Abstract
Background
Stromal-mediated signaling enhances NFκB pathway activity in chronic lymphocytic leukemia B-cells (CLL), leading to cell survival and chemoresistance. Ubiquitination of IκBα may partially account for constitutive activation of NFκB. MLN4924 is an investigational agent that inhibits the Nedd8-activating enzyme, thereby neutralizing Cullin-RING ubiquitin ligases and preventing degradation of their substrates.
Experimental Design
We conducted a pre-clinical assessment of MLN4924 in CLL. Primary CLL cells were co-cultured in vitro with CD40L-expressing stroma to mimic the pro-survival conditions present in lymphoid tissue. The effect of MLN4924 on CLL cell apoptosis, NFκB pathway activity, Bcl-2 family members and cell cycle was assessed by flow cytometry, western blotting, PCR and immunocytochemistry.
Results
CD40L-expressing stroma protected CLL cells from spontaneous apoptosis and induced resistance to multiple drugs, accompanied by NFκB activation and Bim repression. Treatment with MLN4924 induced CLL cell apoptosis and circumvented stroma-mediated resistance. This was accompanied by accumulation of phospho-IκBα, decreased nuclear translocation of p65 and p52 leading to inhibition of both canonical and non-canonical NFκB pathways, and reduced transcription of their target genes, notably chemokines. MLN4924 promoted induction of Bim and Noxa in the CLL cells leading to rebalancing of Bcl-2 family members towards the pro-apoptotic BH3-only proteins. siRNA-mediated knockdown of Bim or Noxa decreased sensitivity to MLN4924. MLN4924 enhanced the antitumor activity of the inhibitors of BCR-associated kinases.
Conclusions
MLN4924 disrupts NFκB activation and induces Bim expression in CLL cells thereby preventing stroma-mediated resistance. Our data provide rationale for further evaluation of MLN4924 in CLL.
Keywords: chronic lymphocytic leukemia, apoptosis, MLN4924, NF-κB, Bim
Introduction
Inefficient B-cell apoptosis is considered the dominant defect in CLL (1, 2). Still, up to 2% of the circulating CLL B-cell pool is renewed daily (3). Gene expression profiling identified lymph nodes as the site of cell activation and proliferation with upregulation of NFκB and B-cell receptor (BCR) signaling (4). Constitutive and BCR-dependent activation of NFκB is an important feature in CLL B-cells and predicts poor disease outcome (5-7). Targeting tyrosine kinases within the BCR-signaling cascade has proven a promising therapeutic strategy with novel inhibitors of Bruton tyrosine kinase (ibrutinib) and phosphoinositide 3-kinase delta (PI-3Kδ) [idelalisib, formerly CAL-101] currently in clinical trials (8, 9). However BCR-independent activation of NFκB may lead to tumor resistance. NFκB activity may be driven via microenvironmental stimuli, examples of which include macrophage-mediated B cell–activating factor of the TNF family/a proliferation-inducing ligand (BAFF/APRIL), T-cell-mediated-CD40L signaling, and Toll-like receptor signaling (7, 10, 11). Thus, neutralization of NFκB is a promising strategy in CLL, as it has the potential to target the proliferative pool of CLL cells and could hypothetically lead to sensitization to both conventional chemotherapy and BCR-targeting agents. The NFκB pathway has been successfully targeted in CLL in vitro (12). However, lack of clinical advances with those agents necessitates development of novel approaches.
MLN4924 is an investigational small molecule inhibitor of NEDD8-activating enzyme (NAE) which has shown promising pre-clinical activity in hematologic malignancies, including acute myeloid leukemia and lymphoma (13, 14). NAE is necessary for activation of Cullin-RING ubiquitin ligases. In vitro MLN4294 leads to accumulation of Cullin-RING E3 ligase (CRL) substrates, including IκBα, Nrf-2, p27 and Cdt1 (13, 15). Disrupted ubiquitination of IκB in the presence of MLN4924 results in inactivation of the NFκB canonical pathway in several tumor types (14, 16). Since the NFκB pathway is predominantly active in the lymphatic tissue, we proposed that its pharmacological inhibition will target CLL cells within their supportive microenvironment. Our pre-clinical work demonstrates for the first time that MLN4924 shows promising ex vivo activity against primary neoplastic B cells derived from patients with CLL. MLN4924 abrogates NFκB pathway activation in CLL cells co-cultured with CD40L-expressing stroma. This results in enhanced expression of the pro-apoptotic BH3-only proteins Bim and Noxa and circumvents stroma-mediated resistance. Furthermore, cooperation between MLN4924 and the BCR-targeting agent CAL-101 warrants exploration of its clinical activity in CLL.
Methods
Patient samples, CLL and stromal cell co-cultures
Following Institutional Review Board approval and provision of written informed consent, peripheral blood was obtained from 42 patients with B-CLL at Dartmouth-Hitchcock Medical Center. The median time from diagnosis to study entry was 4 years; 37 patients (88%) were untreated. Blood was also obtained from 7 healthy volunteers. Standard Ficoll-Hypaque (Amersham, Piscataway, NJ) techniques were used to isolate peripheral blood mononuclear cells (PBMCs). Such CLL samples had more than 90% CD5+/CD19+ cells as determined by flow cytometry. CLL cells were cultured in RPMI 1640 supplemented with 15% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin, 2 mM L-glutamine, 25 mM HEPES, 100 μM minimum essential medium non-essential amino acids and 1 mM sodium pyruvate (Lonza, Walkersville, MD). 10 CLL samples with 17p deletion were obtained from the CLL Center at Dana-Farber Cancer Institute. All experiments were performed with freshly isolated cells except the viability assays involving the latter, which were performed with viably frozen cells.
Mouse fibroblast cell line (L cells) engineered to express CD40L (L4.5) was given to us by Dr. Sonia Neron (Quebec, Canada) (17). Parental L cells were obtained from American Type Culture Collection (Manassas, VA). All were maintained in RPMI 1640 medium with 10% FBS and penicillin-streptomycin. CLL cells were cultured under standardized condition on stroma as previously described (18). Briefly, stromal cells were seeded to achieve 80-100% confluence on the following day when CLL cells were plated at a 50:1 ratio and incubated at 37°C in 5% CO2 in presence or absence of 10 ng/ml IL4 (Cell Signaling, Danvers, MA). For comparison, cells were cultured in suspension (off stroma) at the same density. Cultures were then treated with drugs for the indicated time periods. At harvest, CLL cells were gently washed off the stromal layer. When harvested for protein and mRNA analysis, CLL cells were transferred to a new plate and incubated for an additional 60 minutes for stroma re-attachment to minimize contamination of CLL cells.
For BCR stimulation, CLL cells were seeded at a density of 5×105 per well in 24-well plates pre-coated with 10 μg/well of rabbit anti-human IgM antibody (Jackson Immunoresearch Laboratories, Baltimore, MD).
Cell viability testing and drugs
CLL cell apoptosis was measured in duplicate as previously described using the ApoScreen Annexin V apoptosis Kit (19). Briefly, cells were resuspended in 150 μl of Annexin V binding buffer containing 1 μl of Annexin V-PE, 1 μl of 7-AAD and 1 μl of CD19- or CD3-FITC mAbs (Southern Biotech, Birmingham, AL) followed by flow cytometry on a FACSCalibur (Becton Dickinson, Palo Alto, CA). MLN4924 was provided by Millennium Pharmaceuticals, Inc. (Cambridge, MA). CAL-101, ibrutinib and bortezomib were obtained from Selleck Chemicals (Houston, TX); BMS-345541, fludarabine, chlorambucil, bendamustine and U0126 - from Sigma Aldrich (St. Louis, MO). Survival of the murine stromal cells was analyzed in a caspase-3 activity assay (Cell Signaling).
Immunoblotting
Cells were lysed in RIPA buffer (20 mM Tris, 150 mM NaCl, 1% NP-40, 1 mM NaF, 1 mM Sodium phosphate, 1 mM NaVO3, 1 mM EDTA, 1 mM EGTA, supplemented with protease inhibitor cocktail (Roche, Indianapolis, IN) and 1 mM PMSF). Proteins were analyzed by immunoblotting as previously described (19). The following antibodies were used: Bcl-2, Bcl-xL, Bim, phospho-Bim, phospho-IκBα, cleaved PARP, Mcl-1, p65/RelA, p52/p100, XIAP, FOXO3A (Cell Signaling), Bcl2-A1 (Abcam, Cambridge, MA), NEDD8 (Epitomics, Burlingame, CA), p27 (Santa Cruz, Biotechnology, Santa Cruz, CA), Noxa (Imgenex, San Diego, CA), β-actin (Sigma Aldrich), horseradish peroxidase-conjugated anti-mouse and anti-rabbit antibodies (BioRad).
