Abstract
Enhancers act over many kilobase pairs to activate target promoters, but their activity is constrained by insulator elements that prevent indiscriminate activation of nearby genes. In the July 1, 2009, issue of Genes & Development, Chopra and colleagues (pp. 1505–1509) report that promoters containing a stalled Pol II are activated by enhancers, but these promoters also serve as insulators that block enhancers from reaching more distal genes. This new class of insulators provide critical clues to regulatory mechanisms.
Keywords: Hox, Pol II stalling, insulator, NELF, DSIF, long range
Precise regulation of transcription by RNA polymerase II (Pol II) is critical for the proper growth, development, and survival of an organism. Transcription can be regulated at several stages, and current data increasingly support that the transition from initiation to elongation is frequently exploited as a regulatory target that can govern the level of gene transcription in response to developmental programming (Muse et al. 2007; Wang et al. 2007; Zeitlinger et al. 2007). During this transition, Pol II can pause or stall proximal to the promoter, where it awaits the proper cellular cues and undergoes physical and chemical changes that are necessary for producing a processive elongation complex that is capable of faithfully transcribing the full length of a gene (Saunders et al. 2006; Gilmour 2008). This mechanism has been proposed to provide a number of advantages that result in the rapid, synchronous activation and repression of transcription in response to environmental or developmental stimuli (Lis 1998). It is also proposed to provide a checkpoint that allows time for Pol II to mature into a productive elongation complex, and coordinate multiple pre-mRNA processing events that occur cotranscriptionally. Control of transcription in response to developmental signals is also known to be mediated by short and long-range interactions of promoters with complex cis-element regulatory regions that include enhancers, insulators, and repressive elements (Maeda and Karch 2007).
Interplay of enhancers and insulators
Enhancers are powerful regulatory elements that can reside many thousands of base pairs upstream of or downstream from a target gene and strongly stimulate its transcription. Enhancers are comprised of clusters of short regulatory elements to which a collection of transcription factors bind. In some cases, the mixture of proteins includes not only transcription activators, but also architectural factors that allow the collection of proteins to assemble into a stable structure called the enhanceosome (Merika and Thanos 2001). Enhancers are numerous and distributed broadly over the genome. This broad distribution and ability of enhancers to broadcast their activation signals over long distances is ostensibly a reckless mechanism of gene regulation that potentially could lead to widespread misregulation of gene transcription. Fortunately, enhancers are kept in check by insulator elements that are also numerous and broadly distributed and provide boundaries that confine the action of enhancer to particular gene targets. Insulators block the ability of enhancers to stimulate a promoter when placed between an enhancer and its targeted promoter, and can also function as barrier elements that block the localized spread of active or repressive chromatin (Geyer 1997; Maeda and Karch 2007). Interestingly, the molecular compositions of insulators show considerable variety, and their mechanism of action remains mysterious. The sophisticated and conserved arrangement of these elements in cis at the ancient homeotic gene cluster pays heed to the importance of local chromatin architecture in the regulation of gene transcription. In the July 1, 2009, issue of Genes & Development, Chopra et al. (2009) propose an intimate link between Pol II stalling and enhancer function by demonstrating an intrinsic insulator activity associated with stalled promoters.
A model for precise and responsive transcriptional regulation
The regulation of the Drosophila melanogaster Antennapedia (ANT-C) and bithorax (BX-C) clusters of homeotic genes provide a classic example of the critical need for precise transcription regulation. During early embryogenesis, expression of the genes within the ANT-C and BX-C dictate the identity of the anterior/thoracic and thoracic/posterior parasegments (for reviews, see Akbari et al. 2006; Maeda and Karch 2007). These patterning events occur in a narrow time frame and in segments that are only several cells in width; thus, tight control of gene expression in time and space is crucial for normal development. In the case of the BX-C, promoter regulation is carried out by multiple cis-acting enhancer, repressor, and insulator elements within the 300-kb region of the gene complex. For the well-studied Abd-B gene, gene activation is controlled by a number of iab (infra-abdominal) regions that are situated 3′ of the gene in a linear fashion that mirrors the ordering of the segments within which they activate gene expression (Akbari et al. 2006; Maeda and Karch 2007). The iab regions contain the enhancer and repressor elements, and insulators have been identified that separate the iab regions. The insulating elements can confine both repressor and enhancer activities, and are proposed to interact with each other and/or other elements to create looped chromatin domains (Maeda and Karch 2007). It is believed that these looped domains are responsible for restricting certain IAB–promoter interactions within a domain. Considerable research has focused on determining the molecular mechanisms involved in this intricate system of gene regulation.
