Abstract
Background
Hepatitis C viral (HCV) load detection and quantification is routinely accomplished by HCV RNA measurement, an expensive but essential test, both for the diagnosis and treatment of chronic hepatitis C (CHC). HCV core antigen (Ag) testing has been suggested as an attractive alternative to molecular diagnostics. The aim of the study was to evaluate an automated chemiluminescent immunoassay (CLIA) for HCV core Ag measurement in comparison to quantitative HCV RNA determination.
Methods
HCV Ag was measured in 105 anti-HCV positive patients, from which 89 were HCV RNA positive with CHC and 16 HCV RNA negative after spontaneous HCV clearance. Viral load was quantified with branched DNA (bDNA, Versant, Siemens). Sera were stored at -70°C and then tested with the Architect HCV Ag test (Abbott Laboratories), a two-step CLIA assay, with high throughput and minimal handling of the specimens. Statistical analysis was performed on logarithmically transformed values.
Results
HCV-Ag was detectable and quantifiable in 83/89 and in grey zone in 4/89 HCV RNA positive sera. HCV-Ag was undetectable in all 16 HCV RNA negative samples. The sample with the lowest viral load that tested positive for HCV-Ag contained 1200 IU/mL HCV RNA. There was a positive correlation between HCV RNA and HCV-Ag (r=0.89). The HCV RNA/ HCV Ag ratio varied from 1.5 to 3.25.
Conclusion
The HCV core Ag is an easy test with comparable sensitivity (>90%) and satisfactory correlation with the HCV RNA bDNA assay. Its role in diagnostics and other clinical applications has to be determined based on cost effectiveness.
Keywords: Core antigen, HCV RNA, hepatitis C
Introduction
Hepatitis C virus (HCV) is detected in serum or plasma of chronically infected patients by molecular methods that specifically measure HCV RNA. HCV viral load is routinely quantified by target amplification with polymerase chain reaction (PCR) or signal amplification with branched DNA (bDNA) assays [1].
In view of the fact that HCV RNA quantification is a rather expensive test, that requires technical skills and separate facilities, there have been efforts to establish different accurate, sensitive and cheaper methods for HCV viral load determination. Quantification of HCV core antigen (Ag), as being part of the viral particle, has been suggested as a possible alternative to HCV RNA measurements. In previous years, manual HCV-Ag ELISA tests were available, but they demonstrated significant limitations, mainly in terms of sensitivity [2].
An HCV core Ag automated chemiluminescent immunoassay (CLIA) has been recently developed for the Architect analyzer (Abbott Diagnostics, Wiesbaden, Germany), with enhanced sensitivity and minimal handling of the specimens [3].
The aim of this study was to evaluate the results of the automated HCV-Ag assay compared to quantitative HCV RNA determinations in clinical specimens.
Materials and methods
Materials
Sera from 105 anti-HCV positive patients were included for this study. Among those, 89 were obtained from patients with chronic hepatitis C (CHC) and detectable HCV RNA (36% HCV genotype 1, 6% genotype 2, 37% genotype 3 and 21% genotype 4) and 16 from anti-HCV positive patients with previously diagnosed acute infection and spontaneous HCV RNA clearance. All sera were tested for HCV RNA with a sensitive PCR assay, the Cobas Amplicor v2.0 test HCV RNA PCR (Roche Molecular Systems, Inc., Pleasanton, CA) according to the manufacturer’s recommendations [4].
In HCV RNA positive sera, viral load was quantified by bDNA (Versant 2.0; Siemens Healthcare Diagnostics, Deerfield, IL), an HCV RNA signal amplification method, with detection limit of 615 IU/mL (2.79 log10 IU/mL) [5-7].
HCV Ag detection and quantification
Sera, stored at -70° C, were thawed once and tested with the Architect HCV-Ag assay. Samples were processed automatically, first treated on-board to dissociate antigen-antibody complexes and release the antigen from the viral particles. HCV core Ag is detected with a cut-off value of 3.0 fmol/L, a grey zone of 3-11 fmol/L and an upper limit of 180,000 fmol/L (1.0 fmol/L of HCV Ag equals to 0.02 pg/mL) [3,8].
Statistical analysis
The correlation coefficients between HCV-Ag and HCV RNA were calculated by Spearman’s rank test and a comparison between the groups was carried out using the Mann-Whitney U test. A P value of less than 0.05 was considered statistically significant.