Reverse transcription polymerase chain reaction (RT-PCR)
CLL cells were negatively selected using B-cell Isolation Kit (Miltenyi Biotec #130-093-660, Auburn, CA). This method achieved >99% B-cell purity as determined by flow cytometry. Total RNA was isolated using RNeasy Mini Kit (Qiagen, Valencia, CA). cDNA was synthesized from 500 ng RNA using the iScript cDNA Synthesis Kit (BioRad, Hercules, CA). Quantitative real time PCR (qRT-PCR) was performed in a C1000 Thermal Cycler (BioRad) using Universal PCR Master Mix according to the manufacturer's instructions (Applied Biosystems, Foster City, CA), with template cDNA and gene specific probes. The following probes were used: BIM: Hs00708019_s1; NOXA: Hs00560402_m1, PUMA: Hs00248075_m1; CXCR4: Hs00607978_s1; CXCR5: Hs00540548_s1; BCL2A1: Hs00187845_m1; BCL-xL: Hs00236329_m1; CFLAR: Hs00153439_m1; CCND1: Hs00765553_m1 STAT5A: Hs00234181_m1. Amplification of the sequence of interest was compared to a reference probe (18S RNA, #4308329; all from Life Technologies, Carlsbad, CA).
All samples were analyzed in duplicate. We used the comparative Ct method for relative quantitation (2−ΔΔCt, where ΔΔCt=ΔCtP-ΔCtK; P=Probe and K=reference sample).
Immunocytochemistry
3×105 cells were adhered onto polylysine D-coated coverslips (Sigma Aldrich) during a 45-minute incubation at 37°C, fixed in 10% formalin (Fisher Scientific, Pittsburgh, PA) and permebealized in 1% triton-X 100 in PBS. Coverslips were blocked for 30 minutes in 5% bovine serum albumin (Sigma-Aldrich) in PBS with 0.1% Tween-20, probed with p65/RelA, p52/p100 (Cell Signaling) or cleaved PARP (Thermo Scientific, Lafayette, CO) antibodies and then with Alexa Fluor 594 goat anti-mouse antibodies (Life Technologies). Coverslips were mounted with anti-fading ProLong Gold Solution (Life Technologies) with 4′,6-diamidino-2-phenylindole (for nuclear counterstaining). Fluorescent images were captured with an F-view II monochrome camera (Olympus, U-CMAD3) mounted on an Olympus BX51 microscope.
NFκB activity assay
Cells were treated as described above. Nuclear fractions were isolated and subsequently analyzed for NFκB activity as per manufacturer's instructions using the NFκB p50/p65 Transcription Factor Assay Kit (Cat# ab133128; Abcam).
siRNA-mediated gene silencing
Electroporation of siRNA into CLL cells was performed using Amaxa Human B-cell Nucleofection Kit (Amaxa, Cologne, Germany). 1×107 PBMCs were mixed with 100 μL of Amaxa B-cell nucleofector solution, and 2 μg of siRNA was nucleofected using program X-05. Transfection efficiency, assessed by transfection with 2 μg pMaxGFP plasmid, was 30-60% with cell viability of 50-80% at 24 hours. siRNA oligos were synthesized by Dharmacon (Lafayette, CO). Sense strand against human Bim1: GACCGAGAAGGUAGACAAUUU, Bim2: CUACCUCCCUACAGACAGAUU, Noxa8: GUAAUUAUUGACACAUUUCUU, Noxa9: AGUCGAGUGUGCUACUCAATT, Puma1: GCCUGUAAGAUACUGUAUAUU.
IGHV Mutation status
Following RNA and cDNA isolation (as above), IGHV mutations were analyzed using the IGH Somatic Hypermutation Assay v2.0 (Invivoscribe, San Diego, CA). Briefly, PCR was performed using the supplied master mixes and Amplitaq Gold DNA polymerase (Applied Biosystems) on a Veriti Thermal Cycler Model #9902 (Applied Biosystems) to amplify the IGH sequence fragment between the leader (VHL) and joining (J) regions as per the manufacturer's instructions. To confirm amplification of a single clonal product in the expected size range, an aliquot of each sample was run on a 1.5% agarose gel and separated by gel electrophoresis. Samples were then submitted for DNA sequencing using the supplied sequencing primers. The NCBI IgBLAST tool was used to determine the % divergence of each clonal sequence. Samples which showed less than 2% divergence from germline sequence were deemed to have unmutated IGHV.
Microarray Analysis
The RNA gene expression microarray experiments were carried out by the Dartmouth Genomics & Microarray Laboratory (Lebanon, NH, USA). Beadarrays with probes for all known human genes (Illumina, San Diego, CA) were used for RNA profiling. Reverse transcription using an oligo(dT) primer bearing a T7 promoter and the high yield ArrayScriptTM reverse transcriptase were used to make cDNA. The cDNA was made double-stranded and purified to use as a template for in vitro transcription with T7 RNA Polymerase and the included biotin-NTP mix. The labeled cRNA was purified and 1.5 µg used for hybridization to the beadarrays for 16 hours at 55°C. Following hybridization, the beadarrays were washed and stained with streptavidin-Cy3 (GE Healthcare, Piscataway, NJ). Fluorescent images were obtained with an Illumina 500GX scanner and processed with the BeadScan software (Illumina). Full results are available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE44864.
Statistical analysis
Statistical analysis was performed with Student t test (paired or unpaired) in GraphPad Prism software (LaJolla, CA). p<0.05 was considered to be statistically significant. Microarray data were analyzed for functional significance using Pathway Studio software (Ariadne Genomics/Elsevier, Rockville, MD). Fisher exact test was used to identify ontology groups and pathways statistically enriched in the gene set. Data is presented as mean±SE.
Results
MLN4924 reduces neddylation of cullins and promotes apoptosis in CLL
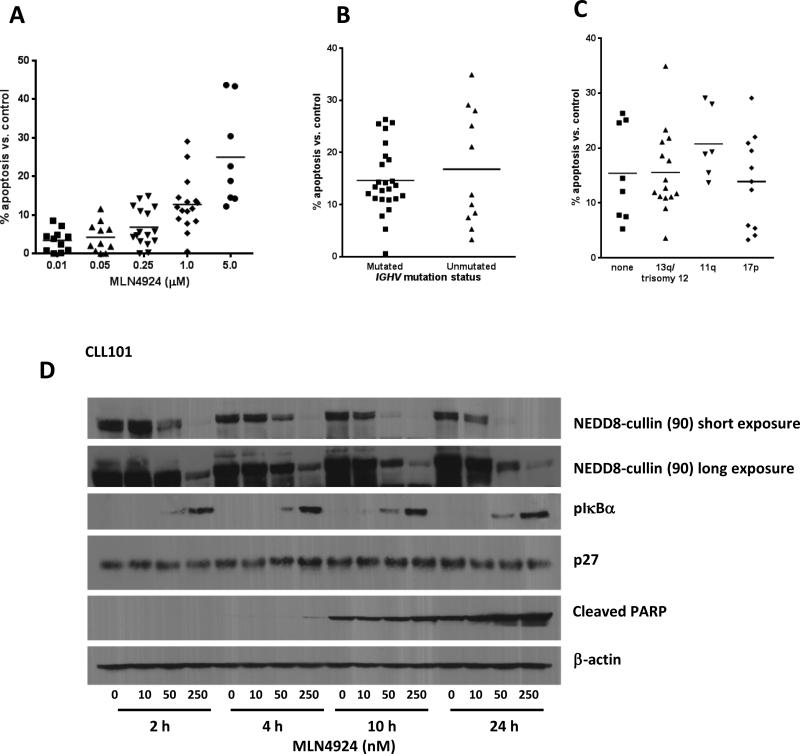
We first investigated whether NAE inhibition induced apoptosis in CLL. CLL cells incubated with 1 μM MLN4924 for 24 hours demonstrated enhanced apoptosis compared to vehicle control (12.8±1.7% Annexin V+ cells; Fig. 1A). While CLL cells exhibited variable sensitivity to MLN4924, we did not find distinct responses depending on either BCR heavy-chain immunoglobulin gene (IGHV) mutational status (Fig. 1B), ZAP-70 or CD38 expression (data not shown) or cytogenetic abnormalities, including 17p deletion (Fig. 1C).
Fig 1. MLN4924 induces apoptosis of the CLL B-cells independent of IGHV mutational status and cytogenetics.
(A) PBMC's from patients with CLL (N=15) were incubated with 0.01 - 5 μM MLN4924 or vehicle control for 24 hours. Horizontal lines represent the mean. (B-C) CLL B-cells were incubated with or without 1 μM MLN4924 for 24 hours. IgHV mutational status was determined as described in the methods. Cytogenetics markers (where available) were determined independent of our laboratory [no marker, 11q, trisomy 12, 13q, 17p]. Horizontal lines represent the mean (p=NS). (A-C) Apoptosis was determined by Annexin V and 7-AAD staining within the CD19+ subset of cells. Since we noted a significant variation in baseline apoptosis between patient samples (range, 4.2 to 44.4%), normalization to the time-matched untreated controls was performed to more clearly reflect the drug-induced apoptosis. (D) MLN4924 inhibits NAE and antagonizes IκB degradation in CLL B-cells. CLL cells (N=4) were incubated with MLN4924 (10 - 250 nM) for 2 - 24 hours. Whole-cell protein lysates were subjected to immunoblotting. A representative image from 1 of 4 experiments is shown.
