediated signaling.
Figure 7. MuV-IA SH inhibited TNF-α activation of NF-κB.
L929 cells were transfected with a reporter plasmid (pκB-TATA-FL) and pCAGGS-MuV-IA SH, pCAGGS-PIV5 SH or pCAGGS-MuV-NP (panel A). At 1 day post-transfection, cells were treated with TNF-α at 10ng/ml for 4 hrs and then assayed for fire fly luciferase activity. Similarly, the effect of the sequence of the SH ORF was examined using a plasmid encoding the SH ORF sequence without expressing the SH polypeptide due to in-frame stop codon insertion downstream of the start codon of the SH ORF (Panel B).
rMuVΔSH was attenuated in vivo
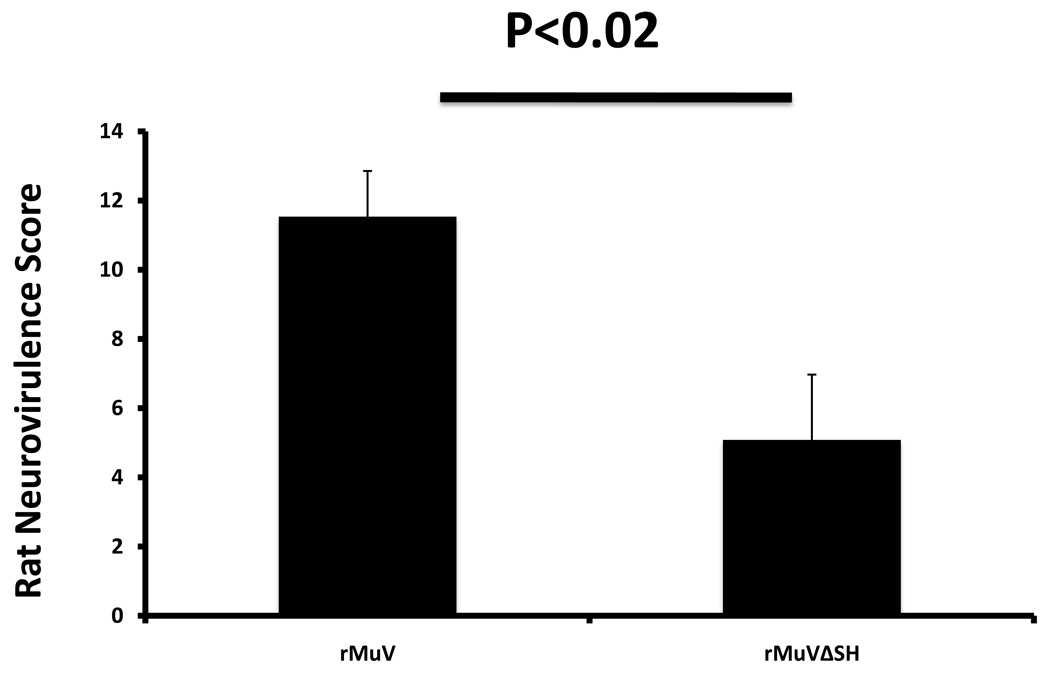
MuV is a human virus and there is no ideal animal model in which to study viral pathogenesis. Intracerebral injection of MuV into newborn rats has been used to compare the relative pathogenecities of different strains of MuV. (Rubin et al., 2005). To compare the neurotoxicity of the viruses, rMuV or rMuVΔSH was injected intracerebrally into brains of newborn rats. Relative neurotoxicity score was calculated based on relative severity of hydrocephalus. As shown in Figure 8, rMuVΔSH had a lower neurotoxicity score than rMuV, indicating that deletion of the SH ORF resulted in attenuation in vivo.
Figure 8. Neurotoxicity of MuVΔSH in vivo.
Newborn rats were infected intracerebrally with rMuV or rMuVΔSH. The animals were sacrificed at 30 days post infection. The brains of the animals were sectioned, stained and neurotoxicity scores were calculated based on relative hydrocephalus score as described in the Materials and Methods.
