Abstract
Mouse selenoprotein P (Sepp1) consists of an N-terminal domain (residues 1-239) that contains 1 selenocysteine (U) as residue 40 in a proposed redox-active motif (-UYLC-) and a C-terminal domain (residues 240-361) that contains 9 selenocysteines. Sepp1 transports selenium from the liver to other tissues by receptor-mediated endocytosis. It also reduces oxidative stress in vivo by an unknown mechanism. A previously uncharacterized plasma form of Sepp1 is filtered in the glomerulus and taken up by renal proximal convoluted tubule (PCT) cells via megalin-mediated endocytosis. We purified Sepp1 forms from the urine of megalin−/− mice using a monoclonal antibody to the N-terminal domain. Mass spectrometry revealed that the purified Urinary Sepp1 consisted of N-terminal Fragments terminating at 11 sites between residues 183 and 208. It was therefore designated Sepp1UF. Because the N-terminal domain of Sepp1 has a thioredoxin fold, Sepp1UF was compared with full-length Sepp1, Sepp1Δ240-361, and Sepp1U40S as a substrate of thioredoxin reductase-1 (TrxR1). All forms of Sepp1 except Sepp1U40S, which contains serine in place of the selenocysteine, were TrxR1 substrates, catalyzing NADPH oxidation when coupled with H2O2 or tert-butyl hydroperoxide as the terminal electron acceptor. These results are compatible with proteolytic cleavage freeing Sepp1UF from full-length Sepp1, the form that has the role of selenium transport, allowing Sepp1UF to function by itself as a peroxidase. Ultimately, plasma Sepp1UF and small selenium-containing proteins are filtered by the glomerulus and taken up by PCT cells via megalin-mediated endocytosis, preventing loss of selenium in the urine and providing selenium for the synthesis of glutathione peroxidase-3.
Keywords: selenoprotein P forms, selenium homeostasis, extracellular peroxidase activity, selenium-containing proteins, megalin
Introduction
Selenoprotein P (Sepp1)1 is an extracellular glycoprotein that is made up of an N-terminal, thioredoxin-fold domain containing a single selenocysteine residue in a redox motif and a smaller C-terminal domain containing multiple selenocysteines—9 in humans and mice. The selenium-rich C-terminal domain endows Sepp1 with a selenium transport function and the N-terminal domain is postulated to function as a redox enzyme (1).
Most research on Sepp1 has focused on the function of its C-terminal domain in regulating whole-body selenium and in transporting selenium from the liver to peripheral tissues (2-4). Genomic analysis and research reports indicate, however, that the N-terminal domain also functions in vivo—probably as an antioxidant enzyme (5-8). It is not clear whether the full-length Sepp1 form exerts both these functions or whether forms of Sepp1 are produced to exert each function.
Zebrafish (and several other species) have two Sepp1 genes—one for the N-terminal domain alone (presumed antioxidant function) and one for the full-length protein (selenium transport function) (9). In most animals, however, including humans and mice, there is only one gene for Sepp1. These observations raise the question of whether the single Sepp1 gene in most species can be the source of two (or more) protein forms of Sepp1.
Over a decade ago, our group detected “isoforms” of rat Sepp1 that terminated at selenocysteine residues (10) This finding raised the possibility that these forms exert distinct functions. In a subsequent in vitro study, however, doubt was cast on the physiological significance of these forms when inefficient read-through of in-frame UGAs in Sepp1 mRNA was shown to produce similar truncated Sepp1 forms (11). Thus, the biological significance of plasma Sepp1 forms that terminate at selenocysteine positions is questionable.
In vitro evidence has been presented that shortened forms of Sepp1 can be produced from the full-length protein. Proteases of the kallikrein family cleaved full-length human SEPP1 into N-terminal and C-terminal fragments (12), raising the possibility that proteolytically generated forms of Sepp1 exist in vivo.
Identification and characterization of naturally occurring Sepp1 forms would likely lead to better understanding of Sepp1 physiology and function. Sepp1 forms are present in vesicles within renal PCT cells, indicating that they are taken up from the glomerular filtrate (13). Moreover, their presence in the glomerular filtrate implies that they are smaller than full-length Sepp1 and might be a modified form of that protein.
Megalin, also known as Lrp2, is a member of the low-density lipoprotein receptor family and is present on the brush border of renal PCT cells. It binds proteins present in the glomerular filtrate and facilitates their endocytosis (14). This process prevents urinary loss of filtered plasma proteins, including Sepp1 forms (15).
Sepp1 has been studied in mice with mutated megalin (15). Most of the ligand-binding region of megalin was deleted in those mice and they excreted Sepp1 forms in their urine as had been expected. Little characterization of those Sepp1 forms was reported, but loss of Sepp1 in the urine was shown to predispose the mice to becoming selenium deficient.
Because characterization of the Sepp1 forms in the urine of megalin−/− mice might provide insight into Sepp1 metabolism and function, we raised mice that lack megalin entirely and characterized N-terminal Sepp1 forms that appeared in their urine. Those forms—like full-length Sepp1—had peroxidase activity when coupled with TrxR1 and NADPH. Thus Sepp1 is processed in vivo to free its enzymatically active N-terminal domain.
Material and methods
Reagents
Oligonucleotides were obtained from core lab facilities at the University of Utah and at Vanderbilt University Medical Center (Molecular Cell Biology Resources Core). 75Se-labeled sodium selenite (specific activity >250 Ci/g selenium) was purchased from the University of Missouri Research Reactor Facility (Columbia, MO). NADPH was purchased from USB Corporation. Glutathione reductase was purchased from Sigma. Recombinant rat TrxR1 (27-28 U/mg) was expressed and purified as described elsewhere (16). Recombinant human thioredoxin-1 (Trx1) was a gift from Dr. Arne Holmgren (Karolinska Institutet, Stockholm, Sweden). All other chemicals were of reagent grade.
Production of Sepp1U40S mice and partial characterization of purified Sepp1U40S
This is the first report of the Sepp1U40S mouse with serine replacing selenocysteine at residue 40. Because the purified protein Sepp1U40S was used in this study to assess the importance of the selenocysteine at residue 40 for the redox activity of Sepp1, we are presenting the methods used to produce the mice and limited characterization of Sepp1U40S protein.
The designation Sepp1U59S is used for the construction of the mutant gene because nucleotide triplets are counted from the start of the 19-amino acid signal peptide. The mice and the resulting protein use the designation Sepp1U40S for consistency with the other mutant of this protein, Sepp1Δ240-361 (17) that does not include the signal peptide in counting the amino acid residues.
Construction of Sepp1U59S targeting vector. To construct the targeting vector, a method described previously was followed (18). Recombineering was used to subclone a 13.1 kb genomic fragment from a BAC clone RP23-41H17, which was obtained from BACPAC resources (http://bacpac.chori.org/). The two oligos used in this step (WS785 and WS786) are shown in Table 1. The resulting plasmid from this step was named pStartK-Sepp1. PCR-based mutagenesis was used to convert the DNA encoding the first selenocysteine from TGA to TCA, which encodes serine. The self-excising neo selection cassette ACN was inserted at the BglI site before the second exon. The resulting plasmid was named pStartK-Sepp1U59SACN. To add a negative selection HSV-tk gene, Gateway recombination was performed to quickly transfer Sepp1U59SACN into an HSV-tk containing vector named pWSTK2. The resulting targeting vector was named pWSTK2-Sepp1U59SACN.
Table 1.