MLN4924 selectively inhibits modification of cullin proteins by NEDD8 thus preventing CRL functional activity and ultimately leading to accumulation of their substrates (15). In the presence of MLN4924, CLL cells demonstrated a dose-dependent decrease in neddylation of cullins as early as 2 hours (Fig. 1D). This effect was more pronounced at later time points when it was also seen at lower drug concentrations. We investigated the effect of MLN4924 on several CRL substrates and observed a concomitant accumulation of phospho-IκBα. CLL cell apoptosis, both spontaneous and drug-induced, was detected at 10 hours and prominent at 24 hours, but was more pronounced in the presence of MLN4924, as evidenced by PARP cleavage. The cell cycle inhibitor p27Kip1 is another CRL substrate and is highly expressed in circulating CLL cells (20). Interestingly, MLN4924 had no effect on p27Kip1 protein levels, pointing to a relatively low turnover of this cell-cycle regulator in resting peripheral blood CLL cells (Fig.1D).
NAE inhibition-mediated abrogation of NFκB reverses the stroma-mediated protection in CLL
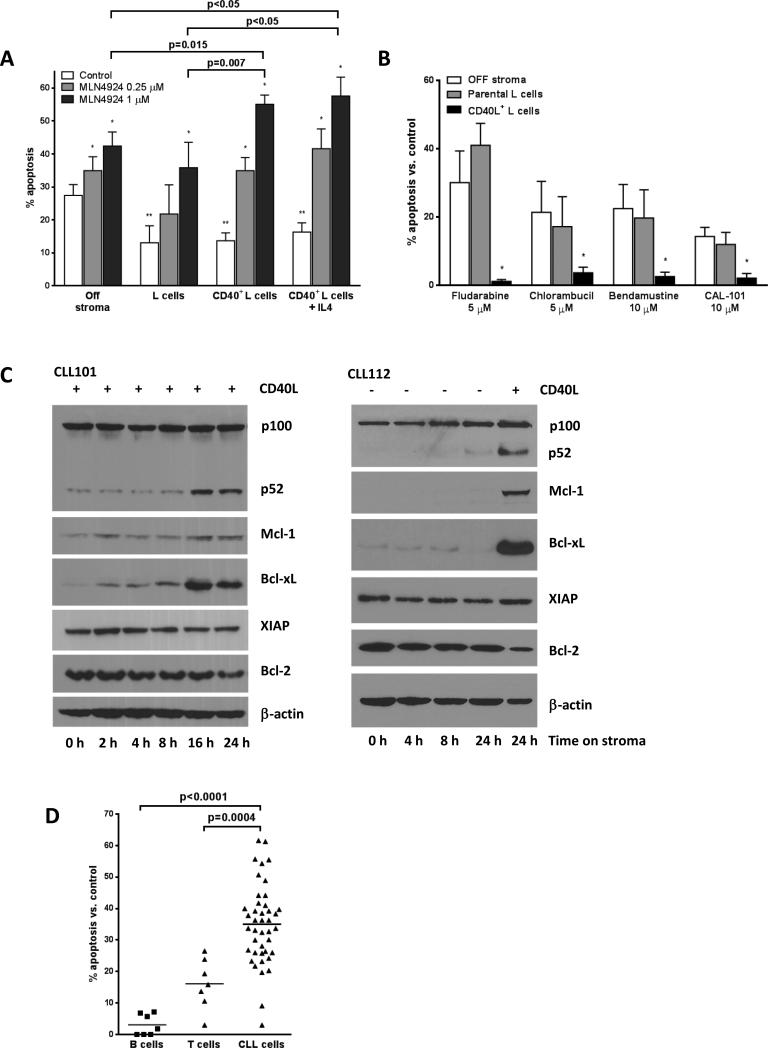
Given the importance of the microenvironment in sustaining CLL cell survival and proliferation in general and the NFκB pathway activity in particular, we further studied the effects of MLN4924 in a stromal co-culture system. Stroma cells present in the lymph node and bone marrow establish direct cell-cell contact with CLL cells engaging multiple pro-survival pathways (18), among which the tumor necrosis factor receptor family, including CD40, plays a prominent role. CD40L+ stroma partially rescued CLL cells from spontaneous apoptosis (Fig. 2A, white bars). We and others have previously shown that CD40L induces drug resistance in CLL (21-23). We confirmed those findings by demonstrating that the CD40L-expressing (but not parental) stroma induced CLL cell resistance to common chemotherapy agents and CAL-101 (Fig. 2B). In agreement with earlier work (24) the CD40L-expressing stroma activated both canonical and non-canonical NFκB pathways in CLL cells after 16-24 hours of co-culture. Activation of the non-canonical NFκB pathway was readily demonstrated by the emergence of p52, a cleaved product of p100 (Fig. 2C). Canonical NFκB pathway activation was confirmed by demonstrating nuclear translocation of p65/RelA, as discussed below. NFκB activation was accompanied by induction of anti-apoptotic proteins Mcl-1 and Bcl-xL on the CD40L-expressing stroma while Bcl-2 levels remained constant (Fig. 2C).
Fig. 2. MLN4924 abrogates the pro-survival effects of CD40L-expressing stroma in CLL cells.
(A) CLL cells (n=20) were cultured on CD40L-expressing or parental L cells for 24 hours, followed by incubation with 0.25 or 1 μM MLN4924 or vehicle control for 48 hours. As a reference, cells were treated off stroma. Cells were also cultured in the presence of 10 ng/ml IL4 (N=8). Apoptosis within CD19+ subset of cells was determined by Annexin V and 7-AAD staining. Data are the mean ± SE. *, p<0.01 compared to untreated control; **, p<0.05 compared to off stroma. (B) CLL cells (N=6) were co-cultured with CD40L-expressing or parental L cells for 24 hours, followed by incubation with the indicated drugs or with vehicle control for 48 hours. As a reference, cells were treated off stroma. Apoptosis within the CD19+ subset of cells was determined by Annexin V and 7-AAD staining and normalized to the untreated controls. Data are the mean ± SE. * - p<0.05 compared to off stroma or parental stroma. (C) CLL cells were co-cultured with CD40L-expressing (left panel) or parental stroma (right panel except for lane 5) for 2 - 24 hours. Whole-cell protein lysates were subjected to immunoblotting. Results from 1 of 3 experiments are shown. (D) Peripheral blood mononuclear cells from patients with CLL or healthy volunteers were cultured on CD40L-expressing stroma for 24 hours, followed by incubation with 1 μM MLN4924 or vehicle control for 48 hours. Apoptosis within CD19+ and CD3+ subset of cells was determined by Annexin V and 7-AAD staining and normalized to the time-matched untreated controls (p<0.0001).
Unexpectedly, we found that CD40L-expressing stroma did not elicit resistance to MLN4924 (Fig. 2A, gray and black bars). Interestingly, 1 μM MLN4924 induced more apoptosis on stroma (55.1±4.2% CLL cells) than off stroma (42.4±2.9%; p=0.015). Thus, CD40L+ (but not parental) L cells also appeared to sensitize CLL cells towards a neddylation inhibitor (Fig. 2A). Similarly, apoptosis occurred irrespective of common genetic features albeit CLL samples carrying deletion 17p were less sensitive to MLN4924 compared to non-17p CLL (Supplementary Fig. 1A-B; p<0.05). The size of a 17p− clone within each sample did not correlate with apoptotic response, indicating its susceptibility to the drug (Supplementary Fig. 1C). Neither protection from spontaneous apoptosis nor sensitivity of CLL cells to MLN4924 was altered by addition of IL4 (Fig. 2A). Importantly, MLN4924 did not lead to caspase activation and did not impair CD40L expression in the stroma (Supplementary Fig. 2). Finally, normal B cells and T cells exhibited decreased sensitivity to MLN4924 compared to CLL cells (Fig. 2D).
