Abstract
It is often discussed whether the biogeography of free-living protists is better explained by the ‘everything is everywhere'(EiE) hypothesis, which postulates that only ecology drives their distribution, or by the alternative hypothesis of ‘moderate endemicity' in which geographic barriers can limit their dispersal. To formally test this, it would be necessary not only to find organisms restricted to a geographical area but also to check for their presence in any other place with a similar ecology. We propose the use of environmental niche models to generate and test null EiE distributions. Here we have analysed the distribution of 18S rDNA variants (ribotypes) of the myxomycete Badhamia melanospora (belonging to the protozoan phylum Amoebozoa) using 125 specimens from 91 localities. Two geographically structured groups of ribotypes congruent with slight morphological differences in the spores can be distinguished. One group comprises all populations from Argentina and Chile, and the other is formed by populations from North America together with human-introduced populations from other parts of the world. Environmental climatic niche models constructed separately for the two groups have significant differences, but show several overlapping areas. However, only specimens from one group were found in an intensively surveyed area in South America where both niche models overlap. It can be concluded that everything is not everywhere for B. melanospora. This taxon constitutes a complex formed by at least two cryptic species that probably diverged allopatrically in North and South America.
Keywords: biogeography of protists, moderate endemicity, Myxomycetes, phylogeography
Introduction
The biogeographical patterns of free-living unicellular eukaryotes are still a subject of debate. The underlying question is to what extent current geographic barriers and historical events have restrained the dispersion of protists, and whether this influence can be traced in the distribution of the organisms that exist today. Some authors (Finlay 2002; Fenchel and Finlay 2004) defend that most of them are cosmopolitan, as described in the ‘everything is everywhere' (EiE) hypothesis. In that case, protist species present in a given location would be a function of only their habitat requirements and not of restricted dispersion. However, others (e.g. Smith and Wilkinson 2007; Vanormelingen et al., 2008) have found evidence in favour of the ‘moderate endemicity' (ME) hypothesis that at least some organisms have geographically restricted distributions (Foissner 1999, 2006; Foissner et al., 2008), not reaching all of their potentially suitable habitats when they disperse.
The advent of phylogeographic methods based on molecular data has improved the resolution for detecting and analysing variability between populations and searching for recent dispersal events. These methods have also shed light on the existence of cryptic species complexes (Amato et al., 2007; Smirnov 2007; Bass et al., 2009; Howe et al., 2009; Morard et al., 2009; Poulíčková et al., 2010; Douglas et al., 2011) that share a common morphology but are genetically distinct.
Clear molecular evidence of geographically restricted 18 S rDNA sequence types (ribotypes) has been found in Foraminifera (Darling et al., 2007; Aurahs et al., 2009), diatoms (Evans et al., 2009; Sorhannus et al., 2010) and Cercozoa (Bass et al., 2007). However, finding a geographic structure does not allow a full rejection of the EiE hypothesis. It is still possible that considered lineages are globally dispersed, but they have different ecological preferences causing them to inhabit distinct areas. One major issue is the limited amount of available information about protists compared with multicellular organisms. In this context, niche models can be a valuable tool for generating null hypotheses of the distribution of the organisms in an EiE scenario from a limited set of data (Aguilar and Lado 2012). Environmental niche models are mathematical functions that describe the probability of the presence of an organism according to the values of a number of environmental variables. They are calculated using the information provided by localities where the organisms were previously found to make an extrapolation to other unexplored areas. These models can be subsequently tested for the presence of barriers to dispersion by checking whether organisms have reached all of their potential habitats.
In this report, we provide a striking example of geographical genetic differentiation in Myxomycetes (Amoebozoa) that has also been analysed taking into account the information provided by niche models. Myxomycetes lend themselves especially well to such studies as their DNA can be extracted and sequenced from fruiting bodies already well preserved in herbaria from many globally widespread locations. B. melanospora Speg is an organism usually found on decaying Cactaceae and other succulent plants. Its fruiting bodies (sporocarps) are easily visible, as they form groups of whitish-grey little balls, approximately 1 mm in diameter, that contain dark-coloured, warted, reticulate spores. The morphology of B. melanospora is highly variable (Lado et al., 2007), but it was not previously known whether this morphological variation is an expression of phenotypic plasticity or is phenotypic evidence of actual genetic divergence. It appears to be mostly restricted to the arid regions of America, where it is very frequent. However, it has never been collected from the Asian arid regions (Novozhilov et al., 2009), and only rarely from other parts of the world, including intensively studied regions like Europe, where it is most frequently found growing on introduced cactae (www.gbif.org).
This study reports an analysis of small subunit ribosomal DNA (SSU rDNA) sequence variation in B. melanospora on its entire known geographical range. For a better understanding of the geographical pattern found, clade-specific habitat preferences have been analysed and compared using niche models. The main character distinguishing clades, that is, the spore morphology, has also been studied using s.e.m. Our results are at odds with the EiE hypothesis, and suggest that B. melanospora constitutes a geographically structured complex formed by at least two cryptic species, which are genetically and morphologically differentiated and display different distributions.
