Abstract
Sphingolipid molecules act as bioactive lipid messengers and exert their actions on the regulation of various cellular signaling pathways. Sphingolipids play essential roles in numerous cellular functions, including controlling cell inflammation, proliferation, death, migration, senescence, tumor metastasis and/or autophagy. Dysregulated sphingolipid metabolism has been also implicated in many human cancers. Macroatuophagy (referred to here as autophagy) “self-eating”, is characterized by nonselective sequestering of cytosolic materials by an isolation membrane, which can be either protective or lethal for cells. Ceramide (Cer), a central molecule of sphingolipid metabolism, has been extensively implicated in the control of autophagy. The increasing evidence suggests Cer is highly involved in mediating two opposing autophagic pathways, which regulate either cell survival or death, autophagy paradox. However, the underlying mechanism that regulates the autophagy paradox remains unclear. Therefore, this review focuses on recent studies with regard to the regulation of autophagy by Cer and elucidate the roles and mechanisms of action of Cer in controlling autophagy paradox.
Keywords: Sphingolipids, ceramide, autophagy, mitophagy, cell death
1. Autophagy
Autophagy (“self-eating” in Greek) is a self-digestive process that targets internal or damaged organelles and misfolded proteins to lysosomal degradation[1]. To date, three types of autophagy have been described, including macroautophagy, microautophagy and chaperone-mediated autophagy (CMA), which distinguish from each other as to the functions and mechanisms[2–4]. Autophagy consists of several distinct processes[5]. It initiates with the formation of an isolation membrane, namely a phagophore, which elongates to engulf cytoplasmic components. With LC3B-II protein (phosphatidyl ethanolamine, PE-conjugated microtubule-associated protein 1 light chain 3b) localized to the isolation membrane, it encloses to form an autophagosome. Fusing of autophagosome with lysosome forms autophagolysosome, in which contents sequestered in autophagosome vesicles are degraded by lysosomal hydrolases (Fig. 1) [6–9]. Autophagy not only results in the removal of damaged proteins and organelles, but also precisely regulates the normal turnover of the intracellular components to ensure the cellular quality control[10]. It primarily acts as a survival mechanism under stress conditions, such as nutrition starvation, via self-cannibalism to provide cellular energy and produce metabolic precursors[8,11,12]. However, under various different stress conditions, dysregulated and/or persistent autophagy may also lead to cell death through different molecular mechanisms[13–15]. Autophagic defects have been implicated in various human diseases, including neurodegenerative diseases and aging, infection, pulmonary and cardiovascular diseases, metabolic stress and cancer[12,16–22].
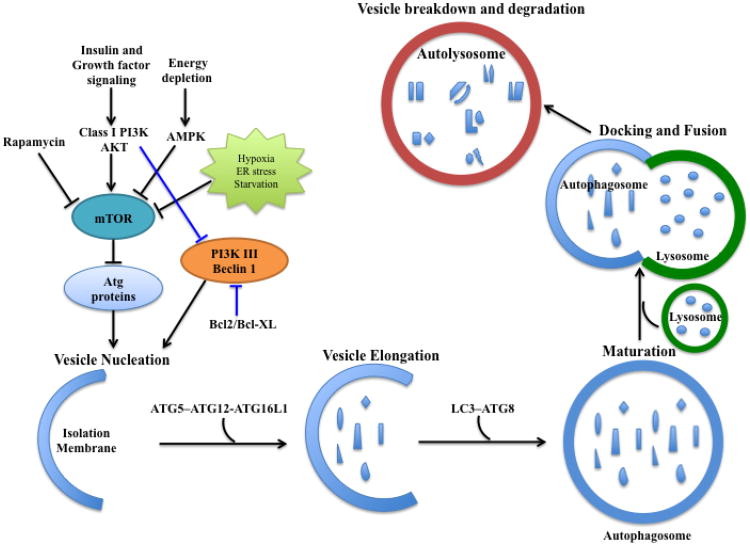
Fig 1. Schematic view of the autophagic pathway.
Autophagic cellular events have distinct stages: vesicle nucleation (formation of the isolation membrane/phagophore), vesicle elongation and maturation, autophagosome-lysosome fusion, and vesicle breakdown and degradation. Autophagy occurs at a basal level and responds to various environmental signals and stress. The well-characterized regulatory modulators include PI3K-AKT-mTOR and Beclin1-Class III PI3K complexes. Insulin and other growth factors activate PI3K-AKT-mTOR, which negatively regulates autophagy. Environmental stress and energy depletion inhibit mTOR, relieving its inhibitory effects on autophagy induction. Both PI3K-AKT and Bcl2/Bcl-XL suppress Beclin1-Class III PI3K complex to inhibit autophagy. Autophagosomal elongation requires ubiquitin-like conjugation system, ATG5-ATG12-ATG16L1 complex. However, the conversion of LC3-I to LC3-II (PE-conjugated form) is a key regulatory step in autophagosome maturation.
2. Autophagy in cell survival and cell death
2.1 Autophagy acts as a pro-survival and cytoprotective mechanism
Basal autophagy primarily regulates cellular homeostasis[8,11]. It usually occurs at a low rate and serves its housekeeping function to remove aggregated proteins and damaged organelles as they are toxic to the cell[23]. In response to most forms of cellular stress, such as metabolic stress or nutrition deprivation, autophagy supplies cells with energy and essential metabolic materials to maintain normal cellular functions[1,10,24]. Mutant mice, in which ATG (Autophagy-related protein)5 or ATG7 was depleted, developed neurodegenerative disease with the accumulation of polyubiquitylated protein aggregates[25,26]. ATG5 deficient mice failed to survive the neonatal period, which showed severe metabolic crisis with low amino acid and ATP levels[27]. Moreover, mitophagy, a form of autophagy that selectively targets damaged mitochondria, is particularly important for the regulation of cell death, as defective mitophagy causes mitochondrial dysfunction, leading to mitochondrial damage and toxic ROS (reactive oxygen species) production, which eventually may result in cell death[28,29]. Moreover, autophagy is also induced under the context of metabolic stress or survival nutrient withdrawal, which stabilized cyclin-dependent kinase inhibitor 27, leading to cell death inhibition[30].
2.2 Autophagy acts as a pro-death mechanism
Autophagic cell death is also known as Type II programmed cell death, in which autophagy per se serves as a cell death mechanism[31]. It is defined as cell death by autophagy, not cell death with autophagy[32]. Autophagic cell death should meet the following standards: (1) the cell death occurs independent of apoptosis; (2) cell death should be rescued by suppression of autophagy via both pharmacological inhibitors (e.g.,3-methyladenine (3-MA)) and genetic approaches, such as gene knockdown/mutation or gene silencing of autophagic regulators; (3) not only the autophagy markers but also autophagy flux is present in the dying cells[33]. On one hand, sustained upregulation of autophagic activity beyond a certain threshold could contribute to irreversible cellular atrophy and cause catastrophic cellular dysfunction due to the huge loss of cytosol and organelles[31]. For example, Bax-/-Bak-/- cells activated autophagy in response to growth factor withdrawal, which caused progressive atrophy and eventually resulted in irreversible cell death[34]. Over-activated autophagy may compromise the recovery ability of a cell if autophagic clearance leads to a complete elimination of an essential organelle[28]. One the other hand, autophagy may also mediate its cell killing effect through the selective degradation of essential proteins in the cell[31]. It was shown that autophagy selectively degraded catalase, which plays an essential role in cellular antioxidant defense. Moreover, catalase deficiency contributed to ROS accumulation in cells, which succumbed to autophagic mediated cell death[35]. Moreover, autophagy may also mediate its cell killing effect through the trigger of apoptosis and/or necrosis[28,31,36]. One prominent example is ATG5, which, under certain conditions, was proteolytically activated to mediate apoptosis via its translocation to mitochondria, triggering mitochondrial outer membrane permeabilization (MOMP)[37]. As previously mentioned, Lenardo and coworkers showed that catalase degradation by autophagy promoted necrotic cell death, which could be prevented by autophagy suppression[35]. Autophagic cell death was also shown to be required in salivary glands during Drosophila development, in which overexpression of ATG1 induced caspase-independent degradation of salivary glands[38]. In addition, Beclin1 mutant, which didn't bind anti-apoptotic protein Bcl-2, resulted in uncontrolled upregulation of autophagy, and accelerated cell death[13].
3 Autophagy and cancer
3.1 Autophagy is a tumor suppressor mechanism
In early stages of cancer development, quality control of autophagy suppresses tumor growth and exerts its anti-carcinogenic function by preventing metabolic or oxidative stress, maintaining normal mitochondrial function and safeguarding against DNA damage and genetic instability[39]. Beclin 1, one essential protein of autophagy, is monoallelically deleted in 40 to 70% of human prostate, ovarian and breast cancers[40–42]. Also, monoallelic disruption of beclin 1 in mice promoted spontaneous tumorigenesis, indicating that Beclin 1 primarily functions as a tumor suppressor[43,44]. Other studies also identified additional autophagy-related proteins (e.g., UVRAG, Bif1 and ATG5) as tumor suppressor proteins[11,12,33]. Moreover, autophagy regulation is closely associated with some oncogenic signaling modulation[33]. For example, PI3K-mTOR-Akt signaling axis is constitutively activated in many human cancers and promotes tumor growth and proliferation, which, by contrast, suppress autophagy[45,46]. Some common oncogenic proteins (e.g., Bcl-2, PI3K and PKB) inhibit autophagy induction; however, several well-known tumor suppressors (e.g., p53, PTEN and TSC1/2) invariably activate autophagy[11].
