Abstract
Most studies of biofilm biology have taken a reductionist approach, where single-species biofilms have been extensively investigated. However, biofilms in nature mostly comprise multiple species, where interspecies interactions can shape the development, structure and function of these communities differently from biofilm populations. Hence, a reproducible mixed-species biofilm comprising Pseudomonas aeruginosa, Pseudomonas protegens and Klebsiella pneumoniae was adapted to study how interspecies interactions affect biofilm development, structure and stress responses. Each species was fluorescently tagged to determine its abundance and spatial localization within the biofilm. The mixed-species biofilm exhibited distinct structures that were not observed in comparable single-species biofilms. In addition, development of the mixed-species biofilm was delayed 1–2 days compared with the single-species biofilms. Composition and spatial organization of the mixed-species biofilm also changed along the flow cell channel, where nutrient conditions and growth rate of each species could have a part in community assembly. Intriguingly, the mixed-species biofilm was more resistant to the antimicrobials sodium dodecyl sulfate and tobramycin than the single-species biofilms. Crucially, such community level resilience was found to be a protection offered by the resistant species to the whole community rather than selection for the resistant species. In contrast, community-level resilience was not observed for mixed-species planktonic cultures. These findings suggest that community-level interactions, such as sharing of public goods, are unique to the structured biofilm community, where the members are closely associated with each other.
Keywords: Klebsiella pneumonia, mixed-species biofilm, Pseudomonas aeruginosa, Pseudomonas protegens, sodium dodecyl sulfate, tobramycin
Introduction
Many studies have demonstrated that biofilms are distinct from their planktonic counterparts. For example, biofilms have been found to be more resistant to antibiotics and host immune responses (Costerton et al., 1999; Mah and O'Toole, 2001; Arciola et al., 2005; Jensen et al., 2007). These resistances are hypothesized to in part reflect the expression of biofilm-specific genes and phenotypes as revealed by transcriptomic and proteomic studies. About 1–2% of the entire genome (79 genes in Escherichia coli and 73 genes in Pseudomonas aeruginosa) and more than 800 proteins were differentially expressed when biofilms and planktonic bacteria were compared (Whiteley et al., 2001; Sauer et al., 2002; Schembri et al., 2003).
Most studies thus far have focused on single-species biofilms to understand the molecular and physiological mechanisms that drive biofilm development (Sauer and Camper, 2001; Sauer et al., 2002). However, biofilms in both natural and engineered environments are highly diverse. For example, 454 pyrosequencing of activated sludge revealed 1183 to 3567 operational taxonomic units (OTUs) (Zhang et al., 2012). In addition, about 100–200 bacterial species were found to colonize and form biofilms on hard and soft tissues of the oral cavity (Paster et al., 2006).
Given the diversity of biofilms in nature, it is likely that interspecies interactions play important roles in determining the development, structure and function of these biofilms. For example, there might be intense competition for resources, as exemplified by the invasion and overgrowth of Hyphomicrobium sp. biofilms by P. putida (Banks and Bryers, 1991). Metabolic cooperation also occurred within biofilms where Burkholderia sp. LB400 feeding on chlorophenyl was observed to excrete chlorobenzoate, which was subsequently metabolized by Pseudomonas sp. B13 (FR1) (Nielsen et al., 2000). Similarly, mixed-species biofilms of Variovarax sp., Comamonas testosteroni and Hyphomicrobium sulfonivorans degraded the phenylurea herbicide, linuron and its metabolic intermediate more efficiently than the comparable single-species biofilms (Breugelmans et al., 2008). These results indicate that interspecies interactions within a biofilm can affect its development, structure and function, and that such interactions are unpredictable from the studies of single-species biofilms.
In this study, a mixed-species biofilm comprising P. aeruginosa, P. protegens (recently renamed from P. fluorescens Pf-5 (Ramette et al., 2011; Lim et al., 2013a)) and Klebsiella pneumoniae was studied. This mixed-species biofilm has previously been shown to be reproducible with respect to structure and viable cell counts (Jackson et al., 2001; Stoodley et al., 2001). In addition, these three species are commonly found in soil habitats and are reported to coexist in metalworking fluids, where the growth of P. protegens is a prerequisite for the growth of P. aeruginosa and K. pneumoniae (Chazal, 1995). These three species were also isolated from the gut of the silk moth, Bombyx mori (Anand et al., 2010), further supporting the relevance of these three species as a mixed-species biofilm model. Here, confocal microscopy and quantitative image analysis of biofilms formed by fluorescent protein-tagged bacteria were used to investigate the development, structure and resilience of this mixed-species biofilm in comparison with single-species biofilms. Development of the biofilms was consistent where the mixed-species biofilm displayed a delayed development and distinct structures that were different from all comparable single-species biofilms. Further, the mixed-species biofilm was more resilient to antimicrobials compared with the single-species biofilms and this community-level protection was shown to be affected by the spatial organization of the community members.
Materials and methods
Bacterial strains and culture media
Bacteria (Table 1) were cultured in either M9 minimal medium (48 mM Na2HPO4; 22 mM KH2PO4; 9 mM NaCl; 19 mM NH4Cl; 2 mM MgSO4; 0.1 mM CaCl2; and 0.04% w/v glucose) supplemented with 0.2% w/v casamino acids (supplemented M9 minimal medium), Luria Bertani broth (LB10) (10 g l−1 NaCl; 10 g l−1 tryptone; 5 g l−1 yeast extract) or Super Optimal Broth (SOB) (10 mM NaCl; 2.5 mM KCl; 10 mM MgCl2; 10 mM MgSO4; 20 g l−1 tryptone; 5 g l−1 yeast extract).
Table 1. List of bacterial strains.
| Species and strain | Genotypic and phenotypic characteristicsa | Source or reference |
|---|---|---|
| Pseudomonas aeruginosa | ||
| PAO1 | Wild type | bATCC BAA-47 |
| cPseudomonas protegens | ||
| Pf-5 | Wild type | bATCC BAA-477 |
| dKlebsiella pneumonia | ||
| KP-1 | Wild type | This project |
| Escherichia coli | ||
| JM109 | endA1 glnV44 thi-1 relA1 gyrA96 recA1 mcrB+ Δ(lac-proAB) e14− [F' traD36 proAB+ lacIq lacZΔM15] hsdR17(rK−mK+) | Yanisch-Perron et al., 1985 |
| HPS1 | F− Δ(lab-proAB) endA1 gyrA96 hsdR17 supE44 relA1 recA1 thi rifR zzx::mini-Tn5Lac4 | Choi et al., 2005 |
| CC118 λpir | Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recAl λ pir | Choi et al., 2005 |
| DH5α λpir | F−,Φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK−, mK+) phoA supE44 thi-1 λ pir | Choi et al., 2005 |
| S17-1 λpir | hsdR recA pro RP4-2 (Tc::Mu; Km::Tn7)(λ pir) | Miller and Mekalanos, 1988 |
| HB101 | F−, hsdS20 (rb−, mb−), supE44, ara14, galK2, lacY1, proA2, rpsL20 (StrR), xyl-5, mtl-1, l-, recA13, mcrA−, mcrB− | Lambertsen et al., 2004 |
StrR: Stretomycin resistance.
American Type Culture Collection (ATCC) number.
Pseudomonas fluorescens has recently been renamed as Pseudomonas protegens (Ramette et al., 2011; Lim et al., 2013a).
K. pneumoniae is an environmental isolate and its genome is currently being sequenced.
Plasmids and plasmid constructions
Plasmids (Table 2) were purified using the Invitrogen PureLink HQ Plasmid Purification Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. R6K replicon-based delivery plasmids were constructed. Briefly, purified ColE1 replicon-based delivery plasmids, pUC18T-mini-Tn7T-Gm-GFPmut3, pUC18T-mini-Tn7T-Gm-eYFP, pUC18T-mini-Tn7T-Gm-eCFP and vector plasmid were sequentially digested with SalI and KpnI restriction endonucleases (New England Biolabs, Ipswich, MA, USA). The 2.6-kbp fragment (containing the gentamicin cassette and fluorescent protein-coding gene) digested from the ColE1 replicon-based delivery plasmid and the 3.0-kbp fragment (containing the R6K ori) digested from the vector plasmid were gel purified using Invitrogen PureLink Quick Gel Extraction Kit (Invitrogen). Purified fragments were ligated with T4 DNA ligase (Promega, Fitchburg, WI, USA). Similarly, the ColE1 replicon-based delivery plasmid, pUC18T-mini-Tn7T-Gm-DsRedExpress, was double digested with SalI and ScaI restriction endonucleases followed by a single digestion using the SapI restriction endonuclease (New England Biolabs), while the vector plasmid was sequentially digested with SalI and SapI restriction endonucleases (New England Biolabs). The 2.9-kbp fragment digested from the ColE1 replicon-based delivery plasmid and the 2.8-kbp fragment digested from the vector plasmid were gel purified and ligated as above. The helper plasmid, pTNS2-ColE1, was constructed by amplifying (primers ColE1_F and ColE1_R, Table 3) the ColE1 ori from a ColE1 replicon-based delivery plasmid and subsequently ligating it into the EcoRI and SmaI restriction endonuclease sites of pTNS2.
