Abstract
In mammals, X- and Y-encoded genes are transcriptionally shut down during male meiosis, but the expression of many of them is (re)activated, after meiosis, in spermatids. Postmeiotic XY gene expression is timely regulated by active epigenetic marks, which are de novo incorporated in the sex chromatin of spermatids, and by repressive epigenetic marks inherited from meiosis; alteration in this process leads to male infertility. In the mouse, postmeiotic XY gene expression is known to depend on genetic information carried by the male specific region of the Y chromosome long arm (MSYq). The MSYq gene Sly has been shown to be a key regulator of postmeiotic sex chromosome gene expression and necessary for the maintenance/recruitment of repressive epigenetic marks on the sex chromatin, but studies suggest that another MSYq gene may be required. The best candidate to date is Ssty, an MSYq multicopy gene of unknown function. Here, we show that SSTY proteins are specifically expressed in round and elongating spermatids and colocalize with the postmeiotic sex chromatin. Moreover, SSTY proteins interact with SLY protein and its X-linked homolog SLX/SLXL1, and may be required for the localization of SLX/SLY proteins in the spermatid nucleus and sex chromatin. Our data suggest that SSTY is a second MSYq factor involved in the control of XY gene expression during sperm differentiation. As Slx/Slxl1 and Sly genes have been shown to be involved in the XY intragenomic conflict which affects the offspring sex-ratio, Ssty might constitute another actor of this conflict.
Keywords: Sex chromosomes, spermatogenesis, chromatin, germ cells, intragenomic conflict
Introduction
The mammalian X and Y chromosomes are enriched in genes which are specifically expressed in the male germ cells and thus predicted to be important for spermatogenesis [1–3]. Their expression is under a precise epigenetic regulation throughout spermatogenesis: the sex chromosomes are transcriptionally active during spermatogonial stem cell proliferation, but expression of most X and Y genes (except microRNA [4]) is completely abolished at the pachytene stage of meiosis by a well-characterized epigenetic process, conserved among mammals, termed Meiotic Sex Chromosome Inactivation (MSCI) [5]. The silencing of the sex chromosomes is initiated by phosphorylation of the histone H2A variant X (H2AFX) by ATR, followed by MDC1-mediated spreading of this signal [6, 7], and subsequent recruitment onto the sex chromatin of repressive epigenetic marks [such as ubiquitinated H2A, trimethylation of the lysine 9 of histone H3 (H3K9me3) and CBX1 (aka HP1β)] which mediate gene silencing (for recent reviews see [8–10]). After meiosis, in haploid germ cells, many X and Y genes are reactivated or specifically activated [2, 11, 12]; their postmeiotic expression is regulated by a conjunction of active RNF8-dependent epigenetic marks and of repressive epigenetic marks inherited from MSCI [13–17].
Deletions of the male specific region of the mouse Y chromosome long arm (MSYq) cause spermiogenesis defects and male infertility associated with an upregulation of X and Y postmeiotic genes [18–24]. We have demonstrated that deficiency in Sly, an MSYq multicopy gene, recapitulates many of the phenotypes observed in MSYq-deleted males (i.e. sperm head abnormalities, decreased sperm motility, sperm DNA damage, abnormal chromatin packaging, etc.), and that SLY protein maintains postmeiotic sex chromosome gene expression at a “low” level [25, 26]. Indeed, in males knocked-down for Sly (thereafter termed Sly-deficient males), the recruitment/maintenance of repressive epigenetic modifications normally associated with the postmeiotic sex chromatin (PMSC) is altered and results in the upregulation of most of the X and Y genes that are (re)activated postmeiotically [25].
Importantly, males without MSYq region exhibit more severe sperm defects than Sly-deficient males, associated with a higher derepression of XY genes and loss of additional epigenetic marks [24–27]. This difference could be due to insufficient knockdown of Sly but we have observed that there is limited amount of SLY protein left in Sly-deficient males [25] (our unpublished data); it is therefore more likely that another MSYq gene contributes to the regulation of postmeiotic sex chromosome gene expression. Owing to the abundance of repetitive sequences on the mouse Y chromosome long arm, the annotation and characterization of MSYq genes is not completed. To date, MSYq is known to encompass four other multicopy genes (Ssty, Srsy, Asty and Orly). Asty and Orly do not have a coding potential [28, 29] (NCBI Gene, http://www.ncbi.nlm.nih.gov/gene) while there is no information on Srsy. The remaining gene, Ssty (Spermatid-specific transcripts Y-encoded), is present in two versions (Ssty1 and Ssty2) and repeated >100 times on MSYq. SSTY proteins are expressed during sperm development [30] and belong to the Spin domain proteins, one member of which has been shown to be a regulator of gene expression and to specifically recognize methylated lysine 4 of histone H3 in vitro [31].
In the present study, we investigate SSTY proteins in more details to gain insight into their function during the differentiation of postmeiotic male germ cells. Our work demonstrates that SSTY proteins are specifically present in round and elongating spermatids, colocalize with the postmeiotic sex chromatin (PMSC) and interact with SLY protein and its X-linked homolog SLX/SLXL1, which are known regulators of PMSC expression [32]. We also provide data suggesting that the localization of SLX/SLY protein to the spermatid nucleus and sex chromatin may depend on the presence of SSTY. All in all, these data are in favor of an important role of SSTY in the control of X and Y gene expression during sperm differentiation, and identify Ssty as a novel potential actor of the intragenomic conflict in which Slx/Sly genes have previously been shown to be involved [32].
Results
SSTY proteins are present in round and elongating spermatids
SSTY proteins are encoded by two highly related multicopy genes located on the Y long arm, Ssty1 and Ssty2 (84% identity at the nucleotide level, Fig. S1A). Ssty1 and Ssty2 genes have previously been estimated to be present on the mouse Yq in ~ 80 and 200 copies, respectively (for M. m. musculus Y chromosome [33]). Our recent Blast search of the NCBI database (National Center for Biotechnology Information, http://blast.ncbi.nlm.nih.gov/Blast.cgi) using Ssty1/2 open reading frames led to the retrieving of 58 and 131 distinct copies of Ssty1 and Ssty2, respectively. We previously showed by western blot analyses that SSTY1 is specifically expressed during sperm development [30]. We decided to revisit the expression of SSTY proteins to determine their precise timing of expression and their intracellular localization. For this, we took advantage of an anti-SSTY1 antibody that has been previously described and we generated as well a new anti-SSTY1/2 antibody recognizing both proteins (Fig. S1); we also made use of the Sly-deficient mouse model which presents a ~2.5 times overexpression of SSTY proteins in spermatids [25].
Firstly, we performed immunohistochemistry on WT testicular sections using anti-SSTY1 and anti-SSTY1/2 antibodies (Figs 1, S2 and S3). Fluorescent immunodetection with anti-SSTY1/2 antibody confirmed that SSTY proteins are expressed in postmeiotic cells. Specifically, SSTY proteins start to be visible in step 2–3 spermatids, their signal increasing in subsequent stages of the round spermatid phase. During spermatid elongation (in step 8 to 11 spermatids), SSTY proteins are still visible while the specific signal is lost in condensing spermatids (i.e. step 12 to 16 spermatids). The specificity of the signal was controlled by performing similar analyses on sections from mutant testes (MSYq−) which do not express SSTY proteins (Figs 1 and S2). A similar pattern of expression was observed on Sly-deficient testicular sections (Fig. S2). As an additional control of specificity, detection on WT and Sly-deficient testicular sections using SSTY1/2 pre-immune serum was performed in parallel (Fig. S2). We also used anti-SSTY1 antibody on both WT and Sly-deficient testicular sections. With a fluorescent immunodetection, anti-SSTY1 signal was observed in step 6 to 11 spermatids of WT and Sly-deficient males (Fig. S3A, B) while using a more sensitive assay (colorimetric immunohistochemistry) a similar pattern of expression to that seen with SSTY1/2 antibody was obtained (i.e. presence in most of the round spermatid stages, in elongating spermatids and absent in condensing spermatids, see Fig. S3C). Finally, this pattern of expression was confirmed by western blot detection of SSTY proteins in fractions enriched in round spermatids or in elongating/condensing spermatids compared with mature spermatozoa collected in the epididymis (Fig. S1C).
