Abstract
There is a concerted understanding of the ability of root exudates to influence the structure of rhizosphere microbial communities. However, our knowledge of the connection between plant development, root exudation and microbiome assemblage is limited. Here, we analyzed the structure of the rhizospheric bacterial community associated with Arabidopsis at four time points corresponding to distinct stages of plant development: seedling, vegetative, bolting and flowering. Overall, there were no significant differences in bacterial community structure, but we observed that the microbial community at the seedling stage was distinct from the other developmental time points. At a closer level, phylum such as Acidobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria and specific genera within those phyla followed distinct patterns associated with plant development and root exudation. These results suggested that the plant can select a subset of microbes at different stages of development, presumably for specific functions. Accordingly, metatranscriptomics analysis of the rhizosphere microbiome revealed that 81 unique transcripts were significantly (P<0.05) expressed at different stages of plant development. For instance, genes involved in streptomycin synthesis were significantly induced at bolting and flowering stages, presumably for disease suppression. We surmise that plants secrete blends of compounds and specific phytochemicals in the root exudates that are differentially produced at distinct stages of development to help orchestrate rhizosphere microbiome assemblage.
Keywords: Arabidopsis thaliana, development, microbiome, rhizosphere, root exudation
Introduction
Plants such as bean, maize, soybean, cowpea, cabbage, cotton and Arabidopsis exhibit age-related resistance (ARR) (Develey-Riviere and Galiana, 2007). For example, susceptibility to Puccinia sorghi (common rust) in maize is manifested in younger plants but as the plants mature, the level of disease resistance augments (Abedon and Tracy, 1996). In Arabidopsis, transitions from the vegetative to the floral phase correlates with resistance to Pseudomonas syringae (Kus et al., 2002; Rusterucci et al., 2005). While ARR at the molecular level has been studied with respect to leaf pathogens, little discussion has focused on root defense strategies and their role in ARR. However, there are some indirect correlations; for example, cotton (Zaki et al., 1998) or bean (Nicoli et al., 2012) plants are more susceptible to root disease (Rizoctonia solani or Fusarium root rot, respectively) at the seedling stage. Recent studies have shown that root secretion of defense-related proteins is enhanced during flowering time (De-la-Pena et al., 2010), suggesting an involvement of plant roots in ARR. Similarly, Chaparro et al. (2013) have shown that Arabidopsis roots release more phenolic-related compounds at later stages of life which might be correlated to defense strategies against pathogens as secondary metabolites are involved in plant immunity against bacterial and fungal pathogens (Rogers et al., 1996; Clay et al., 2009; Millet et al., 2010; An and Mou, 2011; Bednarek, 2012). Thus, there is a need to understand the influence of plant development on microbial associations that might occur naturally in the rhizosphere related to defense but also to other vital plant necessities such as nutrient acquisition.
Under natural conditions, plants tend to require higher quantities of N at later stages of development (Kelly et al., 1995; Rossato et al., 2001; Nazoa et al., 2003; Malagoli et al., 2004) but exactly how this N is obtained under natural conditions is unknown. The classically studied symbiotic relationship between rhizobia and legumes has shown that symbiosis occurs only when the plant is under N-limiting conditions (Davidson and Robson, 1986; Eaglesham, 1989; Zahran, 1999). Similarly, the secretion of flavones and flavonols that initiates rhizobia-legume symbiosis is enhanced under N-limiting conditions (Coronado et al., 1995; Zhang et al., 2009). This combined information suggests that the plant may have some control over the identity and functionality of the rhizosphere microbiome.
Studies have shown that rhizospheric fungal and bacterial communities of a wide range of plants (i.e., Arabidopsis, Medicago, maize, pea, wheat and sugar beet) change according to a plant developmental gradient (Baudoin et al., 2002; Mougel et al., 2006; Houlden et al., 2008; Micallef et al., 2009a). In these studies the microbial communities were assessed through automated ribosomal intergenic spacer analysis or denaturing gradient gel electrophoresis techniques that produce a fingerprint of the community structure but not of its members' identity. While these studies demonstrated that plant microbial communities change in response to plant development they were not able to distinguish how or which microbes contribute to the changes observed. For example, Micallef et al. (2009a) through denaturing gradient gel electrophoresis analysis observed that Arabidopsis rhizosphere microbial communities varied with plant development and that microbial communities in early plant development were more distinct to the bulk soil and that this difference decreased with plant age. Similarly, an assessment of the potato rhizosphere demonstrated that young potato plants showed cultivar-dependent rhizosphere microbial communities but these differences in the microbiomes disappeared as the plants aged (Inceoglu et al., 2011). Soybean rhizosphere microbial communities were also influenced by plant development; early reproductive growth stages of the soybean plant produced more complex microbial communities than late stage soybean plants (Xu et al., 2009). An assessment of the microbial community structure through plant development focusing on the members that make up the community is warranted. Incidentally, the recent characterization of the Arabidopsis thaliana core microbiome provides a tool to decipher the influence of the plant on the rhizosphere microbiome at different stages of development (Bulgarelli et al., 2012; Lundberg et al., 2012).
Evidence demonstrating the close ties root exudates have on the microbial composition of the rhizosphere is mounting (Broeckling et al., 2008; Badri et al., 2009, 2013a; Micallef et al., 2009b; Chaparro et al., 2012, 2013), whereby many chemicals present in root exudates act as substrates, chemotactic or signaling molecules to orchestrate changes in microbial composition (Shaw, 1991; de Weert et al., 2002; Jain and Nainawatee, 2002; Horiuchi et al., 2005; Bais et al., 2006; Badri and Vivanco, 2009; Neal et al., 2012; Badri et al., 2013a). Recently, it was reported that the composition of Arabidopsis root exudates change following a plant developmental gradient (Chaparro et al., 2013). Cumulative secretion levels of sugars and sugar alcohols were higher in early time points and decreased through plant growth. In contrast, the cumulative secretion levels of amino acids and phenolics increased over time. Accordingly, it was hypothesized that seedlings of roots release sugars as substrates for a wide diversity of microbes at early stages of development but as the plant ages it releases specific substrates and potentially antimicrobial compounds in an effort to select for particular microbial inhabitants of the rhizosphere (Badri et al., 2013a; Chaparro et al., 2013). This potential selection of microbes in the rhizosphere as the plant ages might be associated with the ability of beneficial microbes to suppress pathogenic ones (Mendes et al., 2011), trigger induced systemic tolerance to overcome abiotic stress (Selvakumar et al., 2012), increase the plant's innate immunity (Zamioudis and Pieterse, 2012), help in mineral nutrition (Bolan, 1991; van der Heijden et al., 2008) and in overall plant health (Berendsen et al., 2012; Chaparro et al., 2012). Here, we tested this hypothesis by analyzing the rhizosphere microbial composition of Arabidopsis by 454 pyrosequencing at four distinct physiological stages of development: seedling (four–five leaf stage), vegetative (rosette), bolting and flowering. We did not include samples past the flowering stage because previous studies have determined that rhizosphere microbial communities converge past the flowering stage (Micallef et al., 2009a; Lundberg et al., 2012). Further, a metatranscriptomics analysis of the rhizomicrobiome was also conducted to ascertain a relationship between plant growth and microbiome functioning.
Experimental procedures
Soil experiment
Soil where Arabidopsis thaliana genotype (Pna-10) (Li et al., 2010) has naturally grown for more than 8 years was collected in July 2011 from the Michigan Extension Station, Benton Harbor, MI (42° 05′ 34′′ N, 86° 21′ 19′′ W, elevation 630 feet). The soil is described in detail in Broeckling et al. (2008); soil from the same site was used in previous studies (Broeckling et al., 2008; Badri et al., 2009, 2013a). Arabidopsis thaliana seeds were surface-sterilized with Clorox for one minute and subsequently rinsed four times with sterile distilled water. Sterile seeds were placed on Murashige and Skoog (MS) media (Murashige and Skoog, 1962) supplemented with 0.9% bactoagar and 3% sucrose Petri plates. Seeds were incubated in a growth chamber with photoperiod 16 h light/8 h night at 25 °C for seven days. The Arabidopsis seven-day-old seedlings did not introduce any microbes to the system as they were surface sterilized with bleach and no bacterial growth was observed on the MS agar plates even after 7 days of growth. Six replicate pots were maintained for each of the four developmental time points, and one 7-day-old seedling was transplanted to each pot. Individual plants were grown until the following stages were reached: seedling (17 days), vegetative (24 days), bolting (31 days) or flowering (38 days); see Chaparro et al. (2013).