Table 2. List of plasmids used in this study.
| Plasmid/host | Relevant characteristica | Source |
|---|---|---|
| Helper plasmid | ||
| pTNS1/CC118 λpir | helper plasmid, providing the Tn7 transposition function. ApR, R6K ori, ori T | Choi et al. (2005) |
| pTNS2/DH5α λpir | helper plasmid, providing the Tn7 transposition function. ApR, R6K ori, ori T | AY884833b,c |
| pTNS2-ColE1/JM109 | helper plasmid, providing the Tn7 transposition function. ApR, ColE1 ori, ori T | This project |
| ColE1 replicon-based delivery plasmid | ||
| pUC18T-mini-Tn7T-Gm-GFPmut3/HPS1 | pUC18 –based delivery plasmid for mini-Tn7-Gm-GFPmut3. ApR, GmR, ColE1 ori, oriT | DQ493877b,c |
| pUC18T- mini-Tn7T-Gm-DsRedExpress/HPS1 | pUC18 –based delivery plasmid for mini-Tn7-Gm-DsRedExpress. ApR, GmR, ColE1 ori, oriT | DQ493880b,c |
| pUC18T- mini-Tn7T-Gm-eYFP/HPS1 | pUC18 –based delivery plasmid for mini-Tn7-Gm-eYFP. ApR, GmR, ColE1 ori, oriT | DQ493879b,c |
| pUC18T- mini-Tn7T-Gm-eCFP/HPS1 | pUC18 –based delivery plasmid for mini-Tn7-Gm-eCFP. ApR, GmR, ColE1 ori, oriT | DQ493878b,c |
| R6K replicon-based delivery plasmid | ||
| pUC18TR6K-mini-Tn7T-Gm-GFPmut3/S17-1 λpir | pUC18 –based delivery plasmid for mini-Tn7-Gm-GFPmut3. ApR, GmR, R6K ori, oriT | This project |
| pUC18TR6K- mini-Tn7T-Gm-DsRedExpress/S17-1 λpir | pUC18 –based delivery plasmid for mini-Tn7-Gm-DsRedExpress. ApR, GmR, R6K ori, oriT | This project |
| pUC18TR6K- mini-Tn7T-Gm-eYFP/S17-1 λpir | pUC18 –based delivery plasmid for mini-Tn7-Gm-eYFP. ApR, GmR, R6K ori, oriT | This project |
| pUC18TR6K- mini-Tn7T-Gm-eCFP/S17-1 λpir | pUC18 –based delivery plasmid for mini-Tn7-Gm-eCFP. ApR, GmR, R6K ori, oriT | This project |
| Mobilizing plasmid | ||
| pRK600/HB101 | mobilizing plasmid, providing the mobilization ability during conjugation. ApR, CmR, R6K ori | N.A. |
| Vector | ||
| pUC18TR6K-mini-Tn7T1/CC118 λpir | pUC18 –based vector plasmid for construction of R6K replicon-based delivery plasmids in this project. ApR, R6K ori, oriT | AY712953c |
N.A., Not applicable.
ApR: Ampicillin resistance; CmR: Chloramphenicol resistance; GmR: Gentamicin resistance.
Plasmids were generously provided by Herbert P. Schweizer (Choi et al., 2005).
National Center for Biotechnology Information (NCBI) accession number.
Table 3. List of primers used.
| Primer | Sequence | Description |
|---|---|---|
| ColE1_F | 5′AGGATCCCCGGGGATAACGCAGGAAAGAACAT3′ | Primer used during PCR amplification of ColE1 ori. Primer is flanked with SmaI site at 5′ end. |
| ColE1_R | 5′GATTACGAATTCCTGTCAGACCAAGTTTACTC3′ | Primer used during PCR amplification of ColE1 ori. Primer is flanked with EcoRI site at 5′ end. |
| Tn7R | 5′CAGCATAACTGGACTGATTTCAG3′ | Common primer used for checking chromosomal insertion of Tn7. |
| PAglmS-down | 5′GCACATCGGCGACGTGCTCTC3′ | Primer used with Tn7R to check chromosomal insertion of Tn7 in PAO1. |
| PF_tn7_R2 | 5′TGCCGCACATCCACGACATCCTC3′ | Primer used with Tn7R to check chromosomal insertion of Tn7 in Pf-5. |
| KP_tn7_R2 | 5′GTGGCGCCGAACAACGAACTGCT3′ | Primer used with Tn7R to check chromosomal insertion of Tn7 in KP-1. |
Transformation of Pseudomonas species by electroporation
Electrocompetent PAO1 and Pf-5 were prepared according to Choi et al. (2006). During transformation, the ColE1 replicon-based delivery plasmid and the helper plasmid, pTNS1, were added to the electrocompetent cells and electroporated (25 μF, 200 Ω and 2.5 kV cm−1) using a Gene Pulser apparatus (BIO-RAD, Hercules, CA, USA). Transformed cells were recovered by the addition of ice-cold Super Optimal Broth with Catabolite repression (SOC) (SOB supplemented with 2% w/v glucose) and incubated with shaking for 3 h at 37 °C and 30 °C for PAO1 and Pf-5, respectively. Recovered cells were plated onto LB5 agar (5 g l−1 NaCl; 10 g l−1 tryptone; 5 g l−1 yeast extract; 1.5% w/v agar) plates supplemented with 100 μg ml−1 gentamicin.
Transformation of KP-1 by electroporation and conjugation
Electrocompetent KP-1 was prepared as described above with the following modification. KP-1 was grown in SOB supplemented with 0.7 mM of ethylene diamine tetra acetate (EDTA) (Fournet-Fayard et al., 1995). Plasmid, pTNS2-ColE1, was added to electrocompetent KP-1 and electroporated (25 μF, 200 Ω and 1.8 kV cm−1). After recovery for 3 h at 37 °C, the cells were plated onto LB5 agar plates supplemented with 200 μg ml−1 ampicillin.
KP-1 carrying pTNS2-ColE1 was subsequently transformed with R6K replicon-based delivery plasmids by tri-parental conjugation as described (de Lorenzo and Timmis, 1994). Briefly, 100 μl of overnight cultures of KP-1 with pTNS2-ColE1 (recipient), E. coli with pRK600 (mobilizer) and E. coli with R6K replicon-based delivery plasmid (donor) were mixed with 700 μl of 10 mM MgSO4. Cells were collected by centrifugation (7000 g for 2 min), washed twice with 10 mM MgSO4, resuspended in 30 μl of 10 mM MgSO4 and pipetted onto a filter membrane placed on non-selective LB10 agar (LB10 with 1.5% w/v agar) plate. After overnight incubation at 37 °C, cells were recovered and plated onto LB5 agar plates supplemented with 100 μg ml−1 gentamicin.
Determination of Tn7 insertion site
Colony PCR was used to verify chromosomal Tn7 insertion using primers specific for the insertion site (Table 3). The reaction mixture was then run in a C1000 thermal cycler (BIO-RAD) with an initial denaturation at 97 °C for 3 min followed by 35 cycles of amplification (denaturation at 97 °C, 30 s; annealing at 55 °C, 30 s; extension at 72 °C, 1 min) and a final extension at 72 °C for 10 min. The PCR product was visualized on an 1% w/v agarose gel and sequenced.