Promoter-paused/stalled Pol II has insulator activity
In collaboration with the Young and Adelman laboratories, Mike Levine's laboratory (Zeitlinger et al. 2007) demonstrated recently that many homeotic genes likely harbor a promoter-proximal stalled Pol II. Interestingly, of the eight genes contained within the ANT-C and BX-C, only the two promoters that flank each cluster apparently show characteristics of a stalled Pol II. Running with this observation, Chopra et al. 2009 hypothesized that the stalled Pol II could define the functional domains of each hox cluster. To test this, they turned to classic enhancer-blocking assays that use promoter–reporter gene fusions placed in various configurations relative to elements of interest, such as enhancers or insulators. These assays revealed that when any of the four stalled promoters controlling a lacZ reporter gene were placed in between the IAB5 enhancer element and a white reporter gene, only the stalled promoter was activated. In contrast, when a nonstalled promoter was placed in between the enhancer and the white reporter, both genes were activated. These results indicate that the stalled Pol II promoters respond to enhancers and contain an enhancer-blocking activity that prevents enhancers from reaching over these stalled promoters to target other genes (Fig. 1).
Figure 1.
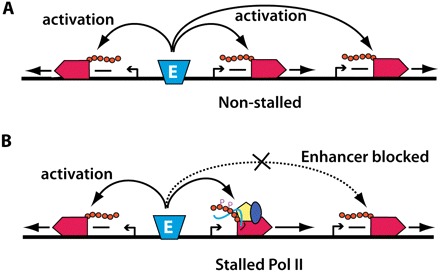
Summary of results from Chopra et al (2009). Chopra et al. (2009) demonstrate enhancer-blocking activity of stalled promoters. (A) Enhancers (blue trapezoid) can activate Pol II (red) at distal as well as nearby genes when the proximal promoter is not stalled. (B) However, distal promoters are insulated from enhancer-directed activation when an intervening promoter contains a stalled Pol II. Components of the stalled complex—NELF (yellow pentagon), DSIF (dark-blue oval), and the short, nascent RNA (blue line)—are shown.
Certain promoter elements can effectively tether an enhancer's activity. For instance, a TATA element has been shown to compete for a functional interaction within an enhancer when an intervening promoter is TATA-less, or if the enhancer is placed in between the two promoters (Ohtsuki et al. 1998). In contrast, whereas stalled promoters are targets of enhancer activity to block downstream activity, they do not inactivate enhancer action on neighboring promoters that reside on the opposite side of the enhancer (Fig. 1). When the IAB5 enhancer was placed in between the Abd-B or Ubx stalled promoters and the white reporter, it was able to activate both promoters. Although a promoter-tethering element was identified recently immediately upstream of the Abd-B gene (Akbari et al. 2008), the entire tethering element was not inculded in the Abd-B promoter construct (VS Chopra, pers. comm.). These results suggest that stalled promoters do not effectively compete with other promoters via a tethering mechanism. Together, these observations suggest that the stalled promoters flanking the hox clusters are important for restricting enhancer function to genes within the Hox cluster and for inhibiting enhancer elements outside the cluster from regulating Hox genes that reside within the complex.
Stalling of Pol II at promoters is controlled by the differential activities of multiple elongation factors. The DSIF and NELF complexes help maintain Pol II in the stalled conformation, while the activity of P-TEFb kinase stimulates the escape of Pol II from the paused state and into productive elongation (Saunders et al. 2006; Gilmour 2008). To test whether the role of these factors in pausing is also important for enhancer-blocking activity, Chopra et al. (2009) then tested the properties of stalled promoters in genetic backgrounds, where each of these factors and an additional elongation factor, Elongin A, were reduced. As expected, reduction in P-TEFb or Elongin A did not alter the enhancer-blocking properties of the stalled Abd-B or Ubx promoters that flank the BX-C. However, reduction of the components in the DSIF or NELF complexes results in a depletion of enhancer blocking. NELF depletion has been shown previously to reduce the level of pausing on several genes in Drosophila. Thus, the above results are consistent with a general requirement for stalling to confer enhancer-blocking properties to a promoter.