DISCUSSION
Immunization against MuV is a part of a 2-dose MMR (mumps, measles and rubella) vaccine regimen that is administrated to children at 1 and 5 years of age in the U.S. Even with a two-dose vaccination schedule, large outbreaks have occurred in vaccinated populations. Here, we describe the rescue of a wild-type mumps virus that is representative of the strain associated with recent outbreaks in the U.S. and Europe. This has enabled us to begin to explore the pathogenesis of MuV using reverse genetics. In this report, we described the potential role of the SH protein in regulating TNF-α, and we found that the deletion of the SH ORF resulted in attenuation in vivo, indicating that SH plays a role in viral pathogenesis. The attenuation of rMuVΔSH in vivo suggests that deleting the SH ORF can be a possible strategy to develop attenuated mumps strains. Recombinant MuVs expressing foreign genes such as GFP and RL have been obtained, and interestingly, the expression level of RL in rMuV-RL in Vero cells remained relatively high after 20 passages (data not shown), indicating that MuV can possibly be used as a vector.
The SH protein of paramyxoviruses was first identified in PIV5-infected cells (Hiebert, Paterson, and Lamb, 1985). A similar gene was predicted basing on sequence analysis of the Enders strain of MuV. However, due to a mutation in the intergenic sequence of the putative SH gene, the SH protein of the Enders strain MuV is not expressed in infected cells (Takeuchi et al., 1991). Thus, the SH protein of MuV has never been detected in MuV-infected cells. Wilson et al (2006) replaced the SH ORF within the genome of PIV5 with the SH ORF of MuV Enders strain and found that the MuV SH can functionally replace the SH ORF of PIV5 (Wilson et al., 2006). Thus, it is thought that the function of MuV SH is the same as the function of the SH ORF of PIV5, a closely related paramyxovirus. In this work, the expression of SH was detected in MuV-infected cells for the first time, confirming the existence of the SH protein in MuV-infected cells. Furthermore, taking advantage of the new reverse genetics system, a recombinant MuV lacking the SH ORF (rMuVΔSH) was obtained and analyzed.
One interesting observation was that rMuVΔSH produced larger plaques. A possible explanation is that the deletion of the SH ORF resulted in a virus that promotes cell-to-cell fusion better than the wild type virus. Because there was no change of total number of ORFs or the overall order of genes, we expect that the relative amounts of viral mRNAs and the expression levels of viral proteins of rMuVΔSH should be similar to those of wild type virus (Figure 4B, C and D). Thus, it is unlikely that the bigger plaque formation by rMuVΔSH was due to a higher level of viral protein expression. Further, we have performed a fusion assay using cells transfected with MuV HN and F in the presence or absence of MuV SH, and did not observe any difference in the extent of cell-to-cell fusion (data not shown), suggesting that the SH does not have a role in promoting cell-to-cell fusion. We speculate that the larger plaques formed by rMuΔSH are due to a higher level of induction of cell death by rMuVΔSH. The viruses infected cells at the same rate; however, the cells infected by rMuVΔSH induced more cell death than rMuV resulting in more rapid cell death at the edge of a plaque.
It is possible that the mRNA of from some ORFs may have biologic functions. For example, the mRNA of the L ORF of PIV5 is capable of activating IFN-β expression (Luthra et al., 2011). In this work, we deleted the ORF of SH and we cannot differentiate the function of the polypeptide encoded by the SH ORF from SH mRNA itself. While the SH polypeptide was needed to block TNF-α mediated signaling, not the sequence of the SH ORF, and we favor a critical role of the SH polypeptide in mumps virus pathogenesis; however, it is possible that the small mRNA potentially expressed from the deleted SH gene could have contributed to the phenotype of rMuVΔSH.
The reduced neurotoxicity of rMuVΔSH in neonatal rat brain indicates that the SH ORF plays a critical role in viral pathogenesis. We propose that infection with rMuVΔSH induced a higher level of pro-inflammatory cytokine expression, resulting in a more rapid resolution of infection, thus limiting damage in the infected brain. A better animal model for studying mumps virus pathogenesis will be highly desirable to understand the exact mechanism of the reduced neurotoxicity.