Results
In HCV RNA positive sera, viral load ranged between 3.06 and 6.9 log10 IU/mL (median 5.48). HCV Ag was undetectable in 2/89 samples (2.25%) with HCV RNA values of 1159 and 1947 IU/mL (3.06 and 3.29 log10 IU/mL). Four samples (4.49%) were found in the HCV Ag grey zone, with values between 7 and 10.6 fmol/L (median 8.72) or in logs0.84 log10 and 1.02 log10 fmol/L (median 0.94). Their respective HCV RNA levels were between 3.07 and 4.9 log10 IU/mL (median 3.39). HCV-Ag was clearly positive and quantifiable in 83 of the 89 HCV RNA positive sera (93.26%) with HCV RNA levels between 3.12 and 6.87 log10 IU/mL (median 5.55). In those samples, positive for both parameters, HCV Ag was found between 1.10 and 4.27 log10 fmol/L (median 3.15) (Table 1). The correlation coefficient (r) of the logarithmic values of HCV RNA and HCV-Ag, in these cases, was 0.89 (y=0.8465x-1.5343) and it was statistically significant (P<0.001) (Fig. 1). The HCV RNA/HCV Ag ratio ranged from 1.5 to 3.25 (median 1.72). The genotype of HCV did not affect HCV Ag detection and did not correlate with HCV RNA/HCV Ag ratios.
Table 1.
Qualitative and quantitative results of hepatitis C virus (HCV) antigen (Ag) in HCV RNA-positive sera

Figure 1.
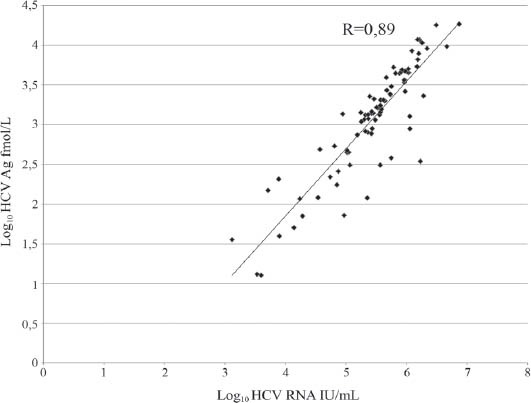
Correlation between hepatitis C virus (HCV) core antigen (HCV-Ag) measured by Architect and HCV-RNA measured by branched DNA (bDNA). The correlation coefficient (R) indicates agreement between logarithmically transformed results
The 16 samples with undetectable HCV RNA also tested negative (100%) by the HCV-Ag assay.
Discussion
Since 1999, an ELISA assay has been introduced for serum HCV-Ag detection and quantification. This method showed marked differences in HCV RNA/core Ag ratios and did not demonstrate the required sensitivity in order to be clinically applied as a substitute for HCV RNA measurements that had become simpler and widely used [2,9-13]. Moreover, HCV-Ag detection has been incorporated in some EIA assays with concomitant anti-HCV antibody detection. This combined type of screening, on one hand leads to a shorter window phase for the detection of HCV, but on the other hand demonstrates lower sensitivity and specificity compared to traditional assays, when HCV RNA is used as a reference [14-17].
The employment of chemiluminescent magnetic particle-based technology and immune complex-dissociating reaction increases the sensitivity of the studied Architect HCV-Ag test 8- to 21-fold from previous HCV core Ag ELISA assays. The analytical sensitivity of the Architect HCV-Ag assay is reported to be between 428 and 2,700 IU/mL HCV RNA, depending on the HCV genotype [18]. In our study we demonstrated sensitivity in clinical specimens equivalent to 1200 IU/mL, corresponding to the serum with the lowest HCV RNA that tested positive for HCV-Ag. This is clear evidence of the enhanced ability of the new HCV core Ag test compared to older ELISAs, to detect low level viremia. In a previous study, in 193 HCV RNA low positive samples (<3.9 log10 IU/mL), HCV-Ag was detected in low levels (median 16 fmol/L) in 81.9% of studied samples. In the same study the sensitivity of HCV-Ag corresponded to 1000 IU/mL of HCV RNA [19], similar to our findings. In this study, we did not observe any influence of the viral genotype on HCV Ag detection. This could be due to the relatively small number of determinations combined with the low prevalence of genotype 2 in the tested population.
The bDNA method that was applied for HCV RNA quantification is a widely used test, with lower sensitivity but higher reproducibility than PCR [20]. Although bDNA lacks in sensitivity compared to PCR, it is an appropriate method to use for HCV RNA correlation with HCV Ag since the latter has even lower analytical sensitivity, according to the manufacturer [18]. It is therefore expected that the vast majority of HCV RNA negative by bDNA, but RT PCR positive samples with viral load between 15 and 615 IU/mL, would be also missed by the HCV Ag test [9]. The quantitative HCV Ag results significantly correlated with HCV RNA measurements by bDNA with a correlation coefficient of 0.89. Higher correlation was reported in a previous study using a more sensitive Cobas-TaqMan RT-PCR assay (r=0.95), but this was achieved with the inclusion of negative results in the calculations. Among 282 samples, 108 were HCV RNA negative and nonreactive for HCV Ag, also demonstrating the observed high specificity of the assay [8]. On the other hand, in a different study [19] the correlation coefficient between HCV-Ag and bDNA was 0.713 and between HCV-Ag and two RT-PCR methods was 0.743 and 0.87, respectively. The latter results were based on calculations with absolute and not logarithmically transformed values. We used logarithmic transformation of HCV RNA values that is common practice in the clinical assessment of chronic hepatitis. Nonetheless, the correlation of HCV Ag with RNA values obtained by RT PCR appears to be slightly higher than that of bDNA. This was also observed in our laboratory in a small number of samples (N=21) that were tested for HCV RNA with an in house RTPCR. In this series the correlation of HCV RNA with HCV Ag was slightly higher, with a correlation coefficient of 0.93 (data not shown) compared to 0.89.