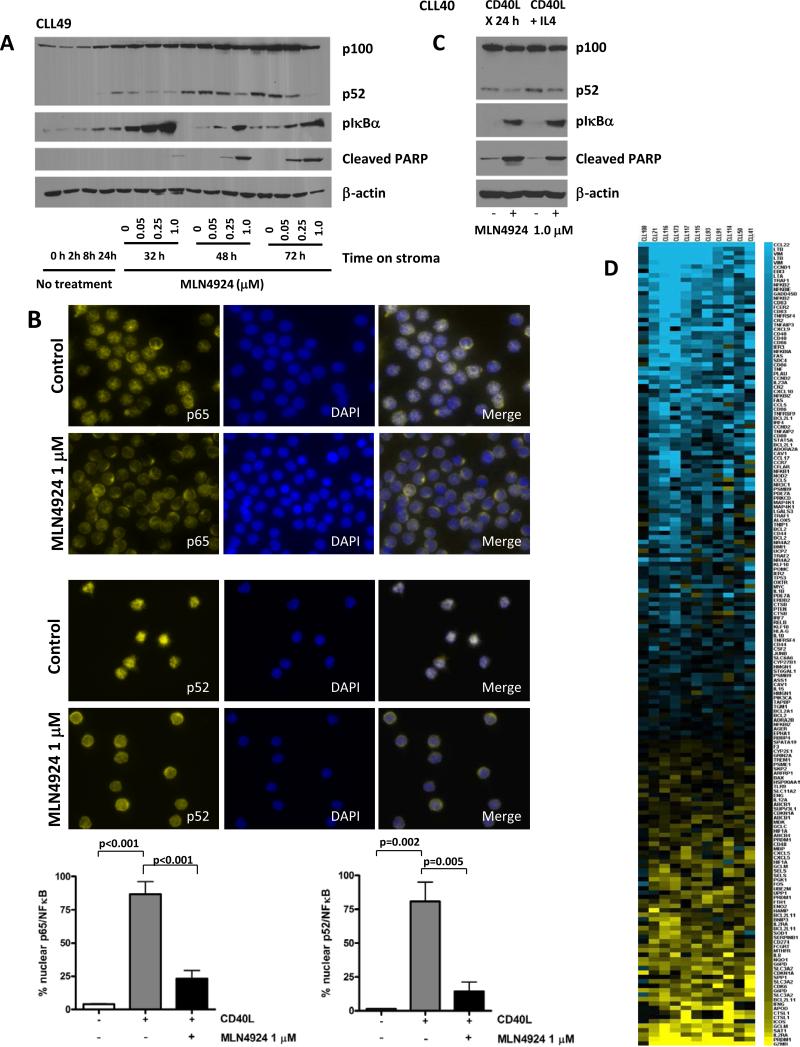
Treatment with MLN4924 in the CD40L+ co-culture system resulted in dose-dependent reversal of p100 cleavage and accumulation of phosphorylated IκBα in CLL cells (Fig. 3A). CLL cells demonstrated strong predominantly nuclear staining for both p65/RelA and p52 upon co-culture with CD40L-expressing stroma, indicating that the NFκB pathway was active (Fig. 3B). However, treatment with MLN4924 resulted in a shift towards cytoplasmic staining for p65 and p100/p52, thus indicating abrogation of both canonical and non-canonical NFκB pathways. Meanwhile, incubation of CLL cells with the parental control stroma did not result in nuclear translocation of either p65 or p52 (Fig. 3B). Addition of IL4 led to a mild increase in p100 processing compared with CD40L alone, yet this was reversed by MLN4924 (Fig. 3C). We have noted no effect on IκBα under those conditions (Fig. 3C). Similarly, upon treatment with MLN4924, the p65/RelA subunit shifted into the cytoplasm in CLL cells exposed to IL4 (Supplementary Fig. 3). We then sought to confirm that CLL cells undergoing apoptosis in response to MLN4924 had lost NFκB activity. Indeed, cells which demonstrated cleaved PARP expression did not show nuclear expression of p65 (Supplementary Fig. 3).
Fig. 3. MLN4924 reverses NFΔB activation in CLL cells.
(A) CLL cells were co-cultured with CD40L-expressing stroma for 24 hours and subsequently incubated with the indicated doses of MLN4924 for 8 - 48 hours. Whole-cell lysates were subjected to immunoblotting. Results from 1 of 4 experiments are shown. (B) CLL cells (N=8) were cultured on CD40L-expressing or parental stroma for 24 hours, followed by incubation with 1 μM MLN4924 for additional 24 hours and immunostained with p65 or p52/p100 antibodies. Nuclei were counterstained with 4,6-diamino-2-phenylindole. A representative case of CLL cells cultured on CD40L-expressing stroma in presence or absence of MLN4924 is shown. 100 cells per sample were scored for expression of p65 or p52 in the nuclear compartment. A comparison of nuclear expression of p65 and p52 in CLL cells co-cultured with either parental or CD40+ L cells, the latter with or without 1 μM MLN4924, is shown in the lower panels. Data are the mean ± SE. (C) CLL cells were co-cultured with CD40L-expressing stroma for 24 hours in the presence or absence of 10 ng/ml IL4. Thereafter, cell were incubated with 1 μM MLN4924 for 24 hours. Whole-cell lysates were subjected to immunoblotting. Results from 1 of 3 experiments are shown. (D) Gene expression profiling reveals a decrease in NFκB-driven gene signature pattern upon treatment with MLN4924. CLL cells were co-cultured with CD40L-expressing stroma for 18 hours and treated or not with 1 μM MLN4924 for additional 24 hours. RNA was isolated from the purified CLL B-cells and microarray analysis was performed as described in the methods. The heat map (left panel) represents a change in expression of the 181 putative NFκB target genes. Blue represents gene downregulation and yellow represents gene upregulation across the individual CLL samples (N=11).
We next employed gene expression profiling to determine which pathways were deregulated by NAE inhibition in CLL and whether the transcriptional targets of NFκB were affected. Of the genes incorporated in the probe set, 7254 were expressed in CLL. Using a cutoff of at least 1.5-fold change we identified 977 genes whose expression was significantly affected by MLN4924 (p<0.01). A set of 559 down regulated genes was analyzed for functional significance. We determined that receptor signaling and expression target pathways involving NFκB were most significantly associated with the downregulated genes (p<0.0001). Of the >400 known NFκB transcriptional targets (http://www.bu.edu/nf-kb/gene-resources/target-genes/) 181 genes were expressed in CLL (Fig. 3D and Supplementary Table) and 79 showed a 50% change in expression (p<0.01). We noted reduced transcription of several groups of NFκB target genes, including anti-apoptotic Bcl-2 family members, genes involved in cell cycle progression, and chemokines (p<0.01). Interestingly, we found a significant downregulation of a number of important cytokine ligands and receptors expressed by the CLL cells which are downstream targets of NFκB, including CCL5, CCL22, CXCR7, CXCR5 and CD40 (Supplementary Table). Chemokine receptors CXCR4 and CXCR5 are vital for CLL cell homing (25). We observed downregulation of the NFκB transcriptional target CXCR5 (26), but not CXCR4, by RT-PCR (Supplementary Fig. 4A). Intriguingly, we detected a 7.7-fold reduction in miR-155, an oncogenic micro-RNA which is predominantly expressed by proliferative CLL cells and is regulated by NFκB (27, 28). Finally, we confirmed that MLN4924 abrogated NFκB activity using an assay which measures p65 and p50 binding to the DNA sequence containing the NFκB response element. In this assay, NFκB activity was reduced in the presence of MLN4924 in a dose-dependent manner (Supplementary Fig. 4B), consistent with previously appreciated effect on IκBα (Fig. 3B). We then analyzed several NFκB transcriptional targets to confirm that the inhibitory effect of MLN4924 was dose-dependent (Supplementary Fig. 4C).
Thus, CLL cells exhibited NFκB activation in stromal co-cultures which was blocked by MLN4924, leading to enhanced apoptosis.
NAE inhibition rebalances Bcl-2 family members towards the pro-apoptotic BH3-only proteins in CLL
While circulating CLL lymphocytes express almost exclusively Bcl-2, cells in the stromal niche have been shown to express other pro-survival Bcl-2 family members. A balance between them and their relative ratio to the pro-apoptotic multi-BH proteins Bax and Bak determines cell fate (29). BH3-only proteins Puma, Noxa, Bim and others also interact with the anti-apoptotic Bcl-2 family members. Recent efforts have seen emergence of Bcl-2 inhibitors and BH3-mimetics (e.g., ABT-263) which have shown promise in treatment of CLL (29). We investigated whether changes in transcription of the anti-apoptotic proteins Bcl-2 and Bcl-xL as identified by gene expression profiling resulted in a significant reduction in the corresponding protein levels to explain enhanced apoptosis in response to MLN4924 on CD40L-expressing stroma.
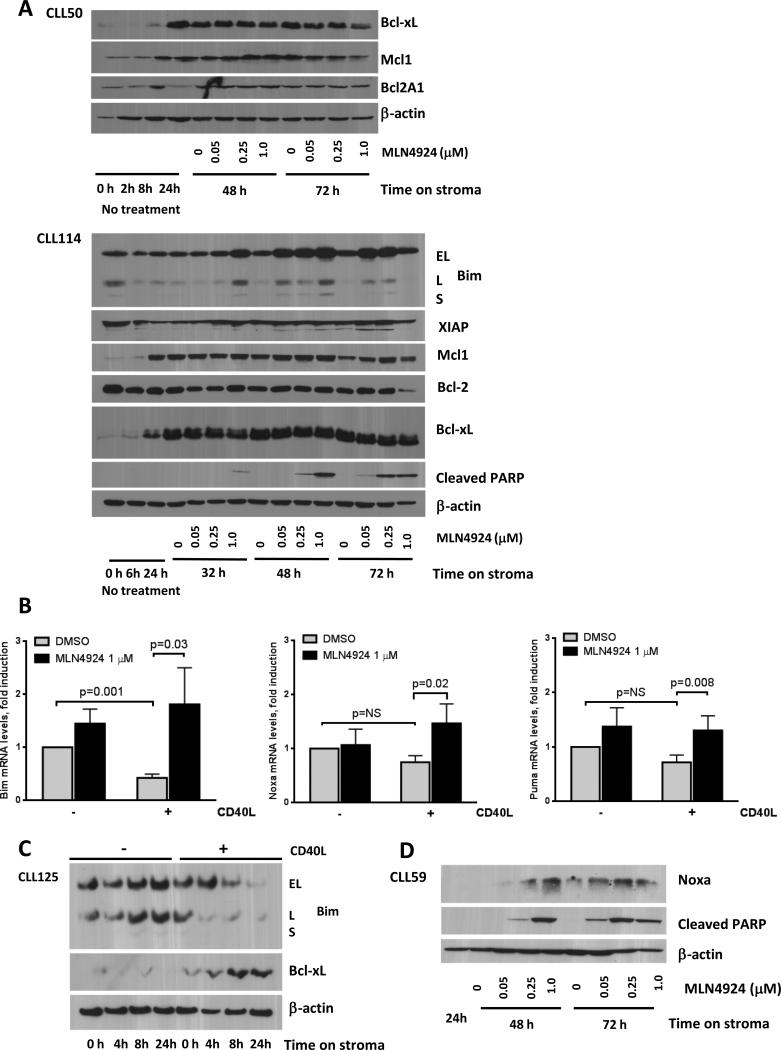
As described above, CD40L-expressing stroma induced Bcl-xL and Mcl-1 in CLL (Fig. 2C and 4A). Additionally, we observed a significant repression of Bim mRNA, but not Noxa or Puma mRNA, in cells cultured on the CD40L+ stroma vs. control stroma (Fig. 4B). Furthermore, CD40L+ but not parental L cells resulted in repression of Bim protein in 4/6 tested CLL samples (Fig. 4C). Consistent with previous reports that unmanipulated L cells do not induce either Mcl-1 or Bcl-xL (30), we observed no change in their expression in CLL cells co-cultured on the parental stroma (Fig. 2C and 4C).