MATERIAL AND METHODS
Plasmids, viruses and cells
All molecular cloning was conducted according to standard procedures as previously described (He et al., 1997). MuV-IA NP, P and L genes were cloned into the pCAGGS expression vector (Niwa, Yamamura, and Miyazaki, 1991). MuV-IA SH gene was cloned into the pCAGGS expression vector. MuV-IA SH (stop codon) was constructed by introducing three continues stop codon sequence into the SH ORF, 6 nucleotides downstream of the start codon. Construction of MuV-IA full-length cDNA in pUC19 was analogous to the PIV5 reverse genetics system (He et al., 1997). To construct pMuVΔSH, the region of the SH ORF from the 4th amino acid to the 57th (156 nt) was substituted with a short 6 nucleotide sequence designed to facilitate subcloning and to maintain the length of the genome a multiple of six (known as the “rule of six”). pMuV-EGFP and pMuV-RL were constructed by inserting either an EGFP or a renilla luciferase gene between F and SH gene flanked by F gene start and SH gene end. Primer sequences, detailed cloning strategies and entire cDNA sequences of MuV are available upon request.
To rescue an infectious virus from cDNA, plasmid (5ug) containing a full-length genome or a mutated MuV genome was co-transfected with plasmids pCAGGS-L (1ug), pCAGGS-NP (1.5ug) and pCAGGS-P (200ng) into BSRT-7 cells. Usually 4 to 7 days post-transfection, syncytia formation could be observed in transfected BSRT-7 cells. Supernatants were plaqued in Vero cells. Plaques could be visualized at 4 to 7 dpi. One or two plaques from each independent rescue were amplified in Vero cells and their genomes were sequenced.
Vero, HeLa, MDBK and L929 cells were maintained in Dulbecco’s modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (P/S)(Mediatech Inc., Holu Hill, FL), BSRT-7 cell were maintained in DMEM supplemented with 10% FBS, 1% P/S and 10% tryptose phosphate broth (TPB) plus G418 at 400 ug/ml. Cells were cultured at 37°C with 5% CO2 and passed the day before infection or transfection at appropriate dilution factors to archived 80–90% confluence the next day. For virus infection, cells were infected with viruses in DMEM plus 1% bovine serum albumin (BSA) at MOI of 0.01, 3 or 5 and incubated for 1 to 2 hours at 37°C with 5% CO2. The culturing medium was then replaced with DMEM supplied with 2% FBS and 1% P/S. For transfection, cells were transfected with plasmids using PLUS™ and Lipofectamine™ reagents from Invitrogen following the manufacturer provided protocol.
MuV-IA (MuV/Iowa.US/2006) was isolated at the Iowa Hygenic Laboartory from a buccal swab obtained from a mumps case during the early phase of the outbreak in 2006. Genotype analysis was performed at the CDC (Rota et al., 2009) and the accession number for the SH sequence is DQ661745. All mumps viruses were grown in Vero cells and were harvested 4 to 7 dpi. Virus titers were measured in Vero cells by plaque assay followed by Giemsa staining as described before (He and Lamb, 1999; He et al., 1997).
Sequencing of viruses
Viral RNA was extracted from cell culture supernatants using QIAamp® viral RNA extraction mini kit from QIAGEN following manufacturer’s protocol. Isolated total RNA was reverse transcribed into cDNA using Super Script® III reverse transcriptase from Invitrogen with random hexamers. Synthesized cDNA was then served as templates for PCR using mumps virus genome specific primers and Taq polymerase from Invitrogen. 15 sets of primers, each contained a forward and a reverse primer, were designed as to divide the genome into 15 overlapped fragments. The primers were used for the subsequent sequencing of the PCR products (Li et al., 2011). Leader and Trailer sequences were sequenced following standard protocol of Rapid Amplification of cDNA Ends (RACE) (Li et al., 2011). Primer sequences are available upon request.
Generation of monoclonal and polyclonal antibodies against mumps NP, P and SH
To generate monoclonal antibodies against MuV-IA, the virus was grown in Vero cells. The medium of infected Vero cells was collected, and clarified with low-speed centrifugation at 3K rpm for 10 minutes. The clarified media containing virus was overlaid onto a 10ml 20% sucrose solution and centrifuged at 40K rpm for 1.5 hrs at 4°C. The pellet was resuspended in 0.5ml 10 X PBS, mixed with 1.3 ml 80% sucrose solution and overlaid by a decreasing sucrose gradient from bottom to top: 1.8 ml 50% sucrose solution and 0.6ml 10% sucrose solution. The sucrose gradient with virus at the bottom was centrifuged at 45K rpm for 3 hrs at 4°C. 1 ml fractions were collected, mixed with 10ml 1X TEN buffer (100mM NaCl, 10mM Tris-base, 1mM EDTA) and spun down at 40K rpm for 1.5 hrs at 4°C. The pellet containing virus was suspended in 50ul of 1 X TEN buffer plus 1% NP-40 and used for generation of mouse hybridoma cells. Mouse hybridoma cells generating monoclonal antibodies against MuV-IA NP and P were engineered by the core facility in the Pennsylvania State University. The hybridomas were culture in D-MEM supplied with sodium pyruvate, with addition of 20%FBS and 0.1% Gentamicin at 37°C with 5% CO2.