Extraction of microbial DNA from soil
In our study we used the classical definition of the rhizosphere which consist of three zones: the endorhizosphere (root tissue area), the rhizoplane (root surface with epidermis) and the ectorhizosphere (soil directly surrounding the root), and we did not distinguish between these zones (Lynch, 1990; Morgan et al., 2005; Brimecombe et al., 2007; Badri and Vivanco, 2009). Rhizosphere soils (or ‘soil' thereafter) for each of the time point's six biological replicates (24 samples; 4 time points) were collected by gently removing the plants from the pots and obtaining the soil attached to the roots (Supplementary Figure S1) and stored at −80 °C for future use. It is worth noting that the removal of the rhizosphere soil was done in such a manner to prevent mechanical root shearing. However, our rhizosphere soil (as per the classical definition) consists of the rhizosphere and the roots present in that soil (Supplementary Figure S1). Once the total RNA was extracted from the soil using the PowerSoil total RNA isolation kit (Mo Bio, Carlsbad, CA, USA) (see Chaparro et al., 2013), total DNA was subsequently extracted using the RNA PowerSoil DNA Elution Accessory Kit (Mo Bio) according to the manufacturer's instructions. The isolated DNA was quantified using a Nanodrop ND-1000 spectrophotometer (Thermo, Waltham, MA, USA). The bacterial hypervariable regions V1-V3 of the 16S rRNA gene were PCR-amplified using individually bar-coded forward primers 27F, 5′-AGAGTTTGATYMTGGCTCAG-3′ and reverse primer 533R, 5′- TTACCGCGGCTGCTGGC -3′. PCR was performed using Taq DNA polymerase (Takara, Mountain View, CA, USA) as previously described (Badri et al., 2013a). Briefly, DNA samples were diluted to a concentration of 5 ng μl−1 and one microliter was used per PCR reaction. The reaction mix (20 μl) contained 0.4 μmoles of each gene-specific primer, 200 μmoles of dNTPs, 1 × reaction buffer and one unit of Taq DNA polymerase (Takara). PCR included 39 cycles of 94 °C for 30 s, 56 °C for 30 s and 72 °C for 1 minute in a thermal cycler (GeneAmp PCR system 2700; Applied Biosystems, Grand Island, NY, USA). After PCR amplification of the 24 soil DNA samples (6 reps per time point), repetitions were pooled in groups leaving three biological replicates per time point (12 samples). Amplicon products were gel purified using Wizard SV gel (Promega, Madison, WI, USA) and PCR clean-up system followed by Agencourt AMPure XP purification kit (Beckman Coulter, Brea, CA, USA). The concentration of DNA in each sample was determined using the Nanodrop ND-1000 spectrophotometer (Thermo). Approximately equal amounts of the 12 purified amplicon products (3 replicates per time point) were pooled and subjected to unidirectional pyrosequencing in 1/8 of a pico titer-plate at the W.M. Keck Center for Comparative and Functional Genomics, Roy J. Carver Biotechnology Center, University of Illinois at Urbana-Champaign on a Roche/454 Genome Sequencer GS-FLX+ instrument (Roche, Branford, CT, USA). Similarly, isolated total RNA (15 μg per sample) was sent to the same facility. Briefly, mRNA isolation using the Ribozero rRNA removal Meta-bacterial kit (Epicentre Biotechnologies, Madison, WI, USA) was performed; using individually barcoded random hexamer primers the isolated mRNA was converted to cDNA. cDNA library normalization was performed using the Trimmer Direct kit (Evrogen, Moscow, Russia), the samples were pooled in equimolar concentrations and pyrosequencing was performed on 1/8 of a pico titer-plate (see Chaparro et al., 2013).
16S rRNA sequencing analysis
Sequence reads were processed using Mothur v. 1.25.1 (Schloss et al., 2009) as previously described (Badri et al., 2013a). Sampling effort was equalized to the depth of the smallest sample (2769 reads) and operational taxonomic units (OTUs) were defined at ⩾97% sequence identity, using the average neighbor algorithm. Reads were classified within Mothur using the naïve Baysian classifier (Wang et al., 2007). Final taxonomic assignment was based on the consensus identification for each OTU (see Supplementary Table S1). Sequences were also assigned to phylotypes using the phylotype command in Mothur. A multivariate data analysis of the OTUs was performed using METAGENassist (Arndt et al., 2012), followed by normalization based on interquantile range (IQR) (Hackstadt and Hess, 2009) and log2-transformation. IQR normalization allows one to increase statistical power by removing sequences that do not fall within the middle 50% of observations and thus reducing the number of statistical tests one has to perform. Principal component analysis (PCA) and significant features were identified for all treatments using METAGENassist (Arndt et al., 2012). The Vegan package (Oksanen et al., 2012) for R was used for community dissimilarity calculations (Bray–Curtis index) and principal coordinate analysis (PCoA).
Metatranscriptomics analysis
Sequence reads were processed using the Metagenomics-RAST (MG-RAST) server (Meyer et al., 2008) and Mothur (Schloss et al., 2009) (see Chaparro et al., 2013). Briefly, 454 pyrosequencing of the isolated mRNA produced 166 250 sequence reads for seedling, vegetative, bolting, flowering and bulk soil samples (see Chaparro et al., 2013). Host-specific species sequences (Arabidopsis thaliana) were removed using the DNA level matching Bowtie algorithm (Langmead et al., 2009) within MG-RAST. Artificially replicated sequences were removed (Gomez-Alvarez et al., 2009). Sequences were also removed if their length differed by more than two standard deviations from the mean length. Additionally, sequences identified via MG-RAST as ribosomal RNAs were removed using Mothur. Once the ribosomal RNAs were removed (6473 rRNA sequences total) from each of the samples they were normalized to 14 740 high-quality sequence reads per time point (Chaparro et al., 2013). Sequence reads were assigned to the Kyoto Encyclopedia of Genes and Genomes (KEGG) protein database (Kanehisa et al., 2004, 2008). Further quality control consisted of selecting sequences with a minimum percent identity cutoff of 70% and an E-value cutoff of 10−5.
Root exudation
Root exudate data was obtained from Chaparro et al. (2013) from in vitro grown Arabidopsis plants that were grown to the designated plant developmental time points (seedling, vegetative, bolting and flowering). Briefly, 7-day old seedlings (see above for seedling growth) were transferred to Magenta boxes containing 10 ml of MS media supplemented with 1% sucrose. Plants were grown until they were 7, 14, 21 or 28 days and were subsequently transferred to new Magenta boxes containing 10 ml of sterilized Millipore water. Root exudation was collected after 3 days of constant secretion (10, 17, 24 or 31 days) where solutions were filtered using nylon filters (0.45 μm; Millipore, MA, USA). Root exudates were freeze-dried, dissolved in 5 ml of 80% methanol, dried under N gas and sent to the Genome Center Core Services at the University of California, Davis for GC–MS analysis on an Agilent 6890 gas chromatograph (see Chaparro et al., 2013).
Statistical analyses
All statistical analyses were done using SAS (ver. 9.3; SAS Institute). The PROC MIXED function was used to implement a two-way ANOVA analysis with a Tukey post-hoc adjustment to determine pairwise differences between the microbial communities at each plant developmental time point. To ensure that the data followed the assumptions of normality, sequences were log2 transformed. To identify if developmentally dependent root exudation influenced the soil microbial community, Pearson correlation analysis was performed with the compounds released as root exudates (identified in Chaparro et al. (2013)) and the corresponding phylogenetic data. To ensure that the data abided by the assumptions of normality, the sequencing data was log2 transformed and the metabolomics data was log transformed. To determine how the functional microbiome changes with plant growth pairwise comparisons between log2 transformed and subsequently standardized early (seedling and vegetative) and late (bolting and flowering) metatranscriptomics data (see Chaparro et al., 2013) were done using a two-sample t-test. Additionally, Pearson correlation analysis was performed with the log transformed root exudate compounds and the transformed and standardized functional genes that significantly changed (81 transcripts; 413 reads) with plant growth to determine if developmentally-dependent root exudation mediates the functions carried out by the rhizosphere microbiome. The interactive pathways explorer 2 (iPATH2) (Yamada et al., 2011) was used to map the functional transcripts that were differentially expressed between early and late plant development (Supplementary Figure S2).
Results
Plant development influences the soil microbial community
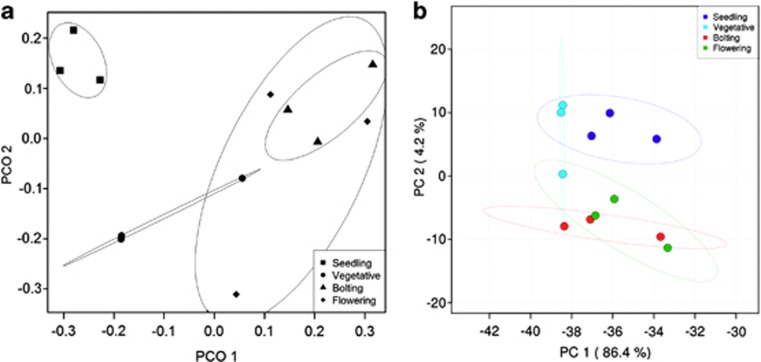
We analyzed the influence of plant development on the rhizosphere microbial community by 454 pyrosequencing and obtained 55 921 high-quality 16S rRNA sequence reads. After equalizing sampling effort, 33 228 reads were retained for analysis. These reads clustered into 7452 OTUs at 3% distance sequence dissimilarity (Supplementary Table S1). We visualized the Bray–Curtis distances between samples using PCoA to determine how dissimilar the soil microbial communities were at each plant developmental time point. The rhizospheric microbial community of seedling was statistically (P<0.05) distinct from that of all other time points (vegetative, bolting and flowering; Figure 1a). On the other hand, the rhizosphere microbial community established at vegetative, bolting and flowering were similar to each other (Figure 1a). We used PCA to identify the factors that influence the soil microbial community at each time point (Figure 1b). The first two principle components explain 90.6% of the variability in the data. Principle component 1 explains 86.4% of the data while principle component 2 explains 4.2% of the data. These data clearly show that the soil microbial community found at seedling was distinct from bolting and flowering. However, we observed that the soil microbial community corresponding to the vegetative stage overlapped with the microbial communities established at seedling and with those at bolting and flowering. This suggests that there is a transition state of the microbial community between seedling and bolting/flowering (Figure 1b).
