Abstract
High-latitude environments, such as the Antarctic McMurdo Dry Valley lakes, are subject to seasonally segregated light–dark cycles, which have important consequences for microbial diversity and function on an annual basis. Owing largely to the logistical difficulties of sampling polar environments during the darkness of winter, little is known about planktonic microbial community responses to the cessation of photosynthetic primary production during the austral sunset, which lingers from approximately February to April. Here, we hypothesized that changes in bacterial, archaeal and eukaryotic community structure, particularly shifts in favor of chemolithotrophs and mixotrophs, would manifest during the transition to polar night. Our work represents the first concurrent molecular characterization, using 454 pyrosequencing of hypervariable regions of the small-subunit ribosomal RNA gene, of bacterial, archaeal and eukaryotic communities in permanently ice-covered lakes Fryxell and Bonney, before and during the polar night transition. We found vertically stratified populations that varied at the community and/or operational taxonomic unit-level between lakes and seasons. Network analysis based on operational taxonomic unit level interactions revealed nonrandomly structured microbial communities organized into modules (groups of taxa) containing key metabolic potential capacities, including photoheterotrophy, mixotrophy and chemolithotrophy, which are likely to be differentially favored during the transition to polar night.
Keywords: ice-covered lake, McMurdo Dry Valleys, microbial diversity, MIRADA-LTERS, network analysis, polar night
Introduction
Microbial diversity and function in aquatic ecosystems are tightly coupled to the physical and geochemical environment (Judd et al., 2006; Galand et al., 2008; Bielewicz et al., 2011). The importance of seasonal succession is increasingly being recognized within the context of geochemically distinct environments (Crump et al., 2003; Andersson et al., 2010; Ghiglione and Murray, 2012; Grzymski et al., 2012). Polar environments are subject to strong seasonal light gradients, where 24-hour daylight drives continual photoautotrophic primary production during the summer, often coinciding with high rates of heterotrophic bacterioplankton production (Takacs and Priscu, 1998; Morán et al., 2001; Alonso-Sáez et al., 2008). Winter sampling is logistically difficult in the polar regions, and thus studies examining microbial dynamics during the darkness of winter and the spring and autumn transition periods are few. Recent studies have shown higher bacterial community richness (Ghiglione and Murray, 2012) and the increased importance of chemolithotrophic Archaea in the Southern Ocean during winter (Grzymski et al., 2012; Williams et al., 2012), whereas others have shown that trophic plasticity is a key survival strategy for protists during the summer–winter transition in Antarctic lakes (Bielewicz et al., 2011).
The perennially ice-covered lakes of the McMurdo (MCM) Dry Valleys, which lie in East Antarctica in the coldest, driest desert on earth, comprise physicochemically stable environments containing microbially dominated ecosystems (Spigel and Priscu, 1998; Takacs and Priscu, 1998; Priscu et al., 1999; Vick and Priscu, 2012). As the sole year-round source of liquid water, such lakes provide the only continuous habitat for aquatic life in the ice-free regions of the Antarctic continent. Permanent ice covers on the lakes severely attenuate the penetration of solar irradiance to between 1% and 2% of incident light (Lizotte and Priscu, 1994) and prohibit wind-driven turbulence, propagating an environment continuously stratified with regard to solar energy and nutrients. A few studies have examined the molecular diversity of bacterial, archaeal (Karr et al., 2005; Glatz et al., 2006; Karr et al., 2006) or protistan (Bielewicz et al., 2011; Kong et al., 2012a) communities in MCM lakes and found that communities are distinctly stratified by depth. The physicochemical stability of these environments makes them excellent locations to examine the impact of seasonal light–dark cycles on microbial community dynamics.
Most studies of the MCM lakes have been confined to summer, when phytoplankton primary production and glacial melt water streams supply >50% of the organic matter supporting heterotrophic growth (Takacs et al., 2001), but a few have examined the activities of bacterioplankton (Takacs and Priscu, 1998; Vick and Priscu, 2012), phytoplankton (Lizotte et al., 1996), flagellates (Thurman et al., 2012) and the diversity of protists (Bielewicz et al., 2011) during the transition periods flanking summer and winter. Mixotrophy, via the combined use of photosynthesis and phagotrophy, is a key adaptive strategy for phytoplankton in MCM lakes that likely allows populations to persist throughout the winter (McKnight et al., 2000; Laybourn-Parry, 2002; Bielewicz et al., 2011; Thurman et al., 2012), whereas metabolic plasticity, such as the ability to switch carbon substrates, is important for heterotrophic bacterioplankton to remain active during winter when phytoplankton-produced organic carbon is in short supply (Vick and Priscu, 2012). Clearly, trophic and metabolic versatilities are vital to the survival of given populations in these lakes and are important to the maintenance of overall ecosystem function.
In addition to physicochemical controls on microbial community structure, recent studies have also shown that microbial co-occurrence patterns can help define ecologically meaningful interactions between species and across domains (Horner-Devine et al., 2007; Fuhrman and Steele, 2008; Steele et al., 2011) and that co-occurring species are often organized into groups, or modules, of functional significance (Chaffron et al., 2010; Barberán et al., 2012). In light of the physicochemical stability of the MCM lakes, we sought to determine the importance of both community succession and operational taxonomic unit (OTU)-level co-occurrence patterns to overall community structure during the summer−autumn transition period (November–March). We used pyrosequencing of the V6 (bacterial and archaeal) and V9 (eukaryotic) hypervariable regions of the small-subunit ribosomal RNA gene on samples from two geochemically distinct MCM lakes, Lake Fryxell (FRX) and the West Lobe of Lake Bonney (WLB), during the austral summer (November) and autumn (March) to analyze microbial community composition, and implemented molecular ecological network analysis to examine inter- and intra-domain co-occurrence patterns.
We provide the first evidence for seasonal shifts in bacterial, archaeal and eukaryotic communities in these Antarctic lakes. Our data also suggest the importance of metabolically plastic taxa in maintaining overall ecosystem function and document the proliferation of several archaeal lineages, which may be important primary producers during the darkness of winter. In combination with studies from the polar oceans (Grzymski et al., 2012; Ghiglione and Murray, 2012; Williams et al., 2012), our results reveal that shifts in community diversity may be characteristic of polar ecosystems during winter and have particular importance in fueling continued biogeochemical cycling in the absence of photosynthetic primary production.
Methods
Sample collection
Duplicate samples for DNA extraction were collected from FRX (depth ∼18 m) and WLB (depth∼38 m) during the Austral summer and autumn at depths of 6 m (bacterial and primary production maximum) and 9 m (chemocline; chlorophyll-a maximum) in FRX (2 November 2007 and 25 March 2008) and 13 m (chemocline; bacterial production, chlorophyll-a, primary production maximum) and 18 m (hypersaline, bottom of trophogenic zone) in WLB (30 November 2007 and 12 March 2008). Corresponding environmental data were collected by the MCM Long-Term Ecological Research program ∼1 week before and 1 week after samples for DNA collection and interpolated to the DNA sample collection date (11 November and 5 December 2007, and 20 and 28 March for FRX; 25 November and 15 December 2007, and 7 and 14 March for WLB). In WLB, environmental data were collected at 17 and 20 or 25 m and interpolated to 18 m. Complete environmental data and methods are available on the MCM Long-Term Ecological Research website (http://www.mcmlter.org/) and are published elsewhere (Vick and Priscu, 2012), and the minimum information about a marker gene sequence-compliant (Yilmaz et al., 2011) environmental data are summarized in supplementary information. Representative vertical profiles of temperature, conductivity and oxygen from each lake are shown in Supplementary Figure S1, and environmental data in Supplementary Table S1. Temperature and conductivity were measured with a SBE 25 Sealogger CTD according to Spigel and Priscu (1998), and dissolved oxygen was measured using the azide modification of the mini-Winkler titration. All water samples were collected through a borehole in the ice cover using a Niskin bottle. Samples for DNA extraction were filtered onto 0.2-μM Sterivex filters (Millipore, Billerica, MA, USA) and stored with 2.0 ml of Puregene lysis buffer at −20 °C until further processing. DNA was extracted as described previously (Amaral-Zettler et al., 2009), and water filtration and DNA extraction protocols can be found at http://amarallab.mbl.edu.