Autophagy may also prevent tumorigenesis as the guardian of the genome[22]. Metabolic stress may lead to ROS accumulation, inducing mutations and DNA strand breaks, which causes tumor suppressor inhibition and oncogene activation[22]. Autophagy acts as a ROS scavenger, maintaining genomic integrity, and prevents tumorigenesis[47,48]. Thus, defective autophagy is highly likely to increase gene mutations and amplification, survive metabolic stress, which susbsequently promotes tumorigenesis[49]. For example, the mitochondrial DNA mutations in autophagy-deficient yeast suggested that autophagy was required to maintain the regular turnover of mitochondria and keep cells from DNA damage and genotoxic stress[8,50]. Failure of p62 clearance or p62 accumulation accounts for another important mechanism of autophagy-mediated tumor suppression[49,51]. Defective autophagy contributed to p62 accumulation, which induced nuclear factor erythroid 2-related factor 2 (NRF2) activation[52,53]. Activated NRF-2 translocated to the nucleus, where it stimulated anti-oxidant defense system and promoted cell survival[51,54]. In addition, p62 is also known as an activator of NF-kB signaling pathway to accelerate tumorigenesis[49,55,56]. In another study, autophagy was significantly elevated in Ras oncogene-induced senescence (OIS) and restricted cell proliferation[57]. Inhibition of autophagy delayed the senescence phenotype[49].
3.2. Autophagy induces tumor promotion/proliferation
In the late stage of oncogenesis or established tumors, autophagy confers tumor cells survival ability by meeting increased metabolic and energetic demands, and executing cellular quality control to eliminate toxic intracellular damages in the aggressive tumor microenvironment[12,33,39]. Pancreatic cancer primary tumors and cell lines showed elevated autophagy under basal conditions[58]. Autophagy inhibition by genetic and pharmacological means (chloroquine treatment) led to elevated DNA damage and impaired mitochondria oxidative phosphorylation, which suppressed pancreatic tumor growth[49]. Autophagy was also shown to be upregulated and promoted the tumorigenicity of the cells expressing activated Ras oncogene[59]. Also, Ras-expressing ATG5-/- and ATG7-/- cells showed reduced tumor growth in mice while Ras-expressing p62-/- cells displayed decreased viability under stress, as well as carcinogenesis inhibition[59]. Moreover, numerous studies showed that autophagy was utilized by cancer cells as an adaptive mechanism to induce chemotherapy resistance and promoted their survival[60]. Thus, inhibition of autophagy by pharmacological inhibitors and genetic approaches may greatly enhance anti-cancer drug cytotoxicity and sensitize cancer cells to various cancer therapies[12,33,49,51]. For example, in a recent study, combination of vinblastine and nanoliposomal C6-ceramide synergistically inhibited cancer growth/progression via attenuation of autophagy maturation, leading to apoptosis and tumor suppression[61]. Autophagy has also been implicated to facilitate the use of cellular glucose toward glycolysis, which is essential for cancer cell transformation and proliferation[62–64].
4. Regulation of autophagy by Cer
4.1 Metabolism of Cer
Sphingolipids are a family of membrane lipids that have structural roles in the regulation of the fluidity and the subdomain structure of the lipid bilayers[65,66]. Cer, a central molecule of sphingolipid metabolism, is composed of a sphingosine base and amide-linked acyl chains varying in length from C14 to C26[67]. Cer then serves as the metabolic and structural precursor for complex sphingolipids, which are composed of hydrophilic head groups, such as sphingomyelin (SM), Cer-1-phosphate and glucosylceramide (GlcCer), which is the precursor for glycolipids and gangliosides[65]. Endogenous Cer can be generated via a de novo pathway, which begins with condensation of serine and palmitoyl CoA by serine-palmitoyl CoA transferase (SPT)[68,69]. Cer can be also generated by the metabolism of other complex sphingolipids, which is tightly regulated by many specialized enzymes[65,66,70]. For example, Cer can be produced by either sphingomyelinases (SMases) mediated sphingomyelin (SM) hydrolysis[71] or cerebrosidase mediated GlcCer and galactosylceramide (GalCer) breakdown[72]. Cer can also be hydrolyzed by ceramidases (CDases) to yield sphingosine[73,74], which can be phosphorylated by sphingosine kinase (SK)1 or SK2, producing sphingosine-1-phosphate (S1P)[75]. Cer is also utilized as a precursor by ceramide kinase (CK), or SM synthase to generate Cer-1-phosphate (C1P) and SM, respectively (Fig. 2) [67,76,77]. Cer is converted into GlcCer by glucosylceramide synthase (GCS), which is Cer transporter (CERT) independent[78]; by contrast, Cer mediated SM synthesis is dependent on CERT, which is responsible for Cer transport from the endoplasmic reticulum (ER) to the Golgi via a non-vesicular mechanism[79–82]. Importantly, non-vesicular transport of GlcCer from its site of synthesis (early Golgi) to distal Golgi compartments is carried out by FAPP2, four-phosphate adaptor protein, controlling the synthesis of glycosphingolipids, which might play crucial roles in determining the lipid composition of the plasma membrane[83].
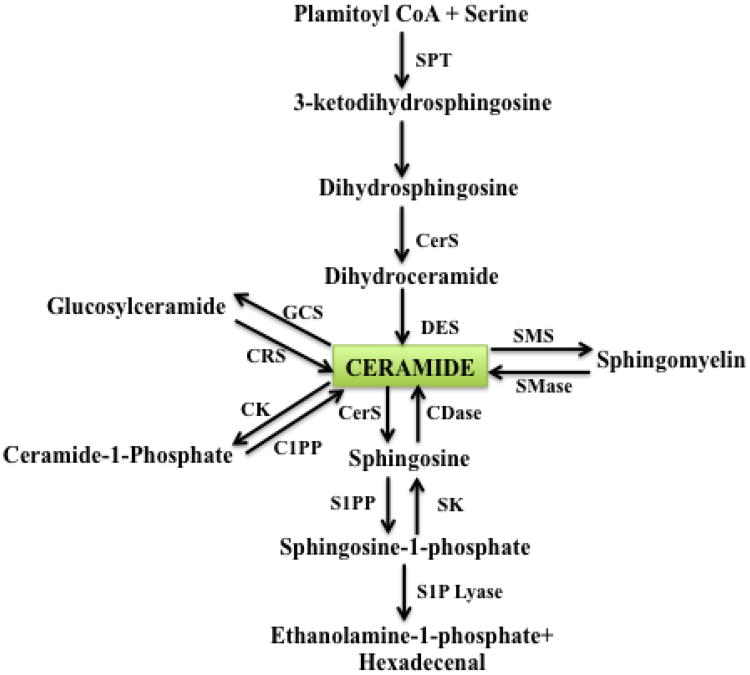
Fig 2. Pathways of sphingolipid metabolism.
Ceramide can be formed de novo or from hydrolysis of sphingomyelin or cerebrosides. Conversely, ceramide can be phosphorylated by ceramide kinase to yield ceramide-1-phosphate, or can serve as a substrate for the synthesis of sphingomyelin or glycolipids. Ceramide can be metabolized by ceramidases (CDases) to yield sphingosine, which in turn is phosphorylated by sphingosine kinases (SKs) to generate sphingosine-1-phosphate (S1P). S1P can be cleared by the action of specific phosphatases that regenerate sphingosine or by the action of a lyase that cleaves S1P into ethanolamine-1-phosphate and a C16-fatty-aldehyde. C1PP, ceramide-1-phosphate phosphatase; CRS, cerebrosidase; CK, ceramide kinase; CerS, ceramide synthase; DES, dihydroceramide desaturase; GCS, glucosylceramide synthase; S1PP, S1P phosphatase; SMS, sphingomyelin synthase; SMase, spingomyelinase; SPT, serine palmitoyl transferase.
4.2 Ceramide synthases(CerS) and de novo generation of Cer
CerS and SPT primarily function to generate Cer de novo[69,84]. CerS was originally identified as the yeast longevity assurance gene 1 (LAG1), known to regulate life-span/longevity in Saccharomyces cerevisiae, and its deletion prolonged the replicative life-span of yeast[85,86]. Moreover, LAC1, a LAG1 homologue, was known to be crucial for yeast Cer synthesis and postulated to perform the same function of CerS[87,88]. The discovery of a mouse homologue of LAG1, also known as LASS1 or the upstream of growth and differentiation factor 1 (UOG1)[87,88], demonstrated that it specifically regulated the synthesis of C18-Cer with a high degree of fatty-acid chain length specificity[86,89]. There exist six LASS proteins (LASS1-6) that were renamed CerS1-6[86]. They all possess a domain of five predicted transmembrane helices termed the TLC (TRAM, LAG1 and CLN8 homology) domain, which contribute to the CerS activity and Cer generation[90]. They also differ in their specificity, and thus produce dihydroceramides(DHCs) with differing chain lengths, which are then desaturated by desaturase(DES) to generate Cer with different fatty acid chain lengths[92,93]. Importantly, CerS1/4 mainly produce C18-Cer, to a lesser extent C20-cermides, whereas CerS5/6 selectively mediates C16-cermide, to a lesser extent C12- and C14-cermides[89]. Moreover, CerS2 generates very long-chain Cers, predominantly C24-cermides and the ultra-long-chain Cers are generated by CerS3[93,94]. There is evidence that Cer with different fatty acid chain lengths produced by CerS1-6 might have distinct functions, and/or mechanism of action in the regulation of cell death and/or autophagy. These recent studies suggest that not all Cers are created equal, and distinct fatty acid chain lengths, subcellular localization/transport, and/or down-stream protein/lipid targets, help define their specific roles in various different stress conditions and/or cell/tissue types. Recently, it has also been postulated that changes in the long chain sphingoid bases of ceramides with 18 versus 16 carbons also play distinct roles in inducing autophagy versus cell death in cardiomyocytes[95].