Targeting vector
| primer designation |
primer sequence (5’-< 3’) | |
|---|---|---|
| pStartK | ||
| WS785 | GTAGTGCTACTTATAAATCCCAACAGAGTCTCTTT TAGCTGTTGCTTCAGCGACTGAATTGGTTCCTTTA AAGCC |
|
| WS786 | ATGTTTATTGCTCACCTAGGCATTATACTAAACAC TCCATGCAAACTACAGCCGCACTCGAGATATCTA GACCCA |
|
| Southern blot probes | ||
| WS869-5F | GAGATCCCAGTAGGTAGGCGACTTG | |
| WS870-5R | CCTGTAACCTTGAGCCAAACTTCCT | |
Generation of Sepp1U40S mice. Standard electroporation of the linearized targeting vector into ES cells was performed as previously described (18). Southern blot analysis was performed to identify correctly targeted ES cell clones. The 5’ Southern probe template (476 bp) was amplified by PCR from the BAC clone RP23-41H17 with primers WS869-5F and WS870-5R (Table 1). DNA isolated from ES cells was digested with BamHI, and run on a 0.9% agarose gel. Southern blot was done with the 5’ probe. The wild-type band is 15.7 kb, and the targeted mutant band is 6.9 kb. The positive targets were further confirmed by Southern blot analysis with XbaI digest using the 5’ probe. Targeted ES cells were injected into blastocysts using our standard protocol. Male chimeric mice were bred with C57BL/6 females to obtain the desired Sepp1U40S allele. Since we used the self-excising neo cassette ACN, the neo was automatically deleted in the F1 generation of heterozygotes. The University of Utah Institutional Animal Care and Use Committee approved animal protocols used to generate the mutant mouse. Adult Sepp1U40S/+ mice (mice heterozygous for the serine mutation allele) were transferred to the animal facility at Vanderbilt University. At Vanderbilt, male and female mice homozygotic for substitution of serine at the first selenocysteine position (Sepp1U40S/U40S) were determined to be fertile. These mice were bred with C57BL/6J mice to produce a congenic Sepp1U40S/U40S breeding colony.
Animal Husbandry
Male C57BL/6-congenic megalin+/− mice (kindly provided by Dr. Joachim Herz of the University of Texas Southwestern Medical Center) were backcrossed 4 times with female FVB/NJ mice (Jackson Lab stock #001800) in order to produce viable megalin−/− mice, as suggested to us by Dr. Ulrich Schweizer of the University of Bonn (Germany). The first 409 pups that survived to be weaned were all genotyped and 4.6% were megalin−/− mice. There were 9 males and 10 females and all of them had a deformity of the forehead as had been observed in megalin−/− mice when they were first produced (19). In subsequent litters, genotyping was carried out only when at least one pup had the forehead deformity. Pups were ear-notched and genotyped by PCR. PCR primer sequences are shown in Table 2. The backcrossed megalin+/− male and female mice were used as breeding pairs and their pups were genotyped. Megalin−/− pups and their megalin+/+ littermates were used for experiments.
Table 2.
PCR primers for genotyping
| primer designation |
primer sequence (5’-< 3’) | PCR product size (pb) |
|
|---|---|---|---|
| Sepp1 | SePS14 | GCCATCAGGGCTCAGTGCAG | wildtype: 600 (SePS14/SePA600) |
| SePA600 | GGTGTTGGGTCGTTTGTTCGGATCG | ||
| NeoS2 | GGATGCCTGCAGAAGACACAAGTA | homozytote: 500 (SePS14/NeoS2) |
|
|
Sepp1 □240-
361 |
MoSePS18 | GCAGGAGCATCTTGGCAGCAGT | mutant: 450 wildtype: 370 |
| MoSePS19 | CACTGACAGGATTCAGTGACTTTCTCCT | ||
| Sepp1U40S | WS1004 | TCCTAGATTGGCAGAGGATAGAATGAA | mutant: 198 wildtype: 116 |
| WS1005 | TCAGAAACACCTTCCAACTGTAATGC | ||
| megalin | G20 | GCACATTTGGCCAGCCAAGG | wildtype: 275 (G20/G21) |
| G21 | CATATCTTGGAAATAAAGCGAC | ||
| S175 | GATTGGGAAGACAATAGCAGGCATGC | homozygote: 325 (G21/S175) |
Sepp1 iJ240-361/+ pairs were mated to produce Sepp1 iJ240-361/iJ240-361 mice (17). This strain and the Sepp1U40S/U40S mice yielded mutant plasma Sepp1 forms that were studied for redox activity.
Mice were housed in plastic cages. The light:dark cycle was 12 h:12 h. Pelleted diet and tap water were provided ad libitum. Experimental diets were formulated by Harlan-Teklad to our specifications (20). The diets were Torula yeast-based and were supplemented with selenium as sodium selenite. The basal (selenium-deficient) form of this experimental diet contained less than 0.01 mg selenium/kg (n=7). Sodium selenite was added to the basal diet during mixing to give added selenium concentrations of 0.25 mg/kg (selenium-adequate or control diet) and 1 mg/kg (high-selenium diet for Sepp1Δ240-361/Δ240-361 mice lacking the selenium-transporting C-terminal domain of Sepp1). The Vanderbilt University Institutional Animal Care and Use Committee approved studies conducted at Vanderbilt.
Study of Sepp1 forms and Gpx3 present in the urine of megalin−/− mice
Megalin−/− and megalin+/+ mice were housed in metabolic cages with free access to food and water. Urine was collected on ice and stored at −80°C. Pooled urine samples from megalin−/− mice and pooled urine samples from megalin+/+ mice were dialyzed overnight against PBS at 4°C to remove small-molecule selenium compounds. Dialyzed urine samples were centrifuged for 15 min at 1200 g and passed through a 0.2 micron syringe filter. The filtrate was applied to a monoclonal antibody (9S4 against Sepp1) column (13) or to a polyclonal-antibody (against Gpx3) column (21). 9S4-binding Sepp1 fragments were eluted from the 9S4 column and Gpx3 was eluted from the anti-Gpx3 column with 0.1 M glycine, pH 2.5. The flow-through fractions that did not bind to the columns were saved for further study.
In a separate experiment, a tracer dose of 75Se-labeled selenite (15 μCi) was injected intraperitoneally into each mouse. 75Se-labeled urine was collected for 24 h following the injection. After urine collection, each mouse was anesthetized with isoflurane and blood was collected from the inferior vena cava and treated with disodium EDTA (1 mg/ml) to prevent coagulation. Plasma was separated by centrifugation and stored at −80°C.
Radioactive urine proteins were separated by SDS-PAGE. The 15% gel was stained with Coomassie blue and dried. The dried gel was exposed to Kodak BioMax film for detection of 75Se-labeled proteins. Non-radioactive urine was separated on a 10% Bis-Tris NuPAGE gel (Invitrogen) for mass spectrometric analysis.
Selenium content of dialyzed urine and of proteins eluted from 9S4 and anti-Gpx3 columns was determined. The protein-containing solutions obtained from the columns were concentrated using an Amicon Ultra centrifugal filter with molecular weight cut off of 3 kDa (Millipore Corporation).
Mass spectrometry of selenoproteins
Native and deglycosylated Sepp1 protein forms that had been purified from urine and plasma were resolved via SDS-PAGE. The gel was stained with colloidal Coomassie and bands of interest were excised from the gel. Bands were sliced into 1 mm3 gel pieces, and proteins were reduced with dithiothreitol, carbamidomethylated with iodoacetamide, and in-gel digested with trypsin or Glu-C using methods described in (22). Also, as previously described (22), extracted peptides were reconstituted in 0.1% formic acid and were analyzed by LC-MS/MS using an Eksigent NanoLC and Autosampler and a Thermo Scientific LTQ Orbitrap XL or LTQ Orbitrap Velos mass spectrometer. Briefly, the instruments were operated using a data-dependent method with dynamic exclusion enabled. Full scan (m/z 300-2000 or 400-2000) spectra were acquired with the Orbitrap as the mass analyzer (resolution 60,000), and the top 5 (LTQ Orbitrap XL) or top 12 (LTQ Orbitrap Velos) most abundant ions in each MS scan were selected for fragmentation in the LTQ. An isolation width of 2 m/z, activation time of 30 ms (LTQ Orbitrap XL) or 10 ms (LTQ Orbitrap Velos), and 35% normalized collision energy were used to generate MS/MS spectra. For peptide identification, tandem mass spectra were searched against a Mus musculus subset of the UniprotKB (www.uniprot.org) protein database. Database searches were performed using a custom version of Sequest (Thermo Scientific) on the Vanderbilt ACCRE Linux cluster and results were assembled in Scaffold 3 (Proteome Software). All searches were configured to include variable modifications of oxidation on methionine and carbamidomethylation on cysteine or selenocysteine. All tandem mass spectra assigned to Sepp1 peptides were manually validated. Plasma Gpx3 was also in-gel digested with trypsin and Asp-N and analyzed by LC-MS/MS as described above.