Sequencing
We amplified V6 hypervariable regions using primers targeting positions (according to the E. coli numbering scheme) 947–1046 (Bacteria) and positions 958–1048 (Archaea) of the 16S ribosomal RNA gene. For Eukarya, amplification of the V9 hypervariable region followed established protocols (Amaral-Zettler et al., 2009). We multiplex-sequenced the resulting amplicons using bar-coded primers (Huber et al., 2007; Amaral-Zettler et al., 2009) on a 454 Genome Sequencer FLX (Roche, Switzerland) using the manufacturer's recommended protocol. The number of reads obtained on a single sample ranged from 1724 to 23 334 (Archaea), 2997 to 15 552 (Bacteria) and 2602 to 12 504 (Eukarya) (Supplementary Table S2).
Sequence processing
Sequences were trimmed, and low-quality reads were removed according to Huse et al., 2007. Sequences were clustered into OTUs using ESPRIT, SLP and mothur to precluster (2%) sequences using single linkage and construct final clusters based on pairwise alignment and average linkage (Huse et al., 2010). Three-percent cluster widths were used for bacterial and archaeal analyses and 6% for eukaryotic analyses. This clustering method is equally effective as ‘denoising' data via methods such as Pyronoise to minimize OTU inflation (Quince et al., 2011). All of our sequence data are minimum information about a marker gene sequence-compliant (Yilmaz et al., 2011) and have been deposited in the National Center for Biotechnology Information normal and Sequence Read Archives under the accession number SRP028879.
Taxonomy assignment
Taxonomic classification was assigned using the Global Alignment for Sequence Taxonomy (GAST) method (Huse et al., 2008). Briefly, hypervariable tag reference sets (V6 or V9) were created from a ribosomal RNA reference database based on the SILVA database (Pruesse et al., 2007), with taxonomy assigned by the RDP Classifier (Wang et al., 2007). We then aligned the tag sequences against the top 100 reference sequences using MUSCLE and considered the best GAST match. Smaller GAST distances equate to better matches. We assigned a tag to a given genus if two-thirds or more of the full-length reference ribosomal RNA sequences containing the exact hypervariable region shared the same genus. If there was no agreement, we moved up the tree one level to family and so on until a consensus was reached.
Diversity calculations
Parametric alpha diversity estimates for Bacteria and Archaea were calculated using CatchAll version 3.2 (Bunge et al., 2012), and eukaryotic nonparametric (Chao2) richness estimates (Chao, 1987) were calculated with the program SPADE (Chao and Shen, 2010). All calculations were performed on pooled sequences from duplicate samples for bacteria and archaea and separate replicated samples for eukaryotes. We performed these calculations using full data sets, as well as normalized data sets in which the number of sequences per sample was made equal through random resampling.
Network analysis
To determine associations between microbial populations and between microbial populations and the environment, we calculated Spearman correlations between environmental data, relative abundances of bacterial and archaeal OTUs and presence–absence of eukaryotic OTUs. Significant correlations (P<0.01; r⩾0.8) were extracted and the resulting matrix of correlation coefficients was loaded into the program Cytoscape (Shannon et al., 2003) for visualization. In total, we included 899 variables in our correlation analysis: 186 eukaryotic, 637 bacterial and 69 archaeal OTUs, as well as 7 environmental variables.
Statistics
Statistics were carried out in R (R Development Core Team, 2008). Beta diversity was examined and plotted using the function nmds (non-metric multidimensional scaling) in the R library labdsv (Roberts, 2010). The importance of environmental factors in partitioning of beta diversity was tested using permutational analysis of variance (Anderson, 2001) with the adonis function, and C-scores were calculated using the oecosimu function with nestedchecker (Stone and Roberts, 1990) and quasiswap (Miklós and Podani 2004) in the R library vegan (Oksanen et al., 2010). Network statistics, including modularity calculations, were carried out using Cytoscape (Shannon et al., 2003; Supplementary Information).
Results and discussion
Seasonal variation in microbial communities
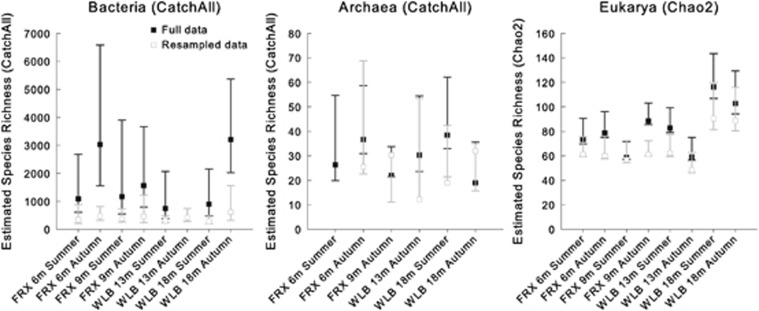
Species richness (alpha diversity) was relatively low across all samples and all three domains, but it followed trends observed in other environments with bacterial richness surpassing archaeal and eukaryotic richness by an order of magnitude (Huber et al., 2007, McCliment et al., 2012). Alpha diversity was generally higher in autumn samples for bacterial and eukaryotic communities in FRX, whereas the opposite was true in WLB (except for 18 m bacterial communities). Archaeal diversity was lower than bacterial and eukaryotic diversity (Figure 1), with coverage (observed/expected diversity) ranging from 53 to 90%. Eukaryotic coverage was highest, ranging from 87 to 98%, whereas bacterial coverage was comparatively low (15 to 50% Supplementary Table S2).
Figure 1.
Alpha diversity estimates with Bonferroni-corrected confidence bounds calculated with CatchAll for Bacteria and Archaea and as the Chao2 index for Eukarya. Archaeal diversity estimates could not be calculated for the FRX 9 m and WLB 13 m summer samples owing to insufficient numbers of reads.
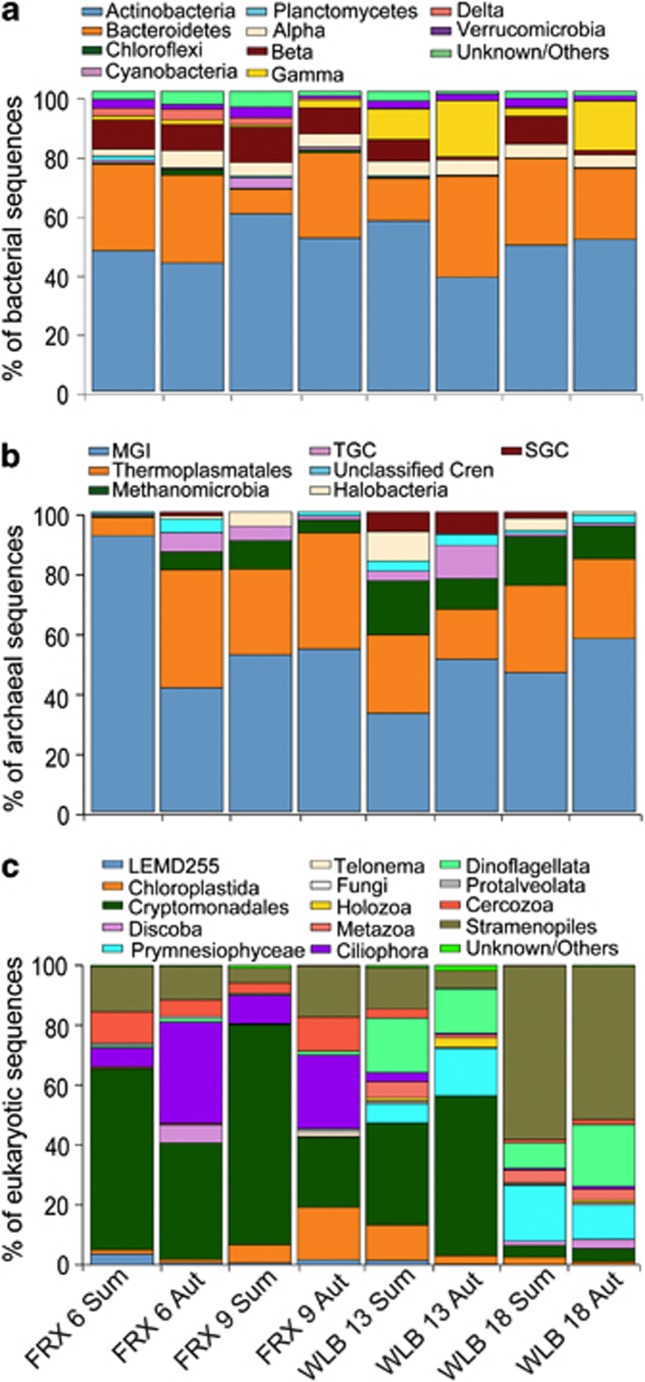
Actinobacteria and Bacteroidetes dominated the bacterial communities of both lakes. This is consistent with reports from other freshwater systems (Newton et al., 2011). The Proteobacteria was the next most abundant phylum in both lakes, with the class Betaproteobacteria dominating FRX and Gammaproteobacteria dominating WLB, a difference that may be explained by the influence of Blood Falls, a Gammaproteobacteria-dominated subglacial feature that flows into the western terminus of WLB (Mikucki and Priscu, 2007). Marine Group I Crenarchaeota dominated the archaeal communities, similar to the upper and intermediate waters of Arctic meromictic Lake A (Comeau et al., 2012), followed by Thermoplasmatales-related Euryarchaeotes and Methanomicrobia. Eukaryotic community composition varied between lakes and depths, with Cryptomonadales and Ciliophora OTUs being the most frequently encountered in FRX, whereas WLB contained OTUs most frequently affiliated with Cryptomonadales, Stramenopiles and Dinoflagellata (Figure 2).