4.3 Cer and autophagy induction
Cer has been implicated in the autophagy induction[96]. Class I PI3K and Akt are well-established autophagy suppressors, and Cer functions to activate PP2A, which blocks Akt activation[97–99]. Various treatments, leading to long-chain Cer accumulation, such as long-chain endogenous Cer generation by exogenous short chain Cer recycling[100]. PDMP (GlcCer synthase inhibitor) or tamoxifen, invariably suppressed Akt activity to stimulate autophagy in human colon (HT-29) and breast cancer (MCF-7) cell lines[98]. Moreover, amino acid deprivation resulted in an increase in Cer levels, which suppressed mTOR activity and induced autophagy in a PP1/PP2A dependent manner. Cer-mediated autophagy induction is also closely linked with its influence on nutrient transporters[101– 103]. Edinger et al. demonstrated that Cer caused a rapid and substantial downregulation of amino acid transporter proteins in a number of cell types[103]. Similarly, Cer suppressed nutrient transporter protein expression, which induced starvation, leading to the AMPK-dependent autophagy induction[104]. Beclin1, the mammalian orthologue of yeast ATG6, plays a central role in autophagy, which is upregulated under stress, and it is mutated or monoallelically deleted in many human cancers[105,106]. Scarlatti et al. showed that exogenous C2-Cer treatment enhances Beclin1 expression and autophagy induction, which was blocked by Cer synthase inhibitor myriocin, indicating Cer regulates Beclin1 expression at the transcriptional or post-transcriptional level[98]. C2-Cer also activated JNK in human cancer cell lines CNE2 and Hep3B, and mediated activation and phosphorylation of c-Jun, a transcription factor, which induced Beclin1 expression at the transcription level[107].
Also, increased levels of Beclin1 can originate from the dissociation of the Beclin1-Bcl2 complex[108]. On the one hand, Cer stimulated stress-activated protein kinase JNK1, leading to Bcl2 phosphorylation, which liberated Beclin1 from its association with Bcl2[109]. On the other hand, Cer mediated activation of Forkhead box protein O3 (FOXO3), which is negatively regulated by Akt, significantly upregulated BH3-only protein BNIP3 expression[96,110]. Elevated BNIP3 expression dissociated Beclin1 from binding to Bcl2 through its competitive binding of Bcl2 and Bcl-xL[111]. Cer-mediated ER stress also contributes to autophagy induction[96]. Disregulated Cer homeostasis in the ER by CerS2 downregulation interfered with intracellular Cer trafficking and distribution[112]. CerS2 downregulation lead to accumulation of long-chain Cers (C14-and C16-Cers) and induces ER-stress related cytoprotective autophagy[112]. Russo et al. demonstrated that dietary myristate oversupply mainly depended on CerS5 to generate Cer, which consequently induced autophagy, resulting in the cardiomyocytes hypertrophy phenotype[113]. Recently, we showed that C18-pyridinium-Cer treatment or endogenous C18-Cer generation by CerS1 expression mediated lethal autophagy induction, independent of apoptosis in human head and neck cancer cells[114]. C18-Cer–induced lethal autophagy was regulated via LC3B-II, and selective targeting of mitochondria by LC3B-II– containing autophagolysosomes (mitophagy) through direct interaction between Cer and LC3B-II upon Drp1-dependent mitochondrial fission[114].
4.4 DHC and autophagy regulation
DHCs are the immediate precursors for de novo synthesis of Cer[65]. Also, DHC is always deemed as biologically inactive molecules[108]. Although its precise physiological role is still controversial and remains to be determined, DHC is shown to be involved in autophagy induction in a number of studies[115– 118]. First, in DU145 prostate cancer cells, autophagy was induced by addition of exogenous C2-DHC while 4-HPR (N-(4-hydroxyphenylretinamide), which elevated DHC level, also induced autophagy, suggesting that a large increase of endogenous DHCs might be responsible for autophagy induction[119]. Second, Signorellli et al. demonstrated that resveratrol-induced autophagy occurred with elevated intracellular DHC levels due to inhibition of DHC desaturase(DEGS) activity and that DHC accumulation led to autophagy induction in human gastric cancer cells HGC-127[120]. Third, under a hypoxia condition, DHC accumulation led to protective autophagy induction, which enhances cell survival and suppress cell proliferation[117]. It was proposed that DHC might function to regulate cell fate by switching from protective autophagy to Cer-mediated apoptosis under stress[96]. Moreover, celecoxib (a selective COX-2 inhibitor) has been recently shown to induce autophagy[121], which predominantly produces C16:0, C24:0 and C24:1 DHCs in human cancer cell lines by inhibiting DEGSs activity[122]. These data suggest that DHCs are involved in autophagy induction, and at least endogenously generated DHCs are not simply inactive sphingolipid molecules[123]. Further studies should dissect and elucidate the signaling pathways that are regulated by DHCs to exert pro-autophagic effects.
5. Cer is involved in the regulation of paradoxical roles of autophagy
5.1 Cer induces cytoprotective autophagy
Cer, especially long chain Cer, is known to be involved in cell death signaling regulation[124]. However, under certain conditions, Cer may induce protective autophagy and utilize autophagic machinery to prevent cells from programmed cell death. Guenther et al. demonstrated that C2-Cer or endogenous Cer starved cells to death by limiting intracellular nutrients subsequent to nutrient transporter downregulation[104]. However, ceramide also triggered homeostatic protective autophagy to prevent cell death as blocking autophagy with chloroquine (CQ) sensitized cells to ceramide exposure[104]. Spassieva et al. demonstrated that down-regulation of CerS2 resulted in no significant decrease of very long chain Cer(VLC) (e.g. C24 or C24:1 Cer), but led to 3-fold increase in long chain Cer (LC) (e.g. C14 or C16 Cer)[112]. SMS-KCNR neuroblastoma cells didn't undergo apoptosis with LC accumulation, which, by contrast, induced autophagy evidenced by LC3B-II lipidation and autophagy-related structures in electron microscope[112]. Mechanistically, CerS2 downregulation stimulated autophagy and UPR (unfolded protein response), which activated the pro-survival IRE1(inositol-requiring element 1), preventing induction of cell death[112]. Also, Park et al. showed that anti-cancer drugs vorinostat and sorafenib activated CD95 via the activation of acid sphingomyelinase(ASMase) and generation of Cer[125]. Cer-CD95 induced autophagy in a PERK dependent manner, which showed cytoprotective effects since suppression of ATG5 expression enhanced sorafenib and vorinostat lethality[125]. Similarly, cyclin-dependent kinase (CDK) inhibitor via Cer-CD95 signaling induced both pro-death/apoptotic signal and pro-survival/autophagic signal in primary hepatocytes[126]. Blockade of protective autophagy induced by Cer-CD95, using 3-MA or siRNA against Atg5, further enhanced cell killing[126]. Tamoxifen, which is known to inhibit GlcCer synthase, elevated endogenous Cer levels and induced autophagic cell death in MCF-7 cells[127], but also triggered protective autophagy to delay cell death in other cancer cells (Fig. 3) [128,129].
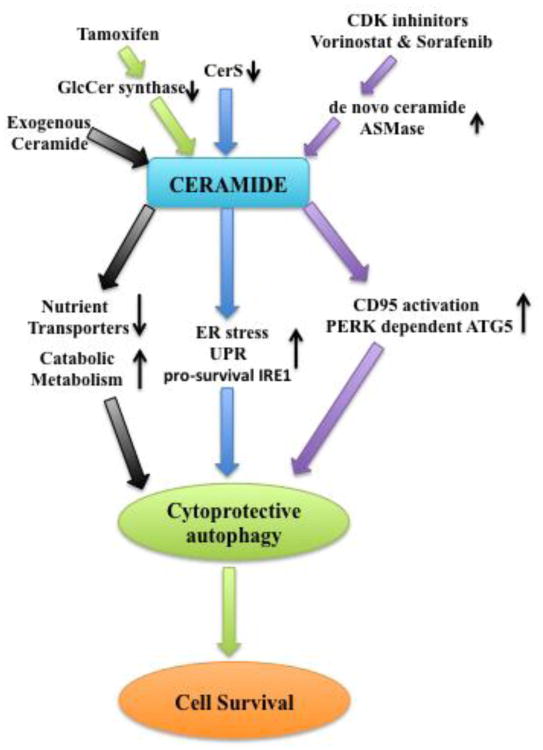
Fig 3. Signaling pathways of protective autophagy that are regulated by ceramide.