Whole-body and tissue selenium determination experiments
At weaning, megalin−/− and megalin+/+ male mice were fed selenium-adequate diet (supplemented with 0.25 mg selenium/kg) and female mice were fed selenium-deficient diet (no selenium supplement). Four weeks after weaning, the male mice were anesthetized with isoflurane and blood was removed from the inferior vena cava with a syringe and needle. Female selenium-deficient mice tolerated the selenium deficiency without observable debility and were studied 12 weeks after weaning. Blood was treated with disodium EDTA (1 mg/ml) to prevent coagulation. An aliquot of whole blood was taken for selenium assay and the remainder of the blood was centrifuged. Plasma was frozen for assay of selenium biomarkers. Liver, kidney, muscle, testis, and brain were harvested and frozen in liquid nitrogen. The remaining carcass was frozen in liquid nitrogen. Plasma and tissues were stored at -80°C. Whole-body selenium concentration was calculated as the sum of blood, tissue, and carcass selenium contents divided by body weight.
Biochemical measurements
Plasma Sepp1 concentration was determined using an ELISA (17). Gpx activity was determined using a coupled assay with 0.25 mM hydrogen peroxide as substrate (23). Selenium was measured using a modification (24) of the fluorometric assay of Koh and Benson (25). The limit of selenium detection by this assay is 1 ng. Protein concentration was carried out using bicinchoninic acid reagents (Pierce) except as otherwise indicated.
Protein purification
A 9S4 monoclonal antibody column was used to purify Sepp1 forms from plasma and urine. The starting material was plasma from C57BL/6-congenic Sepp1+/+, Sepp1 Δ240-361/Δ240-361, and Sepp1U40S/U40S mice (male and female) and urine from megalin−/− and megalin+/+ mice (male and female). Gpx3 was purified from megalin−/− urine using an anti-Gpx3 column. The column-bound proteins were washed sequentially with PBS and 1 M NaCl in PBS. Then proteins were eluted with 0.1 M glycine, pH 2.5. Glycine-eluted samples were immediately neutralized with 1 M Tris, pH 9.0. The samples were concentrated and buffer-exchanged against PBS using Amicon Ultra centrifugal filters with a 30 kDa molecular weight cut off. Protein concentrations of the purified Sepp1 variants were determined by absorbance at 280 nm using theoretical extinction coefficients for the proteins, e.g., 27,765 M−1cm−1 for full-length Sepp1.
Immunofluorescence Microscopy
Mouse kidneys were fixed 1 h at 4°C with 4% formaldehyde in 0.1 M sodium phosphate, pH 7.4. They were then rinsed in phosphate buffer and infiltrated overnight at 4°C in phosphate buffer containing 20% sucrose. The tissue was immersed in Optimal Cutting Temperature Compound (Fisher Scientific), frozen in liquid nitrogen, and stored at −80°C. Cryosections were rinsed with TBST (20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 0.05% Tween 20, 0.025% sodium azide) and blocked with TBST containing 1% bovine serum albumin and 0.1% glycine for 1 hour. Sections were then incubated with rat monoclonal anti-mouse Sepp1 (9S4) and/or rabbit polyclonal anti-megalin primary antibodies (a gift of Dr. Daniel C. Biemesderfer of Yale University) in blocking solution for 1 h at room temperature. Control sections were incubated with equivalent levels of non-immune rat or rabbit IgG. Sections were washed 3 times for 5 minutes each in TBST and incubated for 1 h with affinity-purified secondary antibodies diluted in blocking solution. These included Cy3-conjugated F(ab’)2 mouse anti-rat IgG (Jackson Immunoresearch Laboratories) and Alexa 488-goat anti-rabbit IgG (Invitrogen). Hoechst 33258 was added to the secondary antibody solution. For double antibody staining, additional controls validated that the secondary antibodies only bound to the appropriate primary antibody. Slides were washed 3 times for 5 min with TBST and were mounted in Fluoromount G (Fisher Scientific). Specimens were examined by phase contrast and fluorescence microscopy and images of experimental and control specimens were obtained using identical photographic exposures. The immune-staining experiment was repeated using three animals and the images shown are representative of all replicates.
Sepp1 enzymatic measurements
To determine TrxR1-coupled Sepp1 redox activities, Trx1 or the various forms of Sepp1 (at concentrations of 1 or of 2 μM) were incubated in microwell plates together with 30 nM recombinant rat TrxR1, 200 μM NADPH, and 0.25 or 2.5 mM H2O2, 5 or 10 mM t-BuOOH, or 200 μM insulin as the terminal electron acceptor in a final volume of 100 μL. All assays were performed in 50 mM Tris-HCl, pH 7.5, containing 2 mM EDTA. The reactions were carried out at approximately 22°C and were initiated by the addition of NADPH, whereupon absorbance at 340 nm was followed for either 10 or 30 min. The NADPH extinction coefficient (6,220 M−1cm−1) was used to calculate moles of NADPH consumed. We used the following approximations of molecular weights to convert protein values to μmoles: 40,692 Da for Sepp1, 27,053 Da for Sepp1 Δ240-361, 40,629 Da for Sepp1U40S, and 23,436 Da for Sepp1UF. These values are the theoretically maximal peptide molecular weights for each of the Sepp1 forms. Reaction mixtures containing everything except the Sepp1 form or Trx1 served as background controls and rates obtained with them were subtracted from the rates presented. Each assay was performed at least twice.
Results
Characterization of selenium distribution in megalin−/− mice
In figure 1A, megalin is present on the apical membranes of renal PCT cells and Sepp1 is present inside renal PCT cells near the glomerulus but not in all megalin-containing PCT cells (presumably not in the more distal ones). In megalin−/− mice neither megalin nor Sepp1 can be seen in any PCT cells (Fig 1B). The absence of Sepp1 forms from the more distal PCT cells strongly suggests that Sepp1 forms are completely removed from the glomerular filtrate by megalin-mediated endocytosis and imply that deletion of megalin would lead to excretion of the filtered Sepp1 forms in the urine.
1.
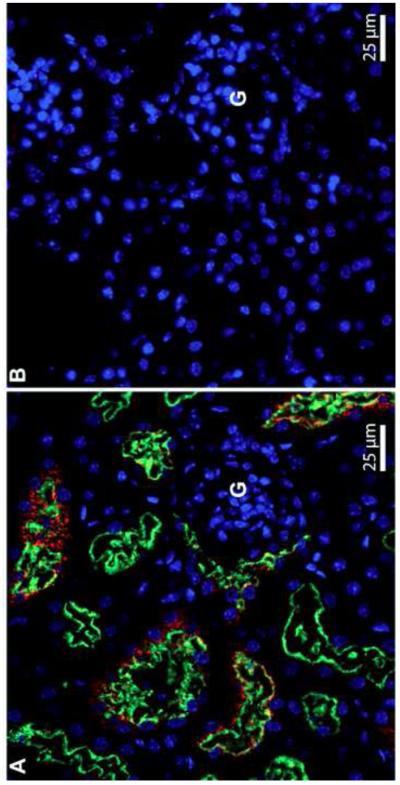
Association of Sepp1 forms (red) with megalin (green) in mouse kidney PCT cells. Panel A depicts megalin+/+ kidney cortex and panel B depicts megalin−/− kidney cortex. G indicates a glomerulus. Cell nuclei are blue.