Figure 2.
Phylum-level diversity of bacterial (a), archaeal (b) and eukaryotic (c) communities in Lakes FRX and WLB during summer (Sum) and autumn (Aut). Bacterial and archaeal OTUs were determined at 97% sequence similarity, and eukaryotic OTUs were determined at 94% sequence similarity. Alpha, Beta, Gamma and Delta refer to the subclasses of Proteobacteria. MGI refers to Marine Group I Crenarchaeota, TGC refers to Terrestrial Group Crenarchaeota and SGC refers to Soil Group Crenarchaeota.
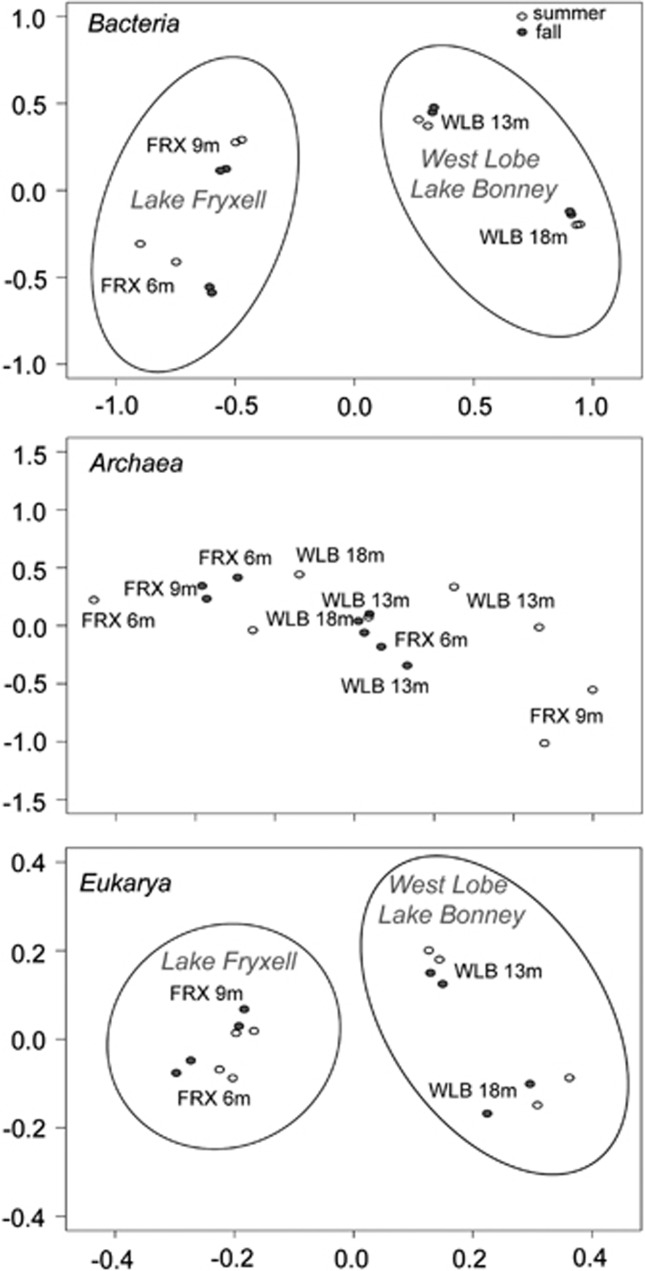
Bacterial and eukaryotic communities both grouped by lake (permutational analysis of variance; P=0.0010 for Bacteria, P=0.0020 for Eukarya) and depth (P=0.0010 for Bacteria and Eukarya). Season (summer vs autumn) was not significant alone, but when included in a model that first accounted for the interaction between lake and depth it explained a significant portion of the variation in the bacterial and eukaryotic communities (P=0.013 and P=0.042, respectively). These results indicate that within lake and depth, bacterial and eukaryotic communities were significantly different between seasons (Figure 3). Archaeal communities did not clearly partition by lake, depth or season, but a model accounting for lake and season indicated that depth was the most important factor explaining the variation between communities (P=0.042; Figure 3). The effects of lake and season were marginally significant for the Archaea (P=0.14 and 0.12, respectively). These results are similar to the seasonal and depth partitioning observed in Arctic meromictic Lake A, where bacterial and eukaryotic phyla varied as a function of both depth and time, and archaeal phylum-level seasonal changes were minor, but depth partitioning was strong (Charvet et al., 2012a, Comeau et al., 2012).
Figure 3.
Non-metric multidimensional scaling of bacterial and archaeal (relative abundance; Bray-Curtis dissimilarity; stress=2.49 and 4.97, respectively) and eukaryotic (presence-absence; Sørensen's similarity; stress=4.68) communities.
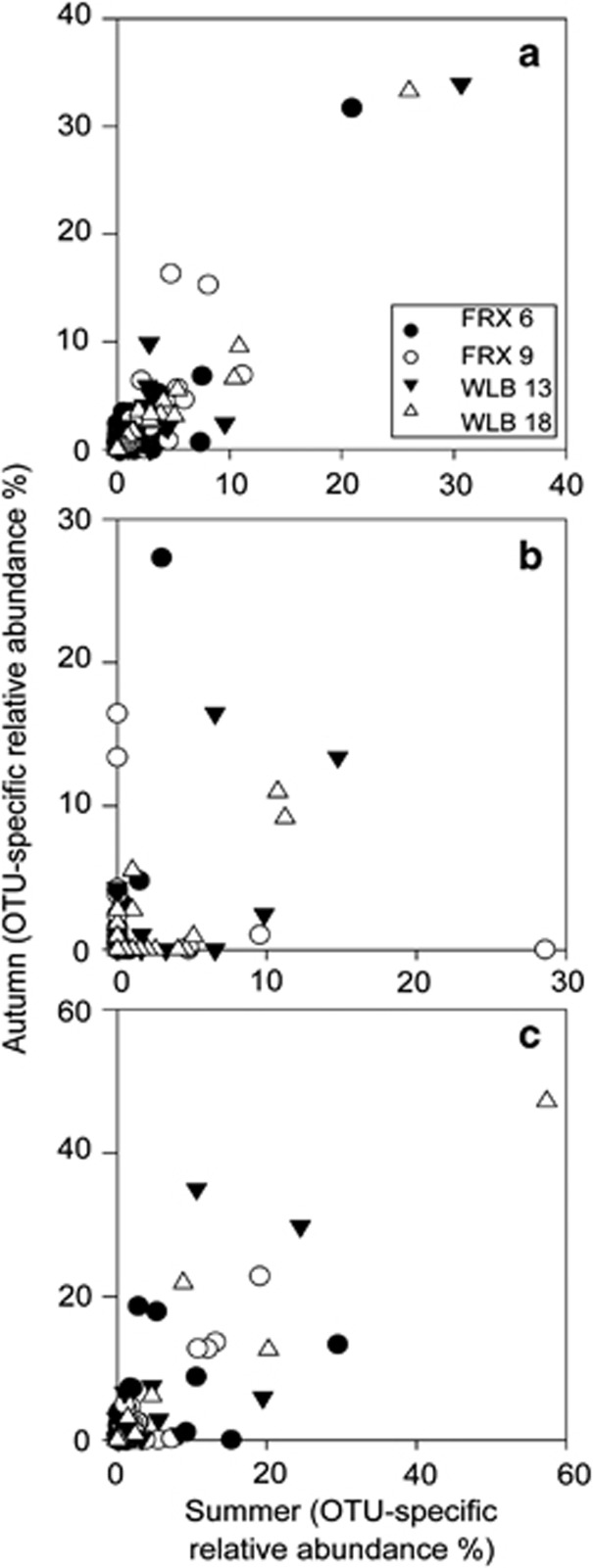
Seasonal changes in community composition were apparent at the OTU level as the percentage of OTUs that went from being rare (<0.1% of community) or absent in summer samples to being abundant (>0.1% of community; Crump et al., 2012) in autumn samples. The changes in composition, shown in Figure 4 as points falling along the y axes, were especially pronounced for the Archaea, describing 28% and 23% of the 6 m and 9 m FRX communities, respectively, and 14% and 10% of the 13 m and 18 m WLB communities, respectively. OTU-level changes in community composition were comparatively small for the Bacteria (0.8 to 5.5% of the communities) and Eukarya (1.7 to 3.0% of OTUs). The use of a higher cutoff (1.0%) for the rare to abundant transition did not change the pattern, although it decreased the percentages (Archaea=4 to 12%, Bacteria=0.2 to 1.0%, Eukarya=0 to 2.0%).
