The current model for conducting genome-wide association studies (GWAS) is primarily phenotype-driven. In this “top-down” approach, the model is the case-control study, where participants are enrolled based on the presence or absence of a clinical phenotype, for example, cardiovascular disease or breast cancer (Pennisi 2007; Wellcome Trust Case Control Consortium 2007). The International HapMap Project has identified a large number of single nucleotide polymorphisms (SNPs) in the human population that enable investigators to genotype subjects for these various polymorphisms and determine associations with a phenotype of interest (Fig. 1A) (International HapMap Consortium et al. 2007).
Figure 1.
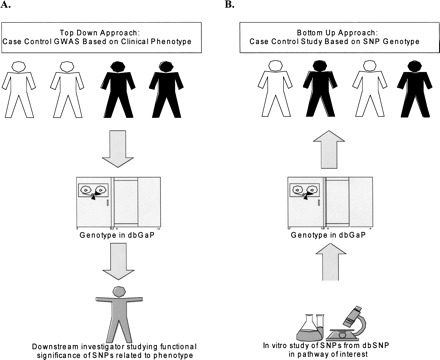
Top-down and bottom-up approaches to genome-wide association studies. (A) In the top-down approach, clinical investigators assemble cases and controls based on a particular phenotype. Genotypic information is then acquired and deposited in dbGaP. (B) In the bottom-up approach, basic investigators identify in vitro phenotypes based on SNP data in dbSNP. They then access clinical populations and controls with whom they are able to conduct “targeted phenotyping.”
Commercially available SNP arrays allow researchers to easily genotype from 100,000 to 1,000,000 SNPs per individual, providing “whole-genome” coverage (Affymetrix GeneChip System, http://www.affymetrix.com/products/system.affx; Illumina, Inc., http://www.illumina.com/pages.ilmn?=39). Typically, only a handful of these SNPs are associated with the particular phenotype under study and are present in only a small fraction of the study population because the minor alleles (variants) tend to be present at low frequencies. The vast majority of SNPs are not associated with the phenotype under study and are ignored because they are not relevant to the phenotype under investigation in the GWAS. For example, the Wellcome Trust Case Control Consortium genotyped 500,000 SNPs in 14,000 cases representing seven common diseases and an additional 3000 controls. From this extensive genotyping (17,000 × 500,000), they identified 24 independent associations (Wellcome Trust Case Control Consortium 2007). The vast majority of the genotyping was not relevant to the seven phenotypes under investigation. The failure to use this “excess” genotypic information, by allowing future investigators to recontact participants and collect hypothesis-driven phenotypic information not collected in the original study, represents a tremendous missed opportunity. This type of “targeted phenotyping” would maximize the utility of data generated in GWAS and stored in existing databases, such as dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) and dbGaP (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap).
Recently, scientists have begun to examine the functional consequences of human SNPs in the laboratory, based on data available from the HapMap Project and other SNP discovery studies, without any a priori knowledge of a human phenotype associated with a particular SNP. Having identified an interesting phenotype in vitro, the next step is to determine the consequence of these SNPs in vivo. Most typically, in a “bottom-up” approach, the researcher will generate an animal model to study the phenotypic consequences of the SNP by performing a case-control study based on genotype. The ability to do a similar case-control study based on genotype in humans would be an extremely powerful approach to understand the consequences of a particular genetic variation. This would involve allowing investigators to identify the usually small number of individuals in any given GWAS who bear a particular variation and then assemble a large cohort of these individuals from multiple GWAS to form the basis of a case group (Fig. 1B). This would be followed by performing hypothesis-driven “targeted phenotyping” from these cases and a randomly selected group of controls to identify particular phenotypes over- or under-represented in the cases relative to controls who do not bear the variation. Such case-control studies based on genotype would prevent the waste of potentially important genotypic information that would otherwise be considered irrelevant based on a lack of association with the phenotype of interest in a phenotype-driven GWAS.