Figure 1.
Multivariate analyses of the rhizosphere microbial community through plant development analyzed by 454 pyrosequencing. (a) Principal Coordinate Analysis (PCoA) for the visualization of pairwise community dissimilarity (Bray–Curtis index) of the rhizosphere microbial community at each plant developmental stage (seedling, vegetative, bolting and flowering). 95% confidence ellipses are shown around each developmental stage. (b) Principal Component Analysis (PCA) of the rhizosphere microbial community at each plant developmental stage. 95% confidence ellipses are shown around each developmental stage.
We also determined the total OTU richness, evenness and diversity of the sequencing data for each time point (seedling, vegetative, bolting and flowering; Table 1). Although there were no statistically significant differences between the time points with respect to overall community characteristics, we observed that vegetative had the largest community richness, diversity and evenness when compared to the other developmental stages, whereas seedling had the lowest diversity and evenness (Table 1). This suggests that while the structure of the overall soil microbial community does not change, specific microbes may be changing through development.
Table 1. Observed species richness (Sobs), Shannon diversity and evenness of the OTU soil microbial community for each plant developmental time point.
| Richness Sobs |
Shannon |
||
|---|---|---|---|
| Evenness | Diversity | ||
| Seedling | 1107.33 | 0.8009 | 5.6136 |
| Vegetative | 1262.67 | 0.8638 | 6.1675 |
| Bolting | 1114.00 | 0.8192 | 5.7484 |
| Flowering | 1040.33 | 0.8245 | 5.7278 |
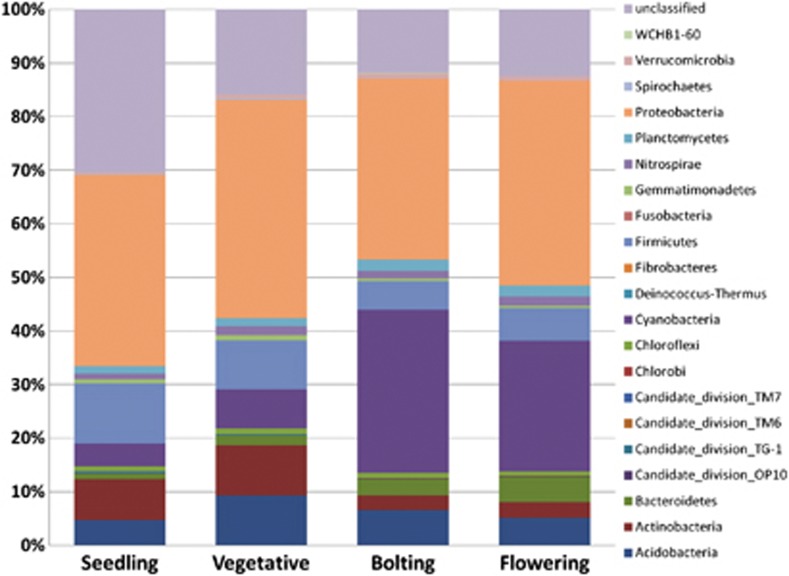
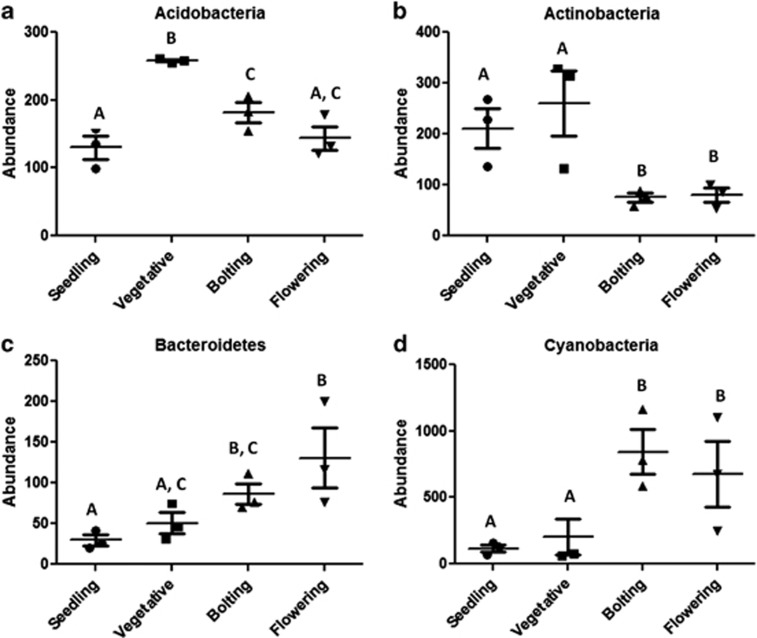
After aligning the OTUs with the SILVA database we classified the soil microbial community into phylotypes consisting of 21 phyla and unclassified (Figure 2). ANOVA analysis with a Tukey post-hoc test identified significant differences between the developmental time points and four phyla (Acidobacteria, Actinobacteria, Bacteroidetes and Cyanobacteria). All other phyla did not significantly change with development. For example, the abundance of Acidobacteria significantly increased (P<0.05) from seedling to vegetative where it reached its peak and then significantly decreased at bolting and flowering to levels similar to those at seedling (Figure 3a). Similarly, the abundance of Actinobacteria was at its highest at the early time points (seedling and vegetative) but then significantly decreased at the later time points (bolting and flowering; Figure 3b). On the other hand, the abundance of Bacteroidetes increased with plant growth reaching its highest abundance at flowering (Figure 3c). Likewise, we observed an increase in the abundance of Cyanobacteria from early time points (seedling and vegetative) to the later time points (bolting and flowering; Figure 3d). These data indicate that while the soil microbial community as a whole did not dramatically change, specific-soil microbial phyla were influenced by plant development.
Figure 2.
Relative abundance (%) of the major bacterial phyla present in the rhizosphere microbial community at each plant developmental stage.
Figure 3.
Bacterial phyla that significantly (P<0.05) change with plant development. (a) Acidobacteria, (b) Actinobacteria, (c) Bacteroidetes and (d) Cyanobacteria. The bars with different letters are significantly different (ANOVA Tukey post-hoc P<0.05) from one another. Each point represents one repetition and graphs show mean±SE.
We further analyzed the soil microbial community at the genus level to determine which genera were influencing the changes observed through development within the phyla Acidobacteria, Actinobacteria, Bacteroidetes and Cyanobacteria. We observed that four genera belonging to Acidobacteria increased in abundance from seedling to vegetative when they reached their peak abundance and then decreased at bolting reaching levels similar to that of seedling during flowering (Supplementary Figure S3A-D). Eight genera belonging to Actinobacteria significantly changed according to plant development (Supplementary Figure S4A-H). For example, Streptomyces increased in abundance from seedling to vegetative when the highest abundance was reached and then significantly decreased at bolting stage to levels below those observed at seedling and remaining at these levels throughout flowering (Supplementary Figure S4E). On the other hand, six genera belonging to the Bacteroidetes phylum significantly increased with plant aging (Supplementary Figures S5A-F). In general, bacteria classified as Cyanobacteria were significantly more abundant at bolting than at seedling and vegetative and then decreased in abundance at flowering (Supplementary Figures S6A-C).
Plant root exudation correlates with rhizosphere microbes through plant development
To determine how developmentally dependent root exudate changes may influence the soil microbiome, Pearson correlation analysis was performed between the compounds released as root exudates (data from Chaparro et al. (2013)) and the rhizosphere bacteria that significantly changed through the life span of the plant (Supplementary Table S2). We observed that Cyanobacteria significantly (P<0.05) correlated with the most root exudate compounds (373 correlations; Table 2) while Bacteroidetes correlated with the least (24 correlations). To determine whether a specific class of root exudate compound was involved in soil microbial community, dynamics root exudates from Chaparro et al. (2013) were categorized as amino acids, phenolics, sugars or sugar alcohols. Phenolic compounds significantly correlated with the soil microbial community of Acidobacteria, Actinobacteria, Bacteroidetes and Cyanobacteria (412 significant correlations; Table 3), followed by amino acids (151), sugars (137) and sugar alcohols (77).
Table 2. Pearson correlation analysis of the OTUs classified as Acidobacteria, Actinobacteria, Bacteroidetes or Cyanobacteria with the compounds released as root exudates.
| Phylum | Number of positive correlations | Number of negative correlations | Total number of correlations |
|---|---|---|---|
| Acidobacteria | 156 | 75 | 231 |
| Actinobacteria | 103 | 46 | 149 |
| Bacteroidetes | 8 | 16 | 24 |
| Cyanobacteria | 130 | 243 | 373 |
The values indicate the number of significant (P<0.05) Pearson correlations for each phyla.