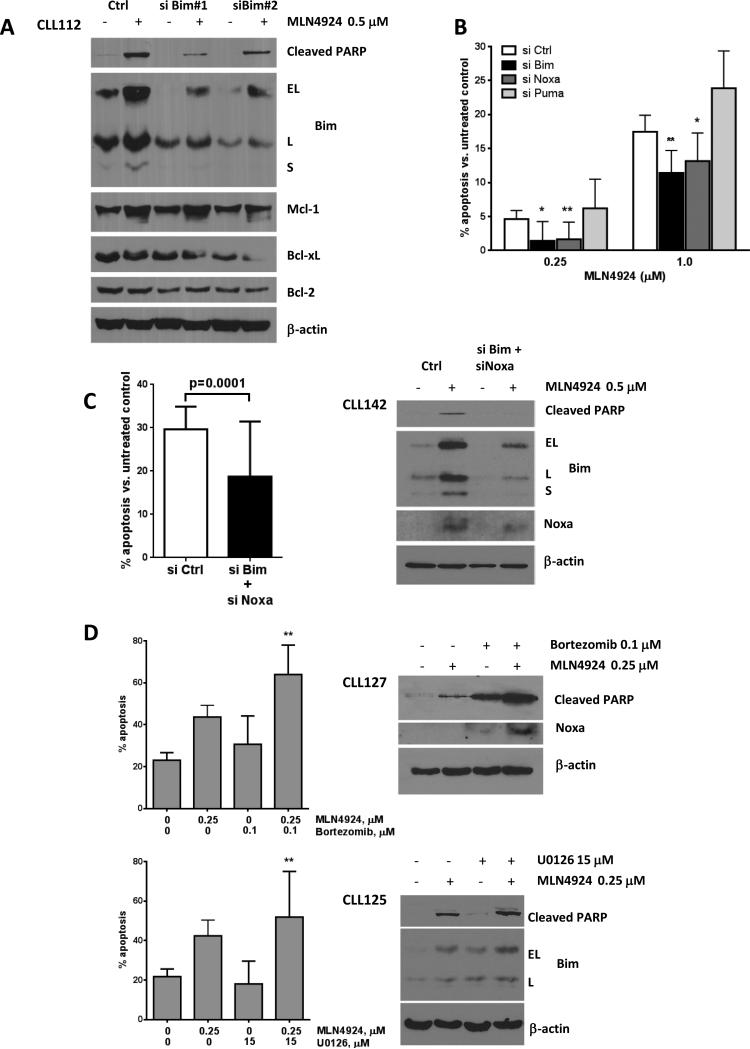
Fig. 4. Treatment of CLL cells with MLN4924 leads to upregulation of BH3-only proteins.
(A) CLL cells were co-cultured with CD40L-expressing L-cells for 24 hours, followed by incubation with 0.05 - 1 μM MLN4924 or vehicle control. Cells were lysed at the indicated time points and subjected to immunoblotting. Two representative images out of 10 independent experiments are shown. (B) CLL cells (N=10) were co-cultured with or CD40L+-expressing or control stroma for 24 hours, followed by incubation with 1 μM MLN4924 or vehicle control for 24 h. Total RNA was isolated from CD19+ CLL B-cells, reverse-transcribed and subjected to real-time PCR with the indicated probes (in duplicates). Results were normalized to 18S levels. Data are the mean ± SE. (C) CLL cells were co-cultured with CD40L-expressing or parental L cells for 4 - 24 hours. Whole-cell protein lysates were subjected to immunoblotting. Results from 1 of 6 experiments are shown. (D) CLL cells were co-cultured with CD40L-expressing stroma for 24 hours, followed by incubation with MLN4924 or vehicle control. Cell were lysed at the indicated time points and subjected to immunoblotting. A representative image out of 5 independent experiments is shown.
Contrary to the microarray findings, Bcl-2 protein expression remained unchanged upon treatment with MLN4924. Meanwhile, of 10 CLL samples tested, modest downregulation of Bcl-xL was detected in 6 samples upon treatment with MLN4924 for 48 hours on CD40L+ stroma, while expression of Mcl-1, XIAP and Bcl2A1 were stable (Fig. 4A). We then studied the effect of MLN4924 on the pro-apoptotic BH3-only proteins. Gene expression profiling experiments revealed a 2-fold upregulation of Bim transcript by MLN4924 (Supplementary Table). We confirmed upregulation of Bim mRNA and protein levels (Fig. 4A and 4B). Bim protein induction was evident by 8 hours of exposure to 1 μM MLN4924. Importantly, Bim induction also occurred in the CLL samples which showed no Bim repression by stroma and no drug-mediated change in anti-apoptotic proteins. BMS-345541, an IκB kinase inhibitor, also led to induction of Bim in CLL cells, confirming the importance of NFκB in Bim regulation (Supplementary Fig. 5). NFκB inhibition was previously shown to upregulate other BH3-only proteins, namely Noxa, in CLL (31). We found that Noxa mRNA and protein as well as Puma mRNA were also induced by MLN4924 in CD40L-expressing stroma co-cultures (Fig. 4B and 4D).
We further sought to confirm the role of BH3-only proteins in MLN4924-mediated apoptosis. siRNA-mediated knockdown yielded consistent reduction in baseline Bim and blunted its upregulation upon treatment with MLN4924 (Fig. 5A). Bim short isoform (S), the most cytotoxic, was no longer detected. This was accompanied by a reduction in apoptosis, despite reduced Bcl-xL expression. These findings were confirmed in an expanded cohort of CLL samples (Fig. 5B). In agreement with earlier data on the involvement of NFκB in regulation of Noxa in CLL, apoptosis was decreased in Noxa-suppressed CLL cells, while manipulation of Puma had no effect (Fig. 5A and Supplementary Fig. 6). Combined knockdown of Bim and Noxa further suppressed CLL cell apoptosis (Fig. 5C).
Fig. 5. BH3-only proteins Bim and Noxa are required for apoptosis induction by MLN4924.
(A) CLL cells were transfected with two individual siRNA's against Bim or control siRNA using Amaxa program X-05. Immediately after nucleofection, cells were cultured on CD40L-expressing stroma for 24 hours, followed by incubation with 0.5 μM MLN4924 or vehicle control for 24 h. Whole-cell lysates were subjected to immunoblotting. A representative blot of three independent experiments is shown. (B-C) CLL cells were transfected with siRNA against Bim (N=7), Noxa (N=7), Puma (N=4), both Bim and Noxa (panel C, N=5) or control siRNA using Amaxa program X-05. Immediately after nucleofection, cells were co-cultured with the CD40L-expressing stroma for 24 hours, followed by incubation with 0.25 or 1 μM MLN4924 or vehicle control for 24 h. Apoptosis within CD19+ subset of cells was determined by Annexin V and 7-AAD staining and normalized to untreated control. Data are the mean ± SE. * - p<0.05 and ** - p<0.01 compared to control siRNA. (D) CLL cells (N=10) were co-cultured with CD40L-expressing stroma for 24 hours, followed by incubation with 0.25 μM MLN4924, 0.1 μM bortezomib, 15 μM U0126, the drugs combined, or with vehicle control for 48 hours. Bortezomib was washed off after 1 hour. Apoptosis was determined as above and normalized to the untreated controls. Data are the mean ± SE. ** - p<0.01 compared to either single drug. For protein detection, cells were incubated with drugs for 24 hours, then lysed and subjected to immunoblotting (representative images out of 3 independent experiments are shown).
Bortezomib was previously shown to induce Noxa in CLL cells (32). Expectedly, treatment of CLL cells with bortezomib resulted in sensitization to MLN4924 (Fig. 5D). Noxa induction was enhanced when MLN4924 was combined with bortezomib compared with either drug alone (Fig. 5D). Meanwhile, CD40L-mediated downregulation of Bim is dependent on activation of extracellular signal-regulated kinase (ERK) (33). While both MLN4924 and U0126 (a mitogen-activated protein kinase inhibitor) induced Bim in CLL cells, the increase in toxicity of the combination was not as pronounced, albeit statistically significant (p<0.01, paired t-test), possibly due to a mild cooperative effect on Bim expression. Thus we found that Bim is frequently downregulated in a CD40L-expressing microenvironment. Meanwhile, NAE inhibition-mediated inactivation of NFκB results in rebalancing of the Bcl-2 family members towards the pro-apoptotic BH3-only proteins Bim and Noxa in CLL cells, which are required for apoptosis induction in this setting.