Figure 1. SSTY proteins are specifically present in round and elongating spermatids.
Representative pictures of immunodetection using SSTY1/2 antibody on testicular sections from WT and MSYq− mice, at stages II–V, VI–VII and VIII–XI. Black and white pictures represent DAPI staining of the nuclei. The different cell types present in testes are indicated: spermatocytes at the pachytene stage (P), round spermatids (RS), elongating spermatids (ES) and condensing spermatids (CS). Anti-SSTY1/2 was detected in green. Lectin-PNA (red) was used to stain acrosomes for staging purposes. The specificity of SSTY1/2 signal was controlled on testicular sections from mutant mice (MSYq−) which do not express SSTY (thus considered as a negative control, ctl-). SSTY proteins are observed in the round spermatids and elongating spermatids, from stage II until stage XI. Scale bar indicates 20μm. See also supplemental Fig. S2 for extended negative control panels and comparison of the pattern of expression of SSTY in WT and shSLY mice.
SSTY proteins are enriched in subnuclear regions and colocalize with the postmeiotic sex chromatin
By immunohistochemistry, SSTY signal was clearly visible in both the spermatid nucleus and cytoplasm (Figs 2 and S1–3). This was confirmed by western blot performed on nuclear and cytosolic fractions of WT and Sly-deficient round spermatids (Fig. S1D). In the nucleus, SSTY was not homogenous but appeared brighter in discrete sub-nuclear regions (Fig. 2). The DAPI staining in the round spermatid nucleus has been well characterized: the most DAPI-dense round-shaped region (usually 1 or 2 per spermatid nucleus) is the chromocenter (i.e. the pericentromeric constitutive heterochromatin), while the less DAPI-dense region near the chromocenter is the postmeiotic sex chromatin (i.e. either the X or Y chromosome) [8]. Interestingly, we observed that some bright foci of SSTY proteins colocalize with the postmeiotic sex chromatin (PMSC, indicated by white arrowheads in Figs 2 and 3) [5, 8]. This signal is stronger in Sly-deficient spermatids than in WT spermatids (Fig. 2), probably due to the increase in the level of SSTY proteins in Sly-deficient mice as we previously reported [25]. In the spermatid nucleus, epigenetic marks known to be associated with transcriptional repression, such as trimethylation of lysine 9 of histone H3 (H3K9me3), are associated with the chromocenter and the PMSC [13–15]. At a higher magnification (Fig. 3), we confirmed that SSTY proteins colocalize with the PMSC, visualized by both the DAPI staining and H3K9me3 signal. While other bright foci of SSTY proteins are observed elsewhere in the nucleus of WT spermatids, there is a clear enrichment in SSTY proteins over the PMSC of Sly-deficient spermatids (Fig. 3).
Figure 2. SSTY proteins are enriched in subnuclear regions of the spermatids.
Representative pictures of immunodetection of SSTY proteins in the nucleus of WT, shSLY and MSYq− spermatids, taken at a higher magnification (x60); (A) using anti-SSTY1 antibody or (B) using anti-SSTY1/2 antibody. As negative controls, both antibodies were used on MSYq− testicular sections (tissue which does not express SSTY) and anti-SSTY1/2 pre-immune serum was used on shSLY sections. Anti-SSTY1/2, anti-SSTY1 or pre-immune serum were detected in green. Lectin-PNA (red) was used to stain acrosomes for staging purpose. Black and white pictures represent DAPI staining of the nuclei. The most DAPI-dense round-shaped structure is the chromocenter; the less DAPI-dense region near the chromocenter is the postmeiotic sex chromatin (indicated by white arrowheads). SSTY protein staining is not diffuse but enriched in some subnuclear regions, among which is the postmeiotic sex chromatin (PMSC, white arrowheads). Scale bar indicates 10μm.
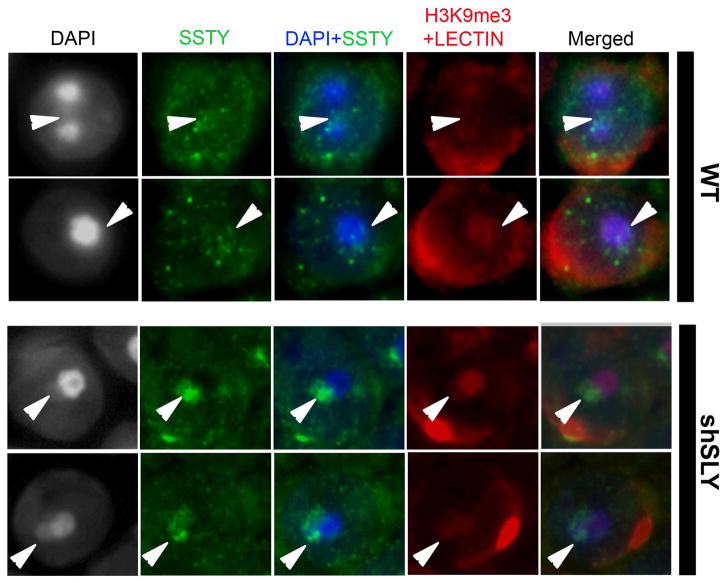
Figure 3. SSTY proteins colocalize with the PMSC and associated epigenetic marks.
Representative pictures of immunodetection of SSTY proteins (green) in the nucleus of WT and shSLY spermatids, taken at a high magnification (x100). In this immunodetection protocol, a permeabilization step was added prior to blocking and incubation with the primary antibody. DAPI (grey or blue) was used to stain nuclei. Lectin-PNA (red) was used to stain acrosomes for staging purposes. The PMSC (indicated by white arrowheads) can be visualized by DAPI staining (grey or blue) and H3K9me3 (red) staining (The most densely stained structure is the chromocenter; the structure less stained near the chromocenter is the PMSC). In WT spermatids, SSTY proteins are visible as bright foci, some of which colocalize with the PMSC. In shSLY spermatids, SSTY protein signal clearly colocalizes with the PMSC.
SSTY proteins interact with SLX/SLY proteins
SLY protein and its X-encoded homologous proteins SLX/SLXL1 have been shown to colocalize with the PMSC and to control its expression [25, 32]. SLY represses PMSC gene expression while SLX/SLXL1 proteins have an antagonistic effect to that of SLY and activate PMSC gene expression [32]. We therefore sought to determine whether SSTY proteins could physically interact with SLY and SLX/SLXL1 proteins. Sly gene encodes two isoforms, Sly1 and Sly2, arising from alternative splicing of exons 5–6 [25] while Slx gene family includes two genes: Slx and Slxl1 [34, 35]. We co-transfected COS-7 with Myc-tagged versions of Ssty1 or Ssty2 together with Flag-tagged Slx, Slxl1, Sly1 or Sly2 genes, and then performed immunoprecipitation experiments using anti-MYC or anti-FLAG antibodies. In these assays, we observed that SSTY1 and SSTY2 proteins pulled down SLY1, SLY2, SLX and SLXL1 proteins (anti-MYC immunoprecipitation) and conversely that SLX/Y proteins pulled down SSTY proteins (anti-FLAG immunoprecipitation) (Fig. 4). No interaction between SLX/SLXL1 and SLY proteins could be detected (data not shown).