Materials and methods
Sampling, DNA extraction and sequencing
Specimens collected by several myxomycetologists – C Lado, S L Stephenson, M Meyer, L H Cavalcanti and R McHugh – comprising more than 10 sporophores were selected for DNA extraction, resulting in a total of 125 herbarium specimens from 91 different localities (Supplementary Table 1). All collections were represented by material that fruited in the field under natural conditions.
DNA was extracted as described elsewhere (Fiore-Donno et al., 2008), and it was amplified by polymerase chain reaction (PCR) using the primers SA' (TGGTTGATCCTGCCAGTAGTGT) and SU19R (TGTCCTCTAATTGTTACTCGA), Mangotaq mix (Bioline, London, UK) and the following cycling parameters: 45 s at 94 °C, 33 × (25 s at 94 °C, 60 s at 42 °C, 3.5 min at 72 °C) and 5 min at 72 °C. To obtain nearly complete SSU rRNA gene sequences, four overlapping sequence fragments were obtained using the primers SA′, SU19R, and S4, S900R, S11.5, SR15, DA2, RB2 (Fiore-Donno et al., 2008) and the same polymerase chain reaction conditions. Purified polymerase chain reaction products (SureClean kit, Bioline) were sequenced directly by Macrogen Korea (Geumcheon-gu, Seoul, South Korea). All new sequences were submitted to GenBank (accession numbers on Supplementary Table 2).
Phylogenetic analyses
Sequences were automatically aligned with Geneious 5.4 and the obtained alignment was corrected by hand. All sites were kept for the analyses. The best available model of molecular evolution was selected with MrModeltest 2.3 (Posada and Crandall 1998), and GTR+Γ+I was the best fit. Phylogenetic trees were primarily constructed using Bayesian inference (BI), with MrBayes 3.1.2 (Huelsenbeck and Ronquist 2001). Two identical searches with ten million generations each (chain temperature=0.2; sample frequency=1000) were conducted. In both runs, probabilities converged on the same stable value approximately after generation 8 000 000. A 50% majority-rule consensus tree was calculated, and posterior probability (PP) was used as an estimate of robustness. All BI analyses were carried out on the freely available Bioportal (www.bioportal.uio.no). Parameter estimates were graphically analysed to assess stability with Tracer 1.0.1. Maximum likelihood trees were calculated with the program RAxML (Stamatakis 2006) using the model GTRGAMMA for nucleotide substitution, the rapid hill-climbing (–fd) option and the rapid bootstrap algorithm (–fa) with 100 bootstrap replicates.
Ribotype networks, representing unique DNA sequences separated by mutational steps, were constructed using statistical parsimony with TCS software.
Scanning electron microscopy
Scanning electron microscopy (s.e.m) images were obtained after the critical-point drying of 32 specimens randomly selected and distributed across the whole phylogeny, using the s.e.m of the Royal Botanic Garden of Madrid, employing a Jeol T 330 A s.e.m, at 10–15 kV. The morphology of 10 spores per specimen (320 spores in total) was studied, and their largest diameter was measured (Supplementary Table 3). Differences in diameter length between genetic groups were tested with a Welsh t-test in R 2.12.2. and a representative spore (close to the average diameter in the specimen, and with typical morphology) from each sample was pictured.
Niche models
Niche models were generated with Maxent 3.3.3a (Phillips and Dudik 2008) using Bioclim variables from WorldClim (Hijmans et al., 2005) with a 2.5 arc-minutes resolution and with collections that were precisely geo-referenced (84 localities). Bioclim variables represent trends in temperature and precipitation along the year, as well as seasonality and extreme or limiting climatic factors. One locality (coordinates −10.022778°, 30.60547°) was excluded from the analyses as it was not covered by some environmental layers. The models were evaluated on the basis of receiver operating characteristic (ROC) analysis, which generates the AUC (area under the curve) score. To check whether models from the genetic groups were significantly different, their outputs were compared using ENMtools (Warren et al., 2010), with D and I indices as measures of niche overlap. The D and I values (Warren et al., 2008) are calculated by comparing two normalized Maxent environmental niche models, and using the estimated values of habitat suitability for each grid. If the niche differences are not caused only by random effects, the obtained D and I values from the two original niche models should display significant differences from the distribution generated by 100 random pseudo-replicates using pooled samples from the two original data groups.
Other statistical analyses
Mantel tests of orthodromic geographic distances versus pairwise raw genetic distances were performed with the package vegan in R 2.12.2, using 1000 permutations and the Pearson product-moment correlation coefficient. Geographic distances between sampling points were previously measured using the package fields, and genetic distances were calculated with the package ape (Paradis et al., 2004). Analyses of molecular variance (AMOVA) were performed with Arlequin 3.5 (Excoffier et al., 2005) and significance was based on 10 000 permutations.