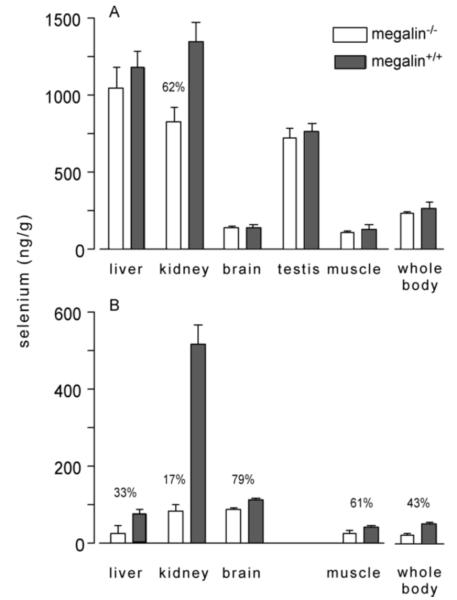
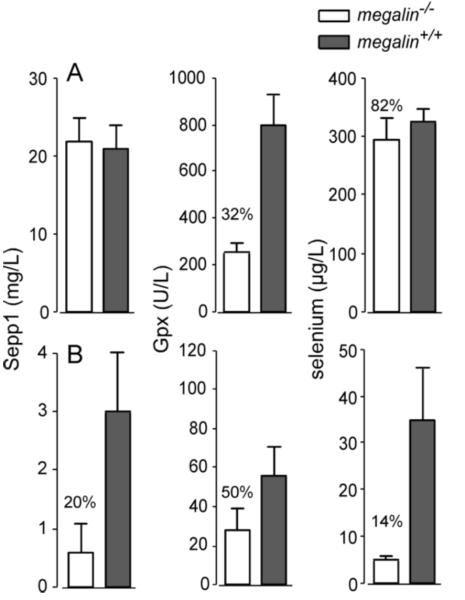
Megalin−/− male mice fed selenium-adequate diet (0.25 mg selenium/kg) maintained a whole-body selenium concentration indistinguishable from that of megalin+/+ mice (Fig 2A). However, kidney selenium concentration fell to 62% of that in megalin+/+ mice and plasma Gpx activity fell to 32% (Fig 3A). These findings indicate that loss of Sepp1 and other selenium-containing proteins in the urine caused by deletion of megalin (shown below) did not lead to selenium deficiency in the mouse when adequate selenium (approximately twice the mouse dietary selenium requirement of 0.1-0.15 mg/kg) was supplied by the diet, but it did cause localized selenium deficiency in the kidney. Gpx3 is synthesized in kidney PCT cells (26) and secreted into the plasma, so loss of Sepp1 uptake by PCT cells in megalin−/− mice likely impaired their ability to synthesize Gpx3 and, thus, lowered plasma Gpx activity.
2.
Whole-body and tissue selenium concentrations in megalin−/− and megalin+/+ mice fed (A) selenium-adequate and (B) selenium-deficient diets. Weanling male mice were fed selenium-adequate diet (supplemented with 0.25 mg selenium/kg) for 4 weeks before tissues were taken for analysis. Weanling female mice were fed selenium-deficient diet for 12 weeks before tissues were taken for analysis. Values are means with 1 S.D. indicated by the half bracket, n=5. Pairs were compared using Student’s t-test and significant differences (p<0.05) are indicated by percentages.
3.
Plasma selenium biomarkers in megalin−/− and megalin+/+ mice fed (A) selenium-adequate diet for 4 weeks and (B) selenium-deficient diet for 12 weeks beginning at weaning. Mice were the same ones presented in figure 2. Values are means with 1 S.D. indicated by half brackets, n=4-5. Pairs were compared using Student’s t-test and significant differences (p<0.05) are indicated by percentages.
To assess the effect of megalin when selenium supply is limiting, we studied selenium deficiency in megalin−/− mice. Female mice were used for this experiment because we were able to produce only a limited number of megalin−/− mice and the male mice that had been produced had been used in other experiments. Figure 2B shows that megalin−/− female mice had a lower whole-body selenium concentration than megalin+/+ female mice when both had been fed selenium-deficient diet for 12 weeks. Selenium concentrations of other tissues were lower in the megalin−/− mice than in the megalin+/+ mice with the kidney level being disproportionately lower than selenium levels in other tissues. All plasma selenium biomarkers were depressed by deletion of megalin in selenium-deficient mice (Fig 3B). Weight gain was not affected in selenium-deficient mice by megalin deletion (not shown) and no obvious neurological impairment was observed in the selenium-deficient megalin−/− mice, although detailed neurological testing was not performed. These findings confirm that loss of megalin, with consequent excretion of Sepp1 forms and other selenium-containing proteins in the urine, exacerbates dietary selenium depletion in tissues (15) with the greatest effect being in kidney.
Appearance of Sepp1 fragments and other selenium-containing proteins in megalin−/− urine
Because PCT cell uptake of Sepp1 from the glomerular filtrate was not detected in megalin−/− mice (Fig 1B), we sought Sepp1 forms in mouse urine after the mice had been labeled with tracer doses of 75Se. Figures 4A and 4B show, respectively, an autoradiogram and a Coomassie blue stain of the same gel of urine samples. Lanes 1 and 3 were loaded with urine from megalin+/+ and megalin−/− mice, respectively. Several bands of 75Se are visible in lane 3 but no 75Se bands are seen in lane 1. The urine samples were dialyzed overnight against PBS to remove small-molecule 75Se and loaded onto lanes 2 and 4 according to the amount of 75Se present in each (see figure legend). In lane 2, no distinct 75Se bands are present but the predominant protein band at ~20 kDa was greatly accentuated (Fig 4B) because a greater amount of sample was applied than in lane 1. That protein band presumably represents ‘mouse urinary protein,’ a normal constituent of mouse urine (27). 75Se bands are present in lane 4 at the same positions as bands in lane 3, indicating that the 75Se in those bands was non-dialyzable. These results confirm that proteins containing selenium were present in the urine of megalin−/− mice but were not detectable in the urine of megalin+/+ mice (15).
4.
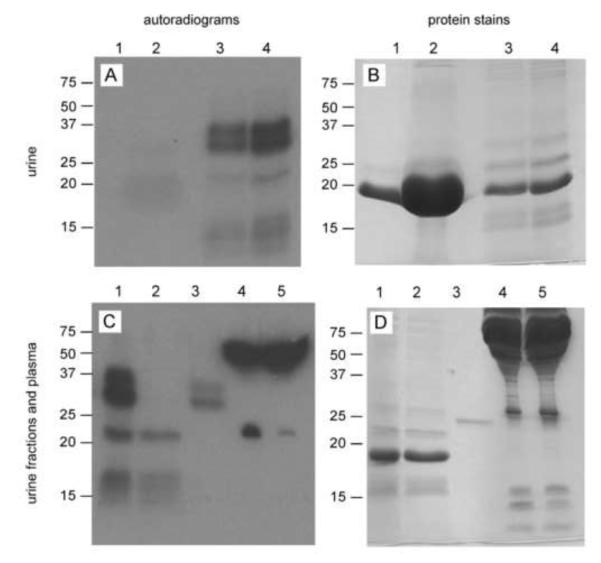
Selenoproteins in the urine of megalin−/− and megalin+/+ mice. Panels A and C are autoradiograms of SDS-PAGE gels of urine and plasma samples from 75Se-labeled megalin−/− and megalin+/+ mice. Panels B and D are the Coomassie stains of the respective gels depicted in panels A and C. Lane 1 in panels A and B was loaded with 1 μl megalin+/+ urine (1660 cpm 75Se) and lane 2 with 20 μl of the same urine after dialysis (1500 cpm 75Se). Lane 3 was loaded with 5 μl megalin−/− urine (2160 cpm 75Se) and lane 4 with 10 μl of the same urine after dialysis (1880 cpm 75Se). Lane 1 in panels C and D was loaded with 10 μl dialyzed megalin−/− urine (2060 cpm 75Se). The dialyzed megalin−/− urine was passed over a monoclonal antibody (9S4) column that binds the N-terminal domain of Sepp1. The flow through was loaded onto lane 2 and the bound fraction was loaded onto lane 3. Lanes 4 and 5 were each loaded with 1 μl plasma from megalin+/+ (1890 cpm 75Se) and megalin−/− (2170 cpm 75Se) mice, respectively.
The nature of the urinary proteins containing 75Se was investigated. Figures 4C and 4D are, respectively, an autoradiogram and a Coomassie blue stain of a gel containing fractions of the 75Se-labeled megalin−/− urine along with plasma from megalin+/+ and megalin−/− mice. Lane 1 contains dialyzed urine from a megalin−/− mouse (same sample as lane 4 in Figs 4A & 4B). The dialyzed megalin−/− mouse urine was passed over a monoclonal antibody (9S4) column that binds the N-terminal domain of Sepp1. The flow-through of that column is seen in lane 2 and the material that bound to the column beads is seen in lane 3. Two 75Se bands are present in lane 3 between the 25 and 37 kDa markers, indicating the presence of N-terminal Sepp1 forms smaller than the full-length plasma Sepp1 seen to migrate at 50 kDa (lanes 4 & 5). A 75Se band is present at 22 kDa in the flow through (lane 2) and corresponds to the 75Se band representing Gpx3 in plasma (lanes 4 & 5). In a separate experiment (not shown), the 22 kDa 75Se band in urine bound to an anti-Gpx3 column. An additional broad 75Se band that did not bind to either antibody column is present between approximately 12 and 17 kDa (lanes 1 & 2, Fig 4C). This band corresponded to visible protein staining (lanes 1 & 2, Fig 4D), indicating that this 75Se is present in a relatively large amount of protein. This 75Se band was not characterized in detail. These findings demonstrate that Sepp1 forms containing the N-terminal domain and Gpx3 forms are present in the urine of megalin−/− mice.