Growth rates and stabilities of the fluorescently tagged bacteria
Fluorescently tagged bacteria and their respective parental strains were grown in supplemented M9 minimal medium (200 r.p.m. at 25±1 °C). Growth was monitored by optical density (OD600) over a 12-h period using an UV spectrophotometer (UV-1800, Shimadzu, Japan). Stability of the fluorescent marker in the absence of antibiotic selection was determined by daily subculture (100 × dilution) in supplemented M9 minimal medium without gentamicin (200 r.p.m. at 25±1 °C) for 7 days. The final culture was diluted to yield approximately 1 × 108 cfu ml−1, stained with 4′,6-diamidino-2-phenylindole (DAPI) as described (Yu et al., 1995), visualized using a confocal laser scanning microscope (CLSM) (LSM 780, Carl Zeiss, Oberkochen, Germany) and the total number of DAPI and fluorescent protein-expressing cells was quantified using ImageJ (http://rsb.info.nih.gov/ij/index.html).
Continuous-culture flow cell and planktonic culture experiments
Biofilms were cultivated in three-channel flow cells (channel dimensions, 1 × 4 × 40 mm3) (Sternberg and Tolker-Nielsen, 2005). The flow cells were supplied with supplemented M9 minimal medium at 9 ml h−1 (mean velocity=0.625 mm s−1) with a Reynolds number of 1.12. Each channel was injected with 0.5 ml of diluted overnight culture containing approximately 1 × 108 cfu ml−1. Mixed-species biofilms were established by inoculating mixed cultures of PAO1, Pf-5 and KP-1 in the ratio of 5:5:1. This ratio was used to account for the faster growth rate of KP-1 and was optimized through previous experiments. Three-day-old biofilms were treated with 0.1% w/v sodium dodecyl sulfate (SDS) in supplemented M9 minimal medium at a flow rate of 9 ml h−1 for 2 h. Tobramycin (10 μg ml−1 in supplemented M9 minimal medium) was added to 2-day-old biofilms at 9 ml h−1 for 24 h.
Mixed-species planktonic culture was prepared by mixing diluted overnight cultures of PAO1, Pf-5 and KP-1 as mentioned above. Subsequently, the mixed culture was further diluted 100 × and allowed to grow (200 r.p.m. at 25±1 °C) for 7 days with daily subculture (100 × dilution). The cfu ml−1 of each species was determined at every 24-h interval.
Quantification using 16S rRNA
Biomass of the mixed-species biofilm was extracted by flushing ice-cold phosphate buffer saline through the flow cell. Total RNA was extracted using the RNeasy Protect Bacteria Mini Kit (Qiagen, Venlo, Netherlands) according to the manufacturer's protocol. Sequencing libraries were prepared from total RNA using the TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, USA) and sequenced on the HiSeq 2500 platform (Illumina) according to the manufacturer's protocols. Subsequently, specific sequence tags for 16S rRNA V4 and V6 regions were used to quantify the relative abundance of the three species. The species-specific tags were derived from the complete genomes of PAO1, Pf-5 and NTUH-K2044 (close relative of KP-1) in the NCBI database (gi number: 110645304, 70728250 and 238892256, respectively). The list of species-specific tag sequences is shown in Supplementary Table 1.
To quantify the relative abundance of the three species, the frequency of the species-specific tag sequences was counted. Relative abundance for the three species in each sample was then expressed as the percentage of the species-specific tag sequence relative to the total number of tag sequences observed.
Microscopy, image and statistical analysis
All microscopic observation and image acquisition were performed by a CLSM (LSM 780, Carl Zeiss) using the multi-track mode to minimize fluorescent bleed-through. The excitation wavelengths for eCFP, eYFP and DsRedExpress were 458, 514 and 561 nm, respectively. The emission wavelengths for eCFP, eYFP and DsRedExpress were 476, 527 and 584 nm, respectively. For each flow cell channel, five image stacks were acquired from the centre of the channel (2 mm from the walls), starting at a distance of 5–10 mm from the inlet (unless otherwise stated) and spaced approximately 5 mm apart (Heydorn et al., 2000). The five image stacks covered a total area of approximately 9 × 105 μm2, which was more than the minimum of 1 × 105 μm2 required for representative data (Korber et al., 1993). For image analysis, three biological replicates (flow cells) with a total of 45 image stacks (15 from each flow cell) were quantified for each biofilm type using IMARIS (Bitplane AG, Belfast, UK). Reproducibility of the biofilms was determined as suggested by Jackson et al. (2001). Statistical analysis was done using IBM SPSS Statistics 20.
Results
Tn7 insertion, its stability and effect on growth
PAO1, Pf-5 and KP-1 were each chromosomally tagged with GFPmut3, eYFP, eCFP or DsRedExpress (data not shown). For optimal discrimination by CLSM, PAO1, Pf-5 and KP-1 tagged with eYFP, eCFP and DsRedExpress, respectively, were used for subsequent experiments. No fluorescent bleed-through was observed for the three fluorescent protein markers used (Supplementary Figure S1). As it has previously been shown that chromosomal insertion and a high level of fluorescent protein expression in the cell could lead to growth defects (Hadjantonakis et al., 2002; Tao et al., 2007), the growth of tagged bacteria was investigated. The growth profiles of tagged PAO1 and Pf-5 were similar to their untagged parental strains while the DsRedExpress-tagged KP-1 exhibited a slight, but not significant reduction in growth (P-value=0.569, one-way ANOVA). In addition, more than 90% of tagged PAO1, Pf-5 and KP-1 cells were shown to maintain their respective fluorescence in the absence of gentamicin selection over a 7-day period (data not shown).