Interestingly, Chopra et al. (2009) also show that negative elongation factors are necessary for the insulator activities of the Fab-7 and Fab-8 elements contained within BX-C. These last results bring forth an interesting question: Why are transcription elongation factors that are necessary for stalling at promoters also necessary for the activity of known insulators? Chopra et al. (2009) speculate that insulators interact with stalled promoters to form higher-order chromatin loop domains similar to known insulator–insulator interactions (Blanton et al. 2003; Cleard et al. 2006; Kyrchanova et al. 2007). Although this idea is yet to be tested directly, physical and functional interactions between the Fab-7 and Fab-8 insulators and the stalled Abd-B promoter have been described (Cleard et al. 2006; Kyrchanova et al. 2007). They further suggest that proteins that bind insulators interact with components of the Pol II complex at stalled genes, which is consistent with the recent observation that the BEAF insulator protein tends to colocalize with NELF (Jiang et al. 2009). The resulting loops may produce restricted structures that prevent cross-talk between domains, but could also be important for targeting enhancers to certain promoters. For instance, the 3′-distal IAB5 and IAB6 enhancers must traverse both the Fab-7 and Fab-8 insulator elements in order to interact with the Abd-B promoter. Perhaps direct interaction of Fab-7 with the Abd-B promoter can specifically target these enhancers to the Abd-B promoter (Cleard et al. 2006).
This new function of stalled promoters raises several interesting questions regarding the mechanisms through which both stalled promoters and insulators contribute to regulation of gene expression. Specifically, it is important to ask why stalled promoters are bestowed with enhancer-blocking activities, while nonstalled promoters are not. Also, a wealth of data point to several equivalencies between insulators and stalled promoters; thus, it is practical to consider the similarities and differences in order to unravel the meaning of promoter–insulator interactions.
Insights from the similarities between promoters and insulators
Insulators can be promoter mimics or interact directly with promoters
Promoters and insulators display similarities in DNase I-hypersensitive site enrichment, GAGA factor (GAF) binding, histone turnover, H3.3 depostition, H2A.Z deposition, H3K4me1,2,3, H3ac, and low H3k27me3 (Mutskov et al. 2002; Barski et al. 2007; Mito et al. 2007; Mavrich et al. 2008). Either very similar mechanisms are occurring at promoters and insulators independently, or these properties are “shared” between the two due to a close proximity brought on by a looping interaction. The former suggests that insulators may block enhancer/repressor interaction with a promoter by acting as a promoter decoy (Geyer 1997). In the promoter decoy or mimicry model, insulators are proposed to recruit components of the transcription machinery that effectively trap enhancers into a nonproductive interaction. Thus, by mimicking a promoter-bound complex, insulators can protect promoters from enhancers when placed in between them.
The shared features of insulators and promoters might also suggest that insulators are specialized promoters. Indeed, several Drosophila insulators have been shown to contain promoters, and the iab regions within the BX-C do support transcription (Geyer 1997; Bae et al. 2002; Drewell et al. 2002). Transcription occurs within and is restricted to the iab domains that will eventually activate transcription of their target gene (Bae et al. 2002; Drewell et al. 2002). Transcription through these regions is proposed to promote an open chromatin organization that allows for the enhancer elements to become functional. Interestingly, removal of an insulator within the BX-C has been shown to abrogate transcription of the neighboring iab domain, resulting in a transformation of the corresponding segment to the anterior one (Drewell et al. 2002). Thus, it is possible that the unidentified promoter is related to the insulator element. However, whether or not intergenic transcription within the BX-C control regions originates from the insulator elements or other cryptic promoters within the iab regions remains to be determined.
The looping domain model proposes that insulators can interact with each other, specialized promoter targeting sequences, and promoters themselves to create looped domains that both target enhancers to specific promoters and restrict enhancer activity to particular domains (Maeda and Karch 2007). This idea is supported by many physical and functional interactions between insulator elements and insulator-binding proteins with promoters (Cleard et al. 2006; Holohan et al. 2007; Kyrchanova et al. 2007). For instance, the insulator-binding protein CTCF has been shown to interact with several known and putative insulator regions within the BX-C, but is also found at the Abd-B promoter (Holohan et al. 2007). It is proposed that CTCF acts as a boundary that restricts the initial activation signal within each iab region, but it is also possible that homo- or hetero-typic interactions between CTCF at insulators and promoters could direct active enhancers to their target.