The selenium content of Sepp1 and Gpx3 forms isolated from megalin−/− urine was determined. After dialysis, centrifugation, and filtration, urine was passed over antibody columns for purification of Sepp1 and Gpx3. In duplicate experiments, eluted Sepp1 fractions contained 17% and 18% of the selenium in pre-column urine (Table 3). Selenium content per protein molecule was calculated to be 0.67 and 0.62 atoms based on Sepp1 molecules with termination after residue 207 (peptide weight of 23,436 Da) (Fig 5). Selenium eluted from the anti-Gpx3 column was below 0.02% of that in pre-column urine (Table 3). These findings indicate that a significant fraction of dialyzed megalin−/− urine selenium—more than 18% when purification yield is taken into consideration—was present as N-terminal Sepp1 forms but that Gpx3 forms, while detected when Gpx3 was labeled by 75Se (Fig 4C), accounted for very little selenium. A further conclusion that can be reached from these experiments is that most of the protein-bound selenium in the megalin−/− urine was present in proteins other than Gpx3 and Sepp1 forms with an intact N-terminal domain.
Table 3.
Selenium in selenoproteins purified from megalin−/− urine (replicates of the experiment)
| seleno- protein |
% of the selenium in dialyzed urine |
ng selenium/mg protein |
nmol selenium/nmol protein (23,436 Da) |
|---|---|---|---|
| Sepp1UF | 17, 18 | 2264, 2090 | 0.67, 0.62 |
| Gpx3 | >0.02, >0.02 | -- | -- |
5.
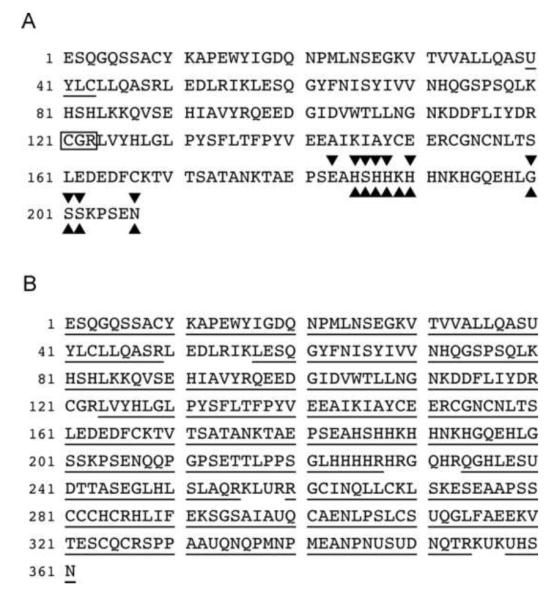
Mass spectrometric analysis of (A) 9S4-binding Sepp1 fragments from megalin−/− urine and (B) Sepp1 purified from plasma. Panel A presents the combined coverage of urinary Sepp1 fragments in the 2 Sepp1 bands between the 37 and 25 kDa markers in lane 3 of figure 4C. The most C-terminal amino acid detected in the upper band was residue 207 and in the lower band it was residue 202. All the amino acid residues shown, except for the 3 in the box, were identified by mass spectrometry. Non-tryptic terminations ( over the terminal residue) in a tryptic digest and non Glu-C terminations (⑩ under the terminal residue) in a Glu-C digest were detected, indicating that the 2 bands were mixtures of Sepp1 fragments. The thioredoxin-like putative active site at residues 40-43 is underlined. Panel B presents underlined coverage (93.6%) of Sepp1 purified from C57BL/6 mouse plasma.
over the terminal residue) in a tryptic digest and non Glu-C terminations (⑩ under the terminal residue) in a Glu-C digest were detected, indicating that the 2 bands were mixtures of Sepp1 fragments. The thioredoxin-like putative active site at residues 40-43 is underlined. Panel B presents underlined coverage (93.6%) of Sepp1 purified from C57BL/6 mouse plasma.
Mass spectrometric characterization of selenoprotein gel bands from megalin−/− urine
The two bands of urinary Sepp1 forms, positions of which are depicted in lane 3 of figure 4C, were analyzed by LC-MS/MS after digestion with trypsin and with Glu-C (Fig 5A). Coverage of the upper band ended with residue 207 and coverage of the lower band ended with residue 202. These collective fragments have been given the designation Sepp1UF (The UF superscript signifies urinary fragments.).
In further analyses of the mass spectrometric results, we sought peptide termination sites not expected to occur with the proteases used for digestion. Figure 5A shows that 10 non-tryptic termination sites were identified in the tryptic digest. They occurred between residues 183 and 208. All the same termination sites were found in the Glu-C digest. However, the non-tryptic termination found after residue 183 was at a Glu-C cleavage site and therefore was expected in the Glu-C digest. In addition, a non Glu-C termination site identified after residue 189 was at a tryptic cleavage site and therefore was expected in the trypsin digest. Thus, the Sepp1 fragments terminate at 11 different sites within a 25-residue stretch. These results indicate that at least 11 N-terminal Sepp1 fragments are excreted in the urine of megalin−/− mice, likely accounting for the heterogeneity of the Sepp1UF observed in figure 4C.
After isolation from C57BL/6 mouse plasma with a monoclonal antibody (9S4) column and SDS-PAGE, native Sepp1 and its deglycosylated form were subjected to LC-MS/MS analysis. Figure 5B shows that 94% coverage was achieved, indicating that full-length Sepp1 was present in mouse plasma. It was not detected in megalin−/− urine, however.
The band identified as Gpx3 by antibody binding was further characterized by LC-MS/MS after trypsin and Asp-N digestions. Coverage of the Gpx3 amino acid sequence was 82%, extending to within 3 residues of the predicted N terminus and 2 residues of the predicted C terminus (Fig 6). These findings suggest that the form of Gpx3 present in the urine was full length.
6.

Mass spectrometric analysis of protein purified from megalin−/− mouse urine using a Gpx3- binding polyclonal antibody column. Digestions were carried out with trypsin and with Asp- N before analysis by mass spectrometry. Amino acids detected by mass spectrometry are underlined.
Purified Sepp1U40S produced 3 bands on SDS-PAGE (lane 3 in Fig 7) compared to one band produced by Sepp1 (lane 1 in Fig 7). The top Sepp1U40S band migrated the same distance as wild-type Sepp1. The other 2 bands migrated farther, suggesting that those proteins had less mass than the protein in band one.
7.
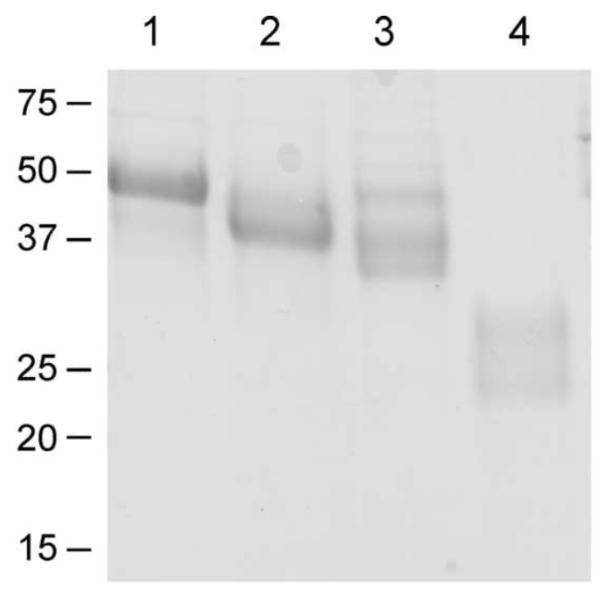
Purified Sepp1 forms used to evaluate redox activity. Sepp1 from C57BL/6 mouse plasma is in lane 1; Sepp1Δ240-361 from Sepp1Δ240-361/Δ240-361 mouse plasma is in lane 2; Sepp1U40S from Sepp1U40S/U40S mouse plasma is in lane 3; and Sepp1UF from megalin−/− mouse urine is in lane 4. All proteins were purified using a Sepp1-binding monoclonal antibody (9S4) column. The SDS-PAGE was performed on a 15% polyacrylamide gel and 4 μg of each protein preparation was loaded. The proteins were stained with Coomassie blue.