Exogenous ceramide induces mild nutrient transporter downregulation, which triggers protective autophagy and enhances catabolic metabolism to maintain cell viability. Ceramide synthase downregulation, resulting in long chain ceramide accumulation, stimulates ER stress and activates pro-survival IRE1 (inositol-requiring element1), which leads to protective autophagy induction and cell death inhibition. Anti-cancer drugs vorinostat and sorafenib, as well as cyclin-dependent kinase (CDK) inhibitor, contribute to upregulation of de novo ceramide generation and acid sphingomyelinase (ASMase) activation, which leads to ceramide accumulation. Ceramide accumulation results in CD95 and PERK-dependent ATG5 activation, leading to protective autophagy induction and inhibition of cell death. Tamoxifen, which is known to inhibit GlcCer synthase, not only elevated endogenous Cer levels, but also triggered protective autophagy to delay cell death.
Although the precise role of DHC is not fully elucidated, a number of studies showed that it is able to induce protective autophagy[115–117,121]. DEGS inhibitor XM462 delayed cell cycle G1/S transition via activation of ER stress and induction of autophagy in gastric carcinoma HCG27 cells[115]. Inhibition of XM462-induced autophagy resulted in significant reduction of metabolic activity measured by MTT assay, indicating DHC induced autophagy promotes survival[115]. Devlin et al. also demonstrated that direct treatment with DHC or indirect augmentation of DHC by siRNA blockade of DEGS1 or 2 decreased cell proliferation[117]. Celecoxib, primarily producing C16:0-, C24:0- and C24:1-DHCs, induced autophagy in human colorectal cancer cells. Inhibition of autophagy using 3-MA and wortmannin or siRNA against ATG8 significantly enhanced Vps34-mediated apoptotic cell death[121].
5.2 Cer induces lethal autophagy
Cer is associated with cell growth arrest and cell death induction[130]. Cer plays a well-established role in triggering programmed cell death in response to growth factor withdrawal, death receptor ligation, hypoxia, and chemotherapeutic drugs[104]. Although many studies confirmed the pivotal role of Cer in mediating lethal autophagy, the precise underlying mechanism is still not completely understood. An initial report from Dbaibo et al. showed that arsenic trioxide (As2O3) induced accumulation of cytotoxic levels of Cer in human leukemia cells through de novo Cer synthesis and inhibition of GluCer synthase activity[131]. Qian et al. demonstrated that As2O3 induced not only apoptosis but also autophagic cell death in leukemia cell lines. The lethal autophagy was attributed to upregulation of Beclin-1 protein and completely prevented by autophagy inhibitor 3-MA[132]. A research study in malignant glioma cells showed that Cer induced autophagic cell death, which was evidenced by occurrence of autophagic vacuoles, acidic vesicular organelles and LC3B-II lipidation[109]. The mechanism depicted was that Cer decreased mitochondrial membrane potential and activated death-inducing mitochondrial protein BNIP3[109]. In human cancer cell lines CNE2(nasopharyngeal carcinoma) and Hep3B(hepatocellular carcinoma), the lethal autophagy was mediated by Cer-induced c-Jun activation through JNK signaling, which transcriptionally upregulated Beclin-1 expression[107]. JNK activity inhibitor SP600125, as well as Beclin-1 siRNA rescued cancer cells from Cer-induced autophagic cell death[107]. Melanoma differentiation associated gene-7(mda-7)/interleukin-24(IL-24) was shown to induce ER stress, which triggered Cer and ROS generation in a PERK-dependent manner[133]. Autophagy-mediated cell death by mda-7/IL-24 depended on Cer and ROS generation, which could be blocked by PERK inhibitor treatment[133]. Patschan et al. exposed human umbilical vein endothelial cells (HUVECs) to glycated collagen I (GC), which led to autophagy induction and ASMase activation, leading to accumulation of ceramide[134], and premature cellular senescence[134]. In human leukemia cells (HL-60) and Chinese hamster ovary cells (CHO), Cer-CAPPs(Cer activated protein phosphatases) have inhibitory effects on Akt-mTOR pathway, which activated autophagy and induced autophagy-mediated cell death, whereas S1P-S1P3 signaling activated Akt-mTOR pathway, which offset autophagy and suppressed lethal autophagy[135]. Hou et al. found that C6-pyridinium Cer preferentially promoted autophagy induction and induced mitochondrial permeabilization, which retarded MCF-7 cells growth and activated apoptosis[136]. Salazar et al. demonstrated that tetrahydrocannabinol (THC), which was known to induce Cer accumulation and eukaryotic translation initiation factor 2α (eIF2) phosphorylation, activated ER stress, leading to autophagy induction via tribbles homolog 3 (TRB3) mediated mTOR inhibition. Cannabinoid-mediated lethal autophagy led to human glioma cell death and contributed to its anti-tumor action in vivo[137]. Our recent data suggested that C18-pyridinium Cer treatment or endogenous C18-Cer generation by CerS1 expression induced lethal autophagy, independent of apoptosis in human head and neck cancer cells[114]. Moreover, knockdown of CerS1 abrogated sodium selenite–induced mitophagy, and stable LC3B-II knockdown protected against CerS1- and C18-Cer–dependent mitophagy and blocked tumor suppression in vivo[114]. Mechanistically, our data suggested that ceramide-LC3-II interaction plays a key role to target autophagolysosomes to damaged mitochondria, which required Drp-1 mediated mitochondrial fission. This, then, results in reduced oxygen consumption rate, decreased ATP production, and subsequent cell death in response to ceramide stress in head and neck cancer cells (Fig. 4) [112].
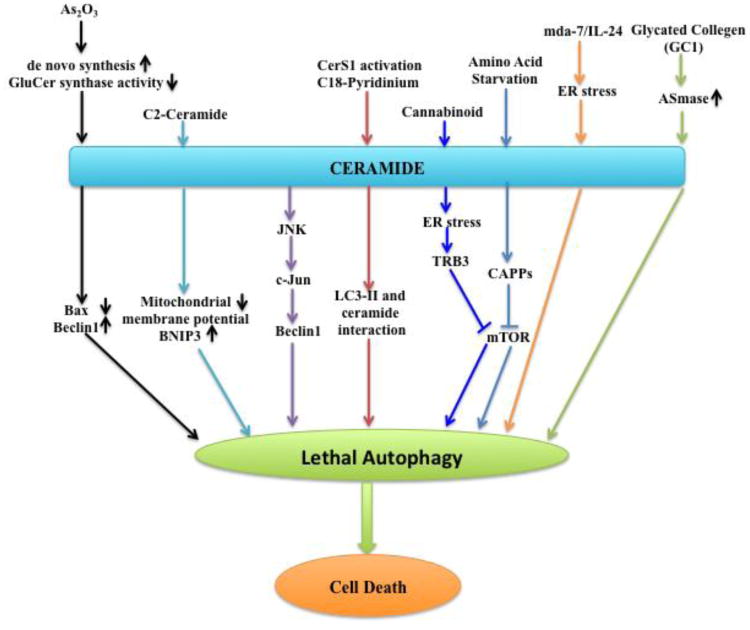
Fig 4. Signaling pathways of lethal autophagy that are regulated by ceramide.
Under most conditions, ceramide leads to lethal autophagy induction and cell death. Arsenic trioxide (As2O3) induces cytotoxic ceramide accumulation via de novo synthesis upregulation and inhibition of GluCer synthase activity, which stimulates Beclin1 expression, leading to lethal autophagy. Ceramide mediated autophagic cell death may be attributed to mitochondrial membrane potential reduction and death-inducing mitochondrial protein BNIP3 activation. Ceramide also induced c-Jun activation through JNK signaling, which transcriptionally upregulated Beclin-1 expression, leading to autophagic cell death. C18-pyridinium ceramide treatment or CerS1 activation induces lethal autophagy, independent of apoptosis, in which ceramide/LC3II interaction plays a key role to target autophagolysosomes to damaged mitochondria. Cannabinoid, which was known to induce ceramide accumulation, activated ER stress, leading to lethal autophagy induction via tribbles homolog 3 (TRB3) mediated mTOR inhibition. Amino acid deprivation also induces CAPPs (ceramide activated protein phosphatases), which have inhibitory effects on Akt/mTOR pathway, resulting in autophagy-mediated cell death. Melanoma differentiation associated gene-7(mda-7)/interleukin-24(IL-24) was shown to induce ER stress, which triggered ceramide-mediated lethal autophagy. Glycated collagen1 (GC1) exposure, which leads to ceramide accumulation and acid sphingomyelinase (ASMase) activation, induces autophagy mediated cellular senescence.
DHC may play a similar role like Cer to induce autophagic cell killing. Gamma-tocotrienol (γTE) treatment led to marked increase of intracellular DHC and dihydrosphingosine and autophagy induction in human prostate PC-3 and LNCaP cancer cells, which caused fatality and decrease cell viability in vitro and inhibited xenograft growth in vivo[118]. This phenomenon was prevented by myriosin, a specific inhibitor of de novo sphingolipid synthesis, supporting the notion that DHC and dihydrosphingosine accumulation could be utilized as a novel anti-cancer mechanism of γTE[118].
6. Conclusion & future perspectives
Autophagy primarily regulates cellular homeostasis[11]. It functions not only to remove aggregated proteins and damaged organelles, but also supplies cells with energy and metabolites to maintain normal cellular functions under stressful conditions[24,39]. However, autophagy has paradoxical functions to regulate the cell fate. It may also induce cell death in two dependent mechanisms: one is caspase-independent (autophagic) cell death, the other is caspase-dependent (iDISC-mediated) cell death[96].