Additionally, our quantitative results yielded an HCV RNA/HCV Ag ratio that varied from one patient to another from 1.5 to 3.25. This finding is considered to be due to the presence of HCV-Ag not only in complete but also in incomplete viral particles and protein structures as has been previously pointed out [2,20,21].
The use of sensitive assays for HCV viral load determination is widely recognized. The assay for HCV-Ag detection by Architect demonstrates high correlation with HCV RNA and improved sensitivity in relation to previous versions of the test. Despite the improvement in sensitivity, there are still obvious limitations of HCV Ag compared to modern molecular methods that can detect as little as 10-15 IU/ mL [22]. However, the high specificity and the simplicity of the test, the rapid turnaround time of the results and its lower price suggest that it could be cost effectively applied in clinical practice. It was demonstrated, in consequence of the high specificity, that HCV Ag positivity is clear proof of viremia and active HCV infection. Therefore, in screening scenarios, only patients with HCV Ag negative results would have to be evaluated for HCV viral load with more sensitive and expensive molecular methods, like RT-PCR, which are definitely essential for the monitoring of treatment. In positive HCV Ag cases, absolute HCV Ag levels need to be carefully interpreted on an individual basis and at this point they cannot replace molecular methods for HCV RNA quantification. More studies, with concomitant consideration of the financial cost of HCV viral load testing, need to be conducted in order to pinpoint the proper applications of HCV Ag testing.
Summary Box
What is ready known:
Hepatitis C virus (HCV) is detected and quantified in serum or plasma by means of HCV RNA measurements, using molecular methods
Quantification of HCV core antigen (HCV-Ag) has been suggested as an attractive alternative to these expensive and demanding measurements
Previous developed HCV-Ag ELISA manual tests demonstrated various limitations
What the new findings are:
Herein, a simple automated HCV core antigen assay was compared to HCV RNA determination by branched DNA (bDNA)
The assay was able to detect an equivalent of at least 1200 IU of the HCV RNA/mL
There was significant correlation between logarithmically transformed values of HCV RNA and HCV-Ag
Variations in HCV RNA/HCV Ag ratio were observed
Biography
Hippokration General Hospital, Athens, Greece
Footnotes
Conflict of Interest: None
References
- 1.Chevaliez S. Virological tools to diagnose and monitor hepatitis C virus infection. Clin Microbiol Infect. 2011;17:116–121. doi: 10.1111/j.1469-0691.2010.03418.x. [DOI] [PubMed] [Google Scholar]
- 2.Bouvier Alias M, Patel K, Dahari H, et al. Clinical utility of total HCV core antigen quantification: a new indirect marker of HCV replication. Hepatology. 2002;36:211–218. doi: 10.1053/jhep.2002.34130. [DOI] [PubMed] [Google Scholar]
- 3.Morota K, Fujinami R, Kinukawa H, et al. A new sensitive and automated chemiluminescent microparticle immunoassay for quantitative determination of hepatitis C virus core antigen. J Virol Methods. 2009;157:8–14. doi: 10.1016/j.jviromet.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Lee SC, Antony A, Lee N, et al. Improved version 2.0 qualitative and quantitative AMPLICOR reverse transcription-PCR tests for hepatitis C virus RNA: calibration to international units, enhanced genotype reactivity, and performance characteristics. J Clin Microbiol. 2000;38:4171–4179. doi: 10.1128/jcm.38.11.4171-4179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alter HJ, Sanchez-Pescador R, Urdea MS, et al. Evaluation of branched DNA signal amplification for the detection of hepatitis C virus RNA. J Viral Hepat. 1995;2:121–132. doi: 10.1111/j.1365-2893.1995.tb00017.x. [DOI] [PubMed] [Google Scholar]