Mass spectrometric analysis of all 3 Sepp1U40S bands showed that serine was present at residue 40 in place of the selenocysteine present in wild-type Sepp1. The bands that migrated farther were forms with C-terminal truncations (results not shown). Thus, replacing the selenocysteine at residue 40 with serine causes truncation of some, but not all, Sepp1U40S molecules. More detailed characterization of the Sepp1U40S mice and the Sepp1U40S protein will be presented in a future report.
Redox activity of Sepp1 forms purified from plasma and megalin−/− urine
Because the putative Sepp1 active site has a thioredoxin-like redox motif with selenocysteine replacing one of its cysteines (Fig 5), all our Sepp1 forms (Fig 7) were studied in a thioredoxin activity assay system (28). NADPH and TrxR1 were used to reduce the Sepp1 form; H2O2, t-BuOOH, and insulin were used as terminal electron acceptors.
The Sepp1 forms with the redox motif intact had significant peroxidase activities that were similar on a molar basis, but Sepp1U40S, the form with its presumed redox motif disrupted, had essentially no activity (Figs 8A & 8B). These findings indicate that the Sepp1 forms present in vivo can serve as substrates of TrxR1. Moreover, these forms have peroxidase activity that is dependent on their selenocysteine-containing redox motif.
8.
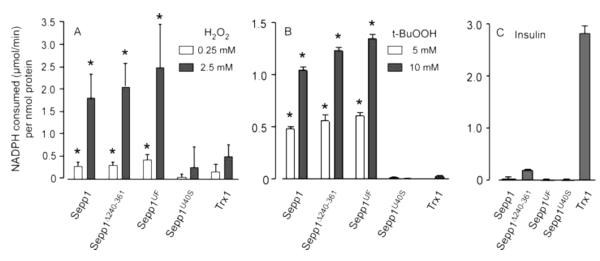
Sepp1 forms containing selenocysteine at residue 40 are substrates of TrxR1 and display peroxidase activity (A & B) but not disulfide reductase activity (C). Bars represent means with 1 S.D. indicated by a bracket. Asterisks indicate values significantly greater (p<0.05) than the respective values (at each H2O2 or t-BuOOH concentration) of the Sepp1U40S protein as assessed by ANOVA followed by Tukey’s post hoc test. The disulfide reductase activities with all forms of Sepp1 (C) were significantly different (p<0.05) from the activity with Trx1.
Trx1, the usual TrxR1 substrate, had low direct peroxidase activity in this assay system (Figs 8A & 8B). However, it reduced disulfides in insulin, a reaction that was not supported by the Sepp1 forms (Fig 8C). Thus, even though the in vivo Sepp1 forms are substrates of TrxR1, their enzymatic activities are different from those of Trx1.
Discussion
This report demonstrates that Sepp1UF is an in vivo form of Sepp1. Its diversion in megalin−/− mice from endocytosis by renal PCT cells to urinary excretion implies that it enters the urinary stream in the glomerulus via filtration of plasma.
Properties and formation of Sepp1UF
Mass spectrometric analysis of Sepp1UF revealed that it consists of 11 N-terminal Sepp1 fragments (Fig 5A). Six of the terminations occur after residues 185-190 and 3 occur after residues 200-202. Thus, 9 of the 11 terminations occur in 2 “hot spots” and the other 2 terminations are nearby. This finding can be explained by proteolytic processing of Sepp1 being the origin of Sepp1UF. It is also possible that some of the termination sites are produced by removal of amino acid residues from longer forms after proteolysis at one or more sites.
Sepp1 has several properties that might influence such proteolytic processing. It binds to cells at 2 sites. One site depends on its heparin-binding property (29,30) and that site is likely to be the heparin-binding site in the N-terminal domain (residues 79-86) (31). The other site is an apoER2-binding site in the C-terminal domain (30). It can be speculated that binding of Sepp1 to cells would make the Sepp1UF termination sites accessible to proteases. Also, because the C-terminal domain associates with the receptor apoER2, proteolytic cleavage might free that domain for endocytosis via apoER2 (32), leaving the N-terminal domain in the extracellular space.
Two highly basic stretches of Sepp1 sequence might also play roles in Sepp1UF production. Seven of the 11 Sepp1UF termination sites are present within or adjacent to a cluster of 8 basic amino acids (within the stretch of residues 185-194) and the other 4 are slightly downstream between residues 200 and 208 (Fig 5A). A second cluster of 10 basic amino acids (within the stretch of residues 223-236) is present downstream of the Sepp1UF termination sites (Fig 5B). Two similar stretches of basic amino acids—mostly histidines—are conserved among species and are potential sites for association of Sepp1 with other proteins, e.g., proteases. This speculative proteolytic process could underlie the presence of Sepp1UF in the urine of megalin−/− mice. Additional work will be necessary to determine the fate of the Sepp1 C-terminus.
A German group observed that the urine of megalin-mutant (Lrp2267/267) mice, in which most of the extracellular domain of megalin was missing, contained Sepp1 forms (15). Using western blots of electrophoresis gels, they observed Sepp1 forms migrating where Sepp1UF migrates. However, they did not characterize those Sepp1 forms further. They also observed a Sepp1 band that migrated with plasma Sepp1. We did not detect such full-length (plasma) Sepp1 in megalin−/− mouse urine (Figs 4 & 5A) and, moreover, would not expect it to be filtered in the glomerulus because of its large size (33). These differences between our findings and those of the German group might stem from a difference between Lrp2267/267 mice and megalin−/− mice, although both were on similar mixed (FVB/NJ and C57BL/6) backgrounds.
Significance of megalin-mediated uptake of Sepp1UF and other selenium-containing proteins by renal PCT cells
Selenium is transferred from the liver to other tissues by receptor-mediated endocytosis of Sepp1. While the receptor apoER2 is responsible for Sepp1 uptake by other tissues (30), kidney PCT cells take up Sepp1 forms from the glomerular filtrate via megalin-mediated endocytosis (13). The present report identifies Sepp1UF as a form of Sepp1 taken up in this manner (Fig 5A).
By facilitating PCT cell uptake of selenium-containing proteins, megalin prevents loss of selenium from the body, supporting tissue selenium levels when dietary selenium intake is low (Fig 2B) (15). Moreover, the PCT cells utilize selenium taken up in this manner to synthesize Gpx3, which is secreted into the plasma (26). Thus, megalin plays a role in whole-body selenium metabolism.
In addition to Sepp1UF, Gpx3 and unidentified selenium-containing polypeptides appeared in the urine of megalin−/− mice (Fig 4). Thus, PCT cells take up a number of selenium-containing proteins, not just Sepp1UF. The unidentified selenium-containing proteins account for most of the selenium in dialyzed urine (Table 3). Therefore, the role of megalin in selenium metabolism is broader than facilitating uptake of Sepp1UF from the glomerular filtrate. It is even possible that C-terminal fragments of Sepp1 are among the selenium-containing proteins that bind megalin and are taken up by PCT cells. Further insights into whole-body selenium metabolism should be gained by identification of additional selenium-containing proteins in megalin−/− urine.
The results of this study allow us to compare and contrast the functions of the Sepp1 receptors apoER2 and megalin. ApoER2 binds to a Sepp1 site in its C terminus and therefore does not bind to Sepp1Δ240-361, which lacks the C-terminal domain (30,34). Megalin, which is a cargo receptor that binds many proteins, appears to bind a number of selenium-containing proteins, including Gpx3 and the Sepp1 N-terminal domain forms Sepp1Δ240-361 (13) and Sepp1UF. It is also possible that megalin binds the C-terminal domain of Sepp1 but more work will be needed to assess that. In addition, we did not assess the possibility that selenium forms bind to cubilin, a ligand-binding protein that is endocytosed through its association with megalin (35).
Peroxidase activity of in vivo Sepp1 forms
Human SEPP1 has been reported to have no Gpx activity with the substrates H2O2 and t-BuOOH and very low activity, relative to Gpx4, with a phospholipid hydroperoxide substrate (36). This finding of limited Gpx activity and our recognition that the putative Sepp1 active site at residues 40-43 resembles a thioredoxin active site led us to examine activities of mouse Sepp1 forms when they were coupled with NADPH and TrxR1 instead of with GSH. We found that both in vivo Sepp1 forms, Sepp1 and Sepp1UF, have significant peroxidase activity with the substrates H2O2 and t-BuOOH in the thioredoxin assay system (Fig 8), raising the possibility that they function as peroxidases in vivo.