Development and structure of mixed-species biofilms relative to those of single-species biofilms
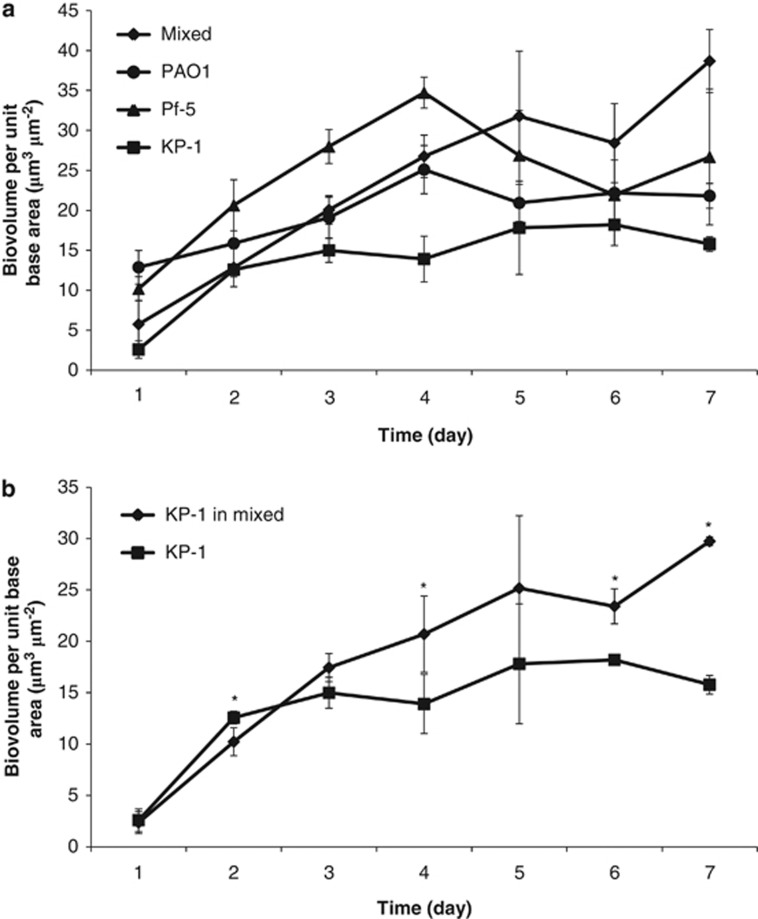
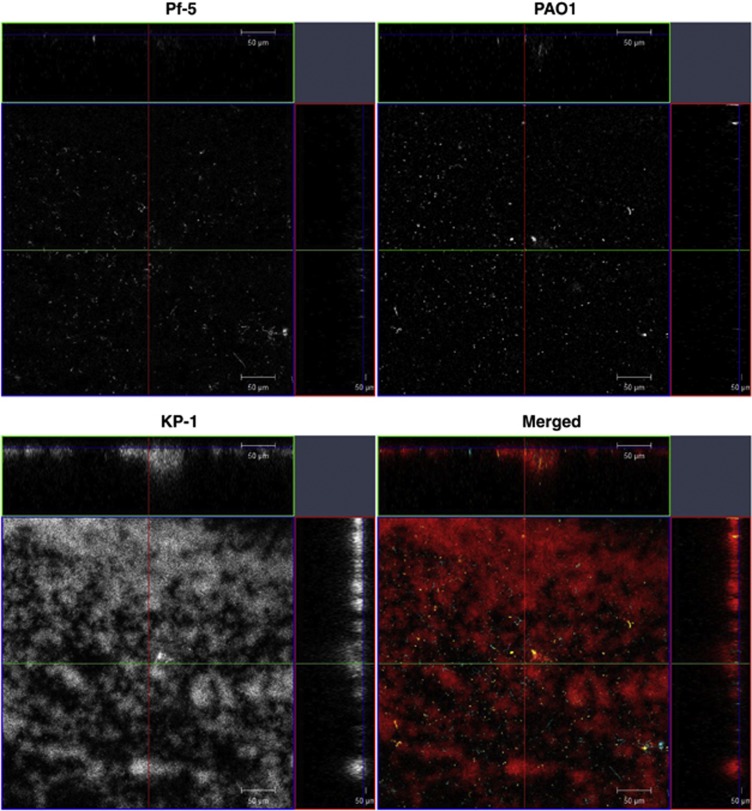
Comparison of the development of single and mixed-species biofilms demonstrated that KP-1 single-species biofilm matured on day 3 when the biovolume per unit base area first peaked at 15.0±1.5 μm3 μm−2 (Figure 1a). The mature KP-1 biofilm was normally flat with no distinct structure (Supplementary Figure S2). By comparison, PAO1 and Pf-5 single-species biofilms first peaked in biomass on day 4, with the appearance of mushroom- and tower-like structures, respectively (Supplementary Figure S2). Their biovolume per unit base area were 25.1±3.0 and 34.7±1.9 μm3 μm−2, respectively, which were both higher than that observed for the KP-1 biofilm (Figure 1a). Biovolume per unit base area of the mixed-species biofilm increased steadily up to day 7, with a slight, but not significant, decrease on day 6. However, this decrease in biomass was consistent across multiple experiments. The biovolume per unit base area of the mature mixed-species biofilm first peaked at 31.8±8.1 μm3 μm−2 (Figure 1a) and exhibited very different structures compared with those exhibited by the single-species biofilms. For example, PAO1 and Pf-5 did not form any mushroom- or tower-like structures in the mixed-species biofilm but were distributed randomly (Figure 2). Surprisingly, KP-1 was observed to form microcolonies that were either shaped as isolated mound-like structures, which measured up to 100 μm in diameter or were similarly thick but flat, presumably due to the merging of multiple microcolonies. As noted above, such distinct structures of KP-1 were seldom observed when KP-1 was grown alone (Supplementary Figure S2).
Figure 1.
Development of single and mixed-species biofilms. (a) Biovolume per unit base area (μm3 μm−2) of PAO1, Pf-5, KP-1 and mixed-species biofilms. Error bars represent standard deviations (n=3). (b) Biovolume per unit base area (μm3 μm−2) of KP-1 biofilm and KP-1 within the mixed-species biofilm. Error bars represent standard deviations (n=3). * denotes significant difference (One-way ANOVA: P-value<0.1).
Figure 2.
Spatial localization and structure of mixed-species biofilm. Confocal microscopy image showing the spatial localization of mixed-species biofilm. The formation of KP-1 (red) microcolonies surrounded by randomly distributed PAO1 (yellow) and Pf-5 (cyan). Top left panel: Pf-5; Top right panel: PAO1; Bottom left panel: KP-1; Bottom right panel: merged image. The top and side images of each panel represent the x-z and y-z planes respectively. The green and red lines indicate the positions corresponding to the x-z and y-z cross sections respectively. The blue line indicates the x-y plane of the main panel. Magnification × 20. Scale bar: 50 μm.
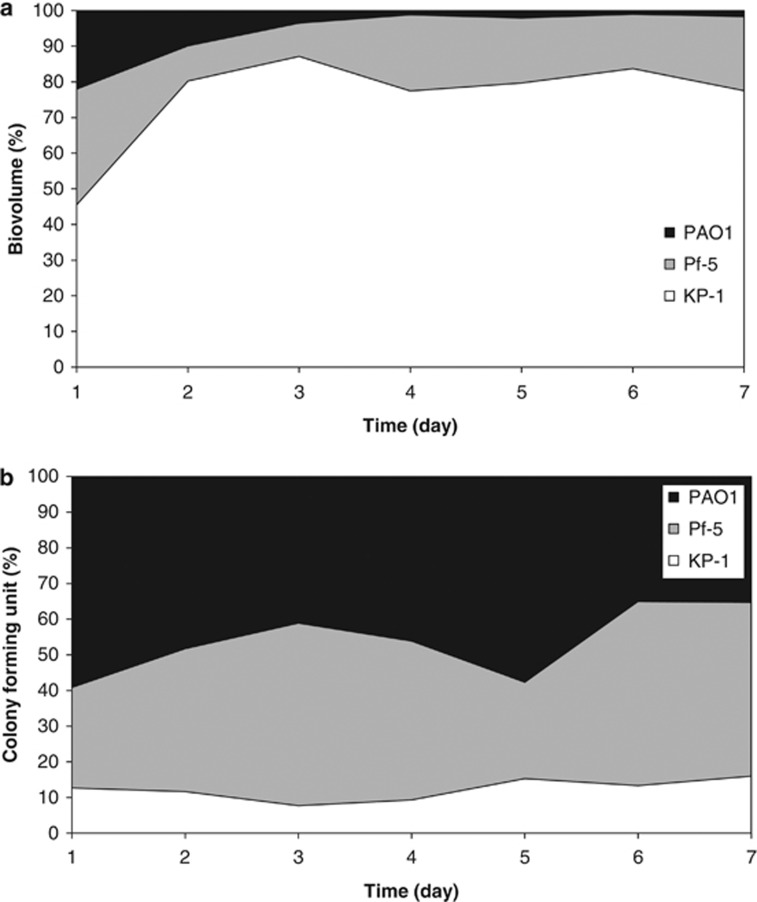
It was interesting to note that the biovolume per unit base area for KP-1 within the mixed-species biofilm was generally higher than the biovolume per unit base area of KP-1 when grown as a single-species biofilm (Figure 1b), even though the amounts of nutrient used for both types of experiment were identical. Quantitative image analysis of the mixed-species biofilms indicated that KP-1 made up 70–80% of the biofilm biomass after day 2, Pf-5 accounted for 10–15% of the biomass and PAO1 was present at 1–5% (Figure 3a). It should be noted that similar quantitative results were obtained irrespective of which fluorescent protein was used to tag the individual species. The biofilm composition was also verified by quantifying the 16S rRNAs extracted from the mixed-species biofilms. The 16S rRNA genes were sequenced from total RNAs extracted from the flow cell grown biofilms on days 4, 5 and 6. The results showed that the mixed-species biofilms mainly comprised KP-1 and small populations of PAO1 and Pf-5 (Supplementary Table 2). One-way analysis of variance (ANOVA) comparisons between the percentages of KP-1, PAO1 and Pf-5 obtained from quantitative image analysis and 16S rRNA sequencing indicated that there was no significant difference between the percentage of each species obtained by the two methods (Supplementary Table 3). Additionally, counts of colony-forming unit in the effluents for both single and mixed-species biofilms were determined (Supplementary Figure 3) and observed to closely reflect the changes in biofilm biomass as determined by quantitative image analysis. Hence, quantitative image analysis by IMARIS can be a useful and non-invasive tool for the analysis of biofilm development and its composition. In contrast to the dominance of KP-1 in the mixed-species biofilms, mixed planktonic cultures of the three species showed that PAO1 was typically present at an average of 46.6±9.2%, Pf-5 at 41.1±9.8% and KP-1 was the lowest at 12.3±2.8% over the 7-day period tested (Figure 3b). The cfu ml−1 levels for PAO1, Pf-5 and KP-1 on day 7 were 1.7 × 108, 2.3 × 108 and 7.7 × 107, respectively.
Figure 3.
Proportions of mixed-species biofilm and planktonic culture. (a) The proportions of the three species within the mixed-species biofilm over the 7-day observation period, derived from quantitative image analysis. (b) The proportions of the three species within the mixed species planktonic culture over the 7-day observation period, derived by quantifying colony-forming units.