To generate polyclonal antibodies against MuV-IA SH, two peptides (N-terminal MPAIQPPLYLTFLLC and C-terminal CYQRSFFHWSFDHSL) were purchased from GenScript Corporation. Two peptides (QFIKQDETGDLIETC and CSRPDNPRGGHRREW) were used to generate polyclonal antibodies against MuV-IA V (GenScript Corporation) in rabbits.
Treatment of infected cells with anti-TNF-α
L929 cells in 6 well plates were infected with rMuVΔSH or rMuV at a MOI of 5 and cultured in DMEM supplemented with 2%FBS and 1% P/S with neutralizing anti-TNF-α antibody or control antibody (BD Pharmingen) at 50 ug/ml for 1 or 2 days (9). At 1 day or 2 dpi, cells were photographed with a microscope with a digital camera, and then collected for MuV-NP staining or TUNEL assay.
Flow cytometry and TUNEL assay
Flow cytometry was performed as previously described (Timani et al., 2008). L929 cell in 6 well plates were infected with rMuVΔSH or rMuV or mock infected at MOI of 3 or 5. At 1 or 2 dpi, attached cells were trypsinized and combined with floating cells in the culture media. Cells were centrifuged and resuspended in 0.5% formaldehyde in phosphate buffered saline (PBS) for 1 hr at 4 °C. The fixed cells were then washed with PBS, permeabilized in 50% FCS-50%DMEM plus three volumes of 70% ethanol overnight. Permeabilized cells were subjected for either TUNEL staining for apoptotic cells according to manufacturer’s protocol or MuV-NP staining for infection rate. When cells were for NP staining, monoclonal MuV-NP antibody was diluted to 1:200 followed by PE anti-mouse secondary antibody staining at a dilution factor of 1:100.
Vero cells were mock infected or infected with rMuVΔSH or rMuV at a MOI of 0.5 or 0.01. At 24 or 48hpi, attached cells were collected in combination with floating cells, fixed. For HN surface staining, cells were directly stained with anti-HN at a dilution factor of 1:50; for total staining of HN and NP staining, fixed cells were permeabilized with 0.1% saponin in PBS and stained with anti-NP at a dilution factor of 1:200 or anti-HN at a dilution factor of 1:50.
Assays for detection of activation of NF-κB
L929 cells on glass cover slips in 6 well plates were infected with rMuVΔSH, or rMuV at MOI of 0.01, or mock infected. At 2 dpi the cover slips were washed with PBS, and fixed in 0.5% formaldehyde. The fixed cells were permeabilized with PBS plus 0.1% saponine and then incubated with mouse anti-P65 (Santa cruz Biotechnology) in PBS with 0.1% saponine followed by secondary FITC labeled goat anti-mouse antibody (Jackson Laboratory). The cells were photographed using a fluorescence microscope with a digital camera.
The NF-κB reporter assay system was performed as described previously (Wilson et al., 2006). L929 cells were plated into 24 well plates and transfected using PLUS™ and Lipofectamine™ reagents with either empty vector, pCAGGS-MuV SH, pCAGGS-MuV SH(stop), pCAGGS-PIV5 SH or pCAGGS-MuV NP, plus a p κ B-TATA-Luc (a reporter plasmid containing a NF-κB promoter region followed by TATA box enhancer and a firefly luciferase gene) and a pCAGGS-RL (a transfection control plasmid expressing renilla luciferase protein). On the second day post transfection, half of the cells were treated with TNF-α (Alexis, San Diego) at a concentration of 10ng/ml in Optima (Invitrogen) for 4 hrs at 37°C with 5% CO2; half of the cells were treated with Optima only. Cells were then lysed with 100ul 1X passive lysis buffer (Promega, Madison, WI) and 10 ul of the lysate were subjected for dual luciferase assay using a dual luciferase assay kit (Promega, Madison, WI). The ratio of TNF-α stimulated cells over no TNF-α stimulation is used as “induction of luciferase activity”.