Figure 4.
Relative abundances (%) of bacterial (a), archaeal (b) and eukaryotic (c) OTUs during summer and autumn. Bacterial and archaeal OTUs were calculated at 97% sequence similarity, and eukaryotic OTUs were calculated at 94% sequence similarity.
Although they were not the dominant members of the communities overall, Stramenopiles (mostly chrysophytes) dominated the eukaryotic OTUs that became abundant during autumn. Charvet et al. (2012b) suggested that the generally small size of chrysophyte cells, relative to other phytoplankton, may account for their dominance in oligotrophic Arctic lakes; decreasing phosphorous concentrations in autumn samples (Vick and Priscu, 2012) may have favored the proliferation of chrysophytes in FRX and WLB. Lizotte et al. (1996) found that chrysophyte communities in the Lake Bonney photic zone were dominated by the mixotrophic genera Ochromonas, which may persist through the seasonal sunset by switching to phagotrophy. The Alphaproteobacteria dominated the autumn proliferation of bacterial OTUs. This contrast with the Actinobacteria- or Bacteroidetes-dominated total communities (summer and autumn together; Figure 2) indicates that the most numerically abundant phyla in the lakes are also capable of persisting through changing environmental conditions, whereas less abundant phyla may opportunistically increase under changing conditions.
Sixty-eight percent of the archaeal OTUs that increased in density during autumn belonged to the Euryarchaeota; half of those grouped with marine or aquatic lineages, whereas the other half grouped with methanogenic clades. The non-methanogenic Euryarchaeota that became abundant during autumn were all members of the Marine Group II, which are known to form seasonal blooms in the surface waters of the North Sea (Pernthaler et al., 2002). One Marine Group II OTU (Archaea_03_5) increased from 3.0% and 0% to 27.3% and 16.5% of the archaeal sequences in the surface and 9 m waters, respectively, of FRX. Thirty-two percent of the autumn archaeal OTUs belonged to the Crenarchaeota, which were dominated by terrestrial and soil groups (42.9%), followed by the Marine Group I Crenarchaeota (28.6%). The increase in euryarchaeal OTUs relative to crenarchaeal OTUs was distinct from the composition of the overall communities, which were dominated by Crenarchaeota rather than Euryarchaeota (Figure 2).
The proliferation of archaeal phylotypes (Grzymski et al., 2012) and proteins associated with chemolithotrophic Archaea (Williams et al., 2012) was reported in Southern Ocean waters during the winter, suggesting that summer communities dominated by photoautotrophy shift to chemolithotrophy during the polar night. Molecular and cultivation studies have revealed the presence of diverse chemolithotrophic microorganisms in FRX and WLB (Priscu et al., 1996, Voytek et al., 1999; Karr et al., 2005; Sattley and Madigan 2006; Kong et al., 2012a), and dark carbon fixation attributed to chemolithotrophs has been measured in these same lakes (Priscu et al., 1996, Vick and Priscu, unpublished data). Kong et al. (2012b) showed that Proteobacteria actively produced RubisCO in WLB during February and March, indicating that chemolithotrophic bacteria were active during the summer–autumn transition. Currently, there are no data regarding the activities of chemolithotrophic archaea in the photic zones of the MCM lakes, but the proliferation of archaeal sequences during autumn suggests that they may be important.
Part of the autumn archaeal ‘bloom' was also owing to the appearance of Terrestrial and Soil Group Crenarchaeota, indicating that allochthonous inputs may affect community structure. Eolian transport is an important dispersal mechanism in the MCM (Šabacká et al., 2012), and the downward migration of lake ice particulate matter (Squyres et al., 1991; Jepsen et al., 2010) may introduce microorganisms (Paerl and Priscu, 1998; Priscu et al., 1998; Gordon et al., 2000) into the water column. Similarly, mid-summer stream-flow is an important source of nutrients, particulate matter (Takacs et al., 2001; Foreman et al., 2004) and perhaps microorganisms (Vincent and Howard-Williams, 1986) to the lakes. Alternatively, the sequences may group with terrestrial lineages, but actually represent native aquatic organisms. Whether these putatively terrestrial sequences are transient, inactive or represent part of the active microbial assemblage is unknown.
Putatively methanogenic lineages (Methanomicrobia and Methanobacteria) accounted for 27% of the autumn proliferation of archaeal OTUs, although the most abundant methanogenic lineages decreased between summer and autumn. All of our samples were taken from oxygenated portions of the water column, but all known Methanomicrobia and Methanobacteria are strict anaerobes. Methanomicrobial sequences in soils surrounding FRX and WLB (Takacs-Vesbach unpublished data) and functional methanogens in the deep waters of FRX (Karr et al., 2006) are possible sources of methanogenic sequences; however, a local BLAST search showed that none of the terrestrial or FRX sequences matched the sequences in our study (data not shown). It is possible that our methanogenic sequences are not functionally methanogens, but their average GAST distances were small (0 to 0.03), indicating >95% accuracy of GAST taxonomic assignments (Huse et al., 2008). Methane production has been documented in oxygenated seawater (Karl et al., 2008, Damm et al., 2010) and an oxygenated oligotrophic lake, where planktonic and phytoplankton-attached Archaea actively transcribed the methyl coenzyme M reductase A gene for methanogenesis (Grossart et al., 2011). Damm et al. (2010) and Grossart et al. (2011) both connected methanogenesis in oxygenated water to phytoplankton activity, and the high concentrations of DMSP in WLB at 13 m (Lee et al., 2004) may provide a substrate pool for methanogenesis through its degradation product methanethiol (Damm et al., 2010). Although the metabolic state of the putatively methanogenic cells in our study is unknown, their presence combined with the supersaturation of methane starting at 12 m in WLB (Priscu and Dore, unpublished data) suggests the possibility of active methanogenesis.
Co-occurrence patterns and the molecular ecological network
Nonrandom community assembly, denoted by nonrandom co-occurrence patterns, is characteristic of assemblages of organisms across domains of life (Gotelli and McCabe, 2002; Horner-Devine et al., 2007). We compared the co-occurrence patterns found in our bacterial, archaeal and eukaryotic sequence data with those of a null distribution, representing random co-occurrence, and used the C-score metric (Stone and Roberts 1990) to determine whether our data differed significantly from a randomly assembled community. We observed nonrandom co-occurrence patterns for our whole data set (C-score=1.56, P=0.01) and for the Bacteria and Eukarya (C-score=1.52, P=0.01 and C-score=1.61, P=0.01, respectively), whereas the C-score for the Archaea alone was marginally significant (C-score=1.41, P=0.19).
