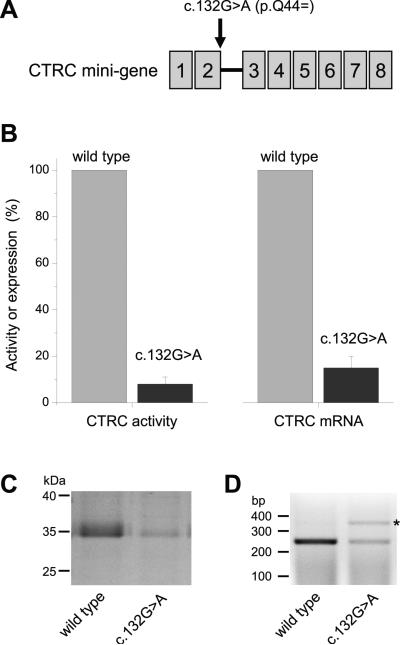
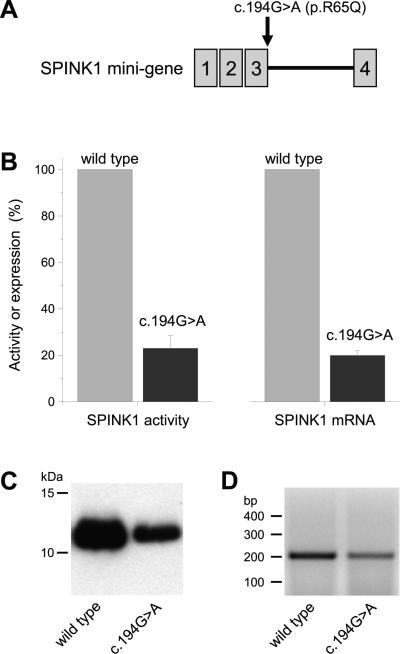
We read with great interest the recent paper by Rosendahl et al.1 describing the incidence of variants in CFTR, SPINK1, CTRC and PRSS1 in a German cohort of chronic pancreatitis cases. The authors conclude that chronic pancreatitis is a complex genetic disease and the net effect of mutations in multiple susceptibility genes underlies increased disease risk. Indeed, in the studied cohort, at least one genetic risk factor was identified in 36.7 % of patients. Surprisingly, however, only 6.5 % of cases carried multiple risk factors, which seems to contradict the complex genetics theory of chronic pancreatitis. While it is likely that more susceptibility genes are yet to be discovered, we submit that known risk genes may have been incompletely characterized resulting in underestimation of the true genetic burden in chronic pancreatitis. Discovery studies tend to focus on exons and exon-intron boundaries and may thus miss many intronic variants. Replication studies often target previously identified variants only. Even if identified, intronic variants other than splice site mutations are often ignored due to inherent difficulties in predicting or experimentally demonstrating functional effects. Synonymous variants in exons are usually presumed to have no functional effects, whereas non-synonymous exonic variants are typically studied in the context of protein function only with the unstated assumption that mRNA expression is unaltered. In fact, exonic mutations, both synonymous and non-synonymous, may have profound effects on pre-mRNA splicing thereby affecting mRNA and protein expression levels. Here we demonstrate, as case-in-point examples, that two exonic variants in CTRC and SPINK1, respectively, markedly decrease mRNA, and consequently protein, levels due to impaired pre-mRNA splicing. Rosendahl et al.1 reported a new synonymous CTRC variant c.132G>A (p.Q44=) found in a patient with chronic pancreatitis. The variation alters the last nucleotide of exon 2. To test the effect of this variant on pre-mRNA splicing, we constructed a CTRC mini-gene with intron 2 placed in the appropriate context within the coding DNA of CTRC. CTRC mini-gene carrying variant c.132G>A exhibited a marked reduction in mRNA and secreted CTRC levels relative to the wild type mini-gene (Figure 1). As expected, when tested in the context of the coding DNA only, the c.132G>A CTRC variant had no effect on mRNA or protein expression (not shown). To extend these observations, we surveyed the known pancreatitis susceptibility genes for previously reported variants that altered the last nucleotide of an exon. We identified the SPINK1 variant c.194G>A (p.R65Q), first reported by Ockenga et al.2 and also described by Rosendahl et al.1 This variant alters the last nucleotide of exon 3 in SPINK1. Previous studies using SPINK1 cDNA constructs indicated that the p.R65Q variant was produced at reduced levels3, which we confirmed and found about a 20-25%, decrease in mRNA and protein expression (not shown). However, when tested in a SPINK1 mini-gene4 containing intron 3, the c.194G>A variant caused a much larger, ~75% reduction in mRNA and protein levels, compared to wild type (Figure 2). These observations indicate that the CTRC c.132G>A (p.Q44=) and the SPINK1 c.194G>A (p.R65Q) variants are pathogenic risk factors for chronic pancreatitis and call for the systematic evaluation of all pancreatitis risk gene variants for possible pre-mRNA splicing defects.