MLN4924 sensitizes CLL cells to the BCR-targeting agents
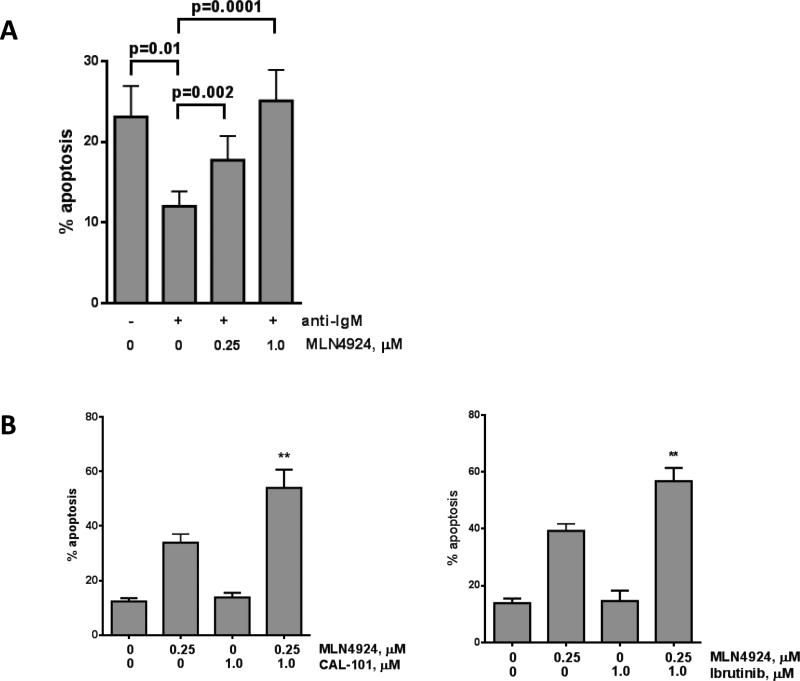
Since NFκB activation is one of the dominant pathways ensuring CLL cell survival in response to BCR signaling, we hypothesized that MLN4924 may enhance the pro-apoptotic effects of the novel BCR kinase inhibitors (9). We first determined whether MLN4924 had an impact on BCR signaling-mediated survival of the CLL cells. As expected, IgM stimulation resulted in CLL cell rescue from spontaneous apoptosis in a subset of patient samples (34), and this was reversed by MLN4924, once again emphasizing the importance of NFκB activation in BCR-mediated survival (Fig. 6A). We then explored the combined effect of MLN4924 and BCR-targeting agents CAL-101 and ibrutinib (8, 9). While CLL cells were resistant to both CAL-101 and ibrutinib in the CD40L-expressing system (but not on parental L cells - not shown), co-incubation with either agent and 0.25 μM MLN4924 led to an increase in cell death compared to MLN4924 alone (Fig. 6B).
Fig. 6. MLN4924 enhances the activity of BCR signaling inhibitor CAL-101 and alkylating agents in CLL.
(A) CLL cells (N=12) were stimulated with immobilized anti-IgM (10 μg/mL) for 1 hour or not and cultured in the presence of 0.25 or 1 μM MLN4924 or vehicle control for 24 hours (in duplicates). (B) CLL cells (N=8) were co-cultured with CD40L-expressing stroma for 24 hours, followed by incubation with 0.25 μM MLN4924, 1 μM CAL-101, 1 μM ibrutinib, the drugs combined, or with vehicle control for 48 hours.
Discussion
Gene expression profiling of peripheral blood CLL cells had initially characterized them as quiescent lymphocytes related to memory cells (2). Subsequently, it was established that, in addition to cell accumulation, proliferation of the neoplastic B-cells provides a significant contribution to the malignant clone (3). Lymph nodes and bone marrow were identified as the preferential sites of activation and proliferation of the CLL cells (4, 5). BCR gene signature along with the NFκB, NFAT and other proliferation/cell cycle functional gene sets are overrepresented in the lymph node, while bone marrow-resident CLL cells show decreased apoptotic priming (4, 22). Signals transduced through the BCR, CD40 and Toll-like receptors converge on NFκB, leading to high NFκB activity in the CLL cells resident in the microenvironment. Hewamana et al. demonstrated that DNA binding of the Rel A subunit is enhanced in CLL compared with normal B-cells, providing evidence for constitutive NFκB activation in CLL (5).
NFκB activation occurs through the canonical and non-canonical pathways (35). In the canonical pathway, the IκB kinase complex phoshorylates IκBα, triggering its ubiquitination and leading to nuclear translocation of the NFκB dimers, predominantly p50/Rel A and p50/c-Rel. Non-canonical activation is based on the proteosome-assisted processing of the precursor protein p100 with nuclear translocation of p52/Rel B (35, 36). Inappropriate degradation of IκBα is among the potential reasons for constitutive activation of NFκB in cancer (37). Thus, blocking proteosomal degradation of IκBα is a promising therapeutic approach. MLN4924 is a recently discovered inhibitor of NAE. MLN4924 binds NAE at its active site forming a covalent MLN4924-NEDD8 adduct thus preventing the modification of cullin proteins by Nedd8 and leading to accumulation of CRL substrates (15). In this work we demonstrate that MLN4924 efficiently reverses cullin neddylation in CLL cells in vitro, a process accompanied by the accumulation of phospho-IκBα and subsequent apoptosis. Accumulation of IκBα in CLL occurred rapidly and at concentrations of MLN4924 sufficient to induce apoptosis (14).
To study the impact of MLN4924 on the microenvironment-mediated NFκB activation, we co-cultured CLL cells with CD40L-expressing stroma. Such a strategy has been shown to activate NFκB and counter the spontaneous apoptosis of the CLL cells in vitro (24, 38). Importantly, CLL cells exposed to CD40L-expressing stroma have decreased “priming” to undergo apoptosis and acquire resistance to both conventional and novel therapeutic agents such as ABT-737 and CAL-101 (21-23, 25). In agreement with earlier reports we demonstrated activation of both canonical and non-canonical NFκB pathways in CLL cells co-cultured with the CD40L-expressing stroma (but not with parental stroma), thus creating a partial recapitulation of the lymph node microenvironment (24). Importantly, we found that CD40L signaling may lead to repression of Bim.
While CLL cells co-cultured with CD40L-expressing stroma were resistant to multiple chemotherapy agents, the protective effects were abrogated by MLN4924. We further observed that NAE inhibition prevented nuclear translocation of the NFκB pathway effectors p65/Rel A and p52 in CLL cells. Attenuated ubiquitination of phospho-IκBα resulted in an increased retention of p65 in the cytoplasm thus leading to inactivation of the canonical pathway. Meanwhile, a non-canonical pathway effector p52 is generated when an E3 ligase induces processing of the NFκB2 precursor protein p100 (36). Since p100 protein expression was not affected by MLN4924 in CLL, while p52 was reduced, it is likely that inactivation of the non-canonical pathway occurred due to E3 ligase inhibition and subsequent reduction in proteosomal processing of p100. It has been previously reported that both canonical and non-canonical NFκB pathways are activated in CLL lymph nodes, pointing to the biological relevance of our findings (4, 24). Interestingly, MLN4924 also induced apoptosis of CLL cells cultured off stroma. Using electrophoretic mobility shift assays, it was shown that peripheral blood CLL cells also demonstrate increased NFκB acitvity compared to normal B cells. It is possible that inhibition of this “baseline” NFκB activity accounts for modest apoptosis in the presence of MLN4924 in this setting (39).
Abrogation of NFκB activity in CLL cells by MLN4924 resulted in decreased transcription of its nuclear targets in a dose-dependent manner. Among those, we found a significant reduction in transcription of genes involved in cell cycle and CLL cell-derived chemokines mediating microenvironment dependence (CXCR5, CCL17, CCL22, etc), some of which are induced via CD40 (40, 41). Furthermore, recent work demonstrates that NFκB upregulates the expression of cytokines and adhesion molecules crucial for CLL survival in the stroma itself (42). Hence MLN4924 may disrupt the chemokine network and cell-cell interactions in the protective microenvironment leading to reduced tissue homing in CLL. In addition, MLN4924 may shift the balance of CD40 signaling towards pro-apoptotic events (43).
While NAE inhibition had minimal effect on expression of the anti-apoptotic Bcl-2 family members, it led to induction of Bim and Noxa. Bim plays an important role in apoptosis regulation in CLL (44). Bim is phosphorylated and ubiquitinated in response to BCR signaling leading to CLL cell survival and disease progression (44). Bim is capable of binding to all Bcl-2 proteins with high affinity, activating the pro-death Bax and/or Bak (45). We found that MLN4924 induced all Bim splice variants (BimEL/L/S) in CLL but the exact mechanism remains unclear. MLN4924 did not induce Forkhead box 3A transcription factor, a regulator of Bim (45) (Supplementary Fig. 7). Earlier reports suggest that NFκB neutralizes Bim via a c-Rel-dependent mechanism in B-cells (46). Additionally, transgenic expression of p52 led to repression of Bim and defective apoptosis of mouse lymphocytes (47). Furthermore, degradation of at least one Bim isoform, BimEL is proposed to occur with the involvement of CRL (48). We found that Bim transcription is upregulated by MLN4924 in CLL cells, implicating de novo synthesis in its induction. While we did not detect Bim phosphorylation in CLL (Supplementary Fig. 7), we did not fully exclude a possibility of attenuated degradation of Bim. . Meanwhile, Noxa has been designated as a putative CRL target and therefore NAE inhibition may abrogate its degradation in CLL (49). Diminished degradation of Bim and Noxa may be contributing to the pro-apoptotic effect of MLN4924 in CLL cells cultured off stroma.