Biofilms were reproducible
The biovolumes per unit base area of all single and mixed-species biofilms were found to decrease from day 3 and later. Hence, 3-day biofilms were selected for subsequent studies to ensure that any change in biovolume and structure was due to the treatment applied. Further, the reproducibility of a 3-day biofilm was tested as suggested by Jackson et al. (2001). Results showed that all the three biological replicates of the single and mixed-species biofilms were reproducible, with almost all of the data falling within the 95% confidence limit (Supplementary Figure S4).
Resource competition and biofilm structure
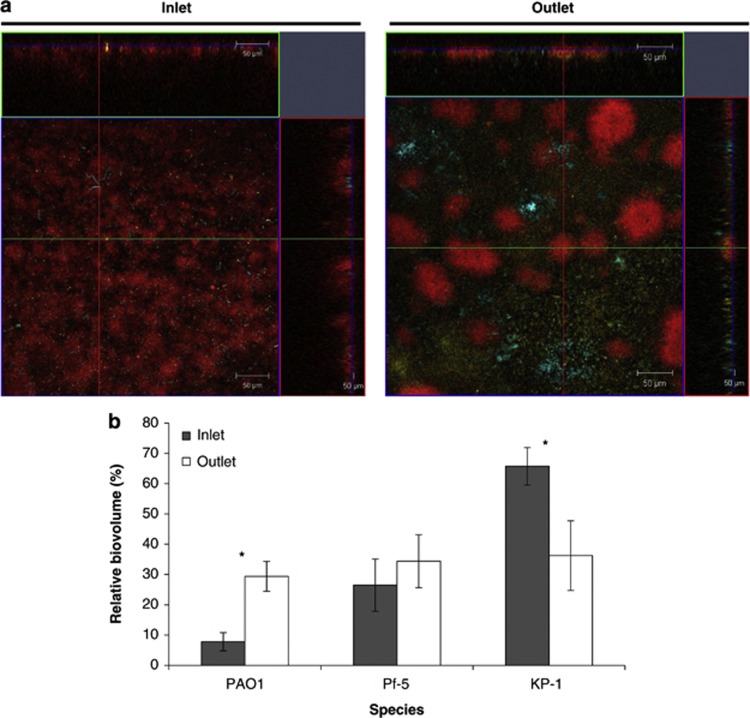
When imaging the mixed-species biofilms, it was observed that biofilms growing at the inlet and outlet ends of a flow cell differed (Figure 4a). The proportion of PAO1 increased from 7.8±3.0% (inlet) to 29.4±4.9% (outlet) whereas the proportion of KP-1 decreased from 65.7±6.2% (inlet) to 36.2±11.5% (outlet) (Figure 4b). Interestingly, the proportions of Pf-5 at the inlet and outlet ends of the flow cells were not significantly different. The specific growth rates for all the three species at different glucose concentrations were thus determined. The growth rate of KP-1 decreased from a maximum of 0.059±0.001 to 0.007±0.002 OD600 h−1 as the glucose concentration decreased from 20 000 to 4 μg ml−1, whereas the growth rates of PAO1 and Pf-5 did not change (Supplementary Figure S5). Given its higher growth rate, it was not surprising that KP-1 dominated the mixed-species biofilm when the glucose concentration was high at the inlet (400 μg ml−1). The glucose concentration in the medium collected from the outlet end was approximately 138±15 μg ml−1 on day 3 (Supplementary Figure S6). Thus, the slower growth of KP-1 at lower glucose concentrations might result in the observed reduction of its biomass at the outlet. When mixed-species biofilms were grown in the presence of 10-fold more glucose (4000 μg ml−1), it was observed that the three species were present in almost equal proportions (data not shown), further illustrating that resource limitation, and thus competition, might affect the composition of a mixed-species biofilm. However, competition for carbon alone may not account for all of the results observed. For example, when grown in pair-wise combinations as planktonic cultures, Pf-5 was observed to outgrow both PAO1 and KP-1 (∼1–2 log10 cfu ml−1 difference) while KP-1 and PAO1 grew together in equal proportions, but achieved a significantly lower growth yield (∼ 2 log10 cfu ml−1 lower) than when grown individually (data not shown). Thus, the accumulation of waste products, oxygen gradients in a biofilm and change in flow dynamics may also impact the assembly of biofilms at the inlet and outlet ends. It should be noted that, for consistency, only image stacks of biofilms grown at the inlet ends were used for the stress experiments (below).
Figure 4.
Differences in compositions and structures of mixed-species biofilms grown at different positions of the flow cell. (a) Confocal microscopy images showing mixed-species biofilms of PAO1 (yellow), Pf-5 (cyan) and KP-1 (red) growing at the inlet end and outlet end of the flow cell. The top and side images represent the x-z and y-z planes respectively. The green and red lines indicate the positions corresponding to the x-z and y-z cross sections respectively. The blue line indicates the x-y plane of the main panel. Magnification × 20. Scale bar: 50 μm. (b) Bar chart depicting the percentages of PAO1, Pf-5 and KP-1 within the mixed-species biofilms grown at the inlet end and outlet end of the flow cell. Error bars represent standard deviations (n=3). * denotes significant difference (One-way ANOVA: P-value<0.05).
Mixed-species biofilm has enhanced stress resistance
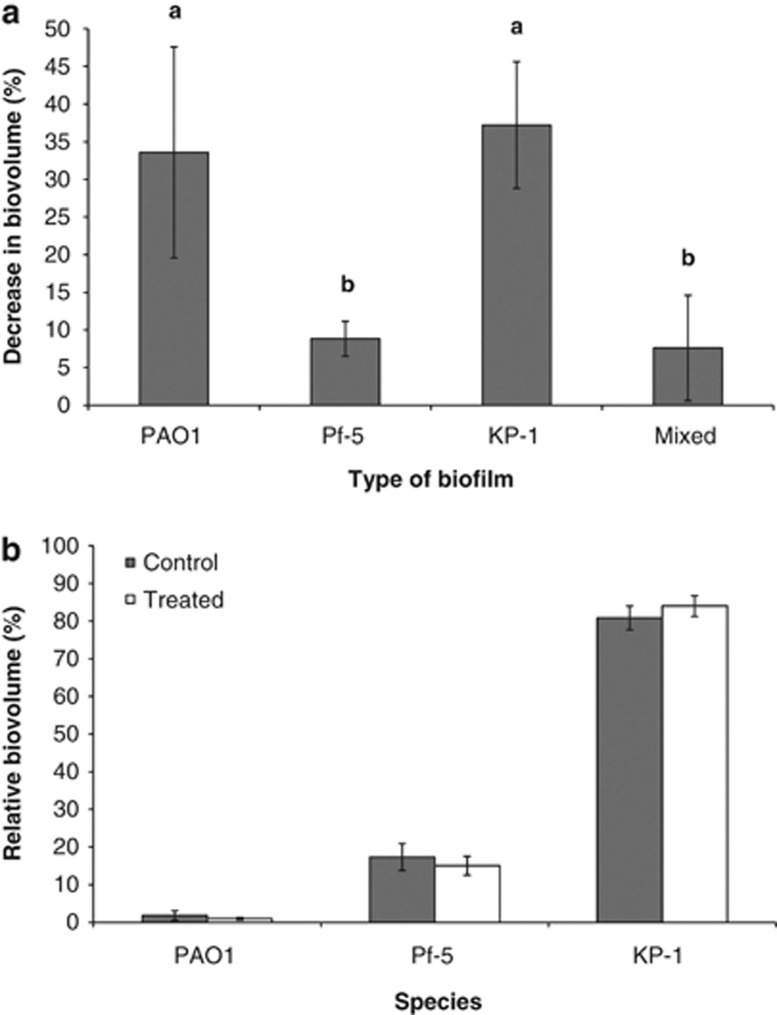
Many studies have shown that biofilms are more resistant to stresses compared with their planktonic counterparts (Nickel et al., 1985; Mah and O'Toole, 2001). However, limited studies have been carried out to determine the relative resistance of mixed-species biofilms compared with single-species biofilms. Here, single and mixed-species biofilms were treated with 10 μg ml−1 tobramycin, an aminoglycoside antibiotic. When grown on their own, single-species biofilms of PAO1, Pf-5 and KP-1 were reduced by 33.6±14.0%, 8.8±2.3% and 37.2±8.4% in biovolume, respectively, while the mixed-species biofilm was reduced by 7.6±7.0% (Figure 5a). This was similarly reflected in the number of colony-forming units in the effluents, where the counts decreased from 1.2 × 108 to 1.1 × 107 (89.4±8.3%) for PAO1, from 8.1 × 107 to 5.6 × 107 (32.8±16.0%) for Pf-5, from 1.0 × 109 to 3.7 × 106 (99.6±0.2%) for KP-1 and from 8.9 × 107 to 7.4 × 107 (19.0±16.3%) for mixed-species biofilm. To determine if the increased tobramycin resistance of the mixed-species biofilm was a consequence of either (a) selection for the tobramycin-resistant Pf-5 or (b) Pf-5 providing protection to the two sensitive species, the proportion of each species was determined for the control and treated mixed-species biofilms. Surprisingly, there was no statistically significant difference between the proportions of PAO1, Pf-5 and KP-1 in the control and treated mixed-species biofilms (Figure 5b). This would suggest that protection against tobramycin for the mixed-species biofilm was extended to all the three species despite the fact that only Pf-5 was resistant to tobramycin. When dual species biofilm of two sensitive species, PAO1 and KP-1, was treated with tobramycin, the biofilm was severely affected with an overall reduction of 42.5±1.4% in biovolume (Supplementary Figure S7), further supporting the notion that Pf-5 might have a protective role in the mixed-species biofilm.