Immunoblotting
Vero cells in 6 well plates at about 90% confluence were infected with mock, MuV-IA, rMuV or rMuVΔSH at a MOI of 0.05. Cells were collected and lysed at 0hr, 24hr, 48hr, or 72hr post-infection in 0.5 ml WCEB buffer (50mM Tris-HCl PH 8.0, 120mM NaCl, 0.5% NP-40, 0.00076% EGTA, 0.2mM EDTA, 10% Glycerol) with a mixture of protease inhibitors as described before (Luthra et al., 2008). Cell lysates were briefly centrifuged to remove cell debris. Cell lysates were loaded into 10% or 17.5% polyacrylamide gel and subjected for SDS-PAGE. Protein were transferred to Immobilon-FL transfer membrane (Millipore), incubated with primary antibody (anti-MuV SH 1:250, anti-MuV V 1:500, anti-MuV NP 1:5000, anti-MuV P 1:2000) and corresponding secondary antibodies conjugated to horseradish peroxidase, and detected by Amersham ECL™ western blotting detection kit (GE Healthcare).
Time course of rMuVΔSH, rMuV and MuV-IA infection in cell culture
Cells in 6cm plates were infected with MuV-IA, rMuV, or rMuVΔSH at MOI of 0.01. 100 µl of supernatant were collected at 0hr, 24hr, 48hr, 72hr postinfection and frozen down at −80°C supplemented with 1% BSA. Virus titers were determined by plaque assay using Vero cells in 24 well plates in triplicates. After 1 to 2 hr’s incubation with the viruses, growth media were changed into semi-solid DMEM with 2% FBS, 1% P/S and 1% low melting point agarose. 4 to 7 dpi, 24 well plates of Vero cells were stained with Giemsa stain and plaques were counted.
Enzyme-linked immunosorbent assay (ELISA) of TNF-α
L929 cells in 6 well plates were infected with mock, rMuV or rMuVΔSH at MOI of 5. Culturing media were collected at 1 dpi, 2 dpi and 3 dpi. The amount of TNF-α secreted into the culturing media was measured using a murine TNF-α detection kit (Amersham Pharmacia) following the procedures described before (Li et al., 2011).
Real time RT-PCR
Vero cells were mock infected or infected with rMuVΔSH or rMuV at a MOI of 0.005. Viral RNA was extracted from infected cells at 4dpi using QIAGEN RNeasy mini kit and reverse transcribed into cDNA using Oligo-dT as primers. MuV F and HN mRNA specific FAM tagged probes were purchased from Applied Biosystems™. Real time PCR was assembled using TaqMan® Gene Expression Master Mix, according to manufacture’s protocol. Ratio between HN mRNA verses F mRNA was calculated using ΔCt.
Examination of MuV neurotoxicity
The rat neurotoxicity test was performed as described before (Rubin et al., 2000). Newborn rats were inoculated intracerebrally with 100 p.f.u. of rMuV (n=36), or rMuVΔSH (n=24) in 20 µl EMEM. Animals were sacrificed at 1 month after injection and the brains were removed, immersionfixed and embedded in paraffin. One 10 µm sagittal section at a constant distance from the anatomical midline from each hemisphere of brain was selected, and stained with haematoxylin and eosin. The neurotoxicity score was calculated based on the cross-sectional area of the brain (excluding the cerebellum) as a percentage of the lateral ventricle on tissue sections from paired brain using Image- Pro Plus image analysis software (Media Cybernetics). The neurotoxicity score was defined as the mean ratio (percentage) of these two measurements on each of the two tissue sections per rat brain. Any rats with signs of pain or distress prior to the planned 1month end point were humanely euthanized immediately and included in analyses. The NIH Guidelines for the Care and Use of Laboratory Animals were strictly adhered to throughout.