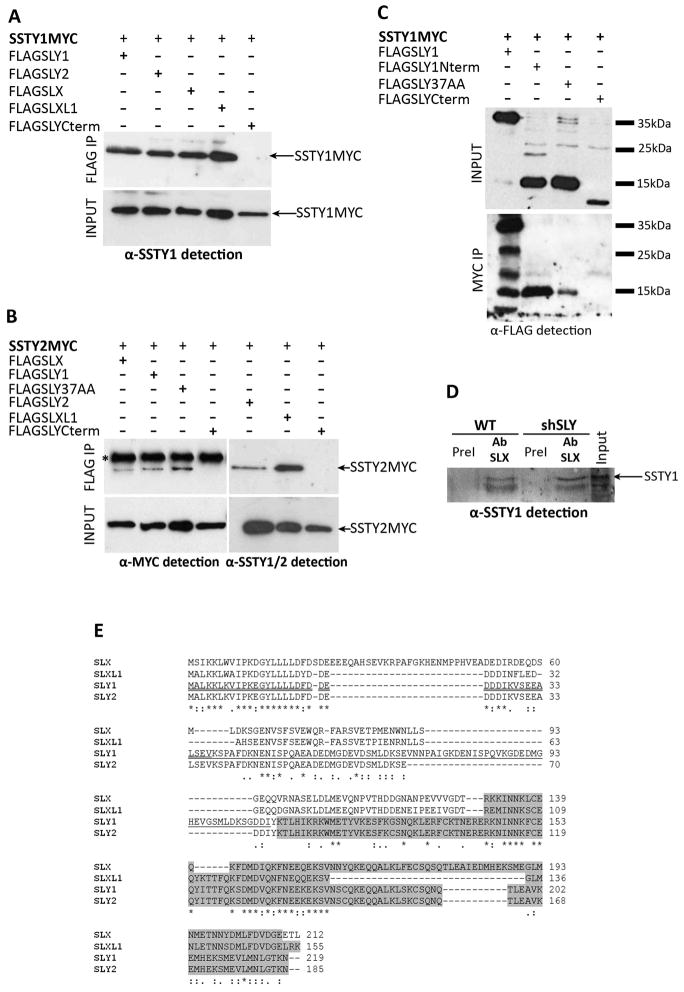
Figure 4. SSTY proteins interact with SLX/SLY proteins.
(A) Anti-SSTY1 antibody detection of protein extracts from COS-7 cells transfected with SSTY1MYC and either FLAGSLY1, FLAGSLY2, FLAGSLX, FLAGSLXL1 or FLAGSLYCterm, before (INPUT) and after immunoprecipitation (IP) with FLAG antibody. SSTY1MYC (indicated by an arrow) is immunoprecipitated by FLAG antibody when co-expressed with FLAGSLY1, FLAGSLY2, FLAGSLX, FLAGSLXL1 but not with FLAGSLYCterm. (B) Anti-MYC antibody and anti-SSTY1/2 antibody detection of protein extracts from COS-7 cells transfected with SSTY2MYC and either FLAGSLX, FLAGSLY1, FLAGFLAGSLY2, FLAGSLXL1, FLAGSLY37AA or FLAGSLYCterm, before (INPUT) and after immunoprecipitation (IP) with FLAG antibody. The star indicates the band corresponding to the light chain of the antibody used for immunoprecipitation. SSTY2MYC (indicated by an arrow) is immunoprecipitated by FLAG antibody when co-expressed with FLAGSLY1, FLAGSLY2, FLAGSLX, FLAGSLXL1, FLAGSLY37AA but not with FLAGSLYCterm. (C) Anti-FLAG detection of protein extracts from COS-7 cells transfected with SSTY1MYC and either FLAGSLY1, FLAGSLY1Nterm, FLAGSLY37AA or FLAGSLYCterm, before (INPUT) and after immunoprecipitation (IP) with MYC antibody. On the right are indicated the bands of the protein molecular weight ladder (35KDa, 25kDa and 15KDa). Anti-FLAG detection shows the presence of FLAGSLY1 protein (~38kDa), FLAGSLY1Nterm (~15kDa), FLAGSLY37AA (~15kDa) and FLAGSLYCterm (~10kDa) peptides (from left to right) before immunoprecipitation (INPUT). Anti-FLAG detection of the immunoprecipitated samples (MYC IP) shows that FLAGSLY1, FLAGSLY1Nterm and FLAGSLY37AA were immunoprecipitated by MYC antibody but not FLAGSLYCterm. In the lane corresponding to FLAGSLY1 immunoprecipitated by MYC antibody, four bands are detected with anti-FLAG antibody. The ~38kDa band corresponds to the intact version of FLAGSLY1, the ~28kDa, ~18kDa and ~15kDa bands likely correspond to shorter version of FLAGSLY1 protein which were produced by proteolysis during the immunoprecipitation process. (D) SSTY1 antibody detection of wild type (WT) and Sly-deficient (shSLY) whole testicular extracts immunoprecipitated either with anti-SLX/SLXL1 antibody or with SLX/SLXL1 pre-immune serum (negative control). SLX/SLXL1 interacts in vivo with SSTY1 (E) CLUSTALW alignment of SLX (GenBank accession number NP_001129948.1), SLXL1 (NP_083457.1), SLY1 (NP_963288.2) and SLY2 (NP_001032837.2) protein sequences. Stars indicate identities and dots similarities (one or two dots indicate strong or weaker conservative changes, respectively), a gap indicates no conservation. The sequence corresponding to SLY1Nterm is underlined once; the sequence corresponding to SLY37AA construct (the minimal domain of interaction with SSTY), which overlaps with part of SLY1Nterm, is underlined twice; the Cor1 domain (as defined by NCBI Conserved Domain), representing SLY1Cterm, is highlighted in grey.
Apart from their Cor1 domain - a domain found in SYCP3 and XLR proteins and thought to mediate interaction with the chromatin (NCBI Conserved Domain Database http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=147120 [36]) - no functional domain has been identified in SLX/SLXL1/SLY proteins. We therefore tried to identify the ‘minimal’ region of SLY that mediates interaction with SSTY proteins. In mouse sex chromosome gene evolution, Sly appeared as a consequence of a chimerism between the 5′ region of Slx and the 3′ of Xlr [29]; the N-terminal region of SLY and SLX/SLXL1 is therefore the most conserved between these proteins, albeit their C-terminal region (which encompasses the Cor1 domain) also bears similarities (Fig. 4E). We produced truncated versions of SLY protein fused to FLAG tag and observed that the construct consisting of the first 108 amino acid residues of SLY1 protein (FLAGSLY1Nterm) pulled down SSTY; while a truncated construct termed FLAGSLYCterm consisting in the last 115 amino acid residues of SLY proteins (i.e. its Cor1 domain) did not. We used other truncated constructs, notably the FLAGSLY37AA, which allowed us to pinpoint the minimal domain of interaction to the 37 first N-terminal residues of SLY (Fig. 4). This region is in fact the most conserved between SLX, SLXL1, SLY1 and SLY2 (Fig. 4E). Interestingly, the FLAGSLY37AA peptide runs at a much higher molecular weight than expected (~15kDa instead of 5.7kDa), which suggests peculiar physicochemical properties.
Moving from the cell model, we investigated SLX/SLY interactions with SSTY in whole testicular extracts by performing immunoprecipitation assays using anti-SLY and anti-SLX/SLXL1 antibodies. These assays allowed us to confirm in vivo interaction between SSTY1 and SLX/SLXL1 proteins in WT and Sly-deficient testes (Fig. 4D) but we did not succeed to immunoprecipitate SSTY with SLY antibody in WT nor in Slx-deficient testes (data not shown). Unfortunately anti-SSTY antibodies did not perform well enough to be used reliably in immunoprecipitation experiments.
