Abstract
Molecular surveys in planktonic marine systems have unveiled a large novel diversity of small protists. A large part of this diversity belongs to basal heterotrophic stramenopiles and is distributed in a set of polyphyletic ribogroups (described from rDNA sequences) collectively named as MAST (MArine STramenopiles). In the few groups investigated, MAST cells are globally distributed and abundant bacterial grazers, therefore having a putatively large impact on marine ecosystem functioning. The main aim of this study is to reevaluate the MAST ribogroups described so far and to determine whether additional groups can be found. For this purpose, we used traditional and state-of-the-art molecular tools, combining 18S rDNA sequences from publicly available clone libraries, single amplified genomes (SAGs) of planktonic protists, and a pyrosequencing survey from coastal waters and sediments. Our analysis indicated a final set of 18 MAST groups plus 5 new ribogroups within Ochrophyta (named as MOCH). The MAST ribogroups were then analyzed in more detail. Seven were typical of anoxic systems and one of oxic sediments. The rest were clearly members of oxic marine picoplankton. We characterized the genetic diversity within each MAST group and defined subclades for the more diverse (46 subclades in 8 groups). The analyses of sequences within subclades revealed further ecological specializations. Our data provide a renovated framework for phylogenetic classification of the numerous MAST ribogroups and support the notion of a tight link between phylogeny and ecological distribution. These diverse and largely uncultured protists are widespread and ecologically relevant members of marine microbial assemblages.
Keywords: 18S rDNA, ecological transitions, MAST, MOCH, pyrotags, SAG
Introduction
Molecular surveys have been instrumental for decoding the wide diversity of microorganisms with crucial ecosystem roles (Falkowski et al., 2008). They have unveiled novel lineages within the three domains of life, bacteria (Achtman and Wagner, 2008), archaea (Robertson et al., 2005) and eukaryotes (Epstein and López-García, 2008), thus opening new avenues for evolutionary and ecological explorations. Within eukaryotes, novel lineages at the highest taxonomic rank were found, such as the Picozoa (Not et al., 2007; Seenivasan et al., 2013) or the rappemonads (Kim et al., 2011), and probably more wait to be unveiled. This new diversity was detected from small subunit rDNA environmental sequences forming monophyletic clades (named as ribogroups) of uncertain affiliation. Then, additional tools were used for their characterization, such as FISH (fluorescence in situ hybridization) (Not et al., 2007), single-cell genomics (Yoon et al., 2011), or isolation in pure culture (Seenivasan et al., 2013). Besides the striking identification of novel high-rank diversity, molecular surveys also identified novelty at all phylogenetic levels, from novel groups within supergroups, to additional diversity within defined taxa (Massana and Pedrós-Alió, 2008).
In molecular surveys of marine planktonic picoeukaryotes, a substantial number of the 18S rDNA sequences formed novel lineages within the supergroups alveolates and stramenopiles, named as MALV (MArine ALveolates) (Guillou et al., 2008) and MAST (Massana et al., 2004). On average, they encompassed 32% and 13% of sequences, respectively, in picoeukaryote surveys (Massana and Pedrós-Alió, 2008). MAST groups were not monophyletic, but formed independent lineages among basal heterotrophic stramenopiles. Twelve MAST groups were first defined (Massana et al., 2004), and seven more were proposed later (Zuendorf et al., 2006; Orsi et al., 2011). Also, a parallel classification was proposed soon after the first study (Richards and Bass, 2005). With only one exception (Cavalier-Smith and Scoble, 2013), this diversity remains uncultured, so determining their cell physiology and ecological attributes is one of the main challenges for future ecological studies. Some groups were investigated in detail, mainly by FISH using group-specific oligonucleotide probes, and MAST cells turned out to be small (2–5 μm) heterotrophic flagellates, widely distributed and active bacterial grazers (Massana et al., 2006; Lin et al., 2012; Piwosz et al., 2013). However, the particular cell size, distribution and activity varied among the investigated groups (Massana et al., 2009; Piwosz and Pernthaler, 2010), revealing a tight link between phylogenetic and functional diversity.
The main aim of this work was to reevaluate the phylogeny, diversity and ecology of MAST ribogroups, using three independent surveys of 18S rDNA sequences derived from traditional and new molecular tools. First, we performed an exhaustive search in the GenBank database for stramenopile sequences derived from both cultures and natural samples obtained through eukaryotic clone libraries. Second, we analyzed a data set of 454 pyrotags from European coastal sites, including oxygenated seawater samples (fractionated as pico-, nano- and microplankton), sediments, and anoxic plankton. This pyrotag data set has already been used to study MAST diversity, distribution and ecology (Logares et al., 2012). Third, we used a collection of single amplified genomes (SAGs) from single-cell protists (Heywood et al., 2011; Martinez-Garcia et al., 2012), which provide a cell-by-cell view of community structure. Our specific objectives were (1) to reevaluate the phylogenetic robustness of described MAST ribogroups, (2) to detect new ribogroups within stramenopiles, (3) to characterize the genetic diversity of each MAST ribogroup and define specific subclades and (4) to derive ecological preferences for the identified ribogroups and subclades by inspecting the samples from where sequences came from. This robust phylogenetic framework is needed for future research addressing the ecology of specific groups, both by using specific probes and by inspecting high-throughput sequencing data sets.