ACKNOWLEDGEMENT
We appreciate comments, suggestions, and technical help from members of the He lab. We are grateful to Dr. Steven Rubin for the neurotoxicity test and ant-HN antibody. This work has been supported by grants from NIH (K02 065795 to B.H.) and National Natural Science Foundation of China Funds for Distinguished Young Scholar—B Plan (No. 30928001 to B.H. and J.W.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Amexis G, Rubin S, Chizhikov V, Pelloquin F, Carbone K, Chumakov K. Sequence diversity of Jeryl Lynn strain of mumps virus: quantitative mutant analysis for vaccine quality control. Virology. 2002;300(2):171–179. doi: 10.1006/viro.2002.1499. [DOI] [PubMed] [Google Scholar]
- Baud V, Karin M. Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 2001;11(9):372–377. doi: 10.1016/s0962-8924(01)02064-5. [DOI] [PubMed] [Google Scholar]
- Carbone KM, Wolinsky JS. Mumps Virus. In: Knipe DM, Howley PM, editors. "Field's Virology". 4 ed. Vol. 1. Philadelphia: Lippincott Williams and Wilkins; 2001. pp. 1381–1400. 2 vols. [Google Scholar]
- Crowley B, Afzal MA. Mumps virus reinfection--clinical findings and serological vagaries. Commun Dis Public Health. 2002;5(4):311–313. [PubMed] [Google Scholar]
- Cusi MG, Santini L, Bianchi S, Valassina M, Valensin PE. Nucleotide sequence at position 1081 of the hemagglutinin-neuraminidasegene in wild-type strains of mumps virus is the most relevant marker of virulence. J Clin Microbiol. 1998;36(12):3743–3744. doi: 10.1128/jcm.36.12.3743-3744.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elango N, Varsanyi TM, Kovamees J, Norrby E. Molecular cloning and characterization of six genes, determination of gene order and intergenic sequences and leader sequence of mumps virus. J Gen Virol. 1988;69(Pt 11):2893–2900. doi: 10.1099/0022-1317-69-11-2893. [DOI] [PubMed] [Google Scholar]
- He B, Lamb RA. Effect of inserting paramyxovirus simian virus 5 gene junctions at the HN/L gene junction: analysis of accumulation of mRNAs transcribed from rescued viable viruses. J. Virol. 1999;73:6228–6234. doi: 10.1128/jvi.73.8.6228-6234.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Leser GP, Paterson RG, Lamb RA. The paramyxovirus SV5 small hydrophobic (SH) protein is not essential for virus growth in tissue culture cells. Virology. 1998;250:30–40. doi: 10.1006/viro.1998.9354. [DOI] [PubMed] [Google Scholar]
- He B, Lin GY, Durbin JE, Durbin RK, Lamb RA. The sh integral membrane protein of the paramyxovirus simian virus 5 is required to block apoptosis in mdbk cells. J Virol. 2001;75(9):4068–4079. doi: 10.1128/JVI.75.9.4068-4079.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Paterson RG, Ward CD, Lamb RA. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology. 1997;237:249–260. doi: 10.1006/viro.1997.8801. [DOI] [PubMed] [Google Scholar]
- Hiebert SW, Paterson RG, Lamb RA. Identification and predicted sequence of a previously unrecognized small hydrophobic protein, SH, of the paramyxovirus simian virus 5. J. Virol. 1985;55:744–751. doi: 10.1128/jvi.55.3.744-751.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiebert SW, Richardson CD, Lamb RA. Cell surface expression and orientation in membranes of the 44 amino acid SH protein of simian virus 5. J. Virol. 1988;62:2347–2357. doi: 10.1128/jvi.62.7.2347-2357.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Rima B, Brown D, Orvell C, Tecle T, Afzal M, Uchida K, Nakayama T, Song JW, Kang C, Rota PA, Xu W, Featherstone D. Proposal for genetic characterisation of wild-type mumps strains: Preliminary standardisation of the nomenclature. Arch Virol. 2005 doi: 10.1007/s00705-005-0563-4. [DOI] [PubMed] [Google Scholar]
