Abstract
A recent meeting (December 2008) regarding chromatin-based epigenetics was hosted by the Banbury Conference Center and Cold Spring Harbor Laboratory. The intent was to discuss aspects of epigenetic control of genomic function, and to arrive at a consensus definition of “epigenetics” to be considered by the broader community. It was evident that multiple mechanistic steps lead to the stable heritance of the epigenetic phenotype. Below we provide our view and interpretation of the proceedings at the meeting.
Keywords: Epigenetics, DNA methylation, noncoding RNA, histone modification, histone variant
Definition: “An epigenetic trait is a stably heritable phenotype resulting from changes in a chromosome without alterations in the DNA sequence.”
The definition of epigenetics proposed here, as with the classical definition (e.g., as proposed by Conrad Waddington in the 1950s), can involve the heritability of a phenotype, passed on through either mitosis or meiosis. Understanding the mechanisms involved in the initiation, maintenance, and heritability of epigenetic states is an important aspect of research in current biology. Several distinct but interconnected molecular pathways have been discovered to date. Below is described a set of operational steps in which such pathways can be placed, in an effort to define the different mechanistic aspects of epigenetic transmission.
Epigenators, Initiators, and Maintainers of the epigenetic process
It is proposed here that there are three categories of signals that culminate in the establishment of a stably heritable epigenetic state: a signal that we propose to call the “Epigenator,” which emanates from the environment and triggers an intracellular pathway; an “Epigenetic Initiator” signal, which responds to the Epigenator and is necessary to define the precise location of the epigenetic chromatin environment; and an “Epigenetic Maintainer” signal, which sustains the chromatin environment in the first and subsequent generations. These classes are depicted in Figure 1 and are explained below.
Figure 1.
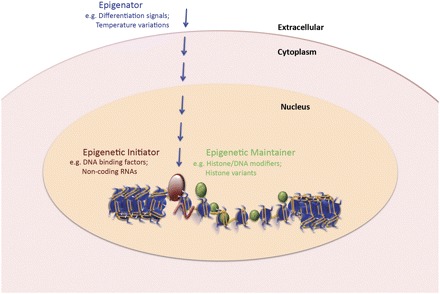
The epigenetic pathway. Three categories of signals are proposed to operate in the establishment of a stably heritable epigenetic state. An extracellular signal referred to as the “Epigenator” (shown in blue) originates from the environment and can trigger the start of the epigenetic pathway. The “Epigenetic Initiator” (shown in red) receives the signal from the “Epigenator” and is capable of determining the precise chromatin location and/or DNA environment for the establishment of the epigenetic pathway. The “Epigenetic Maintainer” (shown in green) functions to sustain the chromatin environment in the initial and succeeding generations. Persistence of the chromatin milieu may require cooperation between the Initiator and the Maintainer. Examples for each category are shown below each heading. Chromatin is depicted in blue.
Epigenator
The epigenetic phenotype is likely triggered by changes in the environment of the cell. Everything occurring upstream of the first event on the chromosome would be part of the Epigenator signal, including an environmental cue or niche and the subsequent signaling pathways leading to the Initiator. Once an Epigenator signal is received, it is converted to an intracellular Epigenator pathway culminating in the “activation” of the Initiator. The Epigenator signaling pathway could be a protein–protein interaction or a modification-based event that unleashes the latent activity of the Initiator. The Epigenator signal will be transient, remaining in the cell long enough to trigger the epigenetic phenotype but not necessary for subsequent events.
Epigenetic Initiator
The Initiator translates the Epigenator signal to mediate the establishment of a local chromatin context at a precise location. Following the priming of the Initiator by the Epigenator signal, the Initiator will define the location on a chromosome where the epigenetic chromatin state is to be established. The Initiator could be a DNA-binding protein, a noncoding RNA, or any other entity that can define the coordinates of the chromatin structure to be assembled. Consequently, some form of sequence recognition must be a feature of this signal. The Initiator will in general be a signal that requires self-reinforcement and self-renewal through positive feedback mechanisms. One operational characteristic of the Initiator is that it may be sufficient to initiate an epigenetic phenotype when introduced into a cell. Also, unlike the Epigenator, the Initiator may not dissipate after its action, but rather may persist with the Maintainer.
Epigenetic Maintainer
The Maintainer sustains the epigenetic chromatin state but is not sufficient to initiate it. This signal involves many different pathways, including DNA methylation, histone modifications, histone variants, nucleosome positioning, and others. Maintainers have the common property that they do not have absolute DNA sequence specificity. Consequently, they could operate at any chromosomal location to which they are recruited by an Initiator. Maintainers may function by carrying an epigenetic signal through the cell cycle or could maintain epigenetic landscapes in terminally differentiated cell types.
The role of one particular class of potential Maintenance signals—i.e., post-translational modifications of histone proteins—requires particular clarification. During the meeting, several examples for an epigenetic role of histone modifications were presented. These included roles of (1) H3K4 and H3K27 methylation, by trithorax and polycomb complexes, respectively, in homeotic gene expression; (2) H3K9 and H4K20 methylation in establishing memory of transcriptional silencing; and (3) H4K16 acetylation in mating-type behavior and aging in Saccharomyces cerevisiae. However, the term “epigenetic” is not always a correct term to define histone modifications. Many modifications play a role in more dynamic processes such as transcriptional induction and DNA repair. Thus, certain histone modifications very likely play a role as Maintainers of epigenetic signals; however, this does not mean that all post-translational modifications of histones are epigenetic in nature.
Biological examples
There are not many well-defined examples of Epigenators. The best example comes from plants, where environmental signals such as temperature affect the epigenetic process of paramutation. Examples of Initiators are noncoding Xist RNA, which is sufficient for silencing the mammalian X chromosome, and DNA-binding factors that lead to reprogramming of differentiated cells into stem cells in metazoans. Maintainers include histones deacetylated by the Sir complex that functions in mating-type switching and sexual differentiation in yeast S. cerevisiae, DNA methylation at CpG islands in plants and some animals, and the histone variant CENPA at centromeres of all eukaryotes.
Final remarks
Epigenetic events in eukaryotic organisms have evolved to provide a more precise and stable control of gene expression and genomic regulation through multiple generations. This is exemplified by the existence of sex-specific dosage compensation or the fine-tuning of allele-specific expression, as seen in imprinted loci. Deregulation of such processes may lead to disease; e.g., misregulation of imprinted genes results in the genesis of Beckwith-Wiedemann and Prader-Willi/Angelman syndromes, whereas the loss of other epigenetic heritance mechanisms results in cellular aging and cancer. In addition, the ability to epigenetically reprogram differentiated cells is becoming of medical importance.
The effort by the meeting participants to define and discuss “epigenetics” was an attempt to add focus and clarity to this exciting and growing area of research.
Acknowledgments
We thank Dr. Terri Grodzicker, Dr. David Stewart, Dr. Jan Witowski, Cold Spring Harbor Laboratory, and the Banbury Conference Center for generously supporting this Epigenetic meeting. We are also grateful to the meeting attendees for stimulating discussion and conversations. Special thanks go to Dr. Bob Kingston for helpful comments and suggestions during the organization of the meeting.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1787609.