Figure 5.
Tobramycin treatment of single and mixed-species biofilms. (a) Percentage decrease in biovolume for PAO1, Pf-5, KP-1 and mixed-species biofilms after treatment with 10 μg ml−1 tobramycin. Error bars represent standard deviations (n=3). a and b denote significant differences (One-way ANOVA: P-value<0.05). (b) Relative biovolume of PAO1, Pf-5 and KP-1 within the control and treated mixed-species biofilms. Error bars represent standard deviations (n=3).
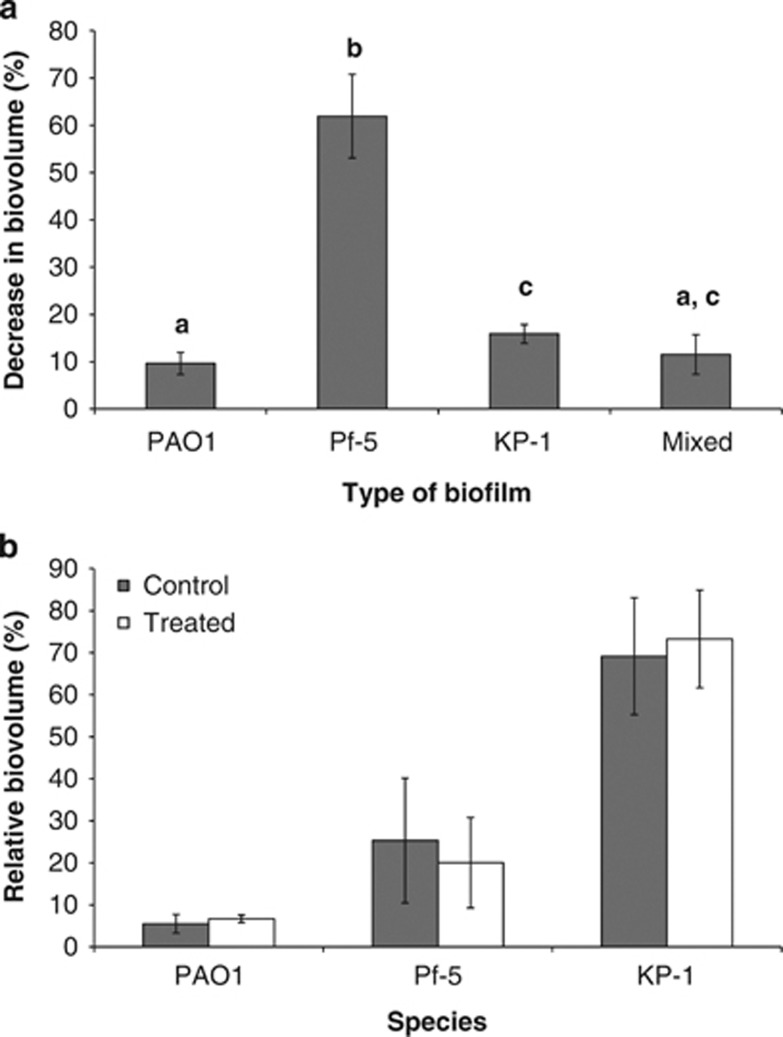
To determine if this protective effect exhibited by the mixed-species biofilm was specific to tobramycin treatment or a more general feature of the mixed-biofilm community, single and mixed-species biofilms were also treated with 0.1% w/v SDS, an anionic surfactant. The single-species biofilms responded differently to SDS compared with tobramycin, where the biovolume of Pf-5 single-species biofilm was significantly reduced (61.9±8.8%) upon SDS exposure, whereas the biovolumes of PAO1 and KP-1 single-species biofilms were only marginally reduced (9.7±2.3% and 15.9±2.0%, respectively) (Figure 6a). However, similar to the result observed for tobramycin treatment, the mixed-species biofilm was resistant to SDS (11.5±4.2% reduction in biovolume). When individual species within the control and treated mixed-species biofilms was again quantified, there was no statistically significant difference between their proportions (Figure 6b), despite Pf-5 being more sensitive to SDS. Dual-species biofilms comprised a sensitive and an insensitive species, i.e. Pf-5 and KP-1 as well as Pf-5 and PAO1, also exhibited increased resistance to SDS compared with Pf-5 single-species biofilms, with a 2.8±1.7% and 46.9±6.0% reduction in biovolumes, respectively (Supplementary Figure S7). Thus, growth as a mixed-species biofilm community is protective against SDS and this protection is inclusive of the most sensitive member of the community. Finally, such community-level resistance is not exclusive to a particular stress but may be a general phenomenon.
Figure 6.
SDS treatment of single and mixed-species biofilms. (a) Percentage decrease in biovolume for PAO1, Pf-5, KP-1 and mixed-species biofilms after treatment with 0.1% w/v SDS. Error bars represent standard deviations (n=3). a, b and c denote significant differences (One-way ANOVA: P-value<0.05). (b) Relative biovolume of PAO1, Pf-5 and KP-1 within the control and treated mixed-species biofilms. Error bars represent standard deviations (n=3).
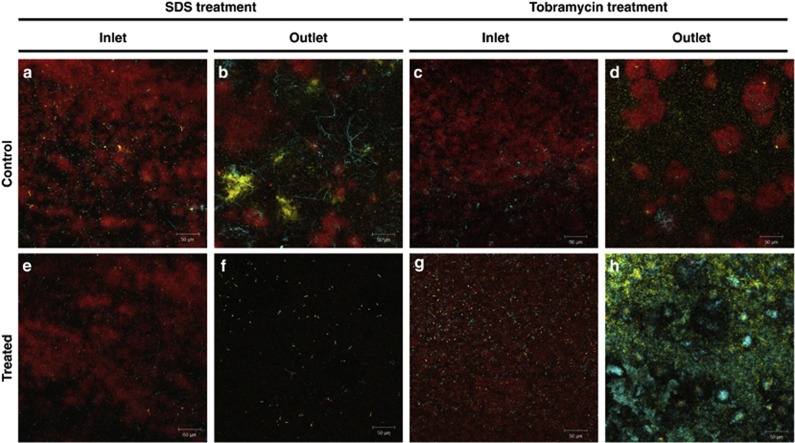
Composition and spatial organization affected resistance of mixed-species biofilm to SDS and tobramycin
As noted above, the proportion of individual species within the mixed-species biofilms growing at the inlet and outlet ends of a flow cell differed. PAO1, Pf-5 and KP-1 in the mixed-species biofilms were also observed to form separate microcolonies at the outlets (Figures 7b and d) rather than being closely associated as seen at the inlets of the flow cells (Figures 7a and c). After treatment with 0.1% w/v SDS, the reduction in biovolume was greater at the outlet (Figure 7f) than at the inlet (Figure 7e). Similarly, treatment with 10 μg ml−1 tobramycin also decreased the biovolume of mixed-species biofilms growing at the inlets of the flow cell (Figure 7g), but resulted in the dominance of PAO1 and Pf-5 at the outlets. Further, PAO1 and Pf-5 also formed prominent microcolonies themselves (Figure 7h) instead of being distributed randomly (Figure 7g). Moreover, when the mixed-planktonic culture of PAO1, Pf-5 and KP-1 was treated with 10 μg ml−1 tobramycin, only the more resistant Pf-5 survived whereas both PAO1 and KP-1 were completely eradicated (Supplementary Figure S8). Collectively, these results suggest that the composition and spatial organization (loose/tight association between the species) of a mixed-species biofilm can impact the survival of the community and its members when challenged with a stress.