- Kolakofsky D, Pelet T, Garin D, Housmann S, Curran J, Roux L. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: The rule of six revisited. J. Virol. 1998;72:891–899. doi: 10.1128/jvi.72.2.891-899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T, Yokosawa N, Yokota S, Fujii N. Association of mumps virus V protein with RACK1 results in dissociation of STAT-1 from the alpha interferon receptor complex. J Virol. 2002;76(24):12676–12682. doi: 10.1128/JVI.76.24.12676-12682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Xu J, Patel J, Fuentes S, Lin Y, Anderson D, Sakamoto K, Wang LF, He B. Function of the small hydrophobic protein of J paramyxovirus. J Virol. 2011;85(1):32–42. doi: 10.1128/JVI.01673-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CS, Chan KP, Goh KT, Chow VT. Hemagglutinin-neuraminidase sequence and phylogenetic analyses of mumps virus isolates from a vaccinated population in Singapore. J Med Virol. 2003;70(2):287–292. doi: 10.1002/jmv.10393. [DOI] [PubMed] [Google Scholar]
- Lin Y, Bright AC, Rothermel TA, He B. Induction of apoptosis by paramyxovirus simian virus 5 lacking a small hydrophobic gene. J Virol. 2003;77(6):3371–3383. doi: 10.1128/JVI.77.6.3371-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Horvath F, Aligo JA, Wilson R, He B. The role of simian virus 5 V protein on viral RNA synthesis. Virology. 2005;338(2):270–280. doi: 10.1016/j.virol.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Luthra P, Sun D, Silverman RH, He B. Activation of IFN-beta expression by a viral mRNA through RNase L and MDA5. Proc Natl Acad Sci U S A. 2011;108(5):2118–2123. doi: 10.1073/pnas.1012409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra P, Sun D, Wolfgang M, He B. AKT1-dependent activation of NF-kappaB by the L protein of parainfluenza virus 5. J Virol. 2008;82(21):10887–10895. doi: 10.1128/JVI.00806-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin M, Quinlisk P, Shimabukuro T, Sawhney C, Brown C, Lebaron CW. Mumps vaccination coverage and vaccine effectiveness in a large outbreak among college students--Iowa, 2006. Vaccine. 2008;26(29–30):3601–3607. doi: 10.1016/j.vaccine.2008.04.075. [DOI] [PubMed] [Google Scholar]
- Matsumoto T. Assembly of paramyxoviruses. Microbiol Immunol. 1982;26(4):285–320. doi: 10.1111/j.1348-0421.1982.tb00180.x. [DOI] [PubMed] [Google Scholar]
- MMWR. Mumps epidemic--United kingdom, 2004–2005. MMWR Morb Mortal Wkly Rep. 2006;55(7):173–175. [PubMed] [Google Scholar]
- MMWR. Update: mumps outbreak - New York and New Jersey, June 2009–January 2010. MMWR Morb Mortal Wkly Rep. 2010;59(5):125–129. [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants by a novel eukaryotic vector. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Okazaki K, Tanabayashi K, Takeuchi K, Hishiyama M, Okazaki K, Yamada A. Molecular cloning and sequence analysis of the mumps virus gene encoding the L protein and the trailer sequence. Virology. 1992;188:926–930. doi: 10.1016/0042-6822(92)90555-4. [DOI] [PubMed] [Google Scholar]
- Otto W, Mankertz A, Santibanez S, Saygili H, Wenzel J, Jilg W, Wieland W, Borgmann S. Ongoing outbreak of mumps affecting adolescents and young adults in Bavaria, Germany, August to October 2010. Euro Surveill. 15(50) [PubMed] [Google Scholar]
- Paterson RG, Lamb RA. RNA editing by G-nucleotide insertion in mumps virus P-gene mRNA transcripts. J. Virol. 1990;64:4137–4145. doi: 10.1128/jvi.64.9.4137-4145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rima BK, Roberts MW, McAdam WD, Martin SJ. Polypeptide synthesis i mumps virus-infected cells. J Gen Virol. 1980;46(2):501–505. doi: 10.1099/0022-1317-46-2-501. [DOI] [PubMed] [Google Scholar]
- Rota JS, Turner JC, Yost-Daljev MK, Freeman M, Toney DM, Meisel E, Williams N, Sowers SB, Lowe L, Rota PA, Nicolai LA, Peake L, Bellini WJ. Investigation of a mumps outbreak among university students with two measles-mumps-rubella (MMR) vaccinations, Virginia, September–December 2006. J Med Virol. 2009;81(10):1819–1825. doi: 10.1002/jmv.21557. [DOI] [PubMed] [Google Scholar]
- Rubin SA, Afzal MA, Powell CL, Bentley ML, Auda GR, Taffs RE, Carbone KM. The rat-based neurovirulence safety test for the assessment of mumps virus neurovirulence in humans: an international collaborative study. J Infect Dis. 2005;191(7):1123–1128. doi: 10.1086/428098. [DOI] [PubMed] [Google Scholar]