Results
Bayesian inference (BI)
To assess the genetic structure of populations of B. melanospora, the largest fragment of the small subunit ribosomal DNA that is free of type I intron insertion sites and displays sufficient variability was amplified. A total of 37 distinct ribotypes with variable length (510–514 bp) were found in the 125 specimens sequenced, and this short and easy-to-sequence gene region showed 87 (16.3%) variable sites, with an average percent identity of 97.2% and an average 53.3% GC content. Most mutations were single nucleotide substitutions. Although there is still very little information about inter- and intraspecific sequence variability in myxomycetes, we have used a 97% similarity percentage as a first rough approximation to a threshold to separate putative cryptic species, under which samples clustered defining 6 groups.
Rooting the tree was difficult owing to the low number of myxomycete accessions in GenBank and because outgroups have longer branches than B. melanospora. An initial phylogenetic analysis with all available nearly complete small subunit ribosomal DNA sequences of the genera Badhamia and Physarum, and newly obtained sequences from seven specimens of B. melanospora and from six other species of Badhamia and Physarum, was carried out (Supplementary Figure 1). Lineages most closely related to B. melanospora were selected as outgroups for subsequent analyses.
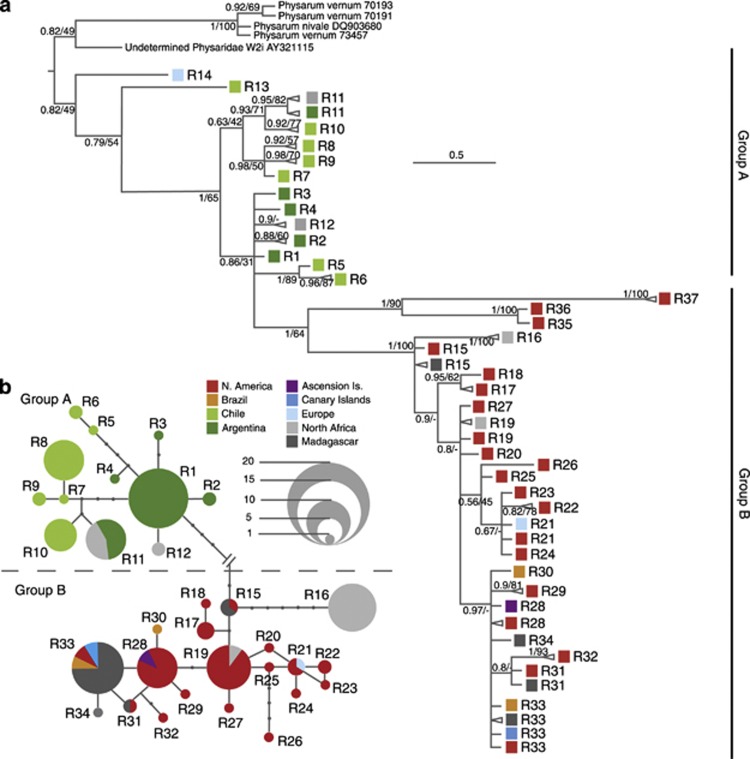
The inferred evolutionary interrelationships among all ribotypes, using the selected outgroups and a final alignment of 519 bp with 164 (31.6%) variable sites, are shown in Figure 1; Supplementary Table 1 gives sample details. One big paraphyletic group (A) of 54 sequences (14 ribotypes) is formed basically by specimens from South America (Chile and Argentina), but also contains sequences from Morocco and France. A well-supported divergent clade (group B) of 71 sequences (23 ribotypes) groups all North American collections, with two from Brazil, and most specimens from other parts of the world (Morocco, Canary Islands, Ascension Island, Europe, Madagascar).
Figure 1.
Phylogenetic analyses. (a): Fifty percent majority-rule rooted consensus tree of a 533 bp fragment of the small subunit rDNA (SSU) of 125 specimens of B. melanospora obtained by Bayesian inference. Triangles represent collapsed groups of samples with the same ribotype and geographic origin. Colours indicate the origin of the specimens. The scale bar represents evolutionary distance in changes per site. Bayesian posterior probabilities and RAxML support values are at each node. (b): Ribotype network of a 533 bp fragment of small subunit rDNA (SSU) of 123 specimens of B. melanospora. Circle size is proportional to the number of specimens within each ribotype, and dots between ribotypes represent unobserved, inferred ancestral ribotypes. Lines between ribotypes represent mutational steps between alleles. Colours denote specimen origin.
Phylogenetic network estimation using statistical parsimony (TCS)
TCS analyses (Figure 1b) were highly congruent with the results yielded by BI. Five highly divergent ribotypes (R13, R14, R35, R36 and R37) did not join the network, but the remaining sequences formed two well-delimited groups of ribotypes coincident with the above-defined groups A and B.