The generality of promoter–insulator interactions
Recent evidence suggests that insulator–promoter interactions are a general means for regulating transcription of a wide variety of genes. Several insulator-binding proteins—CP190, dCTCF, GAF, and BEAF—frequently map by chromatin immunoprecipitation (ChIP) proximal to transcription start sites (TSSs). CP190 and dCTCF sites correlate with active genes, and also mark a boundary between repressive marks (h3k27me3) and active promoters (Mohan et al. 2007). BEAF-binding sites have a higher correlation with TSSs than CP190 or dCTCF sites, and these interactions could be a consequence of both direct binding to promoters and looping between insulators and promoters. Interestingly, the NELF elongation factor, which is important for promoter stalling and insulator function (Chopra et al. 2009), overlaps with ∼40% of identified BEAF sites in a genome-wide analysis (Jiang et al. 2009). In addition, 90% of BEAF-associated genes are contained within the upper half of genes ranked by their level of expression, suggesting that BEAF plays a role in high-level transcription of genes. As demonstrated by Chopra et al. (2009), stalled promoters have insulator function, and it has also been documented that NELF binding (Lee et al. 2008) and stalling/pausing correlate with gene activity levels (Core et al. 2008). These observations suggest a tight link between Pol II stalling, insulator function, known insulator proteins, and gene activity. An intriguing extension of these results is that insulator–promoter interactions could potentially mediate high levels of transcription by creating localized chromatin domain compartments that concentrate transcription factors within regions where high levels of gene expression are needed (Yao et al. 2007). In contrast to what one might expect if insulators interact with stalled promoters, specific interaction of BEAF-containing insulators with stalled promoters is less evident, since only ∼25% of BEAF-occupied sites contain a stalled Pol II. This may be a result of underestimation of the number of stalled genes, but could also be due to differences in the specific composition of certain insulators, evidenced by the incomplete overlap of insulator protein-binding sites (Mohan et al. 2007; Jiang et al. 2009).
Promoters and insulators can act as barriers
Insulators originally were isolated based on their ability to act as barriers that prevent localized spreading of chromatin states associated with activation or insertional position effects (Kellum and Schedl 1991, 1992). It is believed that barrier function comes from the ability of insulators to maintain a localized open chromatin configuration involving nucleosome remodeling and histone modifications. Promoters occasionally have been ascribed with the same properties (Geyer 1997). Moreover, stalling at promoters has been proposed to help maintain promoters in a nucleosome-free state (Shopland et al. 1995; Shopland and Lis 1996), and recent experiments show that depletion of NELF can result in decreased Pol II occupancy and a more repressive chromatin structure at promoters (Muse et al. 2007; Gilchrist et al. 2008). A recent observation that supports this can be found at the 87A heat-shock locus of D. melanogaster. This locus contains two divergently transcribed Hsp70 genes that are flanked by the first insulator elements to be described: scs and scs′ (Kellum and Schedl 1991, 1992). These elements interact to form a looped domain that presumably acts as a barrier to the propagation of changes in chromatin structure. (Blanton et al. 2003). A recent study from our laboratory (Petesch and Lis 2008) has shown that a rapid, transcription-independent loss of nucleosomes across the 87A locus is indeed restricted to the regions within the scs and scs′ elements. However, RNAi depletion of either BEAF or Zw5, which bind the scs and scs′ elements and mediate the looping interaction, does not result in nucleosome loss beyond the insulators for Hsp70 gene activation. Intriguingly, both insulators overlap with promoters that exhibit Pol II enrichment at the promoter (Muse et al. 2007). This leads to the hypothesis that the stalled promoters are capable of enforcing local chromatin barriers in the absence of bona fide insulator-binding activities. It is also possible that insulator-binding proteins could act in concert with a stalled promoter to enhance barrier formation.
GAF ‘makes the connection’
Perhaps the most striking similarity between stalled promoters and insulators is their frequent association with GAF (Maeda and Karch 2007; Lee et al. 2008). GAF is encoded by Trithorax-like (Trl) gene (Farkas et al. 1994), and is named for the (GA)n consensus site that it binds (Adkins et al. 2006). It has three major domains: a single zinc finger DNA-binding domain, a BTB/POZ domain, and a glutamine-rich domain (Q domain). The BTB/POZ domain is found in a wide variety of proteins and is believed to mediate homo- or hetero-protein interactions (Mahmoudi et al. 2002; Adkins et al. 2006). The Q domain is believed to function as a multimerization domain that can also bind to ssDNA (Wilkins and Lis 1999).