SLX/SLXL1 nuclear localization depends on the presence of a Yq gene
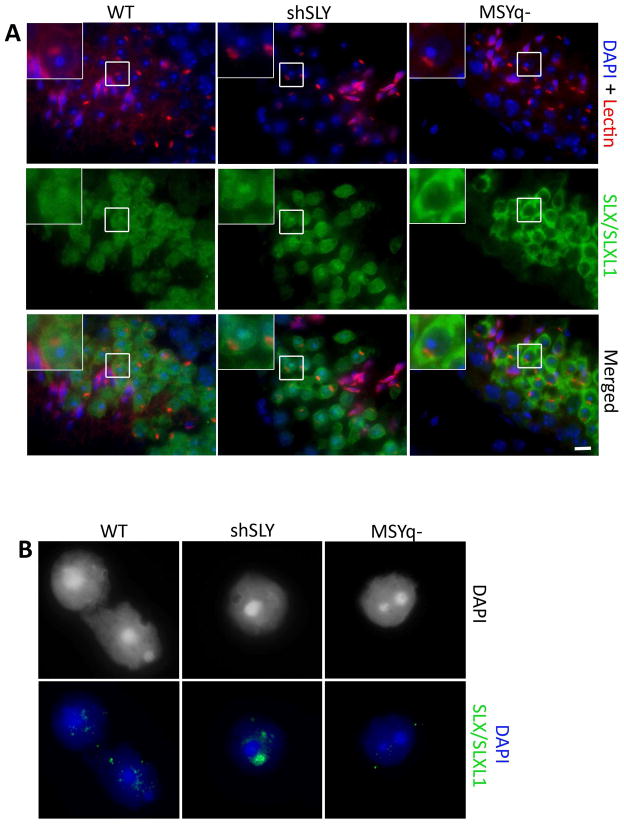
Since we confirmed in vivo interaction between SSTY and SLX/SLXL1 proteins, we next studied in more details SLX/SLXL1 intracellular localization. We have previously shown that, in WT spermatids, SLX/SLXL1 proteins are predominantly cytoplasmic and only faintly detected in the nucleus [34, 35] while, in Sly-deficient spermatids, SLX/SLXL1 proteins are abundant in the nucleus, where they colocalize with the PMSC [32]. Surprisingly, when we looked in MSYq− males, a model deficient for Sly, Ssty and all other MSYq genes, SLX/SLXL1 staining was not in the spermatid nucleus but was restricted to the cytoplasm (Figs 5A and S4). We confirmed these observations by immunofluorescence on surface-spread spermatids (Fig. 5B); in these assays not even a faint SLX/SLXL1 protein signal could be detected in MSYq− spermatid nuclei (0/100) while SLX/SLXL1 were previously detected in the nucleus in ~16% of WT round spermatids and ~76% of Sly-deficient spermatids [32]. These results mean that, in the absence of Ssty (and of other MSYq genes), SLX/SLXL1 does not locate to the spermatid nucleus and PMSC.
Figure 5. SLX/SLXL1 proteins are not visible in the nucleus of MSYq− spermatids.
(A) Detection of SLX/SLXL1 proteins (green) by immunofluorescence in WT, shSLY and MSYq− testis sections. DAPI (blue) was used to stain nuclei and lectin-PNA (red) was used to stain acrosomes. The inset in the upper left corner represents a 2.5 magnification. Scale bar indicates 10μm. (B) Representative pictures of the detection of SLX/SLXL1 proteins (green) by immunofluorescence in WT, shSLY and MSYq− round spermatid nuclei (surface spread technique). DAPI (grey or blue) was used to stain nuclei. No signal can be detected in MSYq− round spermatid nuclei.
Discussion
In the present paper, we studied the Yq-encoded multicopy genes Ssty at the protein level and found that SSTY proteins are present specifically in round and elongating spermatids (step 2–3 to step 11 spermatids). In the round spermatid nucleus, SSTY proteins appear as foci, some of which colocalize with the postmeiotic sex chromatin (PMSC). This pattern of expression is very similar to that of SLX/SLXL1 and SLY proteins, encoded by multicopy genes of the X and Yq chromosomes respectively [28, 34], with the particularity that SLX/SLXL1 locates to the spermatid nucleus and sex chromatin more abundantly when SLY is reduced/absent [32]. Interestingly, the colocalization of SSTY proteins with the sex chromatin is also more apparent in Sly-deficient spermatids; this may be due to the increase in SSTY protein levels in this genotype. Surprisingly, in testes where Sly, Ssty and all other MSYq genes are absent (i.e. in MSYq− males), SLX/SLXL1 proteins are exclusively cytoplasmic. Since SSTY proteins interact with SLX/SLXL1 proteins in a cellular model as well as in vivo, it is plausible that SSTY is the MSYq− encoded factor required for the localization of SLX/SLXL1 proteins in the spermatid nucleus and sex chromatin (PMSC). We also observed that SSTY interacts with SLY proteins in a cellular model but were not able to confirm this interaction in whole testicular extracts, probably due to lack of efficiency of our antibodies. In any case, it remains to be determined if the presence of SLY in the spermatid nucleus and its association with the sex chromatin depends as well on SSTY. The production of a mouse model solely deficient in Ssty should address this question.
SLX/SLXL1 have been shown to be positive regulators of sex chromosome-encoded genes during spermiogenesis while SLY represses partially their expression; in mice, a balanced expression of these two factors is required for normal spermiogenesis to proceed [32]. Although the mechanism by which SLX/SLY protein family regulates sex chromosome gene expression is not elucidated, it is known that they affect the recruitment of repressive epigenetic marks onto the sex chromatin [25, 32]. SLX/SLXL1 and SLY are very acidic proteins with calculated isoelectric points between 4.0 and 4.8 (http://web.expasy.org/protparam/). The minimal domain of interaction of SLX/Y proteins with SSTY is the most conserved region between SLX/SLXL1 and SLY – their N-terminal end – and enriched in acidic amino acids (aspartic acid and glutamic acid). The peptide corresponding to this domain runs in a denaturing polyacrylamide gel electrophoresis at a much higher molecular weight than calculated; this is also the case of SLX/Y proteins and has previously been reported for other acidic proteins/peptides [37]. The acidity of SLX/Y proteins may confer ability to interact with basic DNA-binding proteins. In that aspect, it is worth noting that SSTY1 has a calculated isoelectric point of 9.1 (and SSTY2 of 7.1) and belongs to the Spin-domain proteins. Spindlin1, another member of this protein family which bears 70% of homologies with SSTY proteins, has double-stranded DNA binding activity [38] and has been shown to colocalize with the meiotic spindle during female oogenesis [39]. More recent biochemical and structural studies demonstrated that Spindlin1 binds to the trimethylated histone H3 lysine 4 (H3K4me3) via an aromatic pocket located in its tudor-like domain [31, 40]; the amino acids which form this structure are conserved in SSTY proteins. All these data strongly argue for a role of SSTY in the regulation of postmeiotic sex chromatin expression, together with SLX/Y.
For now, it is difficult to predict whether SSTY would be an activator or a repressor of XY gene expression in spermatids. Given that it appears to preferentially interact with SLX/SLXL1, it would be tempting to presume that SSTY is also an activator of gene expression. However, males carrying large deletions of MSYq genomic region (and consequently deficient for all MSYq genes including Sly and Ssty) have a more dramatic phenotype than Sly ‘only’-deficient males: they present a higher derepression of X and Y genes, loss of additional epigenetic marks [25, 27] associated with more severe spermiogenesis defects, such as an increased incidence of spermhead malformations, of sperm DNA damage and of poorly condensed sperm [26]. Besides, when injected, the sperm of MSYq-deficient males lead to impaired oocyte activation and increased oocyte arrest at pronuclei stage, defects which are not observed in Sly-deficient males [24, 26]. These observations suggest that another MSYq gene is required for sperm differentiation; Ssty remains, to date, the best candidate.