Autophagy is a tumor suppressor mechanism, supported by its role in preventing oxidative stress, maintaining normal mitochondrial function and safeguarding against DNA damage and genetic instability[39]. Autophagy is also a tumor promoting mechanism, which, established cancer cells exploit to meet increased metabolic demands. Also, autophagy is upregulated by cancer cells to resist cancer treatment, suppression of which enhances cytotoxicity of cancer therapy[22,33,49]
Sphingolipids are crucial regulators of autophagy. Various sphingolipids metabolites execute different functions in autophagy regulation. Based on the current literature, S1P-depedent autophagy has been mostly associated with cell survival[135,138–140], which is also comprehensively reviewed by many investigators[67,75,96,108,141,142]. Although Cer-mediated autophagy has also been found to be cytoprotective[104,112], it mainly functions to promote cell death[98,109,114]. Cer utilizes various mechanisms to induce autophagy. It is known to induce autophagy via the blockade of Akt and mTOR signaling[97–99]. It also leads to starvation-induced autophagy by downregulating nutrient transporters[104]. Moreover, it activates JNK/c-Jun pathway to upregulate Beclin1 or dissociates Beclin1 from its binding with Bcl2[98,107,109]. Cer-mediated ER stress is also involved in autophagy induction[96,112]. Our group demonstrated that mitochondrial ceramide directly binds and recruits LC3B-II labeled autophagosomes to damaged mitochondria forlysosomal degradation, also termed lethal mitophagy, [114]. In fact, generation of C18-ceramide by CerS1 and ceramide localization to mitochondria were required for stress-induced lethal autophagy/mitophagy, which was Drp1-mitochondrial fission dependent [114], placing mitochondrial ceramide in the center for selectively inducing lethal mitophagy (Fig. 4). Sphingolipid metabolism and related metabolites are closely linked to mitochondria function[96]. Further studies are needed to dissect the role of Cer in the regulation of lethal mitophagy induction, such as its involvement in manipulating the Parkin/Pink1 pathway. Interestingly, DHC is also involved in autophagy induction[115–117,121]. It may cooperate with Cer to regulate autophagy and determine cell fate. However, the biological activity of DHC is still controversial and needs to be warranted. Finally, the role and mechanisms of action of different Cer species or chain-length specific Cers in autophagy regulation remain unclear, and need to be elucidated[119,123].
Dysregulated Cer metabolism and/or trafficking has been observed in many cancer studies and is closely related to cancer progression and metastasis[143– 146]. Cer has been shown to act as a novel biomarker of cancer therapy response[147], as well as a potential target for the generation of chemotherapeutics[148,149]. In this review, we mainly described the role of Cer in lethal autophagy regulation, which might be an essential mechanism Cer utilize to induce cell killing. In fact, several autophagy-inducing, as well as Cer producing drugs, such as As2O3, tamoxifen and resveratrol, execute their anticancer function via autophagy modulation[108]. Therefore, those observations support the notion that Cer-mediated autophagic cell death may be a promising strategy to inhibit tumor growth. An enormous growth of Cer and DHC research will be anticipated to produce numerous advances in dissecting the roles and mechanisms of Cer and DHC in the regulation of autophagy and cancer pathogenesis. Cer-mediated autophagic cell death pathways may help develop novel therapeutic and/or prevention strategies against various human cancers.
In summary, although there are solid studies to establish a role for Cer in inducing autophagy, more mechanistic studies are needed to clarify how Cer regulates autophagy-mediated cell death versus protection (autophagy paradox) in response to various stress conditions in different cancer types. This regulation seems to be context dependent with regard to subcellular localization of ceramides (mitochondria versus ER), fatty acid chain length composition of ceramides (C18-ceramide versus C16-ceramide), and/or presence/absence of downstream targets of ceramides (Drp1 and LC3B-II) for induction of lethal versus survival autophagy. To this end, it is important to develop new techniques for purification of autophagolysosomes versus lysosomes or mitochondria, and measure their sphingolipid composition using mass spectrometry-based lipidomics, which will help define the roles of specific sphingolipids (and their subcellular localization) in the regulation of autophagy paradox. In addition, defining the molecular details of sphingolipid-protein binding, such as ceramide-LC3B-II complex [114], will be important to understand the mechanisms by which ceramide regulates lethal versus survival autophagy.
Highlights.
Sphingolipids, including ceramide and sphigosine 1-phosphate are key bioactive molecules that regulate autophagy.
This review article focuses on the roles of ceramide on the regulation of autophagy.
Ceramide and dihydro-ceramide induce macroautophagy in various cell types under different stress conditions.
Ceramide-mediated autophagy can be protective or lethal in cancer cells, which is context dependent.
Ceramide-induced autophagy includes lethal mitophagy that results in tumor suppression.
Acknowledgments
This work was supported by grant support from the National Institutes of Health (CA088932, CA173687 and DE016572). We thank Dr. Suzanne Orr (Hollings Cancer Center) for her editorial review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 2.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kon M, Cuervo AM. Chaperone-mediated autophagy in health and disease. FEBS Lett. 2010;584:1399–1404. doi: 10.1016/j.febslet.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 6.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanida I. Autophagy basics. Microbiol Immunol. 2011;55:1–11. doi: 10.1111/j.1348-0421.2010.00271.x. [DOI] [PubMed] [Google Scholar]
- 10.Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13:495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi AMK, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 13.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–2906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- 15.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 16.Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13:805–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubinsztein DC, Mariño G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 18.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizumura K, Cloonan SM, Haspel JA, Choi AMK. The emerging importance of autophagy in pulmonary diseases. Chest. 2012;142:1289–1299. doi: 10.1378/chest.12-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirshenbaum LA. Regulation of autophagy in the heart in health and disease. J Cardiovasc Pharmacol. 2012;60:109. doi: 10.1097/FJC.0b013e31825f6faa. [DOI] [PubMed] [Google Scholar]
- 21.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mah LY, Ryan KM. Autophagy and cancer. Cold Spring Harb Perspect Biol. 2012;4:a008821. doi: 10.1101/cshperspect.a008821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sridhar S, Botbol Y, Macian F, Cuervo AM. Autophagy and disease: always two sides to a problem. J Pathol. 2012;226:255–273. doi: 10.1002/path.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11:709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 26.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 27.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 28.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self- killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 29.Colell A, Ricci JE, Tait S, Milasta S, Maurer U, Bouchier-Hayes L, Fitzgerald P, Guio-Carrion A, Waterhouse NJ, Li CW, Mari B, Barbry P, Newmeyer DD, Beere HM, Green DR. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell. 2007;129:983–997. doi: 10.1016/j.cell.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 30.Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, Slingerland JM, Mills GB. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- 31.Gozuacik D, Kimchi A. Autophagy and cell death. Curr Top Dev Biol. 2007;78:217–245. doi: 10.1016/S0070-2153(06)78006-1. [DOI] [PubMed] [Google Scholar]