GAF has multiple functions. It plays an intricate role in both promoter stalling (Lee et al. 1992, 2008) and interactions between cis-regulatory regions. GAF binding is associated with transcription derepression/activation (Biggin and Tjian 1988; Tsukiyama et al. 1994), repression (Hagstrom et al. 1997; Mishra et al. 2001), insulator activity (Schweinsberg et al. 2004), barrier formation (O'Donnell and Wensink 1994), enhancer blocking (Ohtsuki and Levine 1998), and insulator bypass (Melnikova et al. 2004). GAF's primary role in these processes is thought to be its ability to recruit the ATP-dependent chromosome remodeler NURF, which remodels nucleosomes to create an open chromatin configuration (Adkins et al. 2006). Based on this, the complex and often opposing array of activities listed above likely result from the DNA-binding specificities of protein complexes that bind to sequences exposed by GAF. Two examples of the diverse activities of GAF that are relevant to our discussion are observations that GAF is necessary but not completely sufficient for insulator activity (Melnikova et al. 2004) or promoter-proximal pausing (Lee et al. 1992). However, given the similarities and physical contacts between promoters and insulators and enhancers, the role of GAF in these seemingly separate events deserves re-examination.
In addition to its role in creating an open chromatin architecture, GAF can support bridging interactions between insulators that result in insulator bypass. For example, GAF in one insulator has been shown to interact with the su(Hw) insulator-binding protein Mod(mgd4) and lead to an enhancer bypass of both insulators and activation of a downstream promoter (Fig. 2A; Melnikova et al. 2004). Notably, Mod(mgd4) also has a BTB domain. It is proposed that the interaction loops out the intervening DNA, and somehow allows an interaction between the enhancer and promoter. GAF can also form homo-typic interactions through its BTP/POZ or Q domains that are capable of allowing enhancer-dependent gene activation in both cis and trans when expressed heterologously in yeast (Wilkins and Lis 1999; Mahmoudi et al. 2002; Petrascheck et al. 2005). Given the high coincidence of GAF at promoters and insulators, and its ability to forge homo- or hetero-typic interactions, it seems reasonable to surmise that long-range interactions between insulators and stalled promoters or stalled promoters and enhancers could also be mediated by the presence of GAF at one or both sites.
Figure 2.
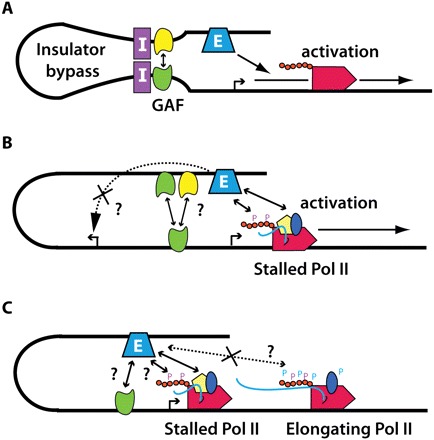
Possible mechanisms of GAF-mediated looping, and possible interactions between enhancers and a stalled Pol II complex. (A) A model depicting GAF-mediated insulator bypass by the pairing of two insulators through interactions between GAF (green) and the Su(Hw) insulator-binding protein Mod(mgd4) (yellow). (B) A speculative model of how interactions between GAF proteins (green) or other proteins (yellow) present within enhancers or insulators could stimulate formation of chromatin domains that could direct enhancers to certain promoters, while bypassing others. These interactions could also be mediated through specific enhancer interactions with stalled Pol II (red). These models have not been tested. (C) It is possible for enhancers (blue trapezoid) to interact with GAF (green) at promoters, components of the stalled complex, or Pol II (red) itself. Specific interactions with a stalled Pol II could be mediated through the C-terminal domain, which is phosphorylated at Ser5 (purple P) of the heptad repeat. Changes in the transcription complex associated with elongation, such as Ser2 phosphorylation of the C-terminal domain (blue P), phosphorylation of Spt5 (blue P), or loss of NELF (yellow pentagon) could prevent enhancers from interacting with nonstalled Pol II.