Finally, Slx/Slxl1 and Sly have recently been shown to be at the basis of an intragenomic conflict occurring between the X and Y chromosomes in the mouse lineage [32]. Indeed, Slx/Slxl1 and Sly have antagonistic effects during spermiogenesis and favor their own transmission to the detriment of the other. A balance exists between Slx/Slxl1 and Sly expression in wild type mice and disruption of this balance causes segregation distortion and male infertility. The conflict in which Slx/Slxl1 and Sly are locked in is thought to have led to the amplification of Slx/Slxl1 and Sly genes. Other X and Y spermiogenic genes would have become amplified in order to compensate the increasing repressive effect of Sly on the PMSC, resulting in the presence of dozen of ampliconic postmeiotic genes on the mouse sex chromosomes [32, 33, 41]. The data we present here suggest that Ssty may also be a regulator of this intragenomic conflict. Ssty genes are located with Sly on the ~500-kb repeat unit that has been massively amplified on the Y chromosome long arm [29]; therefore it seems more likely that Ssty contributes, together with Sly, to the maintenance of PMSC gene expression at a low level. In that aspect, the fact that SSTY protein interacts in vivo with SLX/SLXL1 and appears to favor its nuclear localization is puzzling, and further functional studies will be required to better characterize the individual and global effect of SLX/SLXL1, SLY and SSTY in the postmeiotic control of XY gene expression.
Experimental Procedures
Mice
The Sly-deficient (shSLY), Slx/Slxl1-deficient (shSLX) and MSYq− mice (XSxraY*X) used in this study were produced on a random-bred MF1 albino (National Institute for Medical Research colony) background and maintained as described previously [28, 32]. Non-transgenic male siblings of shSLY or shSLX animals were used as wild-type animals (WT). Animal procedures were subjected to local ethical review (Comité d’Ethique pour l’Expérimentation Animale, Paris Descartes; registration number of the project: CEEA34.JC.114.12). MSYq− males (XY*XSxra) produced on partial C57BL/6 background were also used and gave similar results than XSxraY*X. These mice were maintained as previously described [42] in accordance with the guidelines of the Laboratory Animal Services at the University of Hawaii and the National Research Council’s (NCR) ‘Guide for Care and Use of Laboratory Animals’. The protocol for animal handling and treatment procedures was reviewed and approved by the Animal Care and Use Committee at the University of Hawaii.
Purification and collection of round spermatids, elongating-condensing spermatids, and epididymal spermatozoa
Cells were isolated from 2 month-old mice by Fluorescence-activated cell sorting (FACS) as previously indicated in Bastos H. et al [43] with some modifications. In details, after albuginea dissection, seminiferous tubules were dissociated by using enzymatic digestion with collagenase type I at 120U/ml (Gibco) for 30 min at 36°C in HBSS 1X (Invitrogen) supplemented as previously described [43]. After incubation, a filtration step with 40μm cell strainer was carried out to separate interstitial cells from tubules. Tubules were collected from the surface of the cell strainer, and incubated in Cell Dissociation buffer (Invitrogen) supplemented with 10 μg/ml DNaseI (Sigma) for 15 min at 36°C. The resulting cell suspension was filtered again using a 40μm cell strainer; the flow-through was spun down 10 min at 230g and resuspended at a concentration of 5×106 cell/ml in Incubation buffer (HBSS supplemented with 20 mM HEPES pH7.2, 1.2 mM MgSO4, 1.3 mM CaCl2, 6.6 mM Sodium pyruvate, 0.05% lactate, gluthamine and 1% fetal calf serum). Resuspended cells were then stained with Hoechst 33342 (HO, Invitrogen) at a concentration of 5μg/ml for 45 min at 36°C in the dark. Before FACS analysis, 2 μg/ml propidium iodide (PI) (Sigma) was added to exclude dead cells. Analysis and cell collection was performed at the Cochin Cytometry and Immunobiology Facility using a FACS ARIA III (Becton Dickinson). HO and PI were excited by using a violet laser at 405 nm and cells were separated by using a combination of 605 nm and 450 nm emission spectra. Spermatids were further differentiated into round spermatids (RS) and elongating-condensing spermatids (ES-CS) according to Forward Scatter parameters (FSC) as described previously [43]. A 100 μm nozzle was used with a sample threshold rate of approximately 4000 events/second. Cell purity of the two populations collected by FACS was assessed by DAPI staining, and was shown to be ~95% of round spermatids in the RS fraction and ~88% of elongating and condensing spermatids in the ES-CS fraction.
Sperm were collected from the cauda epididymides. The cauda epididymides were isolated, several incisions were made in the tissue and the sperm were allowed to swim out into PBS, at 37°C, for several minutes. The sperm were then spun down, the supernatant was removed and the pellet was flash-frozen in liquid nitrogen.
Antibodies
To produce anti-SSTY1/2 antibody, a specific peptide (LVGREVQHKFEGKDGSED) was used to immunize two rabbits, followed by booster injections (at two, four and eight weeks) and affinity purification (90 day protocol, Thermo Scientific). For western blot detection, primary antibodies were diluted as follow: rabbit anti-SLY1 antibody [44] 1:2000, rabbit anti-SLX/SLXL1 antibody [34] 1:3000; rabbit anti-SSTY1 antibody (YMT2B) [30] 1:1000; rabbit anti-SSTY1/2 antibody 1:1000; mouse anti-FLAG M2 (Sigma) 1:1000; mouse anti-MYC (Santa Cruz Biotechnology) 1:500. For immunodetection on sections and/or on surface-spread testicular cells, all primary antibodies [anti-SLX/SLXL1 [34], anti-SSTY1 [30], anti-SSTY1/2, anti-H3K9me3 (Millipore)] and pre-immune serum were diluted at 1/50.
Immunofluorescence
For fluorescent or colorimetric (DAB 3, 3′-diaminobenzidine) immunohistochemistry, testes from 2 month-old mice were fixed in 4% buffered paraformaldehyde, then washed in 70% ethanol, dehydrated, and embedded in paraffin. Four-micrometer paraffin sections of testes were mounted on a glass slide and dried overnight at 37°C. Sections were dewaxed in xylene and hydrated in a graded series of alcohols. After washing in de-ionized water, the sections were incubated for 40 min in 0.01 M sodium citrate solution (pH 6) in a water bath at 96°C to allow antigen retrieval. For DAB immunohistochemistry, slides were washed in de-ionized water and incubated in Peroxidase block for 30 min. The subsequent stages were performed as described by the manufacturer (Novolink polymer detection, Leica). For fluorescent immunodetection, slides were washed and blocked for 1 h at room temperature in PBT (PBS, 0.1% Tween, 0.15% BSA). A permeabilization step (10 min with 0.05% Triton X100) was added for immunodetection of H3K9me3. After blocking, slides were incubated overnight at 4°C with primary antibody (or pre-immune serum) diluted at 1/50 in antibody diluent (BD Biosciences). Slides were washed in PBS, incubated in either goat anti-rabbit Alexa 488 (1:500; Molecular Probes) or chicken anti-mouse Alexa 594 (1:500; Molecular Probes) diluted in PBS for 1 h at room temperature, washed in PBS, and mounted in Vectashield DAPI (49,6-diamidino-2-phenylindole) (Vectorlab). Alexa Fluor 594-conjugated peanut agglutinin lectin (Invitrogen) stains the developing acrosome and was used determine the stage of testis tubules and associated spermatids. Briefly, the twelve stages can be differentiated as follow (for more details, see [45–47]): the proacrosomal vesicle appears in stage I and develops into two proacrosomal granules in stages II–III). It then increases in size and flattens (stage IV–V), and starts to surround the spermatid nucleus (stage VI–VII). At stage VIII, the spermatids’ acrosomes face the basal membrane, their nuclei then elongate and flatten (stages IX–X) and gets more and more compact (stages XI–XII). Stage XII shows typical meiotic division figures such as spermatocytes in metaphase.