Group A has 12 different ribotypes, mostly from South American populations – Argentina and Chile – but there are also two from Morocco. One of the Moroccan ribotypes is shared with Argentinean populations; the other is closely related to the most common Argentinean ribotype. It is remarkable that there is no ribotype shared by both Argentina and Chile.
Group B has 20 ribotypes, from North America (USA and Mexico), Brazil and nearly all collections from the Old World – North Africa, Madagascar and Europe – and Atlantic oceanic islands – Canary Islands and Ascension Island. Most ribotypes from the Old World, Atlantic oceanic islands and Brazil group together and are closely related to North American populations.
Statistical analyses
Mantel tests showed that genetic distance is correlated with geographical distance for the American populations (r=0.5486, P<0.001), but it is only weakly correlated when including all the specimens (r=0.09523, P=0.014). Analyses of molecular variance (AMOVA) offered consistent results (Table 1). When comparing South America with North America or with the Old World, there is a higher percentage of variation between populations than within populations, indicating that South American populations are well differentiated. On the other hand, when comparing North America and the Old World the percentage of variation among populations is much lower, showing a higher degree of similarity between them.
Table 1. Analyses of Molecular Variance (AMOVA).
| FST | % AP | % WP | Tot. Var. | |
|---|---|---|---|---|
| NA, SA, OW | 0.51 | 50.69 | 49.31 | 8.83 |
| NA, SA | 0.65 | 65.03 | 34.97 | 10.66 |
| NA, OW | 0.10 | 10.40 | 89.60 | 5.68 |
| SA, OW | 0.53 | 53.12 | 46.88 | 9.29 |
Results obtained after comparing different geographical regions. Abbreviations: NA, North America; SA, South America; OW, Old World; FST, fixation index; % AP, percentage of variation among populations; % WP, percentage of variation within populations; Tot. Var., total variance. Significance tests after 10000 permutations with P<0.001 in all cases.
Scanning electron microscopy (s.e.m.)
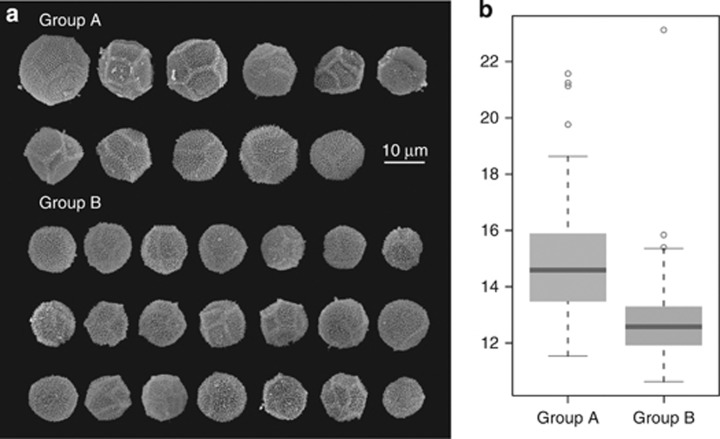
There are significant differences in the diameter of the spores of both ribotype groups (two-tailed Welsh t-test, P<0.001), and also slight changes in the ornamentation of their walls. Although both groups have overlapping sizes, spores from ribotype group A have in general bigger diameters (15.01 μm on average) and, ignoring the outliers, a higher size range than those from group B (12.69 μm on average) (Figures 2a and b). Group A also has more variable wall ornamentations, showing in most cases a very marked reticulum and a polygonal shape, but also less conspicuous ornamentation patterns were observed. Spores from ribotype group B are usually round in outline, with a generally less-marked reticulum.
Figure 2.
Variability in spore morphology. A total of 32 specimens from ribotype groups A and B were randomly selected for morphological studies. Ten spores per specimen were observed and measured (a): (s.e.m) pictures displaying one representative spore from each studied specimen. (b): Box-and-whiskers plot of the diameter of the spores in μm.
Environmental niche models
Separate niche models were generated for each of the two main groups of ribotypes. To predict species occurrence over geographic space, we used a maximum entropy model implemented in Maxent (Phillips and Dudik 2008). Maxent was chosen because it is a powerful tool in comparison with other methods (Elith et al., 2006) even in the presence of small data sets (Hernandez et al., 2006).