- 6.Beld M, Sentjens R, Rebers S, et al. Performance of the New Bayer VERSANT HCV RNA 3.0 assay for quantitation of hepatitis C virus RNA in plasma and serum: conversion to international units and comparison with the Roche COBAS Amplicor HCV Monitor, Version 2.0, assay. J Clin Microbiol. 2002;40:788–793. doi: 10.1128/JCM.40.3.788-793.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elbeik T, Surtihadi J, Destree M, et al. Multicenter evaluation of the performance characteristics of the bayer VERSANT HCV RNA 3.0 assay (bDNA) J Clin Microbiol. 2004;42:563–569. doi: 10.1128/JCM.42.2.563-569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park Y, Lee JH, Kim BS, Kim do Y, Han KH, Kim HS. New automated hepatitis C virus (HCV) core antigen assay as an alternative to real-time PCR for HCV RNA quantification. J Clin Microbiol. 2010;48:2253–2256. doi: 10.1128/JCM.01856-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabrizi F, Lunghi G, Aucella F, et al. Novel assay using total hepatitis C virus (HCV) core antigen quantification for diagnosis of HCV infection in dialysis patients. J Clin Microbiol. 2005;43:414–420. doi: 10.1128/JCM.43.1.414-420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komatsu F, Takasaki K. Determination of serum hepatitis C virus (HCV) core protein using a novel approach for quantitative evaluation of HCV viraemia in anti-HCV-positive patients. Liver. 1999;19:375–380. doi: 10.1111/j.1478-3231.1999.tb00065.x. [DOI] [PubMed] [Google Scholar]
- 11.Maynard M, Pradat P, Berthillon P, et al. Clinical relevance of total HCV core antigen testing for hepatitis C monitoring and for predicting patients’ response to therapy. J Viral Hepat. 2003;10:318–313. doi: 10.1046/j.1365-2893.2003.00430.x. [DOI] [PubMed] [Google Scholar]
- 12.Pradat P, Maynard M, Buti M, et al. The predictive value of core antigen testing for the management of hepatitis C patients receiving pegylated interferon/ribavirin treatment. J Med Virol. 2004;73:392–396. doi: 10.1002/jmv.20104. [DOI] [PubMed] [Google Scholar]
- 13.Veillon P, Payan C, Picchio G, Maniez-Montreuil M, Guntz P, Lunel F. Comparative evaluation of the total hepatitis C virus core antigen, branched-DNA, and amplicor monitor assays in determining viremia for patients with chronic hepatitis C during interferon plus ribavirin combination therapy. J Clin Microbiol. 2003;41:3212–3220. doi: 10.1128/JCM.41.7.3212-3220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alzahrani AJ. Simultaneous detection of hepatitis C virus core antigen and antibodies in Saudi drug users using a novel assay. J Med Virol. 2008;80:603–606. doi: 10.1002/jmv.21075. [DOI] [PubMed] [Google Scholar]
- 15.Laperche S, Le Marrec N, Girault A, et al. Simultaneous detection of hepatitis C virus (HCV) core antigen and anti-HCV antibodies improves the early detection of HCV infection. J Clin Microbiol. 2005;43:3877–3883. doi: 10.1128/JCM.43.8.3877-3883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vermeersch P, Van Ranst M, Lagrou K. Evaluation of the use of a combined HCV antigen/antibody assay in routine laboratory practice. Acta Clin Belg. 2010;65:245–247. doi: 10.1179/acb.2010.053. [DOI] [PubMed] [Google Scholar]
- 17.Yang JF, Lin YY, Hsieh MH, et al. Performance characteristics of a combined hepatitis C virus core antigen and anti-hepatitis C virus antibody test in different patient groups. Kaohsiung J Med Sci. 2011;27:258–263. doi: 10.1016/j.kjms.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Ross RS, Viazov S, Salloum S, Hilgard P, Gerken G, Roggendorf M. Analytical performance characteristics and clinical utility of a novel assay for total hepatitis C virus core antigen quantification. J Clin Microbiol. 2010;48:1161–1168. doi: 10.1128/JCM.01640-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medici MC, Furlini G, Rodella A, et al. Hepatitis C virus core antigen: Analytical performances, correlation with viremia and potential applications of a quantitative, automated immunoassay. J Clin Virol. 2011;51:264–269. doi: 10.1016/j.jcv.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Hadziyannis E, Fried MW, Nolte FS. Evaluation of two methods for quantitation of hepatitis C virus RNA. Mol Diagn. 1997;2:39–46. doi: 10.1054/MODI00200039. [DOI] [PubMed] [Google Scholar]
- 21.Schüttler CG, Thomas C, Discher T, et al. Variable ratio of hepatitis C virus RNA to viral core antigen in patient sera. J Clin Microbiol. 2005;42:1977–1981. doi: 10.1128/JCM.42.5.1977-1981.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.European Association for the Study of the Liver. EASL Clinical Practice Guidelines: management of hepatitis C virus infection. J Hepatol. 2011;55:245–264. doi: 10.1016/j.jhep.2011.02.023. [DOI] [PubMed] [Google Scholar]