- 32.Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, Dawson TM, Dawson VL, El-Deiry WS, Fulda S, Gottlieb E, Green DR, Hengartner MO, Kepp O, Knight RA, Kumar S, Lipton SA, Lu X, Madeo F, Malorni W, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19:107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi KS. Autophagy and cancer. Exp Mol Med. 2012;44:109–120. doi: 10.3858/emm.2012.44.2.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 35.Yu L, Wan F, Dutta S, Welsh S, Liu Z, Freundt E, Baehrecke EH, Lenardo M. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci U S A. 2006;103:4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 38.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sridhar S, Botbol Y, Macian F, Cuervo AM. Autophagy and disease: always two sides to a problem. J Pathol. 2012;226:255–273. doi: 10.1002/path.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito H, Inazawa J, Saito S, Kasumi F, Koi S, Sagae S, Kudo R, Saito J, Noda K, Nakamura Y. Detailed deletion mapping of chromosome 17q in ovarian and breast cancers: 2-cM region on 17q213 often and commonly deleted in tumors. Cancer Res. 1993;53:3382–3385. [PubMed] [Google Scholar]
- 41.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 42.Gao X, Zacharek A, Salkowski A, Grignon DJ, Sakr W, Porter AT, Honn KV. Loss of heterozygosity of the BRCA1 and other loci on chromosome 17q in human prostate cancer. Cancer Res. 1995;55:1002–1005. [PubMed] [Google Scholar]
- 43.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol. 2011;8:528–539. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- 46.Martelli AM, Tazzari PL, Evangelisti C, Chiarini F, Blalock WL, Billi AM, Manzoli L, McCubrey JA, Cocco L. Targeting the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin module for acute myelogenous leukemia therapy: from bench to bedside. Curr Med Chem. 2007;14:2009–2023. doi: 10.2174/092986707781368423. [DOI] [PubMed] [Google Scholar]
- 47.Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eskelinen EL. The dual role of autophagy in cancer. Curr Opin Pharmacol. 2011;11:294–300. doi: 10.1016/j.coph.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 50.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 53.Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Villeneuve NF, Lau A, Zhang DD. Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases. Antioxid Redox Signal. 2010;13:1699–1712. doi: 10.1089/ars.2010.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, Dipaola RS, Karantza-Wadsworth V, White E. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young ARJ, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JFJ, Tavaré S, Arakawa S, Shimizu S, Watt FM, Narita M. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell'Antonio G, Mautner J, Tonon G, Haigis M, Shirihai OS, Doglioni C, Bardeesy N, Kimmelman AC. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JMS, Karantza V, Coller HA, Dipaola RS, Gelinas C, Rabinowitz JD, White E. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen N, Karantza-Wadsworth V. Role and regulation of autophagy in cancer. Biochim Biophys Acta. 2009;1793:1516–1523. doi: 10.1016/j.bbamcr.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adiseshaiah PP, Clogston JD, McLeland CB, Rodriguez J, Potter TM, Neun BW, Skoczen SL, Shanmugavelandy SS, Kester M, Stern ST, McNeil SE. Synergistic combination therapy with nanoliposomal C6-ceramide and vinblastine is associated with autophagy dysfunction in hepatocarcinoma and colorectal cancer models. Cancer Lett. 2013;337:254–265. doi: 10.1016/j.canlet.2013.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 63.Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 64.Thompson CB. Metabolic enzymes as oncogenes or tumor suppressors. N Engl J Med. 2009;360:813–815. doi: 10.1056/NEJMe0810213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ogretmen B, Hannun YA. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat Rev Cancer. 2004;4:604–616. doi: 10.1038/nrc1411. [DOI] [PubMed] [Google Scholar]
- 66.Futerman AH, Hannun YA. The complex life of simple sphingolipids. EMBO Rep. 2004;5:777–782. doi: 10.1038/sj.embor.7400208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ponnusamy S, Meyers-Needham M, Senkal CE, Saddoughi SA, Sentelle D, Selvam SP, Salas A, Ogretmen B. Sphingolipids and cancer: ceramide and sphingosine-1-phosphate in the regulation of cell death and drug resistance. Future Oncol. 2010;6:1603–1624. doi: 10.2217/fon.10.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Merrill AH, Jr, Wang E, Mullins RE. Kinetics of long-chain (sphingoid) base biosynthesis in intact LM cells: effects of varying the extracellular concentrations of serine and fatty acid precursors of this pathway. Biochemistry. 1988;27:340–345. doi: 10.1021/bi00401a051. [DOI] [PubMed] [Google Scholar]
- 69.Kang MS, Ahn KH, Kim SK, Jeon HJ, Ji JE, Choi JM, Jung KM, Jung SY, Kim DK. Hypoxia-induced neuronal apoptosis is mediated by de novo synthesis of ceramide through activation of serine palmitoyltransferase. Cell Signal. 2010;22:610–618. doi: 10.1016/j.cellsig.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 70.Hannun YA, Obeid LM. The Ceramide-centric universe of lipidmediated cell regulation: stress encounters of the lipid kind. J Biol Chem. 2002;277:25847–25850. doi: 10.1074/jbc.R200008200. [DOI] [PubMed] [Google Scholar]
- 71.Tafesse FG, Ternes P, Holthuis JCM. The multigenic sphingomyelin synthase family. J Biol Chem. 2006;281:29421–29425. doi: 10.1074/jbc.R600021200. [DOI] [PubMed] [Google Scholar]
- 72.Stefanić S, Spycher C, Morf L, Fabriàs G, Casas J, Schraner E, Wild P, Hehl AB, Sonda S. Glucosylceramide synthesis inhibition affects cell cycle progression, membrane trafficking, and stage differentiation in Giardia lamblia. J Lipid Res. 2010;51:2527–2545. doi: 10.1194/jlr.M003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morales A, París R, Villanueva A, Llacuna L, García-Ruiz C, Fernández-Checa JC. Pharmacological inhibition or small interfering RNA targeting acid ceramidase sensitizes hepatoma cells to chemotherapy and reduces tumor growth in vivo. Oncogene. 2007;26:905–916. doi: 10.1038/sj.onc.1209834. [DOI] [PubMed] [Google Scholar]
- 74.Park JH, Schuchman EH. Acid ceramidase and human disease. Biochim Biophys Acta. 2006;1758:2133–2138. doi: 10.1016/j.bbamem.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 75.Selvam SP, Ogretmen B. Sphingosine kinase/sphingosine 1-phosphate signaling in cancer therapeutics and drug resistance. Handb Exp Pharmacol. 2013;216:3–27. doi: 10.1007/978-3-7091-1511-4_1. [DOI] [PubMed] [Google Scholar]
- 76.Saddoughi SA, Ogretmen B. Diverse functions of ceramide in cancer cell death and proliferation. Adv Cancer Res. 2013;117:37–58. doi: 10.1016/B978-0-12-394274-6.00002-9. [DOI] [PubMed] [Google Scholar]
- 77.Saddoughi SA, Song P, Ogretmen B. Roles of bioactive sphingolipids in cancer biology and therapeutics. Subcell Biochem. 2008;49:413–440. doi: 10.1007/978-1-4020-8831-5_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nylund M, Kjellberg MA, Molotkovsky JG, Byun HS, Bittman R, Mattjus P. Molecular features of phospholipids that affect glycolipid transfer protein-mediated galactosylceramide transfer between vesicles. Biochim Biophys Acta. 2006;1758:807–812. doi: 10.1016/j.bbamem.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 79.Hanada K, Kumagai K, Tomishige N, Yamaji T. CERT-mediated trafficking of ceramide. Biochim Biophys Acta. 2009;1791:684–691. doi: 10.1016/j.bbalip.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 80.Hanada K, Kumagai K, Tomishige N, Kawano M. CERT and intracellular trafficking of ceramide. Biochim Biophys Acta. 2007;1771:644–653. doi: 10.1016/j.bbalip.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 81.Kumagai K, Yasuda S, Okemoto K, Nishijima M, Kobayashi S, Hanada K. CERT mediates intermembrane transfer of various molecular species of ceramides. J Biol Chem. 2005;280:6488–6495. doi: 10.1074/jbc.M409290200. [DOI] [PubMed] [Google Scholar]
- 82.Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- 83.D'Angelo G, Polishchuk E, Di Tullio G, Santoro M, Di Campli A, Godi A, West G, Bielawski J, Chuang CC, van der Spoel AC, Platt FM, Hannun YA, Polishchuk R, Mattjus P, De Matteis MA. Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature. 2007;449:62–67. doi: 10.1038/nature06097. [DOI] [PubMed] [Google Scholar]
- 84.Bose R, Verheij M, Haimovitz-Friedman A, Scotto K, Fuks Z, Kolesnick R. Ceramide synthase mediates daunorubicin-induced apoptosis: an alternative mechanism for generating death signals. Cell. 1995;82:405–414. doi: 10.1016/0092-8674(95)90429-8. [DOI] [PubMed] [Google Scholar]
- 85.Jazwinski SM, Conzelmann A. LAG1 puts the focus on ceramide signaling. Int J Biochem Cell Biol. 2002;34:1491–1495. doi: 10.1016/s1357-2725(02)00044-4. [DOI] [PubMed] [Google Scholar]
- 86.Pewzner-Jung Y, Ben-Dor S, Futerman AH. When do Lasses (longevity assurance genes) become CerS (ceramide synthases)?: Insights into the regulation of ceramide synthesis. J Biol Chem. 2006;281:25001–25005. doi: 10.1074/jbc.R600010200. [DOI] [PubMed] [Google Scholar]