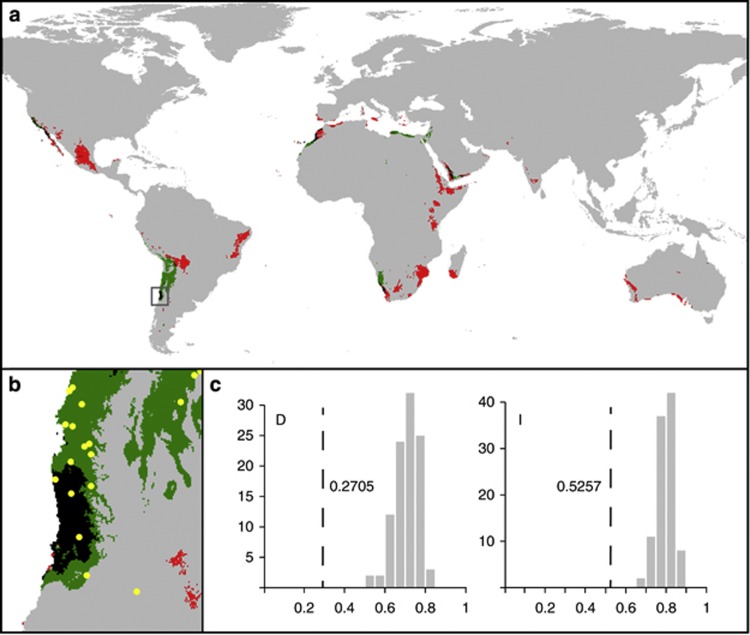
Predicted areas are located in warm arid territories in both cases (Figure 3a). The map obtained for ribotype group A predicts its presence in more restricted areas, situated near the western coasts of the continents. Ribotype group B has a broader niche, which penetrates into more intra-continental localities. In niche-overlapping areas from South America, only samples from group A were collected (hypergeometric distribution P=0.03) (Figure 3b), and in those from North Africa both ribotype groups were found. However, there is no information about overlapping areas in North America.
Figure 3.
Environmental Niche Models. (a): Predictive ecological models based on the Maxent algorithm. Probabilities of presence >0.5 are represented for ribotype groups A (green) and B (red). Overlapping areas are shown in black. (b): Close-up of overlapping niche areas in South America. Yellow dots denote localities where specimens from ribotype group A were found. (c): Niche comparisons of ribotype groups A and B based on D and I as measures of niche overlap. Dotted lines represent niche overlap measures of the original data, and bars show the expected degree of niche overlap when samples are drawn from the same distribution (i.e., pooled samples of occurrence points from the ribotype groups).
Niche models of the ribotype groups are less similar than random. Values of D and I indices obtained from original data (0.339 and 0.597, respectively) were smaller and out of the 95% confidence interval (D: 0.69–0.72, I: 0.78–0.80) of the distribution obtained from random pseudo-replicated models (Figure 3c).
Discussion
This work represents a first step in the study of genetic variation in a complex of cryptic species of myxomycetes in a geographical context. Our data show geographically structured ribotypes, which are congruent with the ME hypothesis. Implementing the use of environmental niche models has revealed that the two major ribotype groups have different habitat preferences, although their models can overlap in some areas. Models can also shed light on many important unresolved biogeographical questions, as the comparison of predicted areas with actual presence data can be a powerful tool for detecting the effect of barriers to dispersion. The absence of specimens from ribotype group B in South American niche-overlapping areas, which is very unlikely only by chance (P=0.03), seems to indicate that there have been impediments to its dispersion to these areas. The future study of niche-overlapping areas in North America from which there is no information could confirm this pattern.
Geographic differentiation among ribotypes
Our results are consistent with data from mating experiments in myxomycete cultures (El Hage et al., 2000; Clark 2000; Clark and Stephenson 2000; Irawan et al., 2000), which found that at least some widespread morphospecies may actually consist of complexes of reproductively isolated clonal lines that can be geographically restricted (Clark 2004). Previous molecular studies on other myxomycete morphospecies did not show any clear relationship between genetic variants and the geographical origin of the specimens (Winsett and Stephenson 2008; Fiore-Donno et al., 2011). However, B. melanospora displays a clear geographic pattern.
The structure found can be interpreted to understand the history of the species. As most basal clades in the BI tree are Argentinean and Chilean populations, B. melanospora has its most probable origin in South America. All North American specimens are part of a well-supported clade that includes most collections from the Old World but does not include any sequence from Argentina or Chile. The TCS analysis shows congruent results, with most closely related sequences from ribotype group A and ribotype group B separated by nine missing ribotypes. Mantel tests and AMOVA also support a North versus South divergence in America, showing a correlation between geographic and genetic distance in this continent, and that, in addition, molecular variance is lower in each subcontinent than in the total data set considering all populations together. The morphological differences we found between ribotype groups support these conclusions. Although the characters studied display overlapping ranges and are not useful as diagnostic characters, they show that genetic differentiation between the groups has had some consequences for morphology.
Furthermore, our data do imply some long distance migration. BI, TCS and AMOVA results suggest quite strongly that North American populations were the main source for populations from the Old World, except for two groups of North African collections and a highly divergent French specimen that were more closely related to South American populations.
Another interesting result is that no ribotype was found on both sides of the Andes – that is, in both Chile and Argentina. The topology of the tree and the TCS network do not show, however, two clearly defined groups of populations from one side and the other. The most likely explanation is that the Andes may have acted as a semi-permeable barrier allowing multiple colonization events across the mountain range.