Since NFκB is among the terminal effectors of BCR signaling, our findings that MLN4924 abolished the protective effect of BCR stimulation suggest that it may overcome the enhanced responsiveness to BCR signaling and the unfavorable prognosis rendered by unmutated IGHV in CLL (50). In CD40L+ stromal co-cultures, CLL cells were rescued from the pro-apoptotic effects of the PI3-Kδ inhibitor CAL-101 and Bruton tyrosine kinase ibrutinib, an effect reversed by MLN4924. Hence, microenvironment-mediated NFκB activation may enhance CLL cell survival independent of BCR signaling and induce resistance to BCR-targeting agents. Remarkable efficacy of MLN4924 in this setting suggests addiction to the NFκB pathway and justifies further investigation of those drug combinations in the clinic.
In summary, we demonstrate that MLN4924 effectively inhibits cullin neddylation in CLL cells. In a model which mimics the lymph node microenvironment, this leads to inactivation of the NFκB pathway, re-expression of Bim and Noxa and prevents stroma-mediated drug resistance. MLN4924 shows cooperation with the BCR-targeting agents . Our data combined with the new knowledge regarding the indispensability of stromal NFκB to CLL cell survival (42) justify further studies of the NAE inhibitor MLN4924 in CLL.
Supplementary Material
Translational Relevance.
Chronic lymphocytic leukemia (CLL) is the most common type of leukemia in the Western Hemisphere. Successful use of purine analogue-containing chemo-immunotherapy regimens has extended survival of younger patients with CLL. However, eventual progression to fludarabine-resistant disease, lack of low-risk curative strategies and limited therapeutic options in the elderly warrant exploration of novel treatment strategies. The tumor microenvironment in the lymphatic tissues provides pro-survival and pro-proliferative stimuli and harbors chemoresistant CLL clones. We demonstrate that MLN4924, an investigational inhibitor of Nedd8-activating enzyme exhibits cytotoxicity towards CLL cells incubated in the microenvironment-mimicking conditions where other drugs are ineffective. MLN4924 disrupts key pathways implicated in progressive disease - NFκB signaling and the anti-apoptotic Bcl-2 family members. Our results justify the development of MLN4924 in CLL.
Acknowledgements
We would like to thank Dr Allison Berger, Ph.D. (Millennium Pharmaceuticals, Inc.) for the helpful discussions and Dr Mark Israel, M.D. for use of the microscope.
Financial support: Support to AVD provided by a National Cancer Institute new faculty award (3P30CA023108-31S4) to the Norris Cotton Cancer Center. AE and AVD are supported by a translational research award from the Leukemia & Lymphoma Society. JRB is supported by the Leukemia Lymphoma Society and the American Cancer Society and is a Scholar in Clinical Research of the Leukemia and Lymphoma Society.
Footnotes
Author contributions
AVD is the principal investigator and takes primary responsibility for the paper. AVD and JCG designed research. JCG, AVD, LAH, OVD, PEK performed experiments. AVD and JCG analyzed data. AE contributed vital reagents. JCG, AE, AVD wrote the paper.
Conflict of Interest: the authors have nothing to disclose.
References
- 1.Danilov AV, Danilova OV, Klein AK, Huber BT. Molecular pathogenesis of chronic lymphocytic leukemia. Curr Mol Med. 2006;6:665–75. doi: 10.2174/156652406778195008. [DOI] [PubMed] [Google Scholar]
- 2.Rosenwald A, Alizadeh AA, Widhopf G, Simon R, Davis RE, Yu X, et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. The Journal of experimental medicine. 2001;194:1639–47. doi: 10.1084/jem.194.11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Messmer BT, Messmer D, Allen SL, Kolitz JE, Kudalkar P, Cesar D, et al. In vivo measurements document the dynamic cellular kinetics of chronic lymphocytic leukemia B cells. J Clin Invest. 2005;115:755–64. doi: 10.1172/JCI23409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herishanu Y, Perez-Galan P, Liu D, Biancotto A, Pittaluga S, Vire B, et al. The lymph node microenvironment promotes B-cell receptor signaling, NF-kappaB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood. 2011;117:563–74. doi: 10.1182/blood-2010-05-284984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hewamana S, Lin TT, Rowntree C, Karunanithi K, Pratt G, Hills R, et al. Rel a is an independent biomarker of clinical outcome in chronic lymphocytic leukemia. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:763–9. doi: 10.1200/JCO.2008.19.1114. [DOI] [PubMed] [Google Scholar]
- 6.Liu Z, Hazan-Halevy I, Harris DM, Li P, Ferrajoli A, Faderl S, et al. STAT-3 activates NF-kappaB in chronic lymphocytic leukemia cells. Mol Cancer Res. 2011;9:507–15. doi: 10.1158/1541-7786.MCR-10-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishio M, Endo T, Tsukada N, Ohata J, Kitada S, Reed JC, et al. Nurselike cells express BAFF and APRIL, which can promote survival of chronic lymphocytic leukemia cells via a paracrine pathway distinct from that of SDF-1alpha. Blood. 2005;106:1012–20. doi: 10.1182/blood-2004-03-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woyach JA, Johnson AJ, Byrd JC. The B-cell receptor signaling pathway as a therapeutic target in CLL. Blood. 2012;120:1175–84. doi: 10.1182/blood-2012-02-362624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevenson FK, Krysov S, Davies AJ, Steele AJ, Packham G. B-cell receptor signaling in chronic lymphocytic leukemia. Blood. 2011;118:4313–20. doi: 10.1182/blood-2011-06-338855. [DOI] [PubMed] [Google Scholar]
- 10.Furman RR, Asgary Z, Mascarenhas JO, Liou HC, Schattner EJ. Modulation of NF-kappa B activity and apoptosis in chronic lymphocytic leukemia B cells. J Immunol. 2000;164:2200–6. doi: 10.4049/jimmunol.164.4.2200. [DOI] [PubMed] [Google Scholar]
- 11.Arvaniti E, Ntoufa S, Papakonstantinou N, Touloumenidou T, Laoutaris N, Anagnostopoulos A, et al. Toll-like receptor signaling pathway in chronic lymphocytic leukemia: distinct gene expression profiles of potential pathogenic significance in specific subsets of patients. Haematologica. 2011;96:1644–52. doi: 10.3324/haematol.2011.044792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pickering BM, de Mel S, Lee M, Howell M, Habens F, Dallman CL, et al. Pharmacological inhibitors of NF-kappaB accelerate apoptosis in chronic lymphocytic leukaemia cells. Oncogene. 2007;26:1166–77. doi: 10.1038/sj.onc.1209897. [DOI] [PubMed] [Google Scholar]
- 13.Swords RT, Kelly KR, Smith PG, Garnsey JJ, Mahalingam D, Medina E, et al. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood. 2010;115:3796–800. doi: 10.1182/blood-2009-11-254862. [DOI] [PubMed] [Google Scholar]
- 14.Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, et al. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-{kappa}B-dependent lymphoma. Blood. 2010;116:1515–23. doi: 10.1182/blood-2010-03-272567. [DOI] [PubMed] [Google Scholar]
- 15.Soucy TA, Smith PG, Rolfe M. Targeting NEDD8-activated cullin-RING ligases for the treatment of cancer. Clin Cancer Res. 2009;15:3912–6. doi: 10.1158/1078-0432.CCR-09-0343. [DOI] [PubMed] [Google Scholar]
- 16.Wei D, Li H, Yu J, Sebolt JT, Zhao L, Lawrence TS, et al. Radiosensitization of human pancreatic cancer cells by MLN4924, an investigational NEDD8-activating enzyme inhibitor. Cancer research. 2012;72:282–93. doi: 10.1158/0008-5472.CAN-11-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neron S, Suck G, Ma XZ, Sakac D, Roy A, Katsman Y, et al. B cell proliferation following CD40 stimulation results in the expression and activation of Src protein tyrosine kinase. Int Immunol. 2006;18:375–87. doi: 10.1093/intimm/dxh377. [DOI] [PubMed] [Google Scholar]
- 18.Kurtova AV, Balakrishnan K, Chen R, Ding W, Schnabl S, Quiroga MP, et al. Diverse marrow stromal cells protect CLL cells from spontaneous and drug-induced apoptosis: development of a reliable and reproducible system to assess stromal cell adhesion-mediated drug resistance. Blood. 2009;114:4441–50. doi: 10.1182/blood-2009-07-233718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Humphries LA, Godbersen JC, Danilova OV, Kaur P, Christensen BC, Danilov AV. Pro-apoptotic TP53 homolog TAp63 is repressed via epigenetic silencing and B-cell receptor signalling in chronic lymphocytic leukaemia. British journal of haematology. 2013;163:590–602. doi: 10.1111/bjh.12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frenquelli M, Muzio M, Scielzo C, Fazi C, Scarfo L, Rossi C, et al. MicroRNA and proliferation control in chronic lymphocytic leukemia: functional relationship between miR-221/222 cluster and p27. Blood. 2010;115:3949–59. doi: 10.1182/blood-2009-11-254656. [DOI] [PubMed] [Google Scholar]