Beyond its role in insulator bypass, we envision several ways that GAF could promote these interactions (Fig. 2B). First, GAF-dependent stalling at promoters could mediate enhancer blocking by allowing efficient enhancer–promoter interaction as a result of the open chromatin configuration. Second, GAF's role in stalling and insulator activity are separable, such that GAF bound at a promoter can, as proposed for dCTCF, form homo- or hetero-typic interactions with components of insulators, independent of the transcription machinery. In this case, the promoter-bound GAF could mediate the formation of loops between cis-acting elements that result in insulator bypass, enhancer blocking, or promoter targeting, depending on the context. Third, a combination of the above models could exist, whereby looping interactions stimulated by GAF could, in concert with its promoter-opening activities, stabilize an interaction between cis-elements and stalled Pol II complexes.
The Levine laboratory (Ohtsuki and Levine 1998) previously uncovered an example of a GAF-mediated enhancer–promoter interaction. GAF-binding sites proximal to the eve promoter block the IAB5 enhancer when inserted between the enhancer and another promoter. Similar to the stalled promoters used in Chopra et al. (2009) the GAF sites were not strictly involved in competition with other promoters. Rather, promoter competition was mediated by the presence of a TATA element in the eve promoter. This last result implies that enhancer–promoter pairing is a function of both their arrangement and local sequence elements; thus, it is important to ask how sequence elements and stalling might control enhancer specificity.
Specificity of stalled promoters for enhancer blocking
Why do stalled promoters act as enhancer blockers, while nonstalled promoters do not? One possibility is that the increased dwell time of Pol II at stalled promoters allows a greater window of opportunity for enhancer–promoter interactions to take place (Core and Lis 2008). This simple interpretation could be combined with the hypothesis that enhancers preferably interact with some component of the stalled complex, or preferably interact with a Pol II that is not yet modified with the characteristics of a productive elongation complex (Fig. 2C). For instance, an enhancer could interact with a stalled Pol II regardless of the modification state of Pol II, or the composition of factors associated with it. A stalling event would then create a nonmoving target that might be bound more easily by an enhancer, since Pol II is not undergoing dynamic conformational changes associated with transcription elongation. If the enhancer–promoter interactions were dependent on the modification state of the polymerase (such as Ser5 phosphorylation of the C-terminal domain), or the composition of factors associated with the Pol II (such as NELF), stalling would again allow more time for the interaction to occur. Conversely, Pol II at nonstalled promoters would be a moving target, and/or spend less time in the state that allows enhancer interaction. Finally, Chopra et al. (2009) demonstrate this preference for several promoters; however, the nonstalled promoters tested appear to be less active or nonactive prior to enhancer function. It will be interesting to see if the specificity of enhancer blocking for stalled promoters holds up in this type of analysis when using a gene that is constitutively active in all cells and shows no signs of stalling in its regulation.
Another possibility that may underlie the mechanisms described above is the inherent selectivity of enhancers for certain types of promoters. Enhancer–promoter specificity has been documented for different types of core promoters. Promoters consist of various core elements that dictate the location and strength with which transcription will initiate (Sandelin et al. 2007; Juven-Gershon et al. 2008b). The most abundant core elements identified thus far include the TATA box, the TFIIB recognition element (BRE), the initiator (Inr), the motif 10 element (MTE), and the downstream promoter element (DPE) (Juven-Gershon et al. 2008b). They adhere to a consensus sequence to varying degrees and all are rarely found together at a single promoter. Some Drosophila enhancers have a preference for activating a promoter containing a DPE (Butler and Kadonaga 2001; Juven-Gershon et al. 2008a), whereas others appear to prefer a promoter with a TATA box, or are not discriminatory (Ohtsuki et al. 1998). A recent analysis identified that stalled promoters are linked with the presence of a DPE or newly identified element, the pause button (Hendrix et al. 2008). The DPE or the pause button appears to synergize with an Inr and GAF sites to provoke stalling at promoters (Hendrix et al. 2008; Lee et al. 2008). These observations, in light of the results from Chopra et al. (2009), may provide clues as to how the promoter DNA sequence elements result in enhancer specificity with promoters. GAF sites might be considered in the future as another element that can dictate some level of promoter specificity by the mechanisms outlined above, or through its known role in insulator bypass. It will be interesting to see if the presence of GAF or a stalled Pol II at promoters are specific targets for different types of enhancers, as seen with several promoter elements. Furthermore, if stalling is involved in promoter/enhancer selectivity, it will be interesting to examine the activators at enhancers in order to gain information about what specific steps of transcription the individual activators stimulate.