Ribotype groups have different environmental niches
Environmental niche models provide evidence for differences in the ecological preferences of B. melanospora ribotype groups, but there are several areas where niches from both groups overlap. Only specimens from ribotype group A have been found in a niche-overlapping area from South America that has been intensively surveyed for myxomycetes (Lado et al., 2012). This exclusion strongly suggests the existence of barriers to dispersion at least between North America and South America, and, unless there is an effect of other unstudied environmental factors, this is prima facie evidence for isolation by geographic distance and inefficient global dispersal. The presence of representatives of both ribotype groups on native plants from the Old World (Euphorbia, Pachypodium) and of group. A individuals on North American plants introduced in the Old World (Opuntia, Agave) (see Supplementary Table 1) indicates that this limited distribution may not be an artefact caused by host plant specificity. One possible limitation of the present analysis is that the modelled niches take account of only climatic factors. Recent studies (Aguilar and Lado 2012; Bates et al., 2013) suggest that climate has a major influence on the distribution of protists at medium to large geographic scales. However, other potentially important environmental variables such as characteristics of the host plant, interaction with other organisms, Ph, soil type, etc may also affect the distribution patterns of B. melanospora. Although the models reasonably predict the distribution of the organisms, it is still possible that a variable not included in the analyses is causing the absence of ribotype group B from the overlapping niche area.
Furthermore, the fact that one ribotype from group B was found only once in Spain and many times in Mexico and another was found once in the Canary Islands and once in Ascension Island but many times in Mexico shows that North American ribotypes can survive in Europe or in ecologically disparate Atlantic islands, suggesting that the absence of most of them from these regions is because of weak dispersal and not because of ecological unsuitability.
It is important to note that there is no information about the actual presence of B. melanospora in several areas with high probability in the models – for example, Southern Africa, Arabia and Australia. Most of these areas have never been surveyed for myxomycetes, and this lack of data makes it harder to interpret our results. It would be worthwhile searching for B. melanospora in these areas; if it were not found, the conclusion that B. melanospora has not been able to reach all ecologically suitable areas during its dispersive processes would be strengthened.
Human introductions
It is striking that Old World strains are nested within the ancestrally New World groups A and B, but in most cases forming a group of closely related ribotypes in group B. AMOVA results also suggest that in most cases Old World populations are more similar to North American populations. This pattern is most simply explained by multiple colonization events from the Americas, especially from North America, to Europe, oceanic islands, Africa and Madagascar.
The four ribotypes found in Morocco do not cluster together; one is almost identical to North American sequences and one to Argentinean sequences, whereas two are distinct Moroccan ribotypes. Environmental niche models show that there are areas in the north-western coast of Africa where niches overlap and ribotypes of both groups coexist. We suggest that there were four colonizations of Morocco from America, either directly or via other unsampled localities in the Old World. Assuming a similar rate of evolutionary change for all strains, two Moroccan ribotypes would be so recent as to be almost identical to their source strains, and the other two would be more ancient – long enough ago for separate ribotypes to have evolved after colonization. Also, we postulate two to three separate colonizations of Madagascar, all most likely from North America. The two Brazilian sequences are also most likely the result of introductions from North America. One of them is of the same ribotype that was apparently introduced into the Canary Islands and Madagascar.
Many specimens collected outside the Americas were found on plants originally introduced from North or Central America (Opuntia, Agave) (see Supplementary Table 1), which quite strongly supports the hypothesis that the dispersion of B. melanospora has been facilitated by human introductions of American succulent plants into the Old World. If these strains had evolved in situ one would expect them to be frequent on native Old World plants.
If our interpretations are correct, then B. melanospora provides the first molecular evidence of human introductions of a myxomycete, but it is possible that at least six other morphospecies of myxomycetes have been introduced in a similar way from America to the Canary Islands (Lado et al 2007). Given the close similarity of most putatively introduced strains, it is unlikely that our results are misled by the absence of data from Africa due to an incomplete sampling. Nonetheless, search for B. melanospora in southern and east Africa is an important test of our human introduction hypothesis.
B. melanospora is probably a cryptic species complex
In the case of B. melanospora, samples clustered forming six groups with a 97% similarity. Furthermore, group B constitutes a well-defined clade, with high support and separated from other sequences by a relatively long branch. This makes it likely that it is reproductively isolated from group A and forms an independent evolutionary line. Groups A and B are genetically, morphologically and geographically strongly differentiated and it is unlikely that there is much, if any, gene flow between them. Very likely they are distinct biological species with different geographic distributions. However, none of the morphological characters studied allow an a priori classification of the specimens in either group, and therefore they cannot be considered as diagnostic characters. It would not be surprising if the two groups each include several cryptic biological species.
Conclusions
Incorporating ecological aspects into biogeographical studies of protists can make it possible to test the EiE hypothesis. In cases where a serious lack of data makes it difficult to carry out both ecological and biogeographic studies, the use of environmental niche models can be of great help, especially when selecting the most appropriate variables for predicting an organism's occurrence. One clear example is myxomycete (mycetozoan) amoebozoans, which are among the most widely distributed of all terrestrial organisms, but also among the least known.