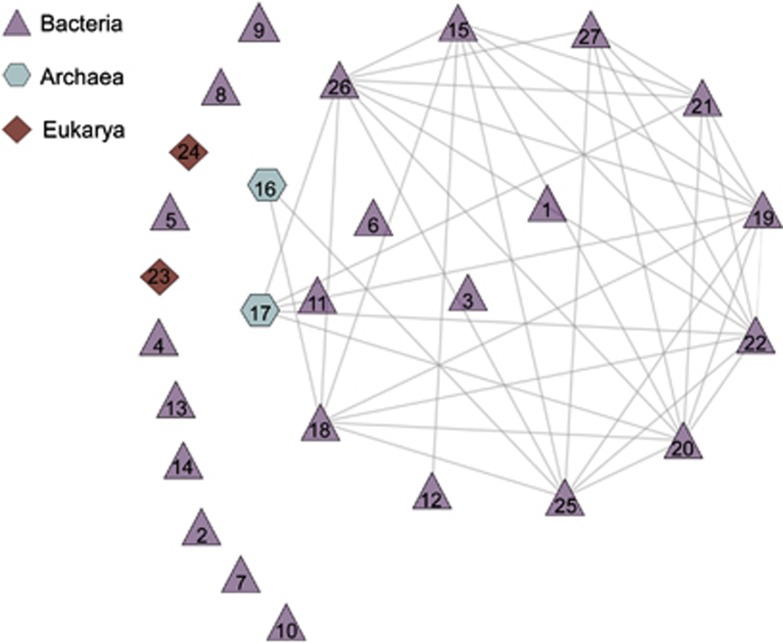
To describe the importance of biotic and abiotic interactions in explaining the nonrandom co-occurrence patterns, we generated a molecular ecological network based on Spearman correlations (P⩽0.01, r⩾0.8) between relative abundances of Bacterial and Archaeal OTUs, presence–absence of Eukaryotic OTUs and discrete values of environmental parameters. In total, we found 20 793 significant correlations between 872 variables (Supplementary Figure S2). We used modularity to detect community structure in our network (Fortunato, 2010; Supplementary Information), resulting in 27 modules containing groups of interconnected nodes (Figure 5; Supplementary Table S3). Each module was designated by a key (the OTU with the highest assignment value to the module) and numbered for convenience in the discussion (Table 1). The modules most important to the network structure were determined based on betweenness centrality (BC). González et al., 2010 showed that nodes with high BC scores were particularly important in maintaining the connectivity of an ecological network, and compared them with keystone species. Nine of 27 modules in our network had BC scores >0 (range 0.005–0.19; Table 1).
Figure 5.
Modules in the molecular ecological network determined by ModuLand. Each module is named according to the node with maximum assignment value to the module.
Table 1. Highest-level taxonomy definition for module keys with the average GAST distance for the reported taxonomy and the betweenness centrality score for each module.
| Module ID | Module Number | Taxonomy | Average GAST Distance | Module Betweenness Centrality Score |
|---|---|---|---|---|
| Acidobacteria_03_11260 | 1 | Acidobacteriaceae | 0.241 | 0 |
| Acidobacteria_03_15770 | 2 | Acidobacteriaceae | 0.258 | 0 |
| Acidobacteria_03_18 | 3 | Geothrix | 0.003 | 0 |
| Acidobacteria_03_1982 | 4 | Acidobacteriaceae | 0.0165 | 0 |
| Acidobacteria_03_231 | 5 | Solibacter | 0.0531 | 0 |
| Acidobacteria_03_5976 | 6 | Acidobacteriaceae | 0.254 | 0 |
| Actinobacteria_03_11036 | 7 | Acidimicrobiaceae | 0.105 | 0 |
| Actinobacteria_03_3 | 8 | Actinobacteria | 0.0026 | 0 |
| Actinobacteria_03_4 | 9 | Sporichthyaceae | 0.0016 | 0 |
| Actinobacteria_03_91 | 10 | Micrococcus | 0.0031 | 0 |
| Alphaproteobacteria_03_13239 | 11 | Rhodospirilliaceae | 0.281 | 0 |
| Alphaproteobacteria_03_2014 | 12 | Rickettsiaceae | 0.0482 | 0 |
| Alphaproteobacteria_03_62 | 13 | Caulobacteraceae | 0.0052 | 0 |
| Alphaproteobacteria_03_6621 | 14 | Sneathiella | 0.0078 | 0 |
| Alphaproteobacteria_03_91 | 15 | Pelagibacter | 0.0496 | 0.19 |
| Archaea_03_117 | 16 | Methanomicrobiales | 0 | 0 |
| Archaea_03_17 | 17 | Thermoplasmatales | 0.0102 | 0 |
| Bacteroidetes_03_1296 | 18 | Flexibacter | 0.0188 | 0.07 |
| Bacteroidetes_03_168 | 19 | Croceibacter | 0.0236 | 0.04 |
| Bacteroidetes_03_262 | 20 | Flavobacterium | 0.0005 | 0.04 |
| Betaproteobacteria_03_143 | 21 | Herbaspirillum | 0.0827 | 0.02 |
| Betaproteobacteria_03_51 | 22 | Methyloversatilis | 0.0063 | 0.04 |
| Eukarya_06_1718 | 23 | Pteridomonas | 0.023 | 0 |
| Eukarya_06_1772 | 24 | Chlorogonium | 0.023 | 0 |
| Gammaproteobacteria_03_4 | 25 | Pseudomonas | 0.0015 | 0.1 |
| Planctomycetes_03_726 | 26 | Planctomyces | 0.0169 | 0.04 |
| Verrucomicrobia_03_119 | 27 | Opitutus | 0.0077 | 0.005 |
We examined the modules with significant BC scores and the modules containing the autumn blooming Archaea in detail, and attempted to assign functions based on the putative physiologies of the organisms present (Supplementary Table S5). Bielewicz et al., (2011) suggested that trophic or metabolic plasticity allows organisms to be more successful in the MCM lakes, and our module analysis supports the importance of innovative energy capture and metabolic flexibility during the transition to polar night.
On the basis of its high BC score (0.19; nearly twice the next highest BC), Module 15 forms the keystone (González et al., 2010) of the MCM network. Five OTUs (40% of the module) were related to taxa that can produce proteorhodopsins (Atamna-Ismaeel et al., 2008, Oh et al., 2011, Huggett and Rappé, 2012), including the module key, Alphaproteobacteria_03_91 (Pelagibacter, GAST=0.049), Bacteroidetes_03_1 (Flavobacteriacea, GAST=0.0029) and Gammaproteobacteria_03_175 (Oceanospirillales, GAST=0.0026). Proteorhodopsins are light-driven proton pumps found mainly in marine and freshwater Alphaproteobacteria, Gammaproteobacteria, Flavobacteria and some Euryarchaeota, which, along with heterotrophic metabolism, can generate energy for growth (‘photoheterotrophy' Giovannoni et al., 2005; Frigaard et al., 2006; Atamna-Ismaeel et al., 2008; DeLong and Béjà, 2010; Steindler et al., 2011) and provide a competitive advantage under conditions of organic carbon or nutrient limitation (Giovannoni et al., 2005), such as those found in the MCM lakes. Fluctuations in the quality and quantity of available organic carbon are thought to affect heterotrophic bacterioplankton metabolism in the MCM lakes during autumn (Vick and Priscu, 2012), and although nutrient limitation is perennial in these lakes phytoplankton activity results in a spring/summer drawdown of N and P (Lizotte et al., 1996). In addition to putatively photoheterotrophic OTUs, which account for 40% of the module, Module 15 contains two OTUs of the genus Hydrogenophaga (GAST=0.0014 and 0.002), which are typically facultatively autotrophic organisms capable of oxidizing hydrogen or using organic carbon to generate energy (for example, Yoon et al., 2008). In total, 53% of the module belongs to groups known to be metabolically flexible, suggesting that oligotrophy and the strong seasonality associated with the MCM lakes favor the ability to shift between energy resources in response to changing environmental conditions. In addition, Module 15's keystone status indicates that these abilities are integral to the MCM lake ecosystem function.