Figure 7.
Composition and spatial organization affected resilience of mixed-species biofilms. (a and c) Confocal microscopy images of control mixed-species biofilms growing at the inlets of the flow cells. (b and d) Confocal microscopy images of control mixed-species biofilms growing at the outlets of the flow cells. (e and g) Confocal microscopy images of treated mixed-species biofilms growing at the inlets of the flow cells. (f and h) Confocal microscopy images of treated mixed-species biofilms growing at the outlets of the flow cells. PAO1 (yellow), Pf-5 (cyan) and KP-1 (red). Magnification: 20 x. Scale bar: 50 μm.
Discussion
Biofilms in the environment typically comprised multiple species living together in communities. While such biofilms can be dominated by a specific species, secondary species are invariably present. For example, while P. aeruginosa dominates biofilms found in the lungs of cystic fibrosis patients, other species such as Abiotrophia adiacens, Bacteroides gracilis, Prevotella salivae, Rothia mucilaginosa and Staphylococcus hominis are also present (Rogers et al., 2004; Lim et al., 2013b). Interactions between different species in the mixed-biofilm communities are believed to affect their development, structures and functions (James et al., 1995; Harrison, 2007; Moons et al., 2009). For instance, P. putida, a p-cresol-degrading organism, was found to surround and protect sensitive Pseudomonas species when the mixed-species biofilms were exposed to p-cresol (Cowan et al., 2000). In contrast, competition was observed when the marine bacterium, Pseudoalteromonas tunicata, produced an antibacterial protein AlpP against other bacteria on the surface of the marine alga Ulva lactuca (Rao et al., 2005). Additionally, type 3 fimbrial adhesin was reported to defend K. pneumoniae against Elastase B secreted by P. aeruginosa in a dual-species biofilm (Childers et al., 2013). Thus, studies on interspecies interactions that occur in mixed-species biofilms will be of utmost importance to understand how such communities function and will ultimately improve strategies for the manipulation of these biofilms.
In this study, the development of a mixed-species biofilm (community) was found to be altered relative to the development of comparable single-species biofilms (population). The biomass of K. pneumoniae KP-1 within the mixed-species biofilm was significantly higher than the biomass of KP-1 single-species biofilm (Figure 1b), despite having access to less nutrients as a consequence of competition with PAO1 and Pf-5. This potentially suggests the presence of metabolic cooperation within the mixed-species biofilm that allows KP-1 to achieve higher biomass with no additional input of carbon. Metabolic cooperation, also known as syntrophy, has been shown where chlorobenzoate, the metabolic waste excreted by Burkholderia sp. LB400, was metabolized by Pseudomonas sp. B13 (FR1) (Nielsen et al., 2000). In doing so, the mixed-species biofilm persisted as the metabolic waste produced by Burkholderia sp. LB400 was constantly removed whereas Pseudomonas sp. B13 (FR1) grew on an alternative nutrient. Such interaction illustrates that a mixed-species biofilm community is better at maximizing and optimizing the use of nutrients to enhance its growth and persistence. Thus, an assembled community can be at a physiological advantage when a single population becomes resource limited. Alternatively, the higher biomass of KP-1 in the mixed-species biofilm can result from physical effects such as increased cohesion mediated by the extracellular matrix provided by either P. aeruginosa PAO1, or P. protegens Pf-5 or both.
Just as biofilms are more resilient than planktonic cells, mixed-species biofilms here were found to be more resilient than single-species biofilms. The increased community-level resistance was observed when treated with tobramycin and SDS, both of which have very different mechanisms of action. Further, the increased community-level resistance was not a result of selection for the insensitive species over the other more-sensitive species, but was rather a cross protection offered by the resistant species to all members in the community. While the specific mechanism of protection is currently unknown, the cross protection can be mediated by the sharing of individually-produced defences. PAO1 encodes SdsA1, a secreted SDS hydrolase, that can degrade and metabolize SDS, thus protecting all the three organisms from the anionic surfactant (Hagelueken et al., 2006). In addition, KP-1 is highly encapsulated and its extensive extracellular matrix may either sequester the SDS, thus reducing its effect on the cells within the biofilm, or may facilitate the retention of defensive enzymes such as SdsA1, which would result in more efficient breakdown of SDS. As a result, the combined effect of SdsA1 and the extracellular matrix may therefore offer protection to the SDS-sensitive Pf-5 within the mixed-species biofilm. SDS-treated dual-species biofilms comprising Pf-5 and KP-1 as well as Pf-5 and PAO1 have also indicated that either KP-1 or PAO1 alone was capable of offering protection to the SDS-sensitive Pf-5 (Supplementary Figure S7). The decreases in biovolumes for both types of dual-species biofilm were significantly lower than the decrease in biovolume observed for Pf-5 single-species biofilm. Nonetheless, the Pf-5 and PAO1 dual-species biofilm was not as resistant to SDS as the Pf-5 and KP-1 dual-species biofilm, and this may, in part, be due to the different mechanism of shared resistance. Hence, it will be interesting to determine the mechanisms of shared resistance to better understand how mixed communities resist stresses.
The community-level resistance to tobramycin can similarly be the consequence of shared defences that are beneficial to the whole community. For example, Pf-5 may produce aminoglycoside-modifying enzymes such as N-acetyltransferase. Aminoglycoside-modifying enzymes catalyze the covalent modification of either amino or hydroxyl functional group, resulting in a modified aminoglycoside that binds inefficiently to ribosomes (Mingeot-Leclercq et al., 1999). The community-level resistance against tobramycin was particularly striking; as Pf-5, which normally contributed to approximately 15% of the total biofilm biomass, was able to protect both PAO1 and KP-1, where only 10% of the mixed-species biofilm was removed. Thus, the role each species can have in a community may not be proportional to its abundance. Nevertheless, close association and interaction between different species within a mixed-species biofilm are essential for the community-level resistance to both tobramycin and SDS (Figure 7). It is clear that community-level resistance is important for bacterial biofilms and similar effect has been observed for a marine-biofilm system, where the mixed-species biofilm was more resistant to oxidative stress and tetracycline exposure than the single-species biofilms (Burmolle et al., 2006).
Intriguingly, when the mixed community was grown in planktonic culture and challenged with 10 μg ml−1 tobramycin, only the tobramycin-resistant Pf-5 survived (Supplementary Figure S8). Hence, the effects on development and resilience are unique to the structured environment of a biofilm, where the presence of extracellular matrix and distinct spatial organization may mediate these effects. In addition, the mixed microbial community may express specific genes and proteins, which are responsible for the protection of a biofilm community, but are either not expressed effectively or not present in a single-species biofilm population. This increased resilience has further implications for the control and manipulation of microbial communities in the medical, industrial and environmental settings since their behaviours cannot be predicted from the studies of single-species biofilms.
In conclusion, this study clearly shows that there are new lessons in microbial cell-cell interactions to be learnt from mixed-biofilm community studies, which are more representative of how bacteria exist in nature and in infections. This model system was shown to be reproducible and demonstrated unique features of a mixed-species biofilm such as altered development and increased resistance to stresses. Hence, it is a suitable model for investigating microbial cell-cell interactions within a mixed-species biofilm, where the individual species can be tracked by fluorescent protein tag and confocal laser scanning microscope. Further, all the three species are genetically tractable and amenable to molecular techniques such as mutagenesis and ‘omics' based approaches. Therefore, this model system offers a unique opportunity to investigate detailed questions on the molecular mechanisms facilitating community assembly and behaviour of a community that is greater than the sum of their members.