Table 3. Pearson correlations analysis of the OTUs classified as Acidobacteria, Actinobacteria, Bacteroidetes or Cyanobacteria with the group of compounds released as root exudates.
| Phylum |
Amino Acids |
Phenolics |
Sugars |
Sugar Alcohols |
||||
|---|---|---|---|---|---|---|---|---|
| Positively | Negatively | Positively | Negatively | Positively | Negatively | Positively | Negatively | |
| Acidobacteria | 35 | 12 | 60 | 38 | 24 | 18 | 37 | 7 |
| Actinobacteria | 3 | 21 | 58 | 25 | 24 | 0 | 18 | 0 |
| Bacteroidetes | 2 | 0 | 4 | 10 | 1 | 5 | 1 | 1 |
| Cyanobacteria | 67 | 11 | 60 | 157 | 0 | 65 | 3 | 10 |
| Total Correlations | 151 | 412 | 137 | 77 | ||||
The values indicate the number of significant (P<0.05) Pearson correlations.
Plant development influences the functional microbiome
To determine the functional genes that were differentially expressed at early (seedling and vegetative) compared to late plant development (bolting and flowering) two-sample t-tests on log2 transformed and standardized data were performed on the metatranscriptomics data from Chaparro et al. (2013). A total of 81 unique transcripts out of 1240 assigned to the KEGG database were significantly (P<0.05) differentially expressed with plant age (Supplementary Table S3). iPATH2 (Yamada et al., 2011) was used to map at what time point (early or late) these 81 functional transcripts were more abundant (Supplementary Figure S2). Of those transcripts, 32 were more abundant during seedling and vegetative stages, while 49 were more numerous during plant bolting and flowering. The majority of the transcripts that were differently expressed between early and late development are involved in metabolism and genetic information processing. Two transcripts are involved in toluene degradation (succinate dehydrogenase iron-sulfur protein) and the transcript involved in nitrotoluene degradation (hydrogenase large subunit) were significantly (P<0.05) more abundant early in plant development. On the other hand, a transcript involved in bacterial chemotaxis (two-component system, chemotaxis family, response regulator) and one involved in streptomycin biosynthesis (dTDP-glucose 4,6-dehydratase) were significantly more abundant in late developmental stages. Among five transcripts involved in nitrogen metabolism, three and two were significantly expressed at early or late plant developmental stages, respectively. For example, the transcript formamidase, carbonic anhydrase and nitric-oxide reductase NorQ protein were highly expressed during plant seedling and vegetative stages. Conversely, the transcripts nitrite reductase (NAD(P)H) large subunit and periplasmic nitrate reductase NapA were more abundant during plant bolting and flowering.
Beneficial microbes are more active during late plant development
The 81 transcripts that were significantly expressed through development were aligned to the BLAT database (Kent, 2002) within MG-RAST (Meyer et al., 2008) to determine which microbes were carrying out the expressed functions (Supplementary Table S3). These transcripts were attributed to bacteria such as Bradyrhizobia, Streptomyces, Azoarcus or Pseudomonas syringae. Accordingly, we categorized these microbes into seven groups depending on their potential activity towards the plant (i.e., symbiotic N fixation, free N fixation, antagonists, PGPRs, pathogens, xenobiotic/metal detoxifiers or unclassified; Table 4). We observed that many of these transcripts aligned to symbiotic N fixers, free N fixers or pathogens (14, 17 or 12 transcripts respectively; Table 4), such as Nitrobacter, Rhodospirillum, Nitrosospira, Mesorhizobium, Cyanobacteria, Azorhizobium or Dickeya dadantii. Interestingly, most of the transcripts that aligned to pathogens were significantly expressed early during development (8 genes) while transcripts that aligned to microbes involved in plant growth promotion or N fixation were more abundant late (23 genes; Table 4). For example, the transcripts involved in bacterial chemotaxis (two-component system, chemotaxis family, response regulator) were significantly more abundant at late time points and aligned to the nitrogen fixing PGPR Methylobacterium extorquens (Ivanova et al., 2001; Sy et al., 2001; Lidstrom and Chistoserdova, 2002). On the other hand, the transcript involved in amoebiasis (Ras-related protein Rab-7A) was significantly more abundant at early time points, and it aligned with the pathogen Moniliophthora perniciosa. Additionally, microbes that are antagonistic and produce fungicides or bactericides such as Streptosporangium, Streptomyces avermitilis and Sorangium cellulosum were transcriptionally active late (Table 4 and Supplementary Table S3) (Burg et al., 1979; Pradella et al., 2002).
Table 4. Taxonomic assignments of the differentially expressed (81) transcripts were categorized based on their activity and whether the corresponding transcript were significantly (t-test P<0.05) expressed early or late in plant development.
| Early | Late | Total | |
|---|---|---|---|
| Symbiotic N fixers | 5 | 9 | 14 |
| Free N fixers | 6 | 11 | 17 |
| Antagonistic | 5 | 6 | 11 |
| Plant growth promoting | 0 | 3 | 3 |
| Pathogen | 8 | 4 | 12 |
| Xenobiotic/metal detoxification | 4 | 5 | 9 |
| Unclassified function | 4 | 11 | 15 |
Plant root exudation correlates with the functional microbiome through plant development
To determine whether root exudation potentially mediates the functions carried out by the rhizosphere microbiome, we correlated the 81 unique and significant transcripts with the compounds released as root exudates (from Chaparro et al. (2013); see Supplementary Table S4). Similar to the 16S rRNA analysis, phenolic compounds appear to mediate the expression of the transcripts (449 significant correlations; Table 5). Additionally, dTDP-glucose 4,6-dehydratase positively correlated with the sugar alcohol myo-Inositol. The transcript two-component system, chemotaxis family, response regulator positively correlated with glycine and xylitol.
Table 5. Pearson correlation analysis of the significantly expressed transcripts at early or late plant development correlated with the group of compounds released as root exudates.
|
Early |
Late |
Overall Total | |||||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Total | Positive | Negative | Total | ||
| Amino Acids | 7 | 160 | 167 | 198 | 1 | 199 | 366 |
| Phenolics | 31 | 139 | 170 | 222 | 57 | 279 | 449 |
| Sugars | 51 | 0 | 51 | 0 | 82 | 82 | 133 |
| Sugar Alcohols | 0 | 47 | 47 | 85 | 6 | 91 | 138 |
The values indicate the number of significant (P<0.05) correlations.
Discussion
Plant developmental changes affect the rhizosphere microbial community
Analysis of the overall bacterial rhizosphere community through plant development revealed that the community did not significantly change with respect to richness, diversity and evenness (Table 1) but Bray–Curtis community dissimilarity analysis revealed that the microbial community at seedling was significantly different from the other developmental stages (vegetative, bolting and flowering; Figure 1a). These results are in agreement with previous reports as the Arabidopsis rhizosphere microbial communities after the bolting stage were not distinct (Micallef et al., 2009a; Lundberg et al., 2012). A more detailed look at the assembled rhizosphere microbial communities through plant development revealed that a core microbiome was established and this constituted bacteria comprising Actinobacteria, Bacteroidetes and Proteobacteria, as was previously observed (Bulgarelli et al., 2012; Lundberg et al., 2012). In addition, the present study demonstrated that Chloroflexi, Firmicutes, Gemmatimonadetes, Nitrospirae, Planctomycetes and Verrucomicrobia were also consistently present throughout plant development (Figure 2). Additionally, Acidobacteria (Figure 3a) and Cyanobacteria (Figure 3d) also comprised the core rhizosphere microbiome but these phyla along with Bacteroidetes (Figure 3c) and Actinobacteria (Figure 3b) significantly changed with plant development suggesting that the plant can select a subset of microbes at different stages of growth. Acidobacteria is one of the most abundant bacterial phyla found in terrestrial ecosystems (Barns et al., 1999) and they have an important role in the carbon cycle due to their ability to degrade complex plant derived polysaccharides such as cellulose and lignin (Ward et al., 2009). Unfortunately, the specific role they play in the soil ecosystem and their role in the rhizosphere (apart from being metabolically active (Lee et al., 2008)) is relatively unknown. Actinobacteria, on the other hand, has recently been associated with disease suppressive soils (Mendes et al., 2011). Streptomyces species, which were significantly more abundant in the vegetative stage of Arabidopsis (Supplementary Figure S4E), are able to increase root nodulation efficiency and promote plant growth of the legume Pisum sativum (Tokala et al., 2002) while simultaneously triggering plant defense in Arabidopsis or apple trees (Cohen et al., 2005; Conn et al., 2008; Lin et al., 2012). On the other hand, Bacteroidetes' role in the rhizosphere has not yet been elucidated but it has been reported that they are important contributors to nutrient turnover in the soil (Yousuf et al., 2012). Bacterial species belonging to Bacteroidetes contain genes involved in denitrification indicating a possible involvement in N cycling (Van Spanning et al., 2005). Cyanobacteria have been shown to colonize plant roots (Gantar et al., 1991; Lundberg et al., 2012), promote plant growth (Prasanna et al., 2009) and are an important plant source for inorganic N due to their ability to fix N (Franche et al., 2009).