Because Sepp1 and Sepp1UF are extracellular proteins and TrxR1 is an intracellular protein, it is not clear how TrxR1 could reduce oxidized Sepp1 forms, thereby enabling them to function as peroxidases. However, evidence has been presented recently that TrxR1 can be induced to migrate to the exofacial aspect of the cell membrane under stress conditions and to reduce substrates outside the cell using reducing equivalents from intracellular NADPH (37). Sepp1 forms bind to cells through their heparin-binding properties (30) and that would position them near the cell membrane, possibly allowing interaction with membrane-associated TrxR1 and its intracellular NADPH supply. Such a mechanism, while still speculative, might serve to protect the cell membrane from injury by peroxides in the extracellular space. Alternatively, Sepp1 forms might also reduce peroxides in a non-catalytic manner—but that type of activity would be inefficient compared to coupling them with TrxR1.
The role of selenocysteine in accelerating thiol/disulfide exchange reactions is complex (38). The presence of a selenocysteine in the active site of one of the redox partners usually accelerates the thiol/disulfide exchange. The reaction between Sepp1 (or Sepp1UF) and TrxR1 involves active sites that both contain selenocysteines; the occurrence of this reaction strongly suggests that a thiol-selenol can reduce a selenenylsulfide in an appropriate microenvironment. The lack of enzymatic activity of Sepp1U40S when coupled with TrxR1 indicates that the selenocysteine residue present in the other Sepp1 variants was involved in catalyzing the observed peroxidase activity.
Conclusions
We identified Sepp1UF as in vivo forms of Sepp1 that are suited to exert an antioxidant function. Sepp1UF lacks the selenium-rich C-terminal domain required for selenium transport but contains the redox-active site and the heparin-binding site. Although important details remain to be clarified, the finding that Sepp1 and Sepp1UF function as peroxidases is a potential explanation for the antioxidant properties of Sepp1. We demonstrated that the selenocysteine-containing redox motif at residues 40-43 is required for this activity and conclude that it is the active site that is reduced by TrxR1 and, in turn, reduces the peroxide substrate.
The demonstration that multiple selenium-containing proteins appear in the urine of megalin−/− mice and that megalin binds the N-terminal domain of Sepp1 forms differentiates megalin function from that of apoER2. Megalin facilitates the endocytosis of many filtered proteins, including several selenium-containing ones. ApoER2 is not known to bind any selenium-containing proteins except Sepp1 forms containing the selenium-rich C-terminal domain. Thus, apoER2 functions to supply selenium to extra-hepatic tissues and megalin functions to prevent the loss of small selenium-containing proteins in the urine.
Highlights.
- Selenoprotein P (Sepp1) has in vivo antioxidant properties by an unknown mechanism.
- A form of Seppl is filtered from plasma in the glomerulus and taken up by megalin.
- SepplUF, ll Seppl N-terminal fragments, is excreted in the urine of megalin−/− mice.
- SepplUF has peroxidase activity when coupled with thioredoxin reductase-l.
- SepplUF might protect cells from injury by extracellular peroxides.
Acknowledgments
The authors are grateful to Dr. Ulrich Schweizer of the University of Bonn (Germany) for advice that helped us to produce viable megalin−/− mice. The authors thank Teri D. Stevenson, Kristin V. Bradshaw (deceased), and Michelle L. Chatterton for animal husbandry.
National Institutes of Health Grants R37 ES002497, R01 DK082813, 5P30 DK058404, and 5P30 ES000267 supported this work. Funding for the Sepp1UF assay experiments carried out in Stockholm came from Karolinska Institutet, the Swedish Cancer Society, and the Swedish Research Council (Medicine). None of the supporting institutions participated in the design, performance, or interpretation of the experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Footnote
- apoER2
- apolipoprotein E receptor-2
- Gpx
- glutathione peroxidase
- LC-MS/MS
- liquid chromatography-coupled tandem mass spectrometry
- PBS
- phosphate-buffered saline
- PCT
- proximal convoluted tubule
- Sepp1
- selenoprotein P
- Sepp1UF
- selenoprotein P urinary fragments
- t-BuOOH
- tert-butyl hydroperoxide
- Trx1
- thioredoxin-1
- TrxR1
- thioredoxin reductase-1
- U
- selenocysteine
References
- 1.Burk RF, Hill KE. Selenoprotein P-Expression, functions, and roles in mammals. Biochim. Biophys. Acta. 2009;1790:1441–1447. doi: 10.1016/j.bbagen.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hill KE, Zhou J, McMahan WJ, Motley AK, Atkins JF, Gesteland RF, Burk RF. Deletion of selenoprotein P alters distribution of selenium in the mouse. J. Biol. Chem. 2003;278:13640–13646. doi: 10.1074/jbc.M300755200. [DOI] [PubMed] [Google Scholar]
- 3.Hill KE, Wu S, Motley AK, Stevenson TD, Winfrey VP, Capecchi MR, Atkins JF, Burk RF. Production of selenoprotein P (Sepp1) by hepatocytes is central to selenium homeostasis. J. Biol. Chem. 2012;287:40414–40424. doi: 10.1074/jbc.M112.421404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schomburg L, Schweizer U, Holtmann B, Flohé L, Sendtner M, Kohrle J. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem. J. 2003;370:397–402. doi: 10.1042/BJ20021853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosschaerts T, Guilliams M, Noel W, Herin M, Burk RF, Hill KE, Brys L, Raes G, Ghassabeh GH, De Baetselier P, Beschin A. Alternatively activated myeloid cells limit pathogenicity associated with African trypanosomiasis through the IL-10 inducible gene selenoprotein P. J. Immunol. 2008;180:6168–6175. doi: 10.4049/jimmunol.180.9.6168. [DOI] [PubMed] [Google Scholar]
- 6.Burk RF, Hill KE, Awad JA, Morrow JD, Kato T, Cockell KA, Lyons PR. Pathogenesis of diquat-induced liver necrosis in selenium-deficient rats. Assessment of the roles of lipid peroxidation by measurement of F2 isoprostanes. Hepatology. 1995;21:561–569. [PubMed] [Google Scholar]
- 7.Lobanov AV, Hatfield DL, Gladyshev VN. Reduced reliance on the trace element selenium during evolution of mammals. Genome Biol. 2008;9:R62. doi: 10.1186/gb-2008-9-3-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinbrenner H, Alili L, Bilgic E, Sies H, Brenneisen P. Involvement of selenoprotein P in protection of human astrocytes from oxidative damage. Free Radic. Biol. Med. 2006;40:1513–1523. doi: 10.1016/j.freeradbiomed.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 9.Kryukov GV, Gladyshev VN. Selenium metabolism in zebrafish: multiplicity of selenoprotein genes and expression of a protein containing seventeen selenocyteine residues. Genes Cells. 2000;5:1049–1060. doi: 10.1046/j.1365-2443.2000.00392.x. [DOI] [PubMed] [Google Scholar]
- 10.Ma S, Hill KE, Caprioli RM, Burk RF. Mass spectrometric characterization of full-length rat selenoprotein P and three isoforms shortened at the C terminus. Evidence that three UGA codons in the mRNA open reading frame have alternative functions of specifying selenocysteine insertion or translation termination. J. Biol. Chem. 2002;277:12749–12754. doi: 10.1074/jbc.M111462200. [DOI] [PubMed] [Google Scholar]