Figure 1.
Effect of the synonymous CTRC variant c.132G>A (p.Q44=) on mRNA and protein expression in transiently transfected HEK 293T cells. (A) Expression plasmids carried a CTRC mini-gene composed of the CTRC coding sequence and intron 2. (B, left panel) Expression of CTRC protein in conditioned medium was measured by activity assay. (B, right panel) Correctly spliced CTRC mRNA in the cell lysate was quantified by real-time PCR. (C) Levels of CTRC protein in the conditioned medium were visualized by SDS-PAGE and Coomassie blue staining. (D) Defective splicing of CTRC mRNA was analyzed by reverse-transcriptase-PCR on ethidium bromide stained agarose gels. Note the reduced levels of the correctly spliced mRNA and the accumulation of the unspliced species (asterisk) in cells transfected with the c.132G>A mini-gene construct.
Figure 2.
Effect of the c.194G>A (p.R65Q) SPINK1 variant on mRNA and protein expression in transiently transfected HEK 293T cells. (A) Expression plasmids carried a SPINK1 mini-gene containing the SPINK1 coding sequence and intron 3. (B, left panel) Expression of SPINK1 protein in conditioned medium was measured by inhibitory activity against human cationic trypsin. (B, right panel) Correctly spliced SPINK1 mRNA in the cell lysate was quantified by real-time PCR. (C) Levels of SPINK1 protein in the conditioned medium were visualized by Western blotting. (D) Defective splicing of SPINK1 mRNA was analyzed by reverse-transcriptase-PCR on ethidium bromide stained agarose gels. Note the reduced levels of the correctly spliced mRNA in cells transfected with the c.194G>A mini-gene construct. Unspliced or misspliced mRNA was not detectable.
ACKNOWLEDGMENTS
These studies were supported by NIH grants R01DK082412, R01DK058088 and R01DK095753 to M.S.-T. The authors thank Dr. Heiko Witt (Technical University, Munich) and Dr. Jonas Rosendahl (University of Leipzig) for stimulating discussions and for sharing unpublished data.
This is a commentary on article Rosendahl J, Landt O, Bernadova J, Kovacs P, Teich N, Bödeker H, Keim V, Ruffert C, Mössner J, Kage A, Stumvoll M, Groneberg D, Krüger R, Luck W, Treiber M, Becker M, Witt H. CFTR, SPINK1, CTRC and PRSS1 variants in chronic pancreatitis: is the role of mutated CFTR overestimated? Gut. 2013 Apr;62(4):582-92.
Footnotes
COMPETING INTEREST STATEMENT
No competing interest to declare.
REFERENCES
- 1.Rosendahl R, Landt O, Bernadova J, et al. CFTR, SPINK1, CTRC and PRSS1 variants in chronic pancreatitis: is the role of mutated CFTR overestimated? Gut. 2013;62:582–592. doi: 10.1136/gutjnl-2011-300645. [DOI] [PubMed] [Google Scholar]
- 2.Ockenga J, Dork T, Stuhrmann M. Low prevalence of SPINK1 gene mutations in adult patients with chronic idiopathic pancreatitis. J Med Genet. 2001;38:243–244. doi: 10.1136/jmg.38.4.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Király O, Wartmann T, Sahin-Tóth M. Missense mutations in pancreatic secretory trypsin inhibitor (SPINK1) cause intracellular retention and degradation. Gut. 2007;56:1433–1438. doi: 10.1136/gut.2006.115725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kereszturi E, Király O, Sahin-Tóth M. Minigene analysis of intronic variants in common SPINK1 haplotypes associated with chronic pancreatitis. Gut. 2009;58:545–549. doi: 10.1136/gut.2008.164947. [DOI] [PMC free article] [PubMed] [Google Scholar]