Plant development influences the functional capacity of the rhizomicrobiome
Metatranscriptomics analyses of mRNA have only recently been used as a means to study microbial communities at a functional level in distinct environments such as the human gut (Gosalbes et al., 2011), the mouse gut (Xiong et al., 2012) and oceans (Poretsky et al., 2009; Baker et al., 2013). In soils, functional microarrays or GeoChip that target specific microbial functions have been successfully used to determine the metabolic potential of the microbial communities (He et al., 2007, 2011; Bai et al., 2013; Zhang et al., 2013). A drawback of the GeoChip is the fact that novel functions and transcripts cannot be identified, and one is biased to the probes present in the chip (Dugat-Bony et al., 2012). Recently, metatranscriptomics in soil has been attempted on the rRNA to identify the active members of the microbial community in the rhizosphere of various crop species (Turner et al., 2013); Urich et al. (2008) described the isolation and sequencing of mRNA from a sandy lawn as a means of describing the microbial transcriptome in the soil. The rhizosphere microbiome plays an essential role in plant health and productivity and it is often referred as the plant's second genome (Berendsen et al., 2012; Chaparro et al., 2012). Accordingly, our metatranscriptomics data permitted a glimpse at the genes that the microbiome was expressing as a whole at each stage of plant development. More transcripts were significantly expressed at late plant developmental time points (Supplementary Table S3) and this may be indicative of the soil microbial community selecting specific functions throughout plant development. For example, dTDP-glucose 4,6-dehydratase involved in streptomycin biosynthesis is more abundant during plant bolting and flowering. Streptomycin is an antibiotic mainly produced by Streptomyces and it is antagonistic against gram-positive and -negative bacteria and has been shown to enhance plant defenses and trigger systemic resistance (Schatz et al., 2005; Conn et al., 2008; Lin et al., 2012). Additionally, we observe that dTDP-glucose-4,6-dehydratase positively correlates with the root exudate compound myo-Inositol which increases streptomycin biosynthesis (Majumdar and Kutzner, 1962; Heding, 1964). Interestingly, we did not find many transcripts that are attributed to plant pathogens at late stages of plant development which is consistent with the increased production of streptomycin and in accordance with ARR. Furthermore, we determined that Sorangium cellulosum which produce bacteriocides and fungicides (Pradella et al., 2002) was also present in late stages of plant development.
The rhizosphere microbiome can also supply the plant with essential nutrients such as nitrogen. Nitrogen is essential for plant growth (Burns 1996; Rossato et al., 2001) and it is usually deficient in soils (Novoa and Loomis, 1981). Thus, in natural environments (as compared to agricultural systems) the plant depends on N fixing and nitrifying bacteria for their N supply. It should be noted that in our study the plants grew in natural soil without external fertilization. In accordance, we observed that throughout plant development functional genes involved in nitrogen metabolism were transcriptionally active (Table 4). Additionally, functions carried out by bacteria involved in N assimilation were prevalent (Table 4 and Supplementary Table S3). This may be indicative of modulation of the microbiome to express specific functions throughout plant growth, that is, differentially express transcripts from beneficial bacteria. For example, transcripts more abundant in early time points align to bacteria involved in providing the plant with N such as Nitrobacter, Rhodospirillum, Nitrosospira, Mesorhizobium or Azorhizobium. Similarly, during bolting and flowering, functional genes expressed in the rhizosphere align to PGPRs such as Bacillus licheniformis (Gutiérrez-Mañero et al., 2001) or Burkholderia ambifaria (Chiarini et al., 2006), free N-fixers such as Cyanothece sp. (Junier et al., 2009), as well as symbiotic N-fixing bacteria (Bradyrhizobium) (Stacey et al., 1995) or Herbaspirillum which is involved in endophytic N fixation (Elbeltagy et al., 2001; Franche et al., 2009). It should be noted that in addition to fixing N through legume symbiosis, Bradyrhizobium promotes plant growth of non-leguminous plants (Antoun et al., 1998). The functional microbiome also expressed genes that aligned to the free N-fixing Cyanobacteria (Supplementary Table S3). These wide changes in the soil bacterial community hint at the soil microbiome shifting and changing according to the needs of the plant and that the rhizosphere functional microbiome can express specific functions at precise stages of plant growth.
Interestingly, we determined that the expression of a functional gene is altered through plant development even when the bacterium performing the function does not change in abundance. For example, Bradyrhizobia classified as Proteobacteria does not significantly change with plant growth (Figure 2) yet transcripts expressed by Bradyrhizobia such as imidazolonepropionase, phenol 2-monooxygenase or glutaminase are more abundant in later stages of plant development (Supplementary Table S3). Alternatively, while Actinobacteria, more specifically Streptomyces, were more abundant during the vegetative stage (Figure 3b, Supplementary Figure S4E) we observed that genes involved in synthesis of the antibiotic streptomycin were higher during plant bolting and flowering (Supplementary Table S3). This indicates that root exudates are presumably able to modulate the expression of specific functional genes without altering the bacterial taxonomic composition of the rhizosphere.
Root exudates act as potential stimulants for rhizomicrobiome functions
While microbial colonization of the rhizosphere has shown an array of benefits to the plant, the exact mechanism by which the plant is able to attract these microbes varies. One such mechanism used is bacterial chemotaxis toward root exudate compounds. Interestingly, we see that the two-component system, chemotaxis family, response regulator involved in bacterial chemotaxis is significantly expressed late in plant development and its expression positively correlates with the root exudates glycine and xylitol (Supplementary Tables S3 and S4). Interestingly, PGPRs and endophytic bacteria show chemotaxis towards glycine (Gaworzewska and Carlile, 1982; de Weert et al., 2002; Gupta Sood, 2003), while the non-symbiotic nitrogen-fixing bacteria Azotobacter vinelandii shows chemotactic behavior towards xylitol (Haneline et al., 1991). This provides an additional mechanism for the plants ability to manipulate and orchestrate the rhizosphere microbiome. While the results presented are only correlative they do highlight the importance of root exudation in rhizosphere plant-microbiome interactions and provide strong evidence to warrant further investigation that would conclusively determine how specific components of the root exudates in the absence of the plant could influence the rhizosphere metatranscriptome. Incidentally, a recent study depicting the microbial communities established from the addition of root exudate blends to the soil demonstrated that different blends produced changes in the microbial community. For example, soil supplemented with ethyl acetate- or water-extracted root exudates generated microbial communities that presumably had the ability to metabolize the pesticide atrazine (Badri et al., 2013a). Similar experiments that add specific compounds (sugars or oxalic acids) to the soil have also shown dramatic shifts in the composition of the microbial community (Eilers et al., 2010; Shi et al., 2011).
The general root exudation patterns reported in Chaparro et al. (2013) do not seem to correlate with general microbiome characteristics as initially hypothesized (i.e., increased root exudation of sugars at early plant development would result in enhanced richness of the rhizosphere microbial community at these early time points). This could be due to the fact that root exudates were collected in vitro while rhizosphere microbial communities were analyzed in vivo; in other words, the root secretion patterns could be different under soil conditions. Accordingly, it has been reported that under in vitro conditions plants grown alone or co-cultured with a microorganism show different patterns of proteins present in the root exudates (De-la-Pena et al., 2008). However, Chaparro et al. (2013) showed evidence that root exudation through development is genetically programmed since there were strong correlations between in vitro root exudation patterns and the ability of the soil microbiome to utilize these compounds in vivo. During the plant vegetative stage (transition state; Figure 1b) we identified the largest OTU richness, diversity and evenness in the rhizosphere (Table 1). This rhizospheric microbial transition state corresponds with a root exudation transition state where the highest diversity of phytochemicals (sugars, sugar alcohols, phenolics and amino acids) was observed (Chaparro et al., 2013). Therefore, it appears that sugars present in the root exudates do not necessarily function as general substrates for soil microbes as is widely reported in the literature (Behera and Wagner, 1974; Jaeger et al., 1999; Fierer et al., 2007; Eilers et al., 2010), whereby compounds such as phenolics or amino acids more readily influence soil rhizosphere microbial communities as well as modulate their transcription (Tables 3 and 5). Recently, Badri et al. (2013a) reported that fractions of root exudates containing phenolic and phenolic-related compounds when applied to the soil (in the absence of the plant) significantly modulated the soil microbiome. In addition, analysis of the rhizosphere microbial community of an Arabidopsis mutant that secretes more phenolic compounds than sugars showed that it cultured more beneficial microbes such as PGPRs and those involved in N-fixation such as Bradyrhizobia and Cyanobacteria when compared to wild type (Badri et al., 2009). Interestingly, the reduction of phenolic exudates by inhibition of phenylalanine ammonia-lyase gene expression in transgenic rice influenced the rhizospheric microbial community, with eight phyla decreasing in abundance in transgenic rice when compared to wild-type plants (Fang et al., 2013). Further studies pinpointing which phenolic compounds are involved in coordinating these microbial interactions are needed.
The data presented here implies and alludes to the fact that the plant through root exudation may be selecting microbes for different functions; however, we would be remiss not to point out other potential mechanisms that may be playing a role. For example, the changes observed in the rhizosphere microbial community could be due to microbial community succession with respect to microbial competition. Unfortunately, presently there is little research regarding microbial succession and stability in soils (Fierer et al., 2010). Recently, it was shown that microbial succession is similar to that of previously described plant and animal succession with respect to species-time relationships (Shade et al., 2013) but what this could mean in the rhizosphere is unclear. There is still much to be explored with respect to plant-microbiome interactions to better understand and decipher the complex patterns and associations that arise in this unique ecological niche.