B. melanospora is a complex case in which limited dispersion, isolation by distance, host specificity and other ecological parameters have acted, giving rise to a set of at least two cryptic species with slight but not completely distinguishable morphologies. In addition, human introduction of host plants may have played an important role in facilitating multiple long distance colonization events from the Americas to the Old World, as well as two putative cases from Mexico to Brazil.
Acknowledgments
We thank S L Stephenson, M Meyer, L H Cavalcanti and R McHugh for contributing specimens and providing comments on the species distribution and morphology, and Fátima Durán, Guillermo Sanjuanbenito, Gemma Andreu, Yolanda Ruiz and Juan Carlos Hernández for technical work. This work was supported by Research Projects CGL2011-22684/BOS and CGL2008-00720/BOS of the Ministry of Science and Innovation of Spain.
Author contributions
MA, AMFD, CL and TCS designed the research; MA and AMFD conducted the research; MA analysed the data; and MA, AMFD, CL and TCS wrote the paper.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Aguilar M, Lado C. Ecological Niche Models reveal the importance of variability in climatic conditions for the biogeography of protosteloid amoebae. ISME J. 2012;6:1506–1514. doi: 10.1038/ismej.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato A, Kooistra WHCF, Levialdi Ghiron JH, Mann DG, Pröschold T, Montresor M. Reproductive isolation among sympatric cryptic species in marine diatoms. Protist. 2007;158:193–207. doi: 10.1016/j.protis.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Aurahs R, Grimm GW, Hemleben V, Hemleben C, Kucera M. Geographical distribution of cryptic genetic types in the planktonic foraminifer Globigerinoides ruber. Mol Ecol. 2009;18:1692–1706. doi: 10.1111/j.1365-294X.2009.04136.x. [DOI] [PubMed] [Google Scholar]
- Bass D, Howe AT, Mylnikov AP, Vickerman K, Chao EE, Smallbone JE, et al. Phylogeny and classification of Cercomonadidae: Cercomonas, Eocercomonas, Paracercomonas, and Cavernomonas gen. n. Protist. 2009;160:483–521. doi: 10.1016/j.protis.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Bass D, Richards TA, Matthai L, Marsh V, Cavalier-Smith T. DNA evidence for global dispersal and probable endemicity of protozoa. BMC Evol Biol. 2007;7:162. doi: 10.1186/1471-2148-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates ST, Clemente JC, Flores GE, Walters WA, Parfrey LW, Knight R, et al. Global biogeography of highly diverse protistan communities in soil. ISME J. 2013;7:652–659. doi: 10.1038/ismej.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J. The species problem in the myxomycetes. Stapfia. 2000;73:39–53. [Google Scholar]
- Clark J. Reproductive systems and taxonomy in the myxomycetes. Syst Geogr Plants. 2004;74:209–216. [Google Scholar]
- Clark J, Stephenson SL. Biosystematics of the myxomycete Physarum melleum. Nova Hedwigia. 2000;71:161–164. [Google Scholar]
- Darling KF, Kucera M, Wade CM. Global molecular phylogeography reveals persistent Arctic circumpolar isolation in a marine planktonic protist. Proc Natl Acad Sci USA. 2007;104:5002–5007. doi: 10.1073/pnas.0700520104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas TE, Kronforst MR, Queller DC, Strassmann JE. Genetic diversity in the social amoeba Dictyostelium discoideum: Population differentiation and cryptic species. Mol Phylogenet Evol. 2011;60:455–462. doi: 10.1016/j.ympev.2011.05.007. [DOI] [PubMed] [Google Scholar]
- El Hage M, Little C, Clark L, Stephenson SL. Biosystematics of Didymium squamulosum complex. Mycologia. 2000;92:54–64. [Google Scholar]
- Elith J, Graham CH, Anderson RP, Dudík M, Ferrier S, Guisan A, et al. Novel methods improve prediction of species' distributions from occurrence data. Ecography. 2006;29:129–151. [Google Scholar]
- Evans KM, Chepurnov VA, Sluiman HJ, Thomas SJ, Spears BM, Mann DG. Highly differentiated populations of the freshwater diatom Sellaphora capitata suggest limited dispersal and opportunities for allopatric speciation. Protist. 2009;160:386–396. doi: 10.1016/j.protis.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Excoffier LG, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fenchel T, Finlay BJ. The ubiquity of small species: patterns of local and global diversity. BioScience. 2004;54:777–784. [Google Scholar]