- 11.Stoytcheva Z, Tujebajeva RM, Harney JW, Berry MJ. Efficient incorporation of multiple selenocysteines involves an inefficient decoding step serving as a potential translational checkpoint and ribosome bottleneck. Mol. Cell. Biol. 2006;26:9177–9184. doi: 10.1128/MCB.00856-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saito Y, Sato N, Hirashima M, Takebe G, Nagasawa S, Takahashi K. Domain structure of bi-functional selenoprotein P. Biochem. J. 2004;381:841–846. doi: 10.1042/BJ20040328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olson GE, Winfrey VP, Hill KE, Burk RF. Megalin mediates selenoprotein P uptake by kidney proximal tubule epithelial cells. J. Biol. Chem. 2008;283:6854–6860. doi: 10.1074/jbc.M709945200. [DOI] [PubMed] [Google Scholar]
- 14.Nykjaer A, Dragun D, Walther D, Vorum H, Jacobsen C, Herz J, Melsen F, Christensen EI, Willnow TE. An endocytic pathway essential for renal uptake and activation of the steroid 25-(OH) vitamin D3. Cell. 1999;96:507–515. doi: 10.1016/s0092-8674(00)80655-8. [DOI] [PubMed] [Google Scholar]
- 15.Chiu-Ugalde J, Theilig F, Behrends T, Drebes J, Sieland C, Subbarayal P, Köhrle J, Hammes A, Schomburg L, Schweizer U. Mutation of megalin leads to urinary loss of selenoprotein P and selenium deficiency in serum, liver, kidneys and brain. Biochem. J. 2010;431:103–111. doi: 10.1042/BJ20100779. [DOI] [PubMed] [Google Scholar]
- 16.Xu J, Arnér ES. Pyrroloquinoline quinone modulates the kinetic parameters of the mammalian selenoprotein thioredoxin reductase 1 and is an inhibitor of glutathione reductase. Biochem. Pharmacol. 2012;83:815–820. doi: 10.1016/j.bcp.2011.12.028. [DOI] [PubMed] [Google Scholar]
- 17.Hill KE, Zhou J, Austin LM, Motley AK, Ham AJ, Olson GE, Atkins JF, Gesteland RF, Burk RF. The selenium-rich C-terminal domain of mouse selenoprotein P is necessary for supply of selenium to brain and testis but not for maintenance of whole-body selenium. J. Biol. Chem. 2007;282:10972–10980. doi: 10.1074/jbc.M700436200. [DOI] [PubMed] [Google Scholar]
- 18.Wu S, Ying G, Wu Q, Capecchi MR. A protocol for constructing gene targeting vectors: generating knockout mice for the cadherin family and beyond. Nature Protocols. 2008;3:1056–1076. doi: 10.1038/nprot.2008.70. [DOI] [PubMed] [Google Scholar]
- 19.Willnow TE, Hilpert J, Armstrong SA, Rohlmann A, Hammer RE, Burns DK, Herz J. Defective forebrain development in mice lacking gp330/megalin. Proc. Natl. Acad. Sci. U.S.A. 1996;93:8460–8464. doi: 10.1073/pnas.93.16.8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill KE, Zhou J, McMahan WJ, Motley AK, Burk RF. Neurological dysfunction occurs in mice with targeted deletion of selenoprotein P gene. J. Nutr. 2004;134:157–161. doi: 10.1093/jn/134.1.157. [DOI] [PubMed] [Google Scholar]
- 21.Olson GE, Whitin JC, Hill KE, Winfrey VP, Motley AK, Austin LM, Deal J, Cohen HJ, Burk RF. Extracellular glutathione peroxidase (Gpx3) binds specifically to basement membranes of mouse renal cortex tubule cells. Am. J. Physiol. Renal Physiol. 2010;298:F1244–F1253. doi: 10.1152/ajprenal.00662.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shonesy BC, Wang X, Rose KL, Ramikie TS, Cavener VS, Rentz T, Baucum AJ, 2nd, Jalan-Sakrikar N, Mackie K, Winder DG, Patel S, Colbran RJ. CaMKII regulates diacylglycerol lipase-alpha and striatal endocannabinoid signaling. Nature Neurosci. 2013;16:456–463. doi: 10.1038/nn.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence RA, Burk RF. Glutathione peroxidase activity in selenium-deficient rat liver. Biochem. Biophys. Res. Commun. 1976;71:952–958. doi: 10.1016/0006-291x(76)90747-6. [DOI] [PubMed] [Google Scholar]
- 24.Sheehan TMT, Gao M. Simplified fluorometric assay of total selenium in plasma and urine. Clin. Chem. 1990;36:2124–2126. [PubMed] [Google Scholar]
- 25.Koh TS, Benson TH. Critical re-appraisal of fluorometric method for determination of selenium in biological materials. J. Assoc. Off. Anal. Chem. 1983;66:918–926. [PubMed] [Google Scholar]
- 26.Avissar N, Ornt DB, Yagil Y, Horowitz S, Watkins RH, Kerl EA, Takahashi K, Palmer IS, Cohen HJ. Human kidney proximal tubules are the main source of plasma glutathione peroxidase. Am. J. Physiol. 1994;266:C367–C375. doi: 10.1152/ajpcell.1994.266.2.C367. [DOI] [PubMed] [Google Scholar]
- 27.Cheetham SA, Smith AL, Armstrong SD, Beynon RJ, Hurst JL. Limited variation in the major urinary proteins of laboratory mice. Physiol. Behavior. 2009;96:253–261. doi: 10.1016/j.physbeh.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Arnér ES, Holmgren A. Measurement of thioredoxin and thioredoxin reductase. Curr. Protoc. Toxicol. 2001 doi: 10.1002/0471140856.tx0704s05. Chapter 7:Unit 7.4.1-7.4.14 DOI: 10.1002/0471140856.tx0704s05; [DOI] [PubMed] [Google Scholar]
- 29.Arteel GE, Franken S, Kappler J, Sies H. Binding of selenoprotein P to heparin: Characterization with surface plasmon resonance. Biol. Chem. 2000;381:265–268. doi: 10.1515/BC.2000.034. [DOI] [PubMed] [Google Scholar]
- 30.Kurokawa S, Hill KE, McDonald WH, Burk RF. Long Isoform Mouse Selenoprotein P (Sepp1) Supplies Rat Myoblast L8 Cells with Selenium via Endocytosis Mediated by Heparin Binding Properties and Apolipoprotein E Receptor-2 (ApoER2) J. Biol. Chem. 2012;287:28717–28726. doi: 10.1074/jbc.M112.383521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hondal RJ, Ma S, Caprioli RM, Hill KE, Burk RF. Heparin-binding histidine and lysine residues of rat selenoprotein P. J. Biol. Chem. 2001;276:15823–15831. doi: 10.1074/jbc.M010405200. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Lu W, Marzolo MP, Bu G. Differential functions of members of the low density lipoprotein receptor family suggested by their distinct endocytosis rates. J. Biol. Chem. 2001;276:18000–18006. doi: 10.1074/jbc.M101589200. [DOI] [PubMed] [Google Scholar]
- 33.Burk RF, Gregory PE. Some characteristics of 75Se-P, a selenoprotein found in rat liver and plasma, and comparison of it with seleno-glutathione peroxidase. Arch. Biochem. Biophys. 1982;213:73–80. doi: 10.1016/0003-9861(82)90441-6. [DOI] [PubMed] [Google Scholar]
- 34.Burk RF, Olson GE, Hill KE, Winfrey VP, Motley AK, Kurokawa S. Maternal-fetal transfer of selenium in the mouse. FASEB J. 2013;27:3249–3256. doi: 10.1096/fj.13-231852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christensen EI, Birn H. Megalin and cubilin: multifunctional endocytic receptors. Nature Reviews. Mol. Cell Biol. 2002;3:256–266. doi: 10.1038/nrm778. [DOI] [PubMed] [Google Scholar]
- 36.Saito Y, Hayashi T, Tanaka A, Watanabe Y, Suzuki M, Saito E, Takahashi K. Selenoprotein P in human plasma as an extracellular phospholipid hydroperoxide glutathione peroxidase. J. Biol. Chem. 1999;274:2866–2871. doi: 10.1074/jbc.274.5.2866. [DOI] [PubMed] [Google Scholar]
- 37.Zhang G, Nitteranon V, Guo S, Qiu P, Wu X, Li F, Xiao H, Hu Q, Parkin KL. Organoselenium compounds modulate extracellular redox by induction of extracellular cysteine and cell surface thioredoxin reductase. Chem. Res. Toxicol. 2013;26:456–464. doi: 10.1021/tx300515j. [DOI] [PubMed] [Google Scholar]
- 38.Hondal RJ, Marino SM, Gladyshev VN. Selenocysteine in thiol/disulfide-like exchange reactions. Antioxid. Redox Signal. 2013;18:1675–1689. doi: 10.1089/ars.2012.5013. [DOI] [PMC free article] [PubMed] [Google Scholar]