Module 17 contained five of the nodes representing the autumn proliferation of archaeal phylotypes (Figure 4, Supplementary Table S3; Archaea_03_17), and it provides another example of competitive energy acquisition in MCM lakes. Two of the nodes belong to the Marine Group I Crenarchaeota and may signify an autumn shift in favor of chemolithotrophic metabolisms similar to that found in the Southern Ocean (Grzymski et al., 2012; Williams et al., 2012). Nodes representing the Marine Group II of the Euryarchaeota also group with Module 17. Currently, there are no cultured representatives of Marine Group II, and thus little is known about their range of metabolic capabilities. However, a complete genome representing the Marine Group II Euryarchaeota was recovered from a Puget Sound metagenome (Iverson et al., 2012), revealing a photoheterotrophic, proteorhodopsin-containing organism. A PCR-based study of Archaea from the North Pacific Subtropical Gyre concluded that approximately 10% of Euryarchaeota contained proteorhodopsin genes (Frigaard et al., 2006). If the putative functions assigned to the Archaea in Module 17 are correct, the module provides further evidence for the importance of metabolic flexibility and suggests that chemolithotrophy may be important in fueling ecosystem production during the polar night.
Protistan organisms often rely on mixotrophic lifestyles to cope with the oligotrophic conditions and seasonal light–dark cycles in MCM lakes (Laybourn-Parry 2002; Bielewicz et al., 2011; Thurman et al., 2012). Phototrophic nanoflagellates in WLB increased their grazing rates on fluorescently labeled bacterial prey throughout the month of March (Thurman et al., 2012), supporting the suggestion of Bielewicz et al. (2011) that cryptophyte populations use phagotrophy as an adaptive strategy during the summer−winter transition. Similarly, our results showed that Chrysophyceae, which are generally dominated by the mixotrophic genus Ochromonas in these lakes (Lizotte et al., 1996), likely increased in abundance during the autumn. Module 22 (BC=0.04, Supplementary Table S3, Betaproteobacteria_03_51) contained 35% of the eukaryotic OTUs that became abundant during autumn, including all of the Chrysophyceae, the heterotrophic nanoflagellate Cryothecomonas and a ciliate.
Module 26 (BC=0.04) contained Actinobacteria (22% of the module), including members of the genus Microthrix (Actinobacteria_03_174; GAST=0.0095). Microthrix and other Actinobacteria generate carbon and energy storage compounds (triacylglcerols), store polyphosphates and possess high-affinity Pst P-uptake systems, all of which may help the organisms compete under conditions of unbalanced growth and P-limiting conditions (McIlroy et al. 2013), such as those found in Lake Bonney (Dore and Priscu, 2001). In addition, Planktophila (Actinobacteria_03_32; GAST=0.0028) are important polysaccharide degraders with the ability to mineralize N-acetylgucosamine, a breakdown product of bacterial cell walls, which may assist in winter survival, and contain actinorhodopsin (Garcia et al., 2013), suggesting a role for photoheterotrophy in Module 26.
Module 21 (BC=0.02; Key=Betaproteobacteria_03_143) contained mostly heterotrophic bacteria and a few Archaea, and a majority of the OTUs in the module increased between the summer and autumn sampling points, especially in the shallower waters of the lakes. Typically, phytoplankton production is thought to draw down nutrient concentrations during the summer, leading to increased nutrient depletion in the already oligotrophic waters of the FRX and WLB photic zones. Members of the Actinobacteria have been shown to proliferate under low-nutrient conditions (reviewed in Newton et al., 2010). The module also contains putative nitrogen-fixing bacteria, which may have increased in response to decreasing nutrient concentrations (Alphaproteobacteria_03_431, Alphaproteobacteria_03_15, Alphaproteobacteria_03_808 and Betaproteobacteria_03_143, the module key).
Taken together, these modules provide evidence for the importance of metabolic and trophic plasticity and nutrient scavenging in the MCM lakes. Other significant modules are discussed in Supplementary Information and provide further examples of the adaptation to oligotrophic or changing environments, along with insights into organic matter processing and eukaryote−prokaryote interactions in lakes FRX and WLB.
Conclusions
Our study comprises the first high-throughput sequencing evaluation of the diversity of Bacteria, Archaea and Eukarya in permanently ice-covered lakes of the Antarctic MCM Dry Valleys. We found that these light- and nutrient-limited systems exhibit low diversity overall, but that the autumn decrease in solar radiation coincides with increases or shifts in microbial diversity across all three domains of life. The statistically significant partitioning of bacterial and eukaryotic communities by season within lake and depth suggests, in agreement with past studies, that these communities are strongly controlled by the vertically stratified water columns of lakes FRX and Bonney, but that they also respond to the change in season. The low archaeal diversity was offset by an autumn ‘bloom' of archaeal OTUs, which likely stemmed from a combination of allochthonous inputs and proliferation of organisms adapted to the winter darkness. Similarly, we found OTUs whose closest relatives are adapted to low-nutrient environments, photoheterotrophic and mixotrophic lifestyles, to be particularly important in the modular community structure in these lakes. We suggest future studies focusing on functional gene analysis, metagenomics or transcription to examine the relationships revealed by our molecular ecological network analysis.
Acknowledgments
We would like to thank the MCM Microbial Observatory, Sukkyun Han, Chao Tang, Amy Chiuchiolo and Marie Šabacká for assistance with sample collection, the 2007–2008 McMurdo Long-Term Ecological Research limnology team for assistance with environmental data collection and Elizabeth McCliment for assistance with sequencing. Funding was provided by NSF DEB-0717390 to Linda A Amaral-Zettler (MIRADA-LTERS) and OPP-1115254, OPP-0838953, OPP-1027284 and OPP- 0839075 to John C Priscu. The Montana Space Grant Consortium provided additional funding for Trista Vick-Majors.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alonso-Sáez L, Sánchez O, Gasol JM, Balagué V, Pedrós-Alio C. Winter-to-summer changes in the composition and single-cell activity of near-surface Arctic prokaryotes. Environ Microbiol. 2008;10:2444–2454. doi: 10.1111/j.1462-2920.2008.01674.x. [DOI] [PubMed] [Google Scholar]
- Amaral-Zettler LA, McCliment EA, Ducklow HW, Huse SM. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One. 2009;4:e6372. doi: 10.1371/journal.pone.0006372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 2001;26:32–46. [Google Scholar]
- Andersson AF, Riemann L, Bertilsson S. Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISME J. 2010;4:171–181. doi: 10.1038/ismej.2009.108. [DOI] [PubMed] [Google Scholar]
- Atamna-Ismaeel N, Sabehi G, Sharon I, Witzel K-P, Labrenz M, Jürgens K, et al. Widespread distribution of proteorhodopsins in freshwater and brackish ecosystems. ISME J. 2008;2:656–662. doi: 10.1038/ismej.2008.27. [DOI] [PubMed] [Google Scholar]
- Barberán A, Bates ST, Casamayor EO, Fierer N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012;6:343–351. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielewicz S, Bell E, Kong W, Friedberg I, Priscu John C, Morgan-Kiss RM. Protist diversity in a permanently ice-covered Antarctic lake during the polar night transition. ISME J. 2011;5:1559–1564. doi: 10.1038/ismej.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunge J, Woodard L, Böhning D, Foster J, Connolly S, Allen H. Estimating population diversity with CatchAll. Bioinformatics. 2012;28:1045–1047. doi: 10.1093/bioinformatics/bts075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffron S, Rehrauer H, Jakob Pernthaler, von Mering C. A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res. 2010;20:947–959. doi: 10.1101/gr.104521.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A. Estimating the population size for capture-recapture data with unequal catchability. Biometrics. 1987;43:783–791. [PubMed] [Google Scholar]
- Chao A, Shen TJ.2010. Program SPADE (Species Prediction And Diversity Estimation). Available at http://chao.stat.nthu.edu.tw/ .