Much like the plant can influence the rhizosphere microbiome the rhizosphere microbiome can also influence the plant. For example, fungal communities have been shown to influence root exudation rates which can in turn influence the rhizosphere microbiome (Meier et al., 2013). Similarly, Lau and Lennon (2011) demonstrated that microbial community structure affects natural plant trait selections. Additionally, it has been established that distinct microbial communities influence plants' ability to tolerate abiotic stress such as drought (Zolla et al., 2013) and even affect leaf metabolome and subsequent insect feeding (Badri et al., 2013b). These examples highlight the multifaceted nature of the interactions in the rhizosphere microbiome.
Conclusions
The conclusions of this study could be summarized as follows: (1) the plant maintains a core rhizosphere microbiome; (2) this core microbiome is likely to express different functions at different stages of plant development; (3) the plant can enhance the expression of a subset of microbial functions at specific times that the core microbiome is not currently expressing; (4) the plant can select a subset of microbes at different stages of development presumably for specific functions that the core microbiome can't express (i.e., N fixation, antibiosis against pathogens, etc.); and (5) the plant secretes blends of compounds and specific phytochemicals in the root exudates that are differentially produced at distinct stages of plant growth to help orchestrate the activities described in 1, 2, 3 and 4. Overall, these concepts suggest that plants and the rhizomicrobiome are in constant communication through the exchange of signals. Experiments targeting some of these ideas are essential to conclusively determine the interactive functionalities that occur in the rhizosphere between crops and their microbiome.
Acknowledgments
Work was supported by a grant from the National Science Foundation to JMV (MCB-0950857). JMC participated in the experimental design, performed the experiments, analyzed the data and wrote the manuscript. DVB participated in the experimental design, helped in data analyses and critical reading of the manuscript. JMV participated in the experimental design, coordinated the studies and wrote the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abedon BG, Tracy WF. Corngrass 1 of Maize (Zea mays L.) Delays Development of Adult Plant Resistance to Common Rust (Puccinia sorghi Schw.) and European Corn Borer (Ostrinia nubilalis Hubner) J Hered. 1996;87:219–223. [Google Scholar]
- An C, Mou Z. Salicylic acid and its function in plant immunity. J Integr Plant Biol. 2011;53:412–428. doi: 10.1111/j.1744-7909.2011.01043.x. [DOI] [PubMed] [Google Scholar]
- Antoun H, Beauchamp C, Goussard N, Chabot R, Lalande R. Potential of Rhizobium and Bradyrhizobium species as plant growth promoting rhizobacteria on non-legumes: effect on radishes (Raphanus sativus L.) Plant Soil. 1998;204:57–67. [Google Scholar]
- Arndt D, Xia J, Liu Y, Zhou Y, Guo AC, Cruz JA, et al. METAGENassist: a comprehensive web server for comparative metagenomics. Nucleic Acids Res. 2012;40:W88–W95. doi: 10.1093/nar/gks497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri DV, Quintana N, El Kassis EG, Kim HK, Choi YH, Sugiyama A, et al. An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol. 2009;151:2006–2017. doi: 10.1104/pp.109.147462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri DV, Vivanco JM. Regulation and function of root exudates. Plant Cell Environ. 2009;32:666–681. doi: 10.1111/j.1365-3040.2008.01926.x. [DOI] [PubMed] [Google Scholar]
- Badri DV, Chaparro JM, Zhang R, Shen Q, Vivanco JM. Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic related compounds predominantly modulate the soil microbiome. J Biol Chem. 2013;288:4502–4512. doi: 10.1074/jbc.M112.433300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri DV, Zolla G, Bakker MG, Manter DK, Vivanco JM. Potential impact of soil microbiomes on the leaf metabolome and on herbivore feeding behavior. New Phytol. 2013;198:264–273. doi: 10.1111/nph.12124. [DOI] [PubMed] [Google Scholar]
- Bai S, Li J, He Z, Van Nostrand JD, Tian Y, Lin G, et al. GeoChip-based analysis of the functional gene diversity and metabolic potential of soil microbial communities of mangroves. Appl Microbiol Biotechnol. 2013;97:7035–7048. doi: 10.1007/s00253-012-4496-z. [DOI] [PubMed] [Google Scholar]
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol. 2006;57:233–266. doi: 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- Baker BJ, Sheik CS, Taylor CA, Jain S, Bhasi A, Cavalcoli JD, et al. Community transcriptomic assembly reveals microbes that contribute to deep-sea carbon and nitrogen cycling. Isme J. 2013;7:1962–1973. doi: 10.1038/ismej.2013.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barns SM, Takala SL, Kuske CR. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl Environ Microbiol. 1999;65:1731–1737. doi: 10.1128/aem.65.4.1731-1737.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudoin E, Benizri E, Guckert A. Impact of growth stage on the bacterial community structure along maize roots, as determined by metabolic and genetic fingerprinting. Appl Soil Ecol. 2002;19:135–145. [Google Scholar]
- Bednarek P. Chemical warfare or modulators of defence responses - the function of secondary metabolites in plant immunity. Curr Opin Plant Biol. 2012;15:407–414. doi: 10.1016/j.pbi.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Behera B, Wagner GH. Microbial Growth Rate In Glucose-amended Soil. Soil Sci Soc Am J. 1974;38:591–594. [Google Scholar]
- Berendsen RL, Pieterse CM, Bakker PA. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Bolan NS. A critical review on the role of mycorrhizal fungi in the uptake of phosphorus by plants. Plant and Soil. 1991;134:189–207. [Google Scholar]
- Brimecombe MJ, De Leij FA, Lynch JM.2007Rhizodeposition and Microbial PopulationsIn: Pinton R, Varanini Z, Nannapaneni P (eds)The Rhizosphere Biochemistry and Organic Substances at the Soil-Plant InterfaceSecond EditionCRC Press: Boca Raton, Fl; 73–109. [Google Scholar]
- Broeckling CD, Broz AK, Bergelson J, Manter DK, Vivanco JM. Root exudates regulate soil fungal community composition and diversity. Appl Environ Microbiol. 2008;74:738–744. doi: 10.1128/AEM.02188-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, Ahmadinejad N, Assenza F, et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488:91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- Burg RW, Miller BM, Baker EE, Birnbaum J, Currie SA, Hartman R, et al. Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob Agents Chemother. 1979;15:361–367. doi: 10.1128/aac.15.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns IG. Nitrogen supply, growth and development. Acta Hort. 1996;428:21–30. [Google Scholar]
- Chaparro JM, Sheflin AM, Manter DK, Vivanco JM. Manipulating the soil microbiome to increase soil health and plant fertility. Biol Fertil Soils. 2012;48:489–499. [Google Scholar]
- Chaparro JM, Badri DV, Bakker MG, Sugiyama A, Manter DK, Vivanco JM. Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS One. 2013;8:e55731. doi: 10.1371/journal.pone.0055731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarini L, Bevivino A, Dalmastri C, Tabacchioni S, Visca P. Burkholderia cepacia complex species: health hazards and biotechnological potential. Trends Microbiol. 2006;14:277–286. doi: 10.1016/j.tim.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323:95–101. doi: 10.1126/science.1164627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MF, Yamasaki H, Mazzola M. Brassica napus seed meal soil amendment modifies microbial community structure, nitric oxide production and incidence of Rhizoctonia root rot. Soil Biol and Biochem. 2005;37:1215. [Google Scholar]
- Conn VM, Walker AR, Franco CM. Endophytic actinobacteria induce defense pathways in Arabidopsis thaliana. Mol Plant Microbe Interact. 2008;21:208–218. doi: 10.1094/MPMI-21-2-0208. [DOI] [PubMed] [Google Scholar]
- Coronado C, Zuanazzi J, Sallaud C, Quirion JC, Esnault R, Husson HP, et al. Alfalfa root flavonoid production is nitrogen regulated. Plant Physiol. 1995;108:533–542. doi: 10.1104/pp.108.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson IA, Robson MJ. Effect of contrasting patterns of nitrate application on the nitrate uptake, N2-fixation, nodulation and growth of white clover. Annals of Botany. 1986;57:331–338. [Google Scholar]
- De-la-Pena C, Badri DV, Lei Z, Watson BS, Brandao MM, Silva-Filho MC, et al. Root secretion of defense-related proteins is development-dependent and correlated with flowering time. J Biol Chem. 2010;285:30654–30665. doi: 10.1074/jbc.M110.119040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-la-Pena C, Lei Z, Watson BS, Sumner LW, Vivanco JM. Root-microbe communication through protein secretion. J Biol Chem. 2008;283:25247–25255. doi: 10.1074/jbc.M801967200. [DOI] [PubMed] [Google Scholar]
- de Weert S, Vermeiren H, Mulders IHM, Kuiper I, Hendrickx N, Bloemberg GV, et al. Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by pseudomonas fluorescens. Mol Plant Microbe Interact. 2002;15:1173–1180. doi: 10.1094/MPMI.2002.15.11.1173. [DOI] [PubMed] [Google Scholar]
- Develey-Riviere MP, Galiana E. Resistance to pathogens and host developmental stage: a multifaceted relationship within the plant kingdom. New Phytol. 2007;175:405–416. doi: 10.1111/j.1469-8137.2007.02130.x. [DOI] [PubMed] [Google Scholar]
- Dugat-Bony E, Peyretaillade E, Parisot N, Biderre-Petit C, Jaziri F, Hill D, et al. Detecting unknown sequences with DNA microarrays: explorative probe design strategies. Environ Microbiol. 2012;14:356–371. doi: 10.1111/j.1462-2920.2011.02559.x. [DOI] [PubMed] [Google Scholar]
- Eaglesham ARJ. Nitrate inhibition of root-nodule symbiosis in doubly rooted soybean plants. Crop science. 1989;29:115–119. [Google Scholar]
- Eilers KG, Lauber CL, Knight R, Fierer N. Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol and Biochem. 2010;42:896. [Google Scholar]