An expanded approach to GWAS will present new challenges in the consent process associated with GWAS so that participants can be contacted in the future by researchers not associated with the original study. Participants may not want to be contacted in the future, and they may not want to know that they bear a SNP that could potentially be related to risk of a particular phenotype (e.g., Alzheimer’s disease or other neurodegenerative disorders). Additionally, they may already be aware of a particular phenotype but be concerned about stigmatization or discrimination related to research on genetic variants associated with that phenotype (e.g., addiction or mental illness). Permission to recontact the participant will, therefore, have to be sought upfront and must be carefully explained and well documented.
The mechanism for recontacting participants will also present challenges. The newly adopted Policy for Sharing of Data Obtained in NIH Supported or Conducted Genome-Wide Association Studies (GWAS) (NOT-OD-07-088, November 16, 2007; http://grants.nih.gov/grants/guide/notice-files/NOT-OD-08-013.html) calls for the deposition of all GWAS data into dbGaP, a restricted database maintained by the National Center for Biotechnology Information (NCBI). Typically, only limited phenotypic data are deposited into dbGaP, all data are coded, and only the depositing investigator maintains a link to personally identifying information about the participant. If a downstream user wants to contact participants to obtain more phenotypic data and to invite them to participate in a follow-up case-control study, then he or she has to contact the primary investigator, who determines whether it is appropriate to follow up with the participant (http://grants.nih.gov/grants/guide/notice-files/NOT-OD-08-013.html). Having a well-defined gatekeeper is important to protect the privacy of research participants and to ensure that they are only invited to participate in those studies that are consistent with their original informed consent. However, individual investigators do not have the time, the resources, or the incentive to assume this responsibility.
One potential solution that has been advocated is to conduct “deep phenotyping” at the outset for all GWAS and then make this information accessible in dbGaP (Tracy 2008). This would limit the need to recontact participants for follow-up studies. However, this would create a tremendous addition of time and expense for the initial GWAS, and the question of upon whom the burden of “deep phenotyping” should fall is not clear. Furthermore, this approach would be limited by the inability to anticipate the nuanced information and measurements necessary to identify all potentially relevant phenotypes, and as clinical phenotypes become more refined, there would still be the need to be able to recontact participants. While investigators should be encouraged to collect a standard set of phenotypic information, permission to recontact and a system for effectuating this is still necessary.
We suggest that an independent governance body assume responsibility for facilitating this type of research (Caulfield et al. 2008). This could be a private entity, such as a centralized ethics review board, or it could be an established government entity, such as the NIH Data Access Committees (DACs). Since the DACs are currently responsible for reviewing and approving applications to dbGaP for data access and use (http://grants.nih.gov/grants/guide/notice-files/NOT-OD-08-013.html), a reasonable first step might be to use this existing infrastructure. DACs would need access to the link to the keycode within the data in order to recontact select participants to inform them of the opportunity to participate in follow-up case-control studies. Although members of the DAC would have access to personally identifying information about participants, they would not be using this information about participants in order to conduct research. Rather, they would be using the information only to contact participants and inform them of additional research opportunities. The members of the DAC would therefore not themselves be engaged in the conduct of research involving human subjects (“Protection of Human Subjects,” 45 Code of Federal Regulations [C.F.R.] section 46, 2003, http://www.hhs.gov/ohrp/humansubjects/guidance/45cfr46.htm). As long as participants are informed upfront that such a committee will have access to their personal information, consent to the overall governance scheme, and agree to be recontacted, no additional oversight should be required.
Typically, participation in the original study is not contingent on consent for recontact. Participants should be given a separate choice about whether they agree to be recontacted and informed of additional research studies. For those who agree, if and when they are contacted, they should be told that these follow-up studies are genotype-driven and what that means, what the genotype and biological pathway of interest are, what the procedures for collecting additional information will be, that half of the participants are controls with no particular genetic variation, and that an invitation to participate is not contingent on the presence of any known phenotype. They will then have to decide whether or not to call the principal investigator for the follow-up study. The extent to which specific consent to the governance scheme is required for the use of previously collected samples and data (where general consent to recontact was obtained) will need to be carefully considered.