- Saito H, Takahashi Y, Harata S, Tanaka K, Sano T, Suto T, Yamada A, Yamazaki S, Morita M. Isolation and characterization of mumps virus strains in a mumps outbreak with a high incidence of aseptic meningitis. Microbiol Immunol. 1996;40(4):271–275. doi: 10.1111/j.1348-0421.1996.tb03346.x. [DOI] [PubMed] [Google Scholar]
- Santak M, Kosutic-Gulija T, Tesovic G, Ljubin-Sternak S, Gjenero-Margan I, Betica-Radic L, Forcic D. Mumps virus strains isolated in Croatia in 1998 and 2005: Genotyping and putative antigenic relatedness to vaccine strains. J Med Virol. 2006;78(5):638–643. doi: 10.1002/jmv.20587. [DOI] [PubMed] [Google Scholar]
- Strohle A, Bernasconi C, Germann D. A new mumps virus lineage found in the 1995 mumps outbreak in western Switzerland identified by nucleotide sequence analysis of the SH gene. Arch Virol. 1996;141(3–4):733–741. doi: 10.1007/BF01718331. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Tanabayashi K, Hishiyama M, Yamada A, Sugiura A. Variation of nucleotide sequences and transcription of the SH gene among mumps virus strains. Virology. 1991;181:364–366. doi: 10.1016/0042-6822(91)90504-5. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Tanabayashi K, Hishiyama M, Yamada YK, Yamada A, Sugiura A. Detection and characterization of mumps virus V protein. Virology. 1990;178:247–253. doi: 10.1016/0042-6822(90)90400-l. [DOI] [PubMed] [Google Scholar]
- Tanabayashi K, Takeuchi K, Okazaki K, Hishiyama M, Yamada A. Expression of mumps virus glycoproteins in mammalian cells from cloned cDNAs: both F and HN proteins are required for cell fusion. Virology. 1992;187:801–804. doi: 10.1016/0042-6822(92)90482-5. [DOI] [PubMed] [Google Scholar]
- Timani KA, Sun D, Sun M, Keim C, Lin Y, Schmitt PT, Schmitt AP, He B. A single amino acid residue change in the P protein of parainfluenza virus 5 elevates viral gene expression. J Virol. 2008;82(18):9123–9133. doi: 10.1128/JVI.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulane CM, Rodriguez JJ, Parisien JP, Horvath CM. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J Virol. 2003;77(11):6385–6393. doi: 10.1128/JVI.77.11.6385-6393.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utz S, Richard JL, Capaul S, Matter HC, Hrisoho MG, Muhlemann K. Phylogenetic analysis of clinical mumps virus isolates from vaccinated and non-vaccinated patients with mumps during an outbreak, Switzerland 1998–2000. J Med Virol. 2004;73(1):91–96. doi: 10.1002/jmv.20064. [DOI] [PubMed] [Google Scholar]
- Watson-Creed G, Saunders A, Scott J, Lowe L, Pettipas J, Hatchette TF. Two successive outbreaks of mumps in Nova Scotia among vaccinated adolescents and young adults. Cmaj. 2006;175(5):483–488. doi: 10.1503/cmaj.060660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxham MN, Server AC, Goodman HM, Wolinsky JS. Cloning and sequencing of the mumps virus fusion protein gene. Virology. 1987;159:381–388. doi: 10.1016/0042-6822(87)90477-6. [DOI] [PubMed] [Google Scholar]
- Whelan J, van Binnendijk R, Greenland K, Fanoy E, Khargi M, Yap K, Boot H, Veltman N, Swaan C, van der Bij A, de Melker H, Hahne S. Ongoing mumps outbreak in a student population with high vaccination coverage, Netherlands, 2010. Euro Surveill. 15(17) doi: 10.2807/ese.15.17.19554-en. [DOI] [PubMed] [Google Scholar]
- Wilson RL, Fuentes SM, Wang P, Taddeo EC, Klatt A, Henderson AJ, He B. Function of Small Hydrophobic Proteins of Paramyxovirus. J Virol. 2006;80(4) doi: 10.1128/JVI.80.4.1700-1709.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosawa N, Yokota S, Kubota T, Fujii N. C-terminal region of STAT-1alpha is not necessary for its ubiquitination and degradation caused by mumps virus V protein. J Virol. 2002;76(24):12683–12690. doi: 10.1128/JVI.76.24.12683-12690.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]