- Elbeltagy A, Nishioka K, Sato T, Suzuki H, Ye B, Hamada T, et al. Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolated from wild rice species. Appl Environ Microbiol. 2001;67:5285–5293. doi: 10.1128/AEM.67.11.5285-5293.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang C, Zhuang Y, Xu T, Li Y, Li Y, Lin W. Changes in rice allelopathy and rhizosphere microflora by inhibiting rice phenylalanine ammonia-lyase gene expression. J Chem Ecol. 2013;39:204–212. doi: 10.1007/s10886-013-0249-4. [DOI] [PubMed] [Google Scholar]
- Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007;88:1354. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- Fierer N, Nemergut D, Knight R, Craine JM. Changes through time: integrating microorganisms into the study of succession. Res Microbiol. 2010;161:635–642. doi: 10.1016/j.resmic.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Franche C, Lindstrom K, Elmerich C. Nitrogen-fixing bacteria associated with leguminous and non-leguminous plants. Plant and Soil. 2009;321:35–59. [Google Scholar]
- Gantar M, Kerby NW, Rowell P, Obreht Z. Colonization of wheat Triticum vulgare L.) by N2-fixing cyanobacteria: I. A survey of soil cyanobacterial isolates forming associations with roots. New Phytologist. 1991;118:477. doi: 10.1111/j.1469-8137.1995.tb04304.x. [DOI] [PubMed] [Google Scholar]
- Gaworzewska ET, Carlile MJ. Positive Chemotaxis Of Rhizobium-Leguminosarum And Other Bacteria Towards Root Exudates From Legumes And Other Plants. J Gen Microbiol. 1982;128:1179–1188. [Google Scholar]
- Gomez-Alvarez V, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from complex microbial communities. Isme J. 2009;3:1314–1317. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- Gosalbes MJ, Durban A, Pignatelli M, Abellan JJ, Jimenez-Hernandez N, Perez-Cobas AE, et al. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS One. 2011;6:e17447. doi: 10.1371/journal.pone.0017447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta Sood S. Chemotactic response of plant-growth-promoting bacteria towards roots of vesicular-arbuscular mycorrhizal tomato plants. FEMS Microbiol Ecol. 2003;45:219–227. doi: 10.1016/S0168-6496(03)00155-7. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Mañero FJ, Ramos-Solano B, An Probanza, Mehouachi J, Tadeo R, Talon FM. The plant-growth-promoting rhizobacteria Bacillus pumilus and Bacillus licheniformis produce high amounts of physiologically active gibberellins. Physiologia Plantarum. 2001;111:206. [Google Scholar]
- Hackstadt AJ, Hess AM. Filtering for increased power for microarray data analysis. BMC Bioinformatics. 2009;10:11. doi: 10.1186/1471-2105-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haneline S, Connelly CJ, Melton T. Chemotactic behavior of azotobacter vinelandii. Appl Environ Microbiol. 1991;57:825–829. doi: 10.1128/aem.57.3.825-829.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Gentry TJ, Schadt CW, Wu L, Liebich J, Chong SC, et al. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. Isme J. 2007;1:67–77. doi: 10.1038/ismej.2007.2. [DOI] [PubMed] [Google Scholar]
- He ZL, Van Nostrand JD, Deng Y, Zhou JZ. Development and applications of functional gene microarrays in the analysis of the functional diversity, composition, and structure of microbial communities. Front Environ Sci Engin China. 2011;5:1–20. [Google Scholar]
- Heding H. Radioactive myoinositol: incorporation into streptomycin. Science. 1964;143:953–954. doi: 10.1126/science.143.3609.953. [DOI] [PubMed] [Google Scholar]
- Horiuchi J, Prithiviraj B, Bais HP, Kimball BA, Vivanco JM. Soil nematodes mediate positive interactions between legume plants and rhizobium bacteria. Planta. 2005;222:848–857. doi: 10.1007/s00425-005-0025-y. [DOI] [PubMed] [Google Scholar]
- Houlden A, Timms-Wilson TM, Day MJ, Bailey MJ. Influence of plant developmental stage on microbial community structure and activity in the rhizosphere of three field crops. FEMS Microbiol Ecol. 2008;65:193–201. doi: 10.1111/j.1574-6941.2008.00535.x. [DOI] [PubMed] [Google Scholar]
- Inceoglu O, Al-Soud WA, Salles JF, Semenov AV, van Elsas JD. Comparative analysis of bacterial communities in a potato field as determined by pyrosequencing. PLoS One. 2011;6:e23321. doi: 10.1371/journal.pone.0023321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova EG, Doronina NV, Trotsenko IuA. Aerobic methylobacteria are capable of synthesizing auxins. Mikrobiologiia. 2001;70:452–458. [PubMed] [Google Scholar]
- Jaeger CH, 3rd, Lindow SE, Miller W, Clark E, Firestone MK. Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and tryptophan. Appl Environ Microbiol. 1999;65:2685–2690. doi: 10.1128/aem.65.6.2685-2690.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain V, Nainawatee HS. Plant flavonoids: signals to legume nodulation and soil microorganisms. J Plant Biochem Biot. 2002;11:1–10. [Google Scholar]
- Junier P, Junier T, Witzel KP, Caru M. Composition of diazotrophic bacterial assemblages in bean-planted soil compared to unplanted soil. Eur J Soil Biol. 2009;45:153–162. [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JT, Bacon RK, Wells BR. Genetic variability in nitrogen utilization at four growth stages in soft red winter wheat. J Plant Nutr. 1995;18:969. [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kus JV, Zaton K, Sarkar R, Cameron RK. Age-related resistance in Arabidopsis is a developmentally regulated defense response to Pseudomonas syringae. Plant Cell. 2002;14:479–490. doi: 10.1105/tpc.010481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau JA, Lennon JT. Evolutionary ecology of plant-microbe interactions: soil microbial structure alters selection on plant traits. New Phytologist. 2011;192:215–224. doi: 10.1111/j.1469-8137.2011.03790.x. [DOI] [PubMed] [Google Scholar]
- Lee SH, Ka JO, Cho JC. Members of the phylum Acidobacteria are dominant and metabolically active in rhizosphere soil. FEMS Microbiol Lett. 2008;285:263–269. doi: 10.1111/j.1574-6968.2008.01232.x. [DOI] [PubMed] [Google Scholar]
- Li X, Bergelson J, Chapple C. The ARABIDOPSIS accession Pna-10 is a naturally occurring sng1 deletion mutant. Mol Plant. 2010;3:91–100. doi: 10.1093/mp/ssp090. [DOI] [PubMed] [Google Scholar]
- Lidstrom ME, Chistoserdova L. Plants in the pink: cytokinin production by methylobacterium. J Bacteriol. 2002;184:1818. doi: 10.1128/JB.184.7.1818.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Ge HM, Yan T, Qin YH, Tan RX. Thaxtomin A-deficient endophytic Streptomyces sp. enhances plant disease resistance to pathogenic Streptomyces scabies. Planta. 2012;236:1849–1861. doi: 10.1007/s00425-012-1741-8. [DOI] [PubMed] [Google Scholar]
- Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JM.(ed). (1990The Rhizosphere Wiley Interscience: Chinchester, UK; p458 [Google Scholar]
- Majumdar SK, Kutzner HJ. Myo-inositol in the biosynthesis of streptomycin by Streptomyces griseus. Science. 1962;135:734. doi: 10.1126/science.135.3505.734. [DOI] [PubMed] [Google Scholar]
- Malagoli P, Laine P, Le Deunff E, Rossato L, Ney B, Ourry A. Modeling nitrogen uptake in oilseed rape cv Capitol during a growth cycle using influx kinetics of root nitrate transport systems and field experimental data. Plant Physiol. 2004;134:388–400. doi: 10.1104/pp.103.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier IC, Avis PG, Phillips RP. Fungal communities influence root exudation rates in pine seedlings. FEMS Microbiol Ecol. 2013;83:585–595. doi: 10.1111/1574-6941.12016. [DOI] [PubMed] [Google Scholar]
- Mendes R, Kruijt M, De Bruijn I, Dekkers E, van der Voort M, JHM Schneider, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. Bmc Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micallef SA, Channer S, Shiaris MP, Colon-Carmona A. Plant age and genotype impact the progression of bacterial community succession in the Arabidopsis rhizosphere. Plant Signal Behav. 2009;4:777–780. doi: 10.4161/psb.4.8.9229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micallef SA, Shiaris MP, Colon-Carmona A. Influence of Arabidopsis thaliana accessions on rhizobacterial communities and natural variation in root exudates. J Exp Bot. 2009;60:1729–1742. doi: 10.1093/jxb/erp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet YA, Danna CH, Clay NK, Songnuan W, Simon MD, Werck-Reichhart D, et al. Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell. 2010;22:973–990. doi: 10.1105/tpc.109.069658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JA, Bending GD, White PJ. Biological costs and benefits to plant-microbe interactions in the rhizosphere. J Exp Bot. 2005;56:1729–1739. doi: 10.1093/jxb/eri205. [DOI] [PubMed] [Google Scholar]
- Mougel C, Offre P, Ranjard L, Corberand T, Gamalero E, Robin C, et al. Dynamic of the genetic structure of bacterial and fungal communities at different developmental stages of Medicago truncatula Gaertn. cv. Jemalong line J5. New Phytol. 2006;170:165–175. doi: 10.1111/j.1469-8137.2006.01650.x. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15:473. [Google Scholar]
- Nazoa P, Vidmar JJ, Tranbarger TJ, Mouline K, Damiani I, Tillard P, et al. Regulation of the nitrate transporter gene AtNRT2.1 in Arabidopsis thaliana: responses to nitrate, amino acids and developmental stage. Plant Mol Biol. 2003;52:689–703. doi: 10.1023/a:1024899808018. [DOI] [PubMed] [Google Scholar]
- Neal AL, Ahmad S, Gordon-Weeks R, Ton J. Benzoxazinoids in root exudates of maize attract pseudomonas putida to the rhizosphere. PLoS One. 2012;7:e35498. doi: 10.1371/journal.pone.0035498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoli A, Zambolim L, TJP Junior, Vieira RF, Teixeira H, JES Carneiro. Resistance of advanced common bean lines to Fusarium root rot. Trop plant pathol. 2012;37:393–398. [Google Scholar]
- Novoa R, Loomis RS. Nitrogen and plant production. Plant and Soil. 1981;58:177–204. [Google Scholar]
- Oksanen J, Blanchet G, Kindt R, Legendre P, Minchin PR, O'Hara RB, et al. 2012vegan: Community Ecology PackageR package version 2.0-4.