Conclusion
In summary, the results from Chopra et al. (2009) unveil an exciting new dimension of how developmental gene programs are regulated. Regulation of long-range enhancer–promoter communication by a stalled Pol II challenges us to rethink how promoter regulation is carried out, and how chromatin domains are formed to affect cis-regulatory trafficking. We attempted here to put the two modes of regulation into context by examining the similarities between the mechanisms at hand in each process. There appear to be several links between the promoter function and regulation of chromatin interactions in cis; namely, paused/stalled Pol II and the presence of GAF and insulator-binding proteins at promoters and distal regulatory sites. These connections can possibly lead to future experiments aimed at determining how stalled Pol II might direct certain chromatin domain formation, or enhancer-specific interactions.
Acknowledgments
We thank Michael Levine, Vivek S. Chopra, Steven J. Petesch, and Abbie Saunders for providing critical feedback on this manuscript. Our research is supported by grants from the NIH.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1827709.
References
- Adkins NL, Hagerman TA, Georgel P. GAGA protein: A multi-faceted transcription factor. Biochem Cell Biol. 2006;84:559–567. doi: 10.1139/o06-062. [DOI] [PubMed] [Google Scholar]
- Akbari OS, Bousum A, Bae E, Drewell RA. Unraveling cis-regulatory mechanisms at the abdominal-A and abdominal-B genes in the Drosophila bithorax complex. Dev Biol. 2006;293:294–304. doi: 10.1016/j.ydbio.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Akbari OS, Bae E, Johnsen H, Villaluz A, Wong D, Drewell RA. A novel promoter-tethering element regulates enhancer-driven gene expression at the bithorax complex in the Drosophila embryo. Development. 2008;135:123–131. [Google Scholar]
- Bae E, Calhoun VC, Levine M, Lewis EB, Drewell RA. Characterization of the intergenic RNA profile at abdominal-A and abdominal-B in the Drosophila bithorax complex. Proc Natl Acad Sci. 2002;99:16847–16852. doi: 10.1073/pnas.222671299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Biggin MD, Tjian R. Transcription factors that activate the ultrabithorax promoter in developmentally staged extracts. Cell. 1988;53:699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- Blanton J, Gaszner M, Schedl P. Protein:protein interactions and the pairing of boundary elements in vivo. Genes & Dev. 2003;17:664–675. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JE, Kadonaga JT. Enhancer–promoter specificity mediated by DPE or TATA core promoter motifs. Genes & Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra VS, Cande J, Hong J-W, Levine M. Stalled Hox promoters as chromosomal boundaries. Genes & Dev. 2009;23:1505–1509. doi: 10.1101/gad.1807309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleard F, Moshkin Y, Karch F, Maeda RK. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using dam identification. Nat Genet. 2006;38:931–935. doi: 10.1038/ng1833. [DOI] [PubMed] [Google Scholar]
- Core LJ, Lis JT. Transcription regulation through promoter–proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewell RA, Bae E, Burr J, Lewis EB. Transcription defines the embryonic domains of cis-regulatory activity at the Drosophila bithorax complex. Proc Natl Acad Sci. 2002;99:16853–16858. doi: 10.1073/pnas.222671199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. The trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- Geyer PK. The role of insulator elements in defining domains of gene expression. Curr Opin Genet Dev. 1997;7:242–248. doi: 10.1016/s0959-437x(97)80134-7. [DOI] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of pol II can enhance gene expression by blocking promoter–proximal nucleosome assembly. Genes & Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DS. Promoter proximal pausing on genes in metazoans. Chromosoma. 2008;118:1–10. doi: 10.1007/s00412-008-0182-4. [DOI] [PubMed] [Google Scholar]
- Hagstrom K, Muller M, Schedl P. A polycomb and GAGA dependent silencer adjoins the fab-7 boundary in the Drosophila bithorax complex. Genetics. 1997;146:1365–1380. doi: 10.1093/genetics/146.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix DA, Hong JW, Zeitlinger J, Rokhsar DS, Levine MS. Promoter elements associated with RNA pol II stalling in the Drosophila embryo. Proc Natl Acad Sci. 2008;105:7762–7767. doi: 10.1073/pnas.0802406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 2007;3:e112. doi: 10.1371/jorunal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Emberly E, Cuvier O, Hart CM. Genome-wide mapping of BEAF binding sites in Drosophila links BEAF to transcription. Mol Cell Biol. 2009 doi: 10.1128/MCB.01748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Kadonaga JT. Caudal, a key developmental regulator, is a DPE-specific transcriptional factor. Genes & Dev. 2008a;22:2823–2830. doi: 10.1101/gad.1698108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter—The gateway to transcription. Curr Opin Cell Biol. 2008b;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- Kellum R, Schedl P. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol. 1992;12:2424–2431. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O, Toshchakov S, Parshikov A, Georgiev P. Study of the functional interaction between mcp insulators from the Drosophila bithorax complex: Effects of insulator pairing on enhancer-promoter communication. Mol Cell Biol. 2007;27:3035–3043. doi: 10.1128/MCB.02203-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol. 2008;28:3290–3300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Kraus KW, Wolfner MF, Lis JT. DNA sequence requirements for generating paused polymerase at the start of hsp70. Genes & Dev. 1992;6:284–295. doi: 10.1101/gad.6.2.284. [DOI] [PubMed] [Google Scholar]
- Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F. Making connections: Boundaries and insulators in Drosophila. Curr Opin Genet Dev. 2007;17:394–399. doi: 10.1016/j.gde.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Mahmoudi T, Katsani KR, Verrijzer CP. GAGA can mediate enhancer function in trans by linking two separate DNA molecules. EMBO J. 2002;21:1775–1781. doi: 10.1093/emboj/21.7.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova L, Juge F, Gruzdeva N, Mazur A, Cavalli G, Georgiev P. Interaction between the GAGA factor and mod(mdg4) proteins promotes insulator bypass in Drosophila. Proc Natl Acad Sci. 2004;101:14806–14811. doi: 10.1073/pnas.0403959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merika M, Thanos D. Enhanceosomes. Curr Opin Genet Dev. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- Mishra RK, Mihaly J, Barges S, Spierer A, Karch F, Hagstrom K, Schweinsberg SE, Schedl P. The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol Cell Biol. 2001;21:1311–1318. doi: 10.1128/MCB.21.4.1311-1318.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- Mohan M, Bartkuhn M, Herold M, Philippen A, Heinl N, Bardenhagen I, Leers J, White RA, Renkawitz-Pohl R, Saumweber H, et al. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J. 2007;26:4203–4214. doi: 10.1038/sj.emboj.7601851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutskov VJ, Farrell CM, Wade PA, Wolffe AP, Felsenfeld G. The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes & Dev. 2002;16:1540–1554. doi: 10.1101/gad.988502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KH, Wensink PC. GAGA factor and TBF1 bind DNA elements that direct ubiquitous transcription of the Drosophila α 1-tubulin gene. Nucleic Acids Res. 1994;22:4712–4718. doi: 10.1093/nar/22.22.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S, Levine M. GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes & Dev. 1998;12:3325–3330. doi: 10.1101/gad.12.21.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S, Levine M, Cai HN. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes & Dev. 1998;12:547–556. doi: 10.1101/gad.12.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrascheck M, Escher D, Mahmoudi T, Verrijzer CP, Schaffner W, Barberis A. DNA looping induced by a transcriptional enhancer in vivo. Nucleic Acids Res. 2005;33:3743–3750. doi: 10.1093/nar/gki689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core proamoters: Insights from genome-wide studies. Nat Rev Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- Schweinsberg S, Hagstrom K, Gohl D, Schedl P, Kumar RP, Mishra R, Karch F. The enhancer-blocking activity of the fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics. 2004;168:1371–1384. doi: 10.1534/genetics.104.029561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopland LS, Lis JT. HSF recruitment and loss at most Drosophila heat shock loci is coordinated and depends on proximal promoter sequences. Chromosoma. 1996;105:158–171. doi: 10.1007/BF02509497. [DOI] [PubMed] [Google Scholar]
- Shopland LS, Hirayoshi K, Fernandes M, Lis JT. HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites. Genes & Dev. 1995;9:2756–2769. doi: 10.1101/gad.9.22.2756. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Becker PB, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- Wang X, Lee C, Gilmour DS, Gergen JP. Transcription elongation controls cell fate specification in the Drosophila embryo. Genes & Dev. 2007;21:1031–1036. doi: 10.1101/gad.1521207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins RC, Lis JT. DNA distortion and multimerization: Novel functions of the glutamine-rich domain of GAGA factor. J Mol Biol. 1999;285:515–525. doi: 10.1006/jmbi.1998.2356. [DOI] [PubMed] [Google Scholar]
- Yao J, Ardehali BM, Fecko CJ, Webb WW, Lis JT. Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol Cell. 2007;28:978–990. doi: 10.1016/j.molcel.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]