- 87.Venkataraman K, Riebeling C, Bodennec J, Riezman H, Allegood JC, Sullards MC, Merrill AH, Jr, Futerman AH. Upstream of growth and differentiation factor 1 (uog1), a mammalian homolog of the yeast longevity assurance gene 1 (LAG1), regulates N-stearoyl-sphinganine (C18-(dihydro)ceramide) synthesis in a fumonisin B1-independent manner in mammalian cells. J Biol Chem. 2002;277:35642–35649. doi: 10.1074/jbc.M205211200. [DOI] [PubMed] [Google Scholar]
- 88.Lee SJ. Expression of growth/differentiation factor 1 in the nervous system: conservation of a bicistronic structure, Proc. Natl Acad Sci U S A. 1991;88:4250–4254. doi: 10.1073/pnas.88.10.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mizutani Y, Kihara A, Igarashi Y. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem J. 2005;390:263–271. doi: 10.1042/BJ20050291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kageyama-Yahara N, Riezman H. Transmembrane topology of ceramide synthase in yeast. Biochem J. 2006;398:585–593. doi: 10.1042/BJ20060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Spassieva S, Seo JG, Jiang JC, Bielawski J, Alvarez-Vasquez F, Jazwinski SM, Hannun YA, Obeid LM. Necessary role for the Lag1p motif in (dihydro)ceramide synthase activity. J Biol Chem. 2006;281:33931–33938. doi: 10.1074/jbc.M608092200. [DOI] [PubMed] [Google Scholar]
- 92.Schulz A, Mousallem T, Venkataramani M, Persaud-Sawin DA, Zucker A, Luberto C, Bielawska A, Bielawski J, Holthuis JCM, Jazwinski SM, Kozhaya L, Dbaibo GS, Boustany RMN. The CLN9 protein, a regulator of dihydroceramide synthase. J Biol Chem. 2006;281:2784–2794. doi: 10.1074/jbc.M509483200. [DOI] [PubMed] [Google Scholar]
- 93.Imgrund S, Hartmann D, Farwanah H, Eckhardt M, Sandhoff R, Degen J, Gieselmann V, Sandhoff K, Willecke K. Adult ceramide synthase 2 (CERS2)-deficient mice exhibit myelin sheath defects, cerebellar degeneration, and hepatocarcinomas. J Biol Chem. 2009;284:33549–33560. doi: 10.1074/jbc.M109.031971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jennemann R, Rabionet M, Gorgas K, Epstein S, Dalpke A, Rothermel U, Bayerle A, van der Hoeven F, Imgrund S, Kirsch J, Nickel W, Willecke K, Riezman H, Gröne HJ, Sandhoff R. Loss of ceramide synthase 3 causes lethal skin barrier disruption. Hum Mol Genet. 2012;21:586–608. doi: 10.1093/hmg/ddr494. [DOI] [PubMed] [Google Scholar]
- 95.Russo SB, Tidhar R, Futerman AH, Cowart LA. Myristate-derived d16:0 Sphingolipids Constitute a Cardiac Sphingolipid Pool with Distinct Synthetic Routes and Functional Properties. J Biol Chem. 2013;288:13397–13409. doi: 10.1074/jbc.M112.428185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Young MM, Kester M, Wang HG. Sphingolipids: regulators of crosstalk between apoptosis and autophagy. J Lipid Res. 2013;54:5–19. doi: 10.1194/jlr.R031278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schubert KM, Scheid MP, Duronio V. Ceramide Inhibits Protein Kinase B/Akt by Promoting Dephosphorylation of Serine 473. J Biol Chem. 2000;275:13330–13335. doi: 10.1074/jbc.275.18.13330. [DOI] [PubMed] [Google Scholar]
- 98.Scarlatti F, Bauvy C, Ventruti A, Sala G, Cluzeaud F, Vandewalle A, Ghidoni R, Codogno P. Ceramide-mediated macroautophagy involves inhibition of protein kinase B and up-regulation of beclin 1. J Biol Chem. 2004;279:18384–18391. doi: 10.1074/jbc.M313561200. [DOI] [PubMed] [Google Scholar]
- 99.Zhou H, Summers SA, Birnbaum MJ, Pittman RN. Inhibition of Akt Kinase by Cell-permeable Ceramide and Its Implications for Ceramide-induced Apoptosis. J Biol Chem. 1998;273:16568–16575. doi: 10.1074/jbc.273.26.16568. [DOI] [PubMed] [Google Scholar]
- 100.Ogretmen B, Pettus BJ, Rossi MJ, Wood R, Usta J, Szulc Z, Bielawska A, Obeid LM, Hannun YA. Biochemical mechanisms of the generation of endogenous long chain ceramide in response to exogenous short chain ceramide in the A549 human lung adenocarcinoma cell line Role for endogenous ceramide in mediating the action of exogenous ceramide. J Biol Chem. 2002;277:12960–12969. doi: 10.1074/jbc.M110699200. [DOI] [PubMed] [Google Scholar]
- 101.Guenther GG, Edinger AL. A new take on ceramide: starving cells by cutting off the nutrient supply. Cell Cycle. 2009;8:1122–1126. doi: 10.4161/cc.8.8.8161. [DOI] [PubMed] [Google Scholar]
- 102.Peralta ER, Edinger AL. Ceramide-induced starvation triggers homeostatic autophagy. Autophagy. 2009;5:407–409. doi: 10.4161/auto.5.3.7809. [DOI] [PubMed] [Google Scholar]
- 103.Edinger AL. Starvation in the midst of plenty: making sense of ceramide-induced autophagy by analysing nutrient transporter expression. Biochem Soc Trans. 2009;37:253–258. doi: 10.1042/BST0370253. [DOI] [PubMed] [Google Scholar]
- 104.Guenther GG, Peralta ER, Rosales KR, Wong SY, Siskind LJ, Edinger AL. Ceramide starves cells to death by downregulating nutrient transporter proteins. Proc Natl Acad Sci U S A. 2008;105:17402–17407. doi: 10.1073/pnas.0802781105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 107.Li DD, Wang LL, Deng R, Tang J, Shen Y, Guo JF, Wang Y, Xia LP, Feng GK, Liu QQ, Huang WL, Zeng YX, Zhu XF. The pivotal role of c-Jun NH2-terminal kinase-mediated Beclin 1 expression during anticancer agents-induced autophagy in cancer cells. Oncogene. 2009;28:886–898. doi: 10.1038/onc.2008.441. [DOI] [PubMed] [Google Scholar]
- 108.Bedia C, Levade T, Codogno P. Regulation of autophagy by sphingolipids. Anticancer Agents Med Chem. 2011;11:844–853. doi: 10.2174/187152011797655131. [DOI] [PubMed] [Google Scholar]
- 109.Daido S, Kanzawa T, Yamamoto A, Takeuchi H, Kondo Y, Kondo S. Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res. 2004;64:4286–4293. doi: 10.1158/0008-5472.CAN-03-3084. [DOI] [PubMed] [Google Scholar]
- 110.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 111.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL. Mitochondrial Autophagy Is an HIF-1-dependent Adaptive Metabolic Response to Hypoxia. J Biol Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 112.Spassieva SD, Mullen TD, Townsend DM, Obeid LM. Disruption of ceramide synthesis by CerS2 down-regulation leads to autophagy and the unfolded protein response. Biochemical Journal. 2009;424:273–283. doi: 10.1042/BJ20090699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Russo SB, Baicu CF, Van Laer A, Geng T, Kasiganesan H, Zile MR, Cowart LA. Ceramide synthase 5 mediates lipid-induced autophagy and hypertrophy in cardiomyocytes. J Clin Invest. 2012;122:3919–3930. doi: 10.1172/JCI63888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sentelle RD, Senkal CE, Jiang W, Ponnusamy S, Gencer S, Panneer Selvam S, Ramshesh VK, Peterson YK, Lemasters JJ, Szulc ZM, Bielawski J, Ogretmen B. Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nat Chem Biol. 2012 doi: 10.1038/nchembio.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gagliostro V, Casas J, Caretti A, Abad JL, Tagliavacca L, Ghidoni R, Fabrias G, Signorelli P. Dihydroceramide delays cell cycle G1/S transition via activation of ER stress and induction of autophagy. The International Journal of Biochemistry & Cell Biology. 2012;44:2135–2143. doi: 10.1016/j.biocel.2012.08.025. [DOI] [PubMed] [Google Scholar]
- 116.Signorelli P, Munoz-Olaya JM, Gagliostro V, Casas J, Ghidoni R, Fabriàs G. Dihydroceramide intracellular increase in response to resveratrol treatment mediates autophagy in gastric cancer cells. Cancer Letters. 2009;282:238–243. doi: 10.1016/j.canlet.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 117.Devlin CM, Lahm T, Hubbard WC, Demark MV, Wang KC, Wu X, Bielawska A, Obeid LM, Ivan M, Petrache I. Dihydroceramide-based Response to Hypoxia. J Biol Chem. 2011;286:38069–38078. doi: 10.1074/jbc.M111.297994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jiang Q, Rao X, Kim CY, Freiser H, Zhang Q, Jiang Z, Li G. Gamma-tocotrienol induces apoptosis and autophagy in prostate cancer cells by increasing intracellular dihydrosphingosine and dihydroceramide. International Journal of Cancer. 2012;130:685–693. doi: 10.1002/ijc.26054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zheng W, Kollmeyer J, Symolon H, Momin A, Munter E, Wang E, Kelly S, Allegood JC, Liu Y, Peng Q, Ramaraju H, Sullards MC, Cabot M, Merrill AH., Jr Ceramides and other bioactive sphingolipid backbones in health and disease: lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim Biophys Acta. 2006;1758:1864–1884. doi: 10.1016/j.bbamem.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 120.Signorelli P, Munoz-Olaya JM, Gagliostro V, Casas J, Ghidoni R, Fabriàs G. Dihydroceramide intracellular increase in response to resveratrol treatment mediates autophagy in gastric cancer cells. Cancer Lett. 2009;282:238–243. doi: 10.1016/j.canlet.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 121.Huang S, Sinicrope FA. Celecoxib-induced apoptosis is enhanced by ABT-737 and by inhibition of autophagy in human colorectal cancer cells. Autophagy. 2010;6:256–269. doi: 10.4161/auto.6.2.11124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schiffmann S, Sandner J, Schmidt R, Birod K, Wobst I, Schmidt H, Angioni C, Geisslinger G, Grösch S. The selective COX-2 inhibitor celecoxib modulates sphingolipid synthesis. J Lipid Res. 2009;50:32–40. doi: 10.1194/jlr.M800122-JLR200. [DOI] [PubMed] [Google Scholar]