Immunofluorescence experiments on surface-spread testicular cells were performed following a protocol adapted from Barlow et al. [48] and previously described [25].
Transfection
The open reading frames (ORF) of SLX, SLXL1, SLY1, SLY2 and truncated forms of SLY (the 37 first amino acids, SLY 37AA; and the 111 last amino acids of the SLY protein sequence corresponding to the COR1 domain, SLY C TERM) were cloned in fusion with a N-Terminal FLAG tag under the control of the CMV promoter of pCDNA3.1 vector (Invitrogen). Similarly, the ORF of SSTY1 and SSTY2 were cloned in fusion with a C-Terminal MYC tag. COS-7 cells were transfected using Lipofectamine 2000 as described by the manufacturer (Invitrogen). Cells were collected for protein extraction 24 to 48 h after transfection.
Protein extracts
Nuclear and cytosolic extracts were obtained from round spermatids following elutriation as described previously [25]. Briefly, a pellet of 1×107 round spermatids was homogenized in a glass pestle with 500μL of lysis buffer [0.6 M Sucrose, 10 mM Hepes pH 7.7, 0.2% NP40, 25 mM KCl, 2 mM EDTA, 0.5 mM EGTA, 1X Protease Inhibitor Cocktail (SIGMA) and 0.2 mM PMSF inhibitors]. After centrifugation for 15 min at 800g, the supernatant was kept as cytoplasmic fraction, while the pellet was washed twice with 1mL of lysis buffer and resuspended in 50μl of nuclear protein extraction buffer [400 mM NaCl, 20 mM Hepes pH 7.7, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol and 1X Protease Inhibitor Cocktail (Sigma) and 0.2 mM PMSF inhibitors]. After 30 min of homogenization at 4°C, the samples were centrifuged for 30 min at 11,000g; the supernatant corresponded to the nuclear protein extract.
Proteins were extracted from transfected cells with ice-cold radioimmunoprecipitation assay (RIPA) buffer [25 mM NaCl, 10 mM Tris-HCl pH 7.5, 5 mM EDTA, 0.1% Nonidet P-40, 1X Protease Inhibitor Cocktail (Sigma) and 0.2 mM PMSF]. Twenty four to 48 h after transfection, cells were pelleted by centrifugation for 5 min at 100g and resuspended with 200μL ice-cold radioimmunoprecipitation assay (RIPA) buffer (25 mM NaCl, 10 mM Tris-HCl pH 7.6, 5 mM EDTA, 0.1% Nonidet P-40, 1X Protease Inhibitor Cocktail and 0.2 mM PMSF), followed by an incubation step at 4°C for 30 min, on a rotating platform to extract the proteins. After centrifugation at 13,000g at 4°C for 30 sec, the supernatant was collected and immediately used for the immunoprecipitation assay.
Flash-frozen testes were grinded and resuspended in 1:9 weight/volume of ice-cold extraction buffer [300 mM NaCl, 20 mM Tris-HCl pH 7.5, 5 mM EDTA, 1% Triton-X-100, 1X Protease Inhibitor Cocktail (Sigma) and 0.2 mM PMSF]. After homogenization and incubation at 4°C for 30 min, cell/tissue lysates were centrifuged at 13,000g at 4°C for 10 min. The supernatant was collected and immediately used for the immunoprecipitation assays.
Immunoprecipitation
For immunoprecipitation assays of MYC-tagged proteins, 50μl of Dynabeads Protein G (Invitrogen) were washed according to the manufacturer’s instructions, resuspended in 250μL of binding buffer (1X PBS with 0.5% Nonidet P-40) containing anti-MYC (Santa Cruz Biotechnology) antibody and incubated overnight at 4°C. Beads were washed in 0.2 M sodium borate (pH 9.0), incubated in 30 mM dimethyl pimelimidate for 30 min at room temperature and washed in 0.2M ethanolamine (pH 8.0) then in binding buffer. Antibody-coupled beads were incubated with cell lysates, overnight at 4°C, and then washed three times in extraction buffer. Elution was performed using 0.1 M Glycine HCl buffer (pH 3.0) which was then neutralized with 1 M Tris buffer (pH 8.8).
For immunoprecipitation assays of FLAG-tagged proteins, 40μL of ANTI-FLAG® M2 Magnetic Beads (SIGMA-ALDRICH) were washed twice in 1X TBS (150 mM NaCl, 50 mM Tris pH 7.4). Beads were incubated with cell lysates, overnight at 4°C, washed three times in 800μL of 1X TBS and resuspended twice in 100μL of 0.1 M Glycine HCl buffer (pH 3.0) for 5 min at room temperature. The pH was neutralized with Triethylammonium bicarbonate buffer (pH 8.4). The samples were concentrated using a speed-vacuum concentrator.
For in vivo immunoprecipitation assays of SLX/SLXL1 proteins, 200μL of Dynabeads® Protein G (Invitrogen) were washed three times in binding buffer (1X PBS with 0.5% Nonidet P-40), resuspended in 800μL of binding buffer with anti-SLX/SLXL1 antibody and incubated overnight at 4°C to allow the binding of the antibody. Beads were washed twice in 1mL of 0.2 M sodium borate (pH 9.0), incubated in 1 mL of 30 mM dimethyl pimelimidate for 30 min at room temperature to covalently couple the antibodies to the beads and washed three times in 1mL of 0.2 M ethanolamine (pH 8.0) and then washed twice in 1mL of binding buffer. Antibody-coupled beads were incubated with 260mg of protein extract for 2 h, washed three times in 500μL of extraction buffer and once in 500μL of 1X TBS with 0.05% Triton X-100. For a non-denaturing elution, beads were resuspended in 60μL of elution buffer (0.1 M Glycine HCl buffer (pH 3.0)) and incubated at room temperature for 5 min. The pH was neutralized with Triethylammonium bicarbonate buffer (pH 8.4).
Western blot
One million of round spermatids, elongating-condensing spermatids, and spermatozoa were denatured in 50μL of 1X LDS Sample Buffer (NuPAGE® LDS Sample Buffer, Life Technologies) with 5% β-Mercaptoethanol and boiled for 25 min at 95°C. The other samples were denatured for 10 min at 95°C in Laemmli denaturing buffer (60 mM Tris pH 6.8, 2% SDS, 10% glycerol, 5% β-Mercaptoethanol, 0.01% Bromophenol blue) and run on a denaturing SDS-polyacrylamide gel. A protein molecular weight ladder (Thermo Scientific PageRuler Prestained Protein Ladder) was loaded and run in parallel. Following transfer, membranes were blocked in 1X PBS, 5% milk powder, 0.1% Tween 20. Incubation with primary antibody was performed overnight at 4°C. Membranes were washed three times in 1X PBS with 0.1% Tween 20, incubated with the corresponding secondary antibody [anti-mouse antibody, 1:3000 (Santa Cruz Biotechnology); anti-rabbit antibody, 1:3000 (Santa Cruz Biotechnology)] coupled to peroxidase and washed three times in 1X PBS with 0.1% Tween 20. The signal was revealed by chemiluminescence according to the manufacturer’s instructions (Pierce® SuperSignal WestPico Chemiluminescent Substrat and Immobilon™ Western Chemiluminescent HRP Substrate, MILLIPORE) and recorded on X-ray films (Kodak® BioMax® XAR Film).