If DACs are going to maintain the link between genomic data and personal identifiers and assume the role of recontacting participants, then strict procedures for maintaining confidentiality will have to be implemented, and a Certificate of Confidentiality should be granted to prevent compelled disclosures (“Protection of Privacy of Individuals Who Are Research Subjects,” 42 United States Code (U.S.C.) section 241(d), 2004, http://caselaw.lp.findlaw.com/scripts/ts_search.pl?title=42&sec=241; NOT-OD-02-037, March 15, 2002, http://grants.nih.gov/grants/guide/notice-files/NOT-OD-02-037.html). Since DACs are housed within a federal agency (i.e., the NIH), an explicit exemption under the Freedom of Information Act (FOIA) should also be granted so that the data do not become publicly available subject to an FOIA request (Freedom of Information Act. 5 United States Code (U.S.C.) section 552(b), 2007, http://www.usdoj.gov/oip/amended-foia-redlined.pdf; Lowrance and Collins 2007). Also, DACs are currently only composed of federal employees. If DACs are to take on this additional responsibility, broader representation from the scientific community, as well as the public, ought to be added.
A major concern with this proposal is that participants may become inundated with invitations to participate in follow-up case-control studies. This may serve as a disincentive, resulting in fewer people agreeing to be recontacted at all. A coordinated system of recontact could also overburden the DACs or whatever governance body takes on the responsibility of facilitating the recontact. An electronic information exchange system may be less intrusive and more efficient. Isaac Kohane and colleagues have proposed a system that would allow investigators to collect longitudinal clinical data from research participants and would enable those who are “tuned in” to obtain information about new research findings that may be relevant to their health (Kohane et al. 2007). If such a communication system were developed and coordinated across GWAS, it could be used by investigators to query the electronic database containing participants’ SNP information, and the system could identify and send a message (electronically or through traditional means) to those participants who have the genotype of interest and a randomly selected control group. The feasibility, efficiency, and cost-effectiveness of developing such a system deserve further consideration. However, even if recontact can be accomplished electronically, oversight will still be required and should be provided by an independent governance body responsible for ensuring adequate protection of research subjects.
Enabling a bottom-up approach to GWAS will significantly advance the pace of human genetic research by expanding the existing mechanisms for studying the functional significance of genetic variation in the human population. It will also maximize the utility of large databanks like dbSNP and dbGaP. The advantages and challenges of using the existing DACs to accomplish this will need to be studied more fully, and there are significant ethical considerations that need to be addressed in the process, but the failure to enable a bottom-up approach will result in a large amount of genomic data that is collected in GWAS being wasted and slow the progress toward personalized genomic medicine.
Acknowledgments
A.L.M.’s work is funded by the Greenwall Foundation Faculty Scholars Program in Bioethics and the NHGRI-ELSI program (NIH 1 R01 HG004333-01). We thank Laurence B. McCullough for thoughtful comments on this manuscript. A.L.M. is a member of the NIH Advisory Committee to the Director (ACD), Working Group on Participant and Data Protection (PDP). The views expressed in this paper are those of the authors and may not be shared by other members of the committee.
Footnotes
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.083584.108.
References
- Caulfield T., McGuire A.L., Cho M., Buchanan J.A., Burgess M.M., Danilczyk U., Diaz C.M., Fryer-Edwards K., Green S.K., Hodosh M.A., et al. Research ethics recommendations for whole-genome research: Consensus statement. PLoS Biol. 2008;6:e73. doi: 10.1371/journal.pbio.0060073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohane I.S., Mandl K.D., Taylor P.L., Holm I.A., Nigrin D.J., Kunkel L.M. Reestablishing the researcher–patient compact. Science. 2007;318:1068. doi: 10.1126/science.1135489. [DOI] [PubMed] [Google Scholar]
- Lowrance W.W., Collins F.S. Identifiability in genomic research. Science. 2007;317:913–914. doi: 10.1126/science.1147699. [DOI] [PubMed] [Google Scholar]
- Pennisi E. Breakthrough of the year: Human genetic variation. Science. 2007;318:1842–1843. doi: 10.1126/science.318.5858.1842. [DOI] [PubMed] [Google Scholar]
- Tracy R.P. ‘Deep phenotyping’: Characterizing populations in the era of genomics and systems biology. Curr. Opin. Lipidol. 2008;19:151–157. doi: 10.1097/MOL.0b013e3282f73893. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]