- 123.Grösch S, Schiffmann S, Geisslinger G. Chain length-specific properties of ceramides. Prog Lipid Res. 2012;51:50–62. doi: 10.1016/j.plipres.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 124.Jana A, Hogan EL, Pahan K. Ceramide and neurodegeneration: susceptibility of neurons and oligodendrocytes to cell damage and death. J Neurol Sci. 2009;278:5–15. doi: 10.1016/j.jns.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Park MA, Zhang G, Martin AP, Hamed H, Mitchell C, Hylemon PB, Graf M, Rahmani M, Ryan K, Liu X, Spiegel S, Norris J, Fisher PB, Grant S, Dent P. Vorinostat and sorafenib increase ER stress, autophagy and apoptosis via ceramide-dependent CD95 and PERK activation. Cancer Biol Ther. 2008;7:1648–1662. doi: 10.4161/cbt.7.10.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang G, Park MA, Mitchell C, Walker T, Hamed H, Studer E, Graf M, Rahmani M, Gupta S, Hylemon PB, Fisher PB, Grant S, Dent P. Multiple cyclin kinase inhibitors promote bile acid-induced apoptosis and autophagy in primary hepatocytes via p53-CD95-dependent signaling. J Biol Chem. 2008;283:24343–24358. doi: 10.1074/jbc.M803444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bursch W, Ellinger A, Kienzl H, Török L, Pandey S, Sikorska M, Walker R, Hermann RS. Active cell death induced by the anti-estrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MCF-7) in culture: the role of autophagy. Carcinogenesis. 1996;17:1595–1607. doi: 10.1093/carcin/17.8.1595. [DOI] [PubMed] [Google Scholar]
- 128.Pattingre S, Bauvy C, Levade T, Levine B, Codogno P. Ceramide-induced autophagy: to junk or to protect cells? Autophagy. 2009;5:558–560. doi: 10.4161/auto.5.4.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Samaddar JS, Gaddy VT, Duplantier J, Thandavan SP, Shah M, Smith MJ, Browning D, Rawson J, Smith SB, Barrett JT, Schoenlein PV. A role for macroautophagy in protection against 4-hydroxytamoxifen-induced cell death and the development of antiestrogen resistance. Mol Cancer Ther. 2008;7:2977–2987. doi: 10.1158/1535-7163.MCT-08-0447. [DOI] [PubMed] [Google Scholar]
- 130.Lavieu G, Scarlatti F, Sala G, Levade T, Ghidoni R, Botti J, Codogno P. Is autophagy the key mechanism by which the sphingolipid rheostat controls the cell fate decision? Autophagy. 2007;3:45–47. doi: 10.4161/auto.3416. [DOI] [PubMed] [Google Scholar]
- 131.Dbaibo GS, Kfoury Y, Darwiche N, Panjarian S, Kozhaya L, Nasr R, Abdallah M, Hermine O, El-Sabban M, de Thé H, Bazarbachi A. Arsenic trioxide induces accumulation of cytotoxic levels of ceramide in acute promyelocytic leukemia and adult T-cell leukemia/lymphoma cells through de novo ceramide synthesis and inhibition of glucosylceramide synthase activity. Haematologica. 2007;92:753–762. doi: 10.3324/haematol.10968. [DOI] [PubMed] [Google Scholar]
- 132.Qian W, Liu J, Jin J, Ni W, Xu W. Arsenic trioxide induces not only apoptosis but also autophagic cell death in leukemia cell lines via up-regulation of Beclin-1. Leuk Res. 2007;31:329–339. doi: 10.1016/j.leukres.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 133.Yacoub A, Hamed HA, Allegood J, Mitchell C, Spiegel S, Lesniak MS, Ogretmen B, Dash R, Sarkar D, Broaddus WC, Grant S, Curiel DT, Fisher PB, Dent P. PERK-dependent regulation of ceramide synthase 6 and thioredoxin play a key role in mda-7/IL-24-induced killing of primary human glioblastoma multiforme cells. Cancer Res. 2010;70:1120–1129. doi: 10.1158/0008-5472.CAN-09-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Patschan S, Chen J, Polotskaia A, Mendelev N, Cheng J, Patschan D, Goligorsky MS. Lipid mediators of autophagy in stress-induced premature senescence of endothelial cells, Am. J Physiol Heart Circ Physiol. 2008;294:H1119–1129. doi: 10.1152/ajpheart.00713.2007. [DOI] [PubMed] [Google Scholar]
- 135.Taniguchi M, Kitatani K, Kondo T, Hashimoto-Nishimura M, Asano S, Hayashi A, Mitsutake S, Igarashi Y, Umehara H, Takeya H, Kigawa J, Okazaki T. Regulation of autophagy and its associated cell death by “sphingolipid rheostat”: reciprocal role of ceramide and sphingosine 1-phosphate in the mammalian target of rapamycin pathway. J Biol Chem. 2012;287:39898–39910. doi: 10.1074/jbc.M112.416552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hou Q, Jin J, Zhou H, Novgorodov SA, Bielawska A, Szulc ZM, Hannun YA, Obeid LM, Hsu YT. Mitochondrially targeted ceramides preferentially promote autophagy, retard cell growth, and induce apoptosis. J Lipid Res. 2011;52:278–288. doi: 10.1194/jlr.M012161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Salazar M, Carracedo A, Salanueva IJ, Hernández-Tiedra S, Lorente M, Egia A, Vázquez P, Blázquez C, Torres S, García S, Nowak J, Fimia GM, Piacentini M, Cecconi F, Pandolfi PP, González-Feria L, Iovanna JL, Guzmán M, Boya P, Velasco G. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest. 2009;119:1359–1372. doi: 10.1172/JCI37948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chang CL, Ho MC, Lee PH, Hsu CY, Huang WP, Lee H. S1P5 is required for sphingosine 1-phosphate-induced autophagy in human prostate cancer PC-3 cells. Am J Physiol Cell Physiol. 2009;297:C451–C458. doi: 10.1152/ajpcell.00586.2008. [DOI] [PubMed] [Google Scholar]
- 139.Lavieu G, Scarlatti F, Sala G, Carpentier S, Levade T, Ghidoni R, Botti J, Codogno P. Regulation of Autophagy by Sphingosine Kinase 1 and Its Role in Cell Survival during Nutrient Starvation. J Biol Chem. 2006;281:8518–8527. doi: 10.1074/jbc.M506182200. [DOI] [PubMed] [Google Scholar]
- 140.Lépine S, Allegood JC, Park M, Dent P, Milstien S, Spiegel S. Sphingosine-1-phosphate phosphohydrolase-1 regulates ER stress-induced autophagy. Cell Death and Differentiation. 2010;18:350–361. doi: 10.1038/cdd.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yester JW, Tizazu E, Harikumar KB, Kordula T. Extracellular and intracellular sphingosine-1-phosphate in cancer. Cancer Metastasis Rev. 2011;30:577–597. doi: 10.1007/s10555-011-9305-0. [DOI] [PubMed] [Google Scholar]
- 142.Van Brocklyn JR, Williams JB. The control of the balance between ceramide and sphingosine-1-phosphate by sphingosine kinase: oxidative stress and the seesaw of cell survival and death. Comp Biochem Physiol B, Biochem Mol Biol. 2012;163:26–36. doi: 10.1016/j.cbpb.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 143.Senkal CE, Ponnusamy S, Bielawski J, Hannun YA, Ogretmen B. Antiapoptotic roles of ceramide-synthase-6-generated C16-ceramide via selective regulation of the ATF6/CHOP arm of ER-stress-response pathways. FASEB J. 2010;24:296–308. doi: 10.1096/fj.09-135087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Karahatay S, Thomas K, Koybasi S, Senkal CE, Elojeimy S, Liu X, Bielawski J, Day TA, Gillespie MB, Sinha D, Norris JS, Hannun YA, Ogretmen B. Clinical relevance of ceramide metabolism in the pathogenesis of human head and neck squamous cell carcinoma (HNSCC): attenuation of C(18)-ceramide in HNSCC tumors correlates with lymphovascular invasion and nodal metastasis. Cancer Lett. 2007;256:101–111. doi: 10.1016/j.canlet.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ryland LK, Fox TE, Liu X, Loughran TP, Kester M. Dysregulation of sphingolipid metabolism in cancer. Cancer Biology & Therapy. 2011;11:138–149. doi: 10.4161/cbt.11.2.14624. [DOI] [PubMed] [Google Scholar]
- 146.Senkal CE, Ponnusamy S, Rossi MJ, Bialewski J, Sinha D, Jiang JC, Jazwinski SM, Hannun YA, Ogretmen B. Role of human longevity assurance gene 1 and C18-ceramide in chemotherapy-induced cell death in human head and neck squamous cell carcinomas. Mol Cancer Ther. 2007;6:712–722. doi: 10.1158/1535-7163.MCT-06-0558. [DOI] [PubMed] [Google Scholar]
- 147.Saddoughi SA, Garrett-Mayer E, Chaudhary U, O'Brien PE, Afrin LB, Day TA, Gillespie MB, Sharma AK, Wilhoit CS, Bostick R, Senkal CE, Hannun YA, Bielawski J, Simon GR, Shirai K, Ogretmen B. Results of a phase II trial of gemcitabine plus doxorubicin in patients with recurrent head and neck cancers: serum C1 8 -ceramide as a novel biomarker for monitoring response. Clin Cancer Res. 2011;17:6097–6105. doi: 10.1158/1078-0432.CCR-11-0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kester M, Kolesnick R. Sphingolipids as therapeutics. Pharmacol Res. 2003;47:365–371. doi: 10.1016/s1043-6618(03)00048-3. [DOI] [PubMed] [Google Scholar]
- 149.Barth BM, Cabot MC, Kester M. Ceramide-based therapeutics for the treatment of cancer. Anticancer Agents Med Chem. 2011;11:911–919. doi: 10.2174/187152011797655177. [DOI] [PubMed] [Google Scholar]