Supplementary Material
Figure S1: Characterization of SSTY antibodies, confirmation of the presence of SSTY proteins in round and elongating spermatids, as well as in the nuclear and cytosolic compartments of spermatids.
Figure S2: SSTY proteins are specifically present in round and elongating spermatids. Supplemental panel to Figure 1.
Figure S3: SSTY1 protein is specifically present in round and elongating spermatids.
Figure S4: Absence of SLX/SLXL1 nuclear signal in MSYq− spermatids. Supplementary panel to Figure 5
Acknowledgments
The authors wish to thank P. Burgoyne, O.A. Ojarikre and A. Rattigan at NIMR, staff of the Cochin mouse house Facility, HistIM Facility, Cellular imaging Facility and Cytometry and Immunobiology Facility. This work was supported by the INSERM, the ANR program (ANR-12-JSV2-0005-01 to JC) and a Marie Curie Fellowship (FP7-PEOPLE-2010-IEF-273143 to JC) and NIH HD072380, NIH RR024206 (Project 2), and HCF13ADVC-60314 to MAW.
Abbreviations
- MSCI
meiotic sex chromosome inactivation
- MSYq
male-specific region of the mouse Y chromosome long arm
- PMSC
postmeiotic sex chromatin
Footnotes
Author Contribution Statement
AC, CM, MES, JA, JC planned experiments, performed experiments and analyzed data; CIR, MAW, AT, DV and JC contributed to essential material; MAW, AT, DV and JC wrote the paper.
References
- 1.Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nature Genetics. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- 2.Mueller JL, Mahadevaiah SK, Park PJ, Warburton PE, Page DC, Turner JM. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nature Genetics. 2008;40:794–799. doi: 10.1038/ng.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Affara NA, Mitchell MJ. The role of human and mouse Y chromosome genes in male infertility. Journal of Endocrinological Investigation. 2000;23:630–645. doi: 10.1007/BF03343787. [DOI] [PubMed] [Google Scholar]
- 4.Song R, Ro S, Michaels JD, Park C, McCarrey JR, Yan W. Many X-linked microRNAs escape meiotic sex chromosome inactivation. Nature Genetics. 2009;41:488–493. doi: 10.1038/ng.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner JM. Meiotic sex chromosome inactivation. Development (Cambridge, England) 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- 6.Royo H, Prosser H, Ruzankina Y, Mahadevaiah SK, Cloutier JM, Baumann M, Fukuda T, Hoog C, Toth A, de Rooij DG, Bradley A, Brown EJ, Turner JM. ATR acts stage specifically to regulate multiple aspects of mammalian meiotic silencing. Genes & Development. 2013;27:1484–1494. doi: 10.1101/gad.219477.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ichijima Y, Ichijima M, Lou Z, Nussenzweig A, Camerini-Otero RD, Chen J, Andreassen PR, Namekawa SH. MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes & Development. 2011;25:959–971. doi: 10.1101/gad.2030811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montellier E, Rousseaux S, Zhao Y, Khochbin S. Histone crotonylation specifically marks the haploid male germ cell gene expression program: Post-meiotic male-specific gene expression. Bioessays. 2011 doi: 10.1002/bies.201100141. [DOI] [PubMed] [Google Scholar]
- 9.van der Heijden GW, Eijpe M, Baarends WM. The X and Y chromosome in meiosis: how and why they keep silent. Asian Journal of Andrology. 2011 doi: 10.1038/aja.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ichijima Y, Sin HS, Namekawa SH. Sex chromosome inactivation in germ cells: emerging roles of DNA damage response pathways. Cell Mol Life Sci. 2012;69:2559–2572. doi: 10.1007/s00018-012-0941-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang PJ, Page DC, McCarrey JR. Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Human Molecular Genetics. 2005;14:2911–2918. doi: 10.1093/hmg/ddi322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendriksen PJM, Hoogerbrugge JW, Themmen APN, Koken MHM, Hoeijmakers JHJ, Oostra BA, van der Lende T, Grootegoed JA. Postmeiotic transcription of X and Y chromosomal genes during spermatogenesis in the mouse. Developmental Biology. 1995;170:730–733. doi: 10.1006/dbio.1995.1252. [DOI] [PubMed] [Google Scholar]
- 13.van der Heijden GW, Derijck AA, Posfai E, Giele M, Pelczar P, Ramos L, Wansink DG, van der Vlag J, Peters AH, de Boer P. Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nature Genetics. 2007;39:251–258. doi: 10.1038/ng1949. [DOI] [PubMed] [Google Scholar]
- 14.Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, Lee JT. Postmeiotic sex chromatin in the male germline of mice. Curr Biol. 2006;16:660–667. doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- 15.Turner JM, Mahadevaiah SK, Ellis PJ, Mitchell MJ, Burgoyne PS. Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Developmental Cell. 2006;10:521–529. doi: 10.1016/j.devcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Sin HS, Barski A, Zhang F, Kartashov AV, Nussenzweig A, Chen J, Andreassen PR, Namekawa SH. RNF8 regulates active epigenetic modifications and escape gene activation from inactive sex chromosomes in post-meiotic spermatids. Genes & Development. 2012;26:2737–2748. doi: 10.1101/gad.202713.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baarends WM, Wassenaar E, Hoogerbrugge JW, Schoenmakers S, Sun ZW, Grootegoed JA. Increased phosphorylation and dimethylation of XY body histones in the Hr6b-knockout mouse is associated with derepression of the X chromosome. J Cell Sci. 2007;120:1841–1851. doi: 10.1242/jcs.03451. [DOI] [PubMed] [Google Scholar]
- 18.Ellis PJI, Clemente EJ, Ball P, Toure A, Ferguson L, Turner JMA, Loveland KL, Affara NA, Burgoyne PS. Deletions on mouse Yq lead to upregulation of multiple X- and Y-linked transcripts in spermatids. Human Molecular Genetics. 2005;14:2705–2715. doi: 10.1093/hmg/ddi304. [DOI] [PubMed] [Google Scholar]
- 19.Moriwaki K, Suh D-S, Styrna J. Genetic factors effecting sperm morphology in the mouse. Mouse News Letter. 1988;82:138. [Google Scholar]
- 20.Styrna J, Klag J, Moriwaki K. Influence of partial deletion of the Y chromosome on mouse sperm phenotype. Journal of Reproduction and Fertility. 1991;92:187–195. doi: 10.1530/jrf.0.0920187. [DOI] [PubMed] [Google Scholar]
- 21.Styrna J, Imai HT, Moriwaki K. An increased level of sperm abnormalities in mice with a partial deletion of the Y chromosome. Genetical Research. 1991;57:195–199. doi: 10.1017/s0016672300029268. [DOI] [PubMed] [Google Scholar]
- 22.Burgoyne PS, Mahadevaiah SK, Sutcliffe MJ, Palmer SJ. Fertility in mice requires X-Y pairing and a Y-chromosomal “spermiogenesis” gene mapping to the long arm. Cell. 1992;71:391–398. doi: 10.1016/0092-8674(92)90509-b. [DOI] [PubMed] [Google Scholar]