Acknowledgments
Kai Wei Kelvin Lee is supported by the National Research Foundation Singapore under the National Research Foundation (NRF) Environmental and Water Technologies (EWT) PhD Scholarship Programme and administered by the Environment and Water Industry Programme Office (EWI).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Anand AA, Vennison SJ, Sankar SG, Prabhu DI, Vasan PT, Raghuraman T, et al. Isolation and characterization of bacteria from the gut of Bombyx mori that degrade cellulose, xylan, pectin and starch and their impact on digestion. J Insect Sci. 2010;10:107. doi: 10.1673/031.010.10701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arciola CR, Campoccia D, Gamberini S, Donati ME, Pirini V, Visai L, et al. Antibiotic resistance in exopolysaccharide-forming Staphylococcus epidermidis clinical isolates from orthopaedic implant infections. Biomaterials. 2005;26:6530–6535. doi: 10.1016/j.biomaterials.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Banks MK, Bryers JD. Bacterial species dominance within a binary culture biofilm. Appl Environ Microbiol. 1991;57:1974–1979. doi: 10.1128/aem.57.7.1974-1979.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breugelmans P, Barken KB, Tolker-Nielsen T, Hofkens J, Dejonghe W, Springael D. Architecture and spatial organization in a triple-species bacterial biofilm synergistically degrading the phenylurea herbicide linuron. FEMS Microbiol Ecol. 2008;64:271–282. doi: 10.1111/j.1574-6941.2008.00470.x. [DOI] [PubMed] [Google Scholar]
- Burmolle M, Webb JS, Rao D, Hansen LH, Sorensen SJ, Kjelleberg S. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl Environ Microbiol. 2006;72:3916–3923. doi: 10.1128/AEM.03022-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazal PM. Pollution of modern metalworking fluids containing biocides by pathogenic bacteria in France. Reexamination of chemical treatments accuracy. Eur J Epidemiol. 1995;11:1–7. doi: 10.1007/BF01719939. [DOI] [PubMed] [Google Scholar]
- Childers BM, Van Laar TA, You T, Clegg S, Leung KP. MrkD(1P) from Klebsiella pneumoniae IA565 allows for co-existence with Pseudomonas aeruginosa and protection from protease-mediated biofilm detachment. Infect Immun. 2013;81:4112–4120. doi: 10.1128/IAI.00521-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K-H, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, et al. A Tn7-based broad-range bacterial cloning and expression system. Nat Meth. 2005;2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- Choi K-H, Kumar A, Schweizer HP. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: Application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Meth. 2006;64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Cowan SE, Gilbert E, Liepmann D, Keasling JD. Commensal interactions in a dual-species biofilm exposed to mixed organic compounds. Appl Environ Microbiol. 2000;66:4481–4485. doi: 10.1128/aem.66.10.4481-4485.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V, Timmis KN. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- Fournet-Fayard S, Joly B, Forestier C. Transformation of wild type Klebsiella pneumoniae with plasmid DNA by electroporation. J Microbiol Meth. 1995;24:49–54. [Google Scholar]
- Hadjantonakis A-K, Macmaster S, Nagy A. Embryonic stem cells and mice expressing different GFP variants for multiple non-invasive reporter usage within a single animal. BMC Biotechnol. 2002;2:11. doi: 10.1186/1472-6750-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagelueken G, Adams TM, Wiehlmann L, Widow U, Kolmar H, Tummler B, et al. The crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, defines a third class of sulfatases. Proc Natl Acad Sci USA. 2006;103:7631–7636. doi: 10.1073/pnas.0510501103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison F. Microbial ecology of the cystic fibrosis lung. Microbiology. 2007;153:917–923. doi: 10.1099/mic.0.2006/004077-0. [DOI] [PubMed] [Google Scholar]
- Heydorn A, Ersboll BK, Hentzer M, Parsek MR, Givskov M, Molin S. Experimental reproducibility in flow-chamber biofilms. Microbiology. 2000;146 (Pt 10:2409–2415. doi: 10.1099/00221287-146-10-2409. [DOI] [PubMed] [Google Scholar]
- Jackson G, Beyenal H, Rees WM, Lewandowski Z. Growing reproducible biofilms with respect to structure and viable cell counts. J Microbiol Meth. 2001;47:1–10. doi: 10.1016/s0167-7012(01)00280-9. [DOI] [PubMed] [Google Scholar]
- James GA, Beaudette L, Costerton JW. Interspecies bacterial interactions in biofilms. J Ind Microbiol. 1995;15:257–262. [Google Scholar]
- Jensen PO, Bjarnsholt T, Phipps R, Rasmussen TB, Calum H, Christoffersen L, et al. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology. 2007;153:1329–1338. doi: 10.1099/mic.0.2006/003863-0. [DOI] [PubMed] [Google Scholar]
- Korber DR, Lawrence JR, Hendry MJ, Caldwell DE. Analysis of spatial variability within mot+ and mot− Pseudomonas fluorescens biofilms using representative elements. Biofouling. 1993;7:339–358. [Google Scholar]
- Lim CK, Hassan KA, Penesyan A, Loper JE, Paulsen IT. The effect of zinc limitation on the transcriptome of Pseudomonas protegens Pf-5. Environ Microbiol. 2013;15:702–715. doi: 10.1111/j.1462-2920.2012.02849.x. [DOI] [PubMed] [Google Scholar]
- Lim YW, Schmieder R, Haynes M, Willner D, Furlan M, Youle M, et al. Metagenomics and metatranscriptomics: windows on CF-associated viral and microbial communities. J Cyst Fibros. 2013;12:154–164. doi: 10.1016/j.jcf.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah T-FC, O'Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- Mingeot-Leclercq MP, Glupczynski Y, Tulkens PM. Aminoglycosides: activity and resistance. Antimicrob Agents Chemother. 1999;43:727–737. doi: 10.1128/aac.43.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moons P, Michiels CW, Aertsen A. Bacterial interactions in biofilms. Crit Rev Microbiol. 2009;35:157–168. doi: 10.1080/10408410902809431. [DOI] [PubMed] [Google Scholar]
- Nickel JC, Ruseska I, Wright JB, Costerton JW. Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob Agents Chemother. 1985;27:619–624. doi: 10.1128/aac.27.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AT, Tolker-Nielsen T, Barken KB, Molin S. Role of commensal relationships on the spatial structure of a surface-attached microbial consortium. Environ Microbiol. 2000;2:59–68. doi: 10.1046/j.1462-2920.2000.00084.x. [DOI] [PubMed] [Google Scholar]
- Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- Ramette A, Frapolli M, Fischer-Le Saux M, Gruffaz C, Meyer JM, Defago G, et al. Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst Appl Microbiol. 2011;34:180–188. doi: 10.1016/j.syapm.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Rao D, Webb JS, Kjelleberg S. Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicata. Appl Environ Microbiol. 2005;71:1729–1736. doi: 10.1128/AEM.71.4.1729-1736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GB, Carroll MP, Serisier DJ, Hockey PM, Jones G, Bruce KD. Characterization of bacterial community diversity in cystic fibrosis lung infections by use of 16S ribosomal DNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol. 2004;42:5176–5183. doi: 10.1128/JCM.42.11.5176-5183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Camper AK. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J Bacteriol. 2001;183:6579–6589. doi: 10.1128/JB.183.22.6579-6589.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schembri MA, Kjaergaard K, Klemm P. Global gene expression in Escherichia coli biofilms. Mol Microbiol. 2003;48:253–267. doi: 10.1046/j.1365-2958.2003.03432.x. [DOI] [PubMed] [Google Scholar]
- Sternberg C, Tolker-Nielsen T. Growing and analyzing biofilms in flow cells. John Wiley and Sons, Inc.: Hoboken, New Jersey; 2005. [Google Scholar]
- Stoodley P, Wilson S, Hall-Stoodley L, Boyle JD, Lappin-Scott HM, Costerton JW. Growth and detachment of cell clusters from mature mixed-species biofilms. Appl Environ Microbiol. 2001;67:5608–5613. doi: 10.1128/AEM.67.12.5608-5613.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W, Evans BG, Yao J, Cooper S, Cornetta K, Ballas CB, et al. Enhanced green fluorescent protein is a nearly ideal long-term expression tracer for hematopoietic stem cells, whereas DsRed-express fluorescent protein is not. Stem Cells. 2007;25:670–678. doi: 10.1634/stemcells.2006-0553. [DOI] [PubMed] [Google Scholar]
- Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, et al. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yu W, Dodds W, Banks M, Skalsky J, Strauss E. Optimal staining and sample storage time for direct microscopic enumeration of total and active bacteria in soil with two fluorescent dyes. Appl Environ Microbiol. 1995;61:3367–3372. doi: 10.1128/aem.61.9.3367-3372.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Shao M-F, Ye L. 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J. 2012;6:1137–1147. doi: 10.1038/ismej.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.