- Poretsky RS, Gifford S, Rinta-Kanto J, Vila-Costa M, Moran MA. Analyzing gene expression from marine microbial communities using environmental transcriptomics. J Vis Exp. 2009;24:e1086. doi: 10.3791/1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradella S, Hans A, Sproer C, Reichenbach H, Gerth K, Beyer S. Characterisation, genome size and genetic manipulation of the myxobacterium Sorangium cellulosum So ce56. Arch Microbiol. 2002;178:484–492. doi: 10.1007/s00203-002-0479-2. [DOI] [PubMed] [Google Scholar]
- Prasanna R, Jaiswal P, Nayak S, Sood A, Kaushik BD. Cyanobacterial diversity in the rhizosphere of rice and its ecological significance. Indian J Microbiol. 2009;49:89–97. doi: 10.1007/s12088-009-0009-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EE, Glazebrook J, Ausubel FN. Mode of action of the Arabidopsis thaliana phytoalexin camalexin and its role in Arabidopsis-pathogen interactions. Mol Plant Microbe In. 1996;9:748–757. doi: 10.1094/mpmi-9-0748. [DOI] [PubMed] [Google Scholar]
- Rossato L, Laine P, Ourry A. Nitrogen storage and remobilization in Brassica napus L. during the growth cycle: nitrogen fluxes within the plant and changes in soluble protein patterns. J Exp Bot. 2001;52:1655–1663. [PubMed] [Google Scholar]
- Rusterucci C, Zhao Z, Haines K, Mellersh D, Neumann M, Cameron RK. Age-related resistance to Pseudomonas syringae pv. tomato is associated with the transition to flowering in Arabidopsis and is effective against Peronospora parasitica. Physiol Mol Plant P. 2005;66:222. [Google Scholar]
- Schatz A, Bugie E, Waksman SA. Streptomycin, a substance exhibiting antibiotic activity against gram-positive and gram-negative bacteria. 1944. Clin Orthop Relat Res. 2005. pp. 3–6. [DOI] [PubMed]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvakumar G, Panneerselvam P, Ganeshamurthy AN, Maheshwari DK.2012Bacterial mediated alleviation of abiotic stress in cropsIn: Maheshwari DK (ed)Bacteria in Agrobiology: Stress Management Springer: New York, NY, USA; 205–224. [Google Scholar]
- Shade A, Gregory Caporaso J, Handelsman J, Knight R, Fierer N. A meta-analysis of changes in bacterial and archaeal communities with time. Isme J. 2013;7:1493–1506. doi: 10.1038/ismej.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw CH. Swimming against the tide: Chemotaxis in Agrobacterium. BioEssays. 1991;13:25–29. doi: 10.1002/bies.950130105. [DOI] [PubMed] [Google Scholar]
- Shi S, Richardson AE, O'Callaghan M, DeAngelis KM, Jones EE, Stewart A, et al. Effects of selected root exudate components on soil bacterial communities. FEMS Microbiol Ecol. 2011;77:600–610. doi: 10.1111/j.1574-6941.2011.01150.x. [DOI] [PubMed] [Google Scholar]
- Stacey G, Sanjuan J, Luka S, Dockendorff T, Carlson RW. Signal exchange in the Bradyrhizobium-soybean symbiosis. Soil Biol and Biochem. 1995;27:473. [Google Scholar]
- Sy A, Giraud E, Jourand P, Garcia N, Willems A, De Lajudie P, et al. Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J Bacteriol. 2001;183:214–220. doi: 10.1128/JB.183.1.214-220.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokala RK, Strap JL, Jung CM, Crawford DL, Salove MH, Deobald LA, et al. Novel plant-microbe rhizosphere interaction involving Streptomyces lydicus WYEC108 and the pea plant (Pisum sativum) Appl Environ Microbiol. 2002;68:2161–2171. doi: 10.1128/AEM.68.5.2161-2171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TR, Ramakrishnan K, Walshaw J, Heavens D, Alston M, Swarbreck D, et al. Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. Isme J. 2013;7:2248–2258. doi: 10.1038/ismej.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urich T, Lanzen A, Qi J, Huson DH, Schleper C, Schuster SC. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One. 2008;3:e2527. doi: 10.1371/journal.pone.0002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden MGA, Bardgett RD, Van Straalen NM. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett. 2008;11:296–310. doi: 10.1111/j.1461-0248.2007.01139.x. [DOI] [PubMed] [Google Scholar]
- Van Spanning RJM, Delgado MJ, Richardson DJ.2005The nitrogen cycle: denitrification and its relationship to N2 fixationIn: Werner D, Newton WE (eds)Nitrogen Fixation in Agriculture, Forestry, Ecology, and the Environment Springer: Netherlands; 277–342. [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. App Environ Microb. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward NL, Challacombe JF, Janssen PH, Henrissat B, Coutinho PM, Wu M, et al. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl Environ Microbiol. 2009;75:2046–2056. doi: 10.1128/AEM.02294-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Frank DN, Robertson CE, Hung SS, Markle J, Canty AJ, et al. Generation and analysis of a mouse intestinal metatranscriptome through Illumina based RNA-sequencing. PLoS One. 2012;7:e36009. doi: 10.1371/journal.pone.0036009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wang G, Jin J, Liu J, Zhang Q, Liu X. Bacterial communities in soybean rhizosphere in response to soil type, soybean genotype, and their growth stage. Soil Biol Biochem. 2009;41:919. [Google Scholar]
- Yamada T, Letunic I, Okuda S, Kanehisa M, Bork P. iPath2.0: interactive pathway explorer. Nucleic Acids Res. 2011;39:W412–W415. doi: 10.1093/nar/gkr313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousuf B, Keshri J, Mishra A, Jha B. Application of targeted metagenomics to explore abundance and diversity of CO(2)-fixing bacterial community using cbbL gene from the rhizosphere of Arachis hypogaea. Gene. 2012;506:18–24. doi: 10.1016/j.gene.2012.06.083. [DOI] [PubMed] [Google Scholar]
- Zahran HH. Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol Mol Biol Rev. 1999;63:968–989. doi: 10.1128/mmbr.63.4.968-989.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki K, Misaghi IJ, Heydari A. Control of Cotton Seedling Damping-off in the Field by Burkholderia (Pseudomonas) cepacia. Plant Disease. 1998;82:291–293. doi: 10.1094/PDIS.1998.82.3.291. [DOI] [PubMed] [Google Scholar]
- Zamioudis C, Pieterse CM. Modulation of host immunity by beneficial microbes. Mol Plant Microbe Interact. 2012;25:139–150. doi: 10.1094/MPMI-06-11-0179. [DOI] [PubMed] [Google Scholar]
- Zhang J, Subramanian S, Stacey G, Yu O. Flavones and flavonols play distinct critical roles during nodulation of Medicago truncatula by Sinorhizobium meliloti. Plant J. 2009;57:171–183. doi: 10.1111/j.1365-313X.2008.03676.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lu Z, Liu S, Yang Y, He Z, Ren Z, et al. Geochip-based analysis of microbial communities in alpine meadow soils in the Qinghai-Tibetan plateau. BMC Microbiol. 2013;13:72. doi: 10.1186/1471-2180-13-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla G, Badri DV, Bakker MG, Manter DK, Vivanco JM. Soil microbiomes vary in their ability to confer drought tolerance to Arabidopsis. App Soil Ecol. 2013;68:1. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.