- 23.Touré A, Szot M, Mahadevaiah SK, Rattigan A, Ojarikre OA, Burgoyne PS. A new deletion of the mouse Y chromosome long arm associated with loss of Ssty expression, abnormal sperm development and sterility. Genetics. 2004;166:901–912. doi: 10.1534/genetics.166.2.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamauchi Y, Riel JM, Stoytcheva Z, Burgoyne PS, Ward MA. Deficiency in mouse Y chromosome long arm gene complement is associated with sperm DNA damage. Genome Biology. 2010;11:R66. doi: 10.1186/gb-2010-11-6-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cocquet J, Ellis PJ, Yamauchi Y, Mahadevaiah SK, Affara NA, Ward MA, Burgoyne PS. The multicopy gene Sly represses the sex chromosomes in the male mouse germline after meiosis. PLoS Biology. 2009;7:e1000244. doi: 10.1371/journal.pbio.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riel JM, Yamauchi Y, Sugawara A, Li HY, Ruthig V, Stoytcheva Z, Ellis PJ, Cocquet J, Ward MA. Deficiency of the multi-copy mouse Y gene Sly causes sperm DNA damage and abnormal chromatin packaging. J Cell Sci. 2013;126:803–813. doi: 10.1242/jcs.114488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynard LN, Turner JM. Increased sex chromosome expression and epigenetic abnormalities in spermatids from male mice with Y chromosome deletions. J Cell Sci. 2009;122:4239–4248. doi: 10.1242/jcs.049916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toure A, Clemente EJ, Ellis P, Mahadevaiah SK, Ojarikre OA, Ball PA, Reynard L, Loveland KL, Burgoyne PS, Affara NA. Identification of novel Y chromosome encoded transcripts by testis transcriptome analysis of mice with deletions of the Y chromosome long arm. Genome Biology. 2005;6:R102. doi: 10.1186/gb-2005-6-12-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellis PJ, Ferguson L, Clemente EJ, Affara NA. Bidirectional transcription of a novel chimeric gene mapping to mouse chromosome Yq. BMC Evol Biol. 2007;7:171. doi: 10.1186/1471-2148-7-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Touré A, Grigoriev V, Mahadevaiah SK, Rattigan A, Ojarikre OA, Burgoyne PS. A protein encoded by a member of the multicopy Ssty gene family located on the long arm of the mouse Y chromosome is expressed during sperm development. Genomics. 2004;83:140–147. doi: 10.1016/s0888-7543(03)00216-7. [DOI] [PubMed] [Google Scholar]
- 31.Wang W, Chen Z, Mao Z, Zhang H, Ding X, Chen S, Zhang X, Xu R, Zhu B. Nucleolar protein Spindlin1 recognizes H3K4 methylation and stimulates the expression of rRNA genes. EMBO Rep. 2011;12:1160–1166. doi: 10.1038/embor.2011.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cocquet J, Ellis PJ, Mahadevaiah SK, Affara NA, Vaiman D, Burgoyne PS. A genetic basis for a postmeiotic X versus Y chromosome intragenomic conflict in the mouse. PLoS Genet. 2012;8:e1002900. doi: 10.1371/journal.pgen.1002900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellis PJ, Bacon J, Affara NA. Association of Sly with sex-linked gene amplification during mouse evolution: a side effect of genomic conflict in spermatids? Human Molecular Genetics. 2011;20:3010–3021. doi: 10.1093/hmg/ddr204. [DOI] [PubMed] [Google Scholar]
- 34.Reynard LN, Turner JM, Cocquet J, Mahadevaiah SK, Toure A, Hoog C, Burgoyne PS. Expression analysis of the mouse multi-copy X-linked gene Xlr-related, meiosis-regulated (Xmr), reveals that Xmr encodes a spermatid-expressed cytoplasmic protein, SLX/XMR. Biology of Reproduction. 2007;77:329–335. doi: 10.1095/biolreprod.107.061101. [DOI] [PubMed] [Google Scholar]
- 35.Cocquet J, Ellis PJ, Yamauchi Y, Riel JM, Karacs TP, Rattigan A, Ojarikre OA, Affara NA, Ward MA, Burgoyne PS. Deficiency in the multicopy Sycp3-like X-linked genes Slx and Slxl1 causes major defects in spermatid differentiation. Mol Biol Cell. 2010;21:3497–3505. doi: 10.1091/mbc.E10-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsuchida J, Nishina Y, Wakabayashi N, Nozaki M, Sakai Y, Nishimune Y. Molecular cloning and characterization of meichroacidin (male meiotic metaphase chromosome-associated acidic protein) Developmental Biology. 1998;197:67–76. doi: 10.1006/dbio.1998.8885. [DOI] [PubMed] [Google Scholar]
- 38.Zhao Q, Qin L, Jiang F, Wu B, Yue W, Xu F, Rong Z, Yuan H, Xie X, Gao Y, Bai C, Bartlam M, Pei X, Rao Z. Structure of human spindlin1. Tandem tudor-like domains for cell cycle regulation. The Journal of Biological Chemistry. 2007;282:647–656. doi: 10.1074/jbc.M604029200. [DOI] [PubMed] [Google Scholar]
- 39.Oh B, Hwang S-Y, Solter D, Knowles B. Spindlin, a major maternal transcript expressed in the mouse during the transition from oocyte to embryo. Development (Cambridge, England) 1997;124:493–503. doi: 10.1242/dev.124.2.493. [DOI] [PubMed] [Google Scholar]
- 40.Yang N, Wang W, Wang Y, Wang M, Zhao Q, Rao Z, Zhu B, Xu RM. Distinct mode of methylated lysine-4 of histone H3 recognition by tandem tudor-like domains of Spindlin1. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17954–17959. doi: 10.1073/pnas.1208517109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Good JM. The conflict within and the escalating war between the sex chromosomes. PLoS Genet. 2012;8:e1002955. doi: 10.1371/journal.pgen.1002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamauchi Y, Riel JM, Wong SJ, Ojarikre OA, Burgoyne PS, Ward MA. Live offspring from mice lacking the Y chromosome long arm gene complement. Biology of Reproduction. 2009;81:353–361. doi: 10.1095/biolreprod.109.076307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bastos H, Lassalle B, Chicheportiche A, Riou L, Testart J, Allemand I, Fouchet P. Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytometry A. 2005;65:40–49. doi: 10.1002/cyto.a.20129. [DOI] [PubMed] [Google Scholar]
- 44.Reynard LN, Cocquet J, Burgoyne PS. The multi-copy mouse gene Sycp3-like Y-linked (Sly) encodes an abundant spermatid protein that interacts with a histone acetyltransferase and an acrosomal protein. Biology of Reproduction. 2009;81:250–257. doi: 10.1095/biolreprod.108.075382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russell LD, Hikim APS, Ettlin RA, Clegg ED. Cache River Press, editor . Histological and Histopathological Evaluation of the Testis. Clearwater, FL: 1990. [Google Scholar]
- 46.Kotaja N, Kimmins S, Brancorsini S, Hentsch D, Vonesch JL, Davidson I, Parvinen M, Sassone-Corsi P. Preparation, isolation and characterization of stage-specific spermatogenic cells for cellular and molecular analysis. Nat Methods. 2004;1:249–254. doi: 10.1038/nmeth1204-249. [DOI] [PubMed] [Google Scholar]
- 47.Ahmed EA, de Rooij DG. Staging of mouse seminiferous tubule cross-sections. Methods Mol Biol. 2009;558:263–277. doi: 10.1007/978-1-60761-103-5_16. [DOI] [PubMed] [Google Scholar]
- 48.Barlow AL, Benson FE, West SC, Hultén MA. Distribution of the RAD51 recombinase in human and mouse spermatocytes. EMBO Journal. 1997;16:5207–5215. doi: 10.1093/emboj/16.17.5207. COCQ. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Characterization of SSTY antibodies, confirmation of the presence of SSTY proteins in round and elongating spermatids, as well as in the nuclear and cytosolic compartments of spermatids.
Figure S2: SSTY proteins are specifically present in round and elongating spermatids. Supplemental panel to Figure 1.
Figure S3: SSTY1 protein is specifically present in round and elongating spermatids.
Figure S4: Absence of SLX/SLXL1 nuclear signal in MSYq− spermatids. Supplementary panel to Figure 5