- Charvet S, Vincent W, Comeau A, Lovejoy C. Pyrosequencing analysis of the protist communities in a High Arctic meromictic lake: DNA preservation and change. Front Microbiol. 2012;3:422. doi: 10.3389/fmicb.2012.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet S, Vincent WF, Lovejoy C. Chrysophytes and other protists in High Arctic lakes: molecular gene surveys, pigment signatures and microscopy. Polar Biol. 2012;35:733–748. [Google Scholar]
- Comeau A, Harding T, Galand P, Vincent WF, Lovejoy C. Vertical distribution of microbial communities in a perennially stratified Arctic lake with saline, anoxic bottom waters. Sci Rep. 2012;2:604. doi: 10.1038/srep00604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump BC, Kling GW, Bahr M, Hobbie JE. Bacterioplankton community shifts in an Arctic lake correlate with seasonal changes in organic matter source. Appl Environl Microbiol. 2003;69:2253–2268. doi: 10.1128/AEM.69.4.2253-2268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump BC, Amaral-Zettler L, Kling G. Microbial diversity in arctic freshwaters is structured by inoculation of microbes from soils. ISME J. 2012;6:1629–1639. doi: 10.1038/ismej.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm E, Helmke E, Thoms S, Schauer U, Nöthig E, Bakker K, et al. Methane production in aerobic oligotrophic surface water in the central Arctic Ocean. Biogeosciences. 2010;7:1099–1108. [Google Scholar]
- DeLong EF, Béjà O. The light-driven proton pump proteorhodopsin enhances bacterial survival during tough times. PLoS Biol. 2010;8:e1000359. doi: 10.1371/journal.pbio.1000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore J, Priscu J. Phytoplankton phosphorus deficiency and alkaline phosphatase activity in the McMurdo Dry Valley Lakes, Antarctica. Limnol Oceanogr. 2001;46:1331–1346. [Google Scholar]
- Foreman CM, Wolf CF, Priscu JC. Impact of episodic warming events on the physical, chemical, and biological relationships of Lakes in the McMurdo Dry Valleys, Antarctica. Aq Geochem. 2004;10:239–368. [Google Scholar]
- Fortunato S. Community detection in graphs. Phys Rep. 2010;486:75–174. [Google Scholar]
- Frigaard N-U, Martinez A, Mincer TJ, DeLong Edward F. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature. 2006;439:847–850. doi: 10.1038/nature04435. [DOI] [PubMed] [Google Scholar]
- Fuhrman J, Steele J. Community structure of marine bacterioplankton: patterns, networks, and relationships to function. Aquatic Microbial Ecology. 2008;53:69–81. [Google Scholar]
- Glatz R, Lepp P, Ward B, Francis C. Planktonic microbial community composition across steep physical/chemical gradients in permanently ice-covered Lake Bonney, Antarctica. Geobiology. 2006;4:53–67. [Google Scholar]
- Galand PE, Lovejoy C, Pouliot J, Garneau M, Vincent WF. Microbial community diversity and heterotrophic production in a coastal Arctic ecosystem: a stamukhi lake and its source waters. Limnol Oceangr. 2008;53:813–823. [Google Scholar]
- Garcia SL, McMahon KD, Martinez-Garcia M, Srvastava A, Sczyrba A, Stepanauskas R, et al. Metabolic potential of a single cell belonging to one of the most abundant lineages in freshwater bacterioplankton. ISME J. 2013;7:137–147. doi: 10.1038/ismej.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiglione JF, Murray A.E. Pronounced summer to winter differences and higher wintertime richness in coastal Antarctic marine bacterioplankton. Environ Microbiol. 2012;14:617–629. doi: 10.1111/j.1462-2920.2011.02601.x. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Bibbs L, Cho J, Stapels M, Desiderio R, Vergin K, et al. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature. 2005;438:82–85. doi: 10.1038/nature04032. [DOI] [PubMed] [Google Scholar]
- González M, Dalsgaard A, Olesen JB. Centrality measures and the importance of generalist species in pollination networks. Ecol Complex. 2010;7:36–43. [Google Scholar]
- Gordon DA, Priscu J, Giovannoni S. Origin and phylogeny of microbes living in permanent Antarctic lake ice. Microb Ecol. 2000;39:197–202. doi: 10.1007/s002480000016. [DOI] [PubMed] [Google Scholar]
- Gotelli NJ, McCabe DJ. Species co-occurrence: a meta-analysis of J. M. Diamond's assembly rules model. Ecology. 2002;83:2091–2096. [Google Scholar]
- Grossart HP, Frindte K, Dziallas C, Eckert W, Tang KW. Microbial methane production in oxygenated water column of an oligotrophic lake. Proc Natl Acad Sci USA. 2011;108:19657–19661. doi: 10.1073/pnas.1110716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzymski JJ, Riesenfeld CS, Williams TJ, Dussaq AM, Ducklow H, Erickson M, et al. A metagenomic assessment of winter and summer bacterioplankton from Antarctica Peninsula coastal surface waters. ISME J. 2012;2:1–15. doi: 10.1038/ismej.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner-Devine MC, Silver JM, Leibold MA, Bohannan BJM, Colwell RK, Fuhrman JA, et al. A comparison of taxon co-occurrence patterns for macro- and microorganisms. Ecology. 2007;88:1345–1353. doi: 10.1890/06-0286. [DOI] [PubMed] [Google Scholar]
- Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA, et al. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- Huggett M, Rappé M. Genome sequence of strain HIMB30, a novel member of the marine Gammaproteobacteria. J Bacteriol. 2012;194:723–733. doi: 10.1128/JB.06506-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Dethlefsen L, Huber JA, Mark Welch D, Relman DA, Sogin ML. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson V, Morris RM, Frazar CD, Berthiaume CT, Morales RL, Armbrust EV. Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science. 2012;335:587–590. doi: 10.1126/science.1212665. [DOI] [PubMed] [Google Scholar]
- Jepsen S, Adams E, Priscu JC. Sediment melt-migration dynamics in perennial Antarctic lake ice. Arctic Antarctic Alpine Res. 2010;42:57–66. [Google Scholar]
- Judd KE, Crump Byron C, Kling George W. Variation in dissolved organic matter controls bacterial production and community composition. Ecology. 2006;87:2068–2079. doi: 10.1890/0012-9658(2006)87[2068:vidomc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Karl DM, Beversdorf L, Björkman KM, Church MJ, Martinez A, Delong EF. Aerobic production of methane in the sea. Nat Geoscience. 2008;7:473–478. [Google Scholar]
- Karr EA, Ng JM, Belchik SM, Sattley WM, Madigan MT, Achenbach LA. Biodiversity of methanogenic and other Archaea in the permanently frozen Lake Fryxell, Antarctica. Appl Environ Microbiol. 2006;72:1663–1666. doi: 10.1128/AEM.72.2.1663-1666.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karr EA, Sattley WM, Madigan MT, Achenbach LA. Diversity and distribution of sulfate-reducing bacteria in permanently frozen Lake Fryxell, McMurdo Dry Valleys, Antarctica. Appl Environ Microbiol. 2005;71:6353–6359. doi: 10.1128/AEM.71.10.6353-6359.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W, Dolhi JM, Chiuchiolo A, John Priscu, Morgan-Kiss RM. Evidence of form II RubisCO (cbbM) in a perennially ice-covered Antarctic lake. FEMS Microbiol Ecol. 2012;82:491–500. doi: 10.1111/j.1574-6941.2012.01431.x. [DOI] [PubMed] [Google Scholar]
- Kong W, Ream DC, Priscu JC, Morgan-Kiss RM. Diversity and expression of RubisCO genes in a perennially ice-covered Antarctic lake during the polar night transition. Appl Environ Microbiol. 2012;78:4358–4366. doi: 10.1128/AEM.00029-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laybourn-Parry J. Survival mechanisms in Antarctic lakes. Philos Trans R Soc London Ser B Biol Sci. 2002;357:863–869. doi: 10.1098/rstb.2002.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PA, Priscu JC, Ditullio GR, Riseman SF, Tursich N, DeMora SJ. Elevated levels of dimethylated-sulfur compounds in Lake Bonney, a poorly ventilated Antarctic lake. Limnol Oceanogr. 2004;49:1044–1055. [Google Scholar]
- Lizotte MP, Priscu JC. Natural fluorescence and quantum yields in vertically stationary phytoplankton from perennially ice-covered lakes. Limnol Oceanogr. 1994;39:1399–1410. [Google Scholar]
- Lizotte MP, Sharp TR, Priscu JC. Phytoplankton dynamics in the stratified water column of Lake Bonney, Antarctica: biomass and productivity during the winter-spring transition. Polar Biol. 1996;16:155–162. [Google Scholar]
- McCliment EA, Nelson CE, Carlson CA, Alldredge AL, Witting J, Amaral-Zettler LA. An all-taxon microbial inventory of the Moorea coral reef ecosystem. ISME J. 2012;6:309–319. doi: 10.1038/ismej.2011.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlroy SJ, Kristiansen R, Albertsen M, Karst SM, Rossetti S, Nielsen JL, et al. Metabolic model for the filamentous ‘Candidatus Microthrix pavicella' based on genomic and Metagenomic analyses. ISME J. 2013;7:1161–1172. doi: 10.1038/ismej.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight DM, Howes BL, Taylor CD, Goehringer DD. Phytoplankton dynamics in a stably stratified Antarctic lake during winter darkness. J Phycology. 2000;36:852–861. [Google Scholar]
- Miklós I, Podani J. Randomization of presence–absence matrices: comments and new algorithms. Ecology. 2004;85:86–92. [Google Scholar]
- Mikucki JA, Priscu JC. Bacterial diversity associated with Blood Falls, a subglacial outflow from the Taylor Glacier, Antarctica. Appl Environ Microbiol. 2007;73:4029–4039. doi: 10.1128/AEM.01396-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morán X, Gasol J, Pedrós-Alió C, Estrada M. Dissolved and particulate primary production and bacterial production in offshore Antarctic waters during austral summer: coupled or uncoupled. Mar Ecol Prog Ser. 2001;222:25–39. [Google Scholar]
- Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev. 2011;75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh HM, Kang I, Lee K, Jang Y, Lim S, Cho J. Complete genome sequence of strain IMCC9063, belonging to SAR11 subgroup 3, isolated from the Arctic Ocean. J Bacteriol. 2011;193:3379–3380. doi: 10.1128/JB.05033-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Guillaume Blanchet F, Kindt R, Legendre P, O'Hara RB, Simpson GL, et al. 2010. vegan: Community ecology package. R Package version 1.17-3.