Materials and methods
Stramenopile sequences derived from GenBank
A reference data set of 370 near complete 18S rDNA sequences was compiled to represent all stramenopiles lineages. This data set was used to identify similar sequences in the NCBI-GenBank database (Release 183.0; April 2011) using a local BLAST search (Altschul et al., 1990) run with relaxed parameters (-evalue 0.0001 -max_target_seqs 10 000 -perc_identity 90). This yielded 6373 sequences between 700 and 2000 bp in size and with a bit score of >1000. Non-stramenopile sequences were detected using KeyDNATools (Guillou et al., 2013) and phylogeny; only a few were kept as outgroups. The GenBank retrieved data set was aligned with mothur (Schloss et al., 2009) based on the SILVA 108 template alignment (Quast et al., 2013) and chimeras (∼350 sequences) were identified by ChimeraSlayer (Haas et al., 2011). Studies reporting protist molecular surveys were checked to confirm that all stramenopile diversity was represented in this data set, and a few divergent lineages were added (such as MH-IX from Richards and Bass, 2005). Finally, sequences shorter than 1100 bp were removed, resulting in a final GenBank data set of ∼4000 entries.
Stramenopile sequences derived from SAGs
During the Tara-Oceans expedition (Karsenti et al., 2011), surface and deep chlorophyll maximum samples were collected at several sites in the Mediterranean Sea and the Indian Ocean, cryopreserved with glycine betaine, and shipped to the Bigelow Laboratory for Ocean Sciences. Two single-cell sorts by flow cytometry were done per sample, a first based on pigmented cells and a second based on DNA staining of unpigmented cells larger than bacteria (Heywood et al., 2011). Single plastidic or aplastidic cells were deposited into 384-well plates, subjected to MDA (multiple displacement amplification), and the genomic DNA was PCR screened with the 18S rDNA eukaryote primers 528f and EukB (Heywood et al., 2011). Sequences (∼1200 bp) were obtained from 903 SAGs and identified by BLAST (complete analysis in Sieracki et al., in preparation). Here, we considered 568 SAGs affiliating to stramenopiles.
Stramenopile sequences derived from BioMarKs pyrotags
During the BioMarKs project (http://www.biomarks.eu/), samples were collected in six European coastal sites at different water column depths (surface and deep chlorophyll maximum) in three size fractions (picoplankton (0.8–3 μm), nanoplankton (3–20 μm) and microplankton (20–2000 μm)), and at sediments (Logares et al., 2012). DNA and RNA extracts were obtained from the samples, the later reverse transcribed to cDNA, and protist diversity was assessed by pyrosequencing the PCR products obtained using V4 18S rDNA primers (Stoeck et al., 2010). Sequences thus obtained were named as pyrotags. The data set was curated by removing short or low quality reads and chimeras (Logares et al., 2012). High-quality pyrotags (∼1.5 million) were clustered at 97% similarity with USEARCH (Edgar, 2010) to obtain the operational taxonomic unit (OTU) table (pyrotags per sample). OTUs were compared with reference data sets (including the MAST data set generated here), and assigned to a taxonomic group when they had an e-value of below 10−100 against a reference sequence. This assignation was validated by phylogeny. Metazoan pyrotags were removed from the OTU table to calculate the contributions of different groups.
Phylogenetic analysis
Maximum likelihood trees with the aligned GenBank retrieved data set were done using RAxML (v7.2.8; Stamatakis, 2006) with the GTR+G evolutionary model under rapid hill climbing mode. Alternative trees were run to select the best likelihood tree and bootstrap was calculated with pseudoreplicates. Bootstrap values were added to the best tree with RAxML and the final trees were edited with Archaeopterix (Han and Zmasek, 2009). A series of preliminary trees (100 replicates for topology and bootstrap) were done to curate the data set by individual BLAST inspection of sequences forming long branches. Sometimes, similar sequences shorter than 1100 bp were identified and incorporated to the data set to break up the long branches (∼50 cases). In other instances, long-branch sequences were removed because they were chimeras (∼30 cases), had introns that disturbed the alignment (∼20 cases), or still remained unique (closest sequence below 92% similarity; ∼20 cases). The curated data set was merged with the 568 stramenopile sequences from SAGs and run with USEARCH to keep the longest sequence that represent OTUs clustered at 98% similarity. The final set of 1226 sequences, including 20 alveolate and rhizaria outgroups, was aligned de novo with MAFFT v6.903b (Katoh et al., 2002) using the E-INS-i algorithm, slightly edited at both ends (2720 final positions) and used to construct an ML tree with 1000 replicates for topology and bootstrap. Phylogenies were run in the Mare Nostrum supercomputer cluster (http://www.bsc.es/marenostrum-support-services).
Final curated data set and intra-group analyses
Once the ribogroups were well defined, a final check on NCBI release 193.0 (December 2012) was done to obtain the final list of 1762 MAST and 36 MOCH (Marine OCHrophyta) sequences (generally larger than 800 bp). Basic properties of the sites from which sequences originated, such as the habitat (marine or freshwater), substrate (plankton or sediment), oxygen content and marine region, were taken from the GenBank entry. Phylogenetic trees pointed to intragroup divergent sequences, which were often slightly edited by removing variable positions at the ends, likely due to sequencing errors or to short chimeric fragments (506 sequences edited). Then, the final MAST data set was exhaustively analyzed group by group. First, sequences were aligned by MAFFT using the E-INS-i algorithm and ML trees were done with 100 replicates for topology and bootstrap. Second, MAST groups with a substantial genetic diversity were subdivided into subclades supported by high bootstrap values. Third, USEARCH was used to cluster group sequences in OTUs defined at distinct similarity thresholds. And fourth, ML trees were done after adding the pyrotags to previous alignments with MAFFT, in order to assign the pyrotags to subclades. The final data sets of MAST and MOCH sequences are shown in Supplementary Tables S1 and S2.
Results
High-rank phylogeny of stramenopiles