- Finlay BJ. Global dispersal of free-living microbial eukaryotic species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- Fiore-Donno A-M, Meyer M, Baldauf SL, Pawlowski J. Evolution of dark-spored myxomycetes (slime-molds): molecules versus morphology. Mol Phylogenet Evol. 2008;46:878–889. doi: 10.1016/j.ympev.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Fiore-Donno A-M, Novozhilov YK, Meyer M, Schnittler M. Genetic structure of two protist species (Myxogastria, Amoebozoa) suggests asexual reproduction in sexual Amoebae. PLoS One. 2011;6:e22872. doi: 10.1371/journal.pone.0022872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissner W. Protist diversity: estimates of the near imponderable. Protist. 1999;150:363–368. doi: 10.1016/S1434-4610(99)70037-4. [DOI] [PubMed] [Google Scholar]
- Foissner W. Biogeography and dispersal of microorganisms: a review emphasising protists. Acta Protozool. 2006;45:111–136. [Google Scholar]
- Foissner W, Chao A, Katz LA. Diversity and geographic distribution of ciliates (Protista: Ciliophora) Biodivers Conserv. 2008;17:329–343. [Google Scholar]
- Hernandez PA, Graham CH, Master LL, Albert DL. The effect of sample size and species characteristics on performance of different species distribution modeling methods. Ecography. 2006;29:773–785. [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology. 2005;25:1965–1978. [Google Scholar]
- Howe AT, Bass D, Vickerman K, Chao EE, Cavalier-Smith T. Phylogeny, taxonomy, and astounding genetic diversity of Glissomonadida ord. nov., the dominant gliding zooflagellates in soil (Protozoa: Cercozoa) Protist. 2009;160:159–189. doi: 10.1016/j.protis.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Irawan B, Clark J, Stephenson SL. Biosystematics of the Physarum compressum morphospecies. Mycologia. 2000;92:884–893. [Google Scholar]
- Lado C, Estarda-Torres A, Stephenson SL. Myxomycetes collected in the first phase of a north-south transect of Chile. Fungal Divers. 2007;25:81–101. [Google Scholar]
- Lado C, Mosquera J, Estrada A, Beltrán E, Wrigley de Basanta D. Description and culture of a new succulenticolous Didymium (Myxomycetes) Mycologia. 2007;99:602–611. doi: 10.3852/mycologia.99.4.602. [DOI] [PubMed] [Google Scholar]
- Lado C, Wrigley de Basanta D, Estrada-Torres A, Stephenson SL. The biodiversity of myxomycetes in central Chile. Fungal Divers. 2012;59:3–32. [Google Scholar]
- Morard R, Quillévéré F, Escarguel G, Ujiie Y, Garidel-Thoron T, Norris RD, et al. Morphological recognition of cryptic species in the planktonic foraminifer Orbulina universa. Mar Micropaleontol. 2009;71:148–165. [Google Scholar]
- Novozhilov YK, Schnittler M, Stephenson SL. Species and Communities in Extreme Environments Festschrift towards the 75th Anniversary and a Laudatio in Honour of Academician Yuri Ivanovich Chernov. Pensoft Publishers & KMK Scientific Press: Sofia-Moscow; 2009. [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Phillips SJ, Dudik M. Modelling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography. 2008;31:161–175. [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Poulíčková A, Vesela J, Neustupa J, Skaloud P. Pseudocryptic diversity versus cosmopolitanism in diatoms: a case study on Navicula cryptocephala Kutz. (Bacillariophyceae) and morphologically similar taxa. Protist. 2010;161:353–369. doi: 10.1016/j.protis.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Smirnov AV. Cryptic freshwater amoeba species in the bottom sediments of Nivå Bay (Øresund, Baltic Sea) Eur J Protistol. 2007;43:87–94. doi: 10.1016/j.ejop.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Smith HG, Wilkinson DM. Not all free-living microorganisms have cosmopolitan distributions—the case of Nebela (Apodera) vas Certes (Protozoa, Amoebozoa, Arcellinida) J Biogeogr. 2007;34:1822–1831. [Google Scholar]
- Sorhannus U, Ortiz JD, Wolf M, Fox MG. Microevolution and speciation in Thalassiosira weissflogii (Bacillariophyta) Protist. 2010;161:237–249. doi: 10.1016/j.protis.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Vanormelingen P, Verleyen E, Vyverman W. The diversity and distribution of diatoms: from cosmopolitanism to narrow endemism. Biodivers Conserv. 2008;17:393–405. [Google Scholar]
- Warren DL, Glor RE, Turelli M. Environmental Niche Equivalency versus Conservatism: Quantitative Approaches to Niche Evolution. Evolution. 2008;62:2868–2883. doi: 10.1111/j.1558-5646.2008.00482.x. [DOI] [PubMed] [Google Scholar]
- Warren DL, Glor RE, Turelli M. ENMTools: a toolbox for comparative studies of environmental niche models. Ecography. 2010;33:607–611. [Google Scholar]
- Winsett KE, Stephenson SL. Using ITS sequences to assess intraspecific genetic relationships among geographically separated collections of the myxomycete Didymium squamulosum. Rev Mex Micol. 2008;27:59–65. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.