- Paerl HW, Priscu JC. Microbial phototrophic, heterotrophic, and diazotrophic activities associated with aggregates in the permanent ice cover of Lake Bonney, Antarctica. Microb Ecol. 1998;36:221–230. doi: 10.1007/s002489900109. [DOI] [PubMed] [Google Scholar]
- Pernthaler A, Preston CM, Pernthaler J, DeLong EF, Amann R. Comparison of fluorescently labeled oligonucleotide and polynucleotide probes for the detection of pelagic marine bacteria and archaea. Appl Environ Microbiol. 2002;68:661–667. doi: 10.1128/AEM.68.2.661-667.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priscu JC, Downes MT, McKay CP. Extreme supersaturation of nitrous oxide in a poorly ventilated Antarctic lake. Limnol Oceanogr. 1996;41:1544–1551. doi: 10.4319/lo.1996.41.7.1544. [DOI] [PubMed] [Google Scholar]
- Priscu JC, Fritsen CH, Adams EE, Giovannoni SJ, Paerl HW, McKay CP, et al. Perennial Antarctic lake ice: an oasis for life in a polar desert. Science. 1998;280:2095–2098. doi: 10.1126/science.280.5372.2095. [DOI] [PubMed] [Google Scholar]
- Priscu JC, Wolf CF, Takacs CD, Fritsen CH, Laybourn-Parry J, Roberts EC, et al. Carbon transformations in a perennially ice-covered Antarctic lake. Bioscience. 1999;49:997–1008. [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2008R: a language and environment for statistical computing. Vienna, Austria. , www.R-project.org . [Google Scholar]
- Roberts D.2010. labdsv: ordination and multivariate analysis for ecology package. R Package version 1.4-1.
- Šabacká M, Priscu JC, Basagic HJ, Fountain AG, Wall DH, Virginia RA, et al. Aeolian flux of biotic and abiotic material in Taylor Valley, Antarctica. Geomorphology. 2012;155-156:102–111. [Google Scholar]
- Sattley WM, Madigan MT. Isolation, characterization, and ecology of cold-active, chemolithotrophic, sulfur-oxidizing bacteria from perennially ice-covered Lake Fryxell, Antarctica. Appl Environ Microbiol. 2006;72:5562–5568. doi: 10.1128/AEM.00702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spigel RH, Priscu JC.1998Physical limnology of the McMurdo Dry Valley lakesIn: Priscu JC, (ed.)Ecosystem Dynamics in a Polar Desert: the McMurdo Dry Valleys, Antarctica. Antarctic Research Series Vol. 72American Geophysical Union: Washington D.C.153–187. [Google Scholar]
- Squyres S, Andersen D, Nedell S, Wharton R. Lake Hoare, Antarctica: sedimentation through a thick perennial ice cover. Sedimentology. 1991;38:363–379. doi: 10.1111/j.1365-3091.1991.tb01265.x. [DOI] [PubMed] [Google Scholar]
- Steele JA, Countway PD, Xia L, Vigil PD, Beman JM, Kim DY, et al. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J. 2011;5:1414–1425. doi: 10.1038/ismej.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler L, Schwalbach MS, Smith DP, Chan F, Giovannoni SJ. Energy starved Candidatus Pelagibacter ubique substitutes light-mediated ATP production for endogenous carbon respiration. PLoS One. 2011;6:e19725. doi: 10.1371/journal.pone.0019725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone L, Roberts A. The checkerboard score and species distributions. Oecologia. 1990;85:74–79. doi: 10.1007/BF00317345. [DOI] [PubMed] [Google Scholar]
- Takacs CD, Priscu JC. Bacterioplankton dynamics in the McMurdo Dry Valley lakes, Antarctica: production and biomass loss over four seasons. Microb. Ecol. 1998;36:239–250. doi: 10.1007/s002489900111. [DOI] [PubMed] [Google Scholar]
- Takacs CD, Priscu JC, McKnight DM. Bacterial dissolved organic carbon demand in McMurdo Dry Valley Lakes, Antarctica. Limnol Oceanogr. 2001;46:1189–1194. [Google Scholar]
- Thurman J, Parry J, Hill PJ, Priscu John C, Vick TJ, Chiuchiolo A, et al. Microbial dynamics and flagellate grazing during transition to winter in Lakes Hoare and Bonney, Antarctica. FEMS Microbiol Ecol. 2012;82:449–458. doi: 10.1111/j.1574-6941.2012.01423.x. [DOI] [PubMed] [Google Scholar]
- Vick TJ, Priscu JC. Bacterioplankton productivity in lakes of the Taylor Valley, Antarctica during the polar night transition. Aquat Microb Ecol. 2012;68:77–90. [Google Scholar]
- Vincent W, Howard-Williams C. Antarctic stream ecosystems: physiological ecology of a blue-green algal epilithon. Freshwater Biol. 1986;16:219–233. [Google Scholar]
- Voytek MA, Priscu JC, Ward BB. The distribution and relative abundance of ammonia-oxidizing bacteria in lakes of the McMurdo Dry Valley, Antarctica. Hydrobiologia. 1999;401:113–130. [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TJ, Long E, Evans F, Demaere MZ, Lauro FM, Raftery MJ, et al. A metaproteomic assessment of winter and summer bacterioplankton from Antarctic Peninsula coastal surface waters. ISME J. 2012;2:1–18. doi: 10.1038/ismej.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz P, Kottmann R, Field D, Knight R, Cole JR, et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MixS) specifications. Nat Biotechnol. 2011;29:415–420. doi: 10.1038/nbt.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KS, Tsukada N, Sakai Y, Ishii M, Igarashi Y, Nishihara H. Isolation and characterization of a new facultatively autotrophic hydrogen-oxidizing Betaproteobacterium, Hydrogenophaga sp. AH-24. FEMS Microbiol Lett. 2008;278:94–100. doi: 10.1111/j.1574-6968.2007.00983.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.