- 21.Soderquist R, Bates DJ, Danilov AV, Eastman A. Gossypol overcomes stroma-mediated resistance to the BCL2 inhibitor ABT-737 in chronic lymphocytic leukemia cells ex vivo. Leukemia. 2013;27:2262–4. doi: 10.1038/leu.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davids MS, Deng J, Wiestner A, Lannutti BJ, Wang L, Wu CJ, et al. Decreased mitochondrial apoptotic priming underlies stroma-mediated treatment resistance in chronic lymphocytic leukemia. Blood. 2012;120:3501–9. doi: 10.1182/blood-2012-02-414060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogler M, Butterworth M, Majid A, Walewska RJ, Sun XM, Dyer MJ, et al. Concurrent up-regulation of BCL-XL and BCL2A1 induces approximately 1000-fold resistance to ABT-737 in chronic lymphocytic leukemia. Blood. 2009;113:4403–13. doi: 10.1182/blood-2008-08-173310. [DOI] [PubMed] [Google Scholar]
- 24.Tromp JM, Tonino SH, Elias JA, Jaspers A, Luijks DM, Kater AP, et al. Dichotomy in NF-kappaB signaling and chemoresistance in immunoglobulin variable heavy-chain-mutated versus unmutated CLL cells upon CD40/TLR9 triggering. Oncogene. 2010;29:5071–82. doi: 10.1038/onc.2010.248. [DOI] [PubMed] [Google Scholar]
- 25.Burger JA, Ghia P, Rosenwald A, Caligaris-Cappio F. The microenvironment in mature B-cell malignancies: a target for new treatment strategies. Blood. 2009;114:3367–75. doi: 10.1182/blood-2009-06-225326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu R, Zhao X, Gurney TA, Landau NR. Functional analysis of the proximal CCR5 promoter. AIDS Res Hum Retroviruses. 1998;14:1509–19. doi: 10.1089/aid.1998.14.1509. [DOI] [PubMed] [Google Scholar]
- 27.Wang M, Tan LP, Dijkstra MK, van Lom K, Robertus JL, Harms G, et al. miRNA analysis in B-cell chronic lymphocytic leukaemia: proliferation centres characterized by low miR-150 and high BIC/miR-155 expression. J Pathol. 2008;215:13–20. doi: 10.1002/path.2333. [DOI] [PubMed] [Google Scholar]
- 28.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–9. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 29.Billard C. Design of novel BH3 mimetics for the treatment of chronic lymphocytic leukemia. Leukemia. 2012;26:2032–8. doi: 10.1038/leu.2012.88. [DOI] [PubMed] [Google Scholar]
- 30.Cosimo E, McCaig AM, Carter-Brzezinski LJ, Wheadon H, Leach MT, Le Ster K, et al. Inhibition of NF-kappaB-mediated signaling by the CDK inhibitor CR8 overcomes pro-survival stimuli to induce apoptosis in chronic lymphocytic leukemia cells. Clin Cancer Res. 2013 doi: 10.1158/1078-0432.CCR-12-2170. [DOI] [PubMed] [Google Scholar]
- 31.Tromp JM, Geest CR, Breij EC, Elias JA, van Laar J, Luijks DM, et al. Tipping the Noxa/Mcl-1 balance overcomes ABT-737 resistance in chronic lymphocytic leukemia. Clin Cancer Res. 2012;18:487–98. doi: 10.1158/1078-0432.CCR-11-1440. [DOI] [PubMed] [Google Scholar]
- 32.Smit LA, Hallaert DY, Spijker R, de Goeij B, Jaspers A, Kater AP, et al. Differential Noxa/Mcl-1 balance in peripheral versus lymph node chronic lymphocytic leukemia cells correlates with survival capacity. Blood. 2007;109:1660–8. doi: 10.1182/blood-2006-05-021683. [DOI] [PubMed] [Google Scholar]
- 33.Hallaert DY, Jaspers A, van Noesel CJ, van Oers MH, Kater AP, Eldering E. c-Abl kinase inhibitors overcome CD40-mediated drug resistance in CLL: implications for therapeutic targeting of chemoresistant niches. Blood. 2008;112:5141–9. doi: 10.1182/blood-2008-03-146704. [DOI] [PubMed] [Google Scholar]
- 34.Deglesne PA, Chevallier N, Letestu R, Baran-Marszak F, Beitar T, Salanoubat C, et al. Survival response to B-cell receptor ligation is restricted to progressive chronic lymphocytic leukemia cells irrespective of Zap70 expression. Cancer research. 2006;66:7158–66. doi: 10.1158/0008-5472.CAN-06-0085. [DOI] [PubMed] [Google Scholar]
- 35.Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 36.Sun SC. The noncanonical NF-kappaB pathway. Immunol Rev. 2012;246:125–40. doi: 10.1111/j.1600-065X.2011.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo JL, Kamata H, Karin M. IKK/NF-kappaB signaling: balancing life and death--a new approach to cancer therapy. J Clin Invest. 2005;115:2625–32. doi: 10.1172/JCI26322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cuni S, Perez-Aciego P, Perez-Chacon G, Vargas JA, Sanchez A, Martin-Saavedra FM, et al. A sustained activation of PI3K/NF-kappaB pathway is critical for the survival of chronic lymphocytic leukemia B cells. Leukemia. 2004;18:1391–400. doi: 10.1038/sj.leu.2403398. [DOI] [PubMed] [Google Scholar]
- 39.Hewamana S, Alghazal S, Lin TT, Clement M, Jenkins C, Guzman ML, et al. The NF-kappaB subunit Rel A is associated with in vitro survival and clinical disease progression in chronic lymphocytic leukemia and represents a promising therapeutic target. Blood. 2008;111:4681–9. doi: 10.1182/blood-2007-11-125278. [DOI] [PubMed] [Google Scholar]
- 40.Davids MS, Burger JA. Cell Trafficking in Chronic Lymphocytic Leukemia. Open J Hematol. 2012:3. doi: 10.13055/ojhmt_3_s1_03.120221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scielzo C, Apollonio B, Scarfo L, Janus A, Muzio M, Ten Hacken E, et al. The functional in vitro response to CD40 ligation reflects a different clinical outcome in patients with chronic lymphocytic leukemia. Leukemia. 2011;25:1760–7. doi: 10.1038/leu.2011.149. [DOI] [PubMed] [Google Scholar]
- 42.Lutzny G, Kocher T, Schmidt-Supprian M, Rudelius M, Klein-Hitpass L, Finch AJ, et al. Protein Kinase C-beta-Dependent Activation of NF-kappaB in Stromal Cells Is Indispensable for the Survival of Chronic Lymphocytic Leukemia B Cells In Vivo. Cancer Cell. 2013;23:77–92. doi: 10.1016/j.ccr.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Messmer D, Kipps TJ. CD154 gene therapy for human B-cell malignancies. Ann N Y Acad Sci. 2005;1062:51–60. doi: 10.1196/annals.1358.008. [DOI] [PubMed] [Google Scholar]
- 44.Paterson A, Mockridge CI, Adams JE, Krysov S, Potter KN, Duncombe AS, et al. Mechanisms and clinical significance of BIM phosphorylation in chronic lymphocytic leukemia. Blood. 2012;119:1726–36. doi: 10.1182/blood-2011-07-367417. [DOI] [PubMed] [Google Scholar]
- 45.Gillings AS, Balmanno K, Wiggins CM, Johnson M, Cook SJ. Apoptosis and autophagy: BIM as a mediator of tumour cell death in response to oncogene-targeted therapeutics. FEBS J. 2009;276:6050–62. doi: 10.1111/j.1742-4658.2009.07329.x. [DOI] [PubMed] [Google Scholar]
- 46.Banerjee A, Grumont R, Gugasyan R, White C, Strasser A, Gerondakis S. NF-kappaB1 and c-Rel cooperate to promote the survival of TLR4-activated B cells by neutralizing Bim via distinct mechanisms. Blood. 2008;112:5063–73. doi: 10.1182/blood-2007-10-120832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, Zhang B, Yang L, Ding J, Ding HF. Constitutive production of NF-kappaB2 p52 is not tumorigenic but predisposes mice to inflammatory autoimmune disease by repressing Bim expression. J Biol Chem. 2008;283:10698–706. doi: 10.1074/jbc.M800806200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dehan E, Bassermann F, Guardavaccaro D, Vasiliver-Shamis G, Cohen M, Lowes KN, et al. betaTrCP- and Rsk1/2-mediated degradation of BimEL inhibits apoptosis. Mol Cell. 2009;33:109–16. doi: 10.1016/j.molcel.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jia L, Yang J, Hao X, Zheng M, He H, Xiong X, et al. Validation of SAG/RBX2/ROC2 E3 ubiquitin ligase as an anticancer and radiosensitizing target. Clin Cancer Res. 2010;16:814–24. doi: 10.1158/1078-0432.CCR-09-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lanham S, Hamblin T, Oscier D, Ibbotson R, Stevenson F, Packham G. Differential signaling via surface IgM is associated with VH gene mutational status and CD38 expression in chronic lymphocytic leukemia. Blood. 2003;101:1087–93. doi: 10.1182/blood-2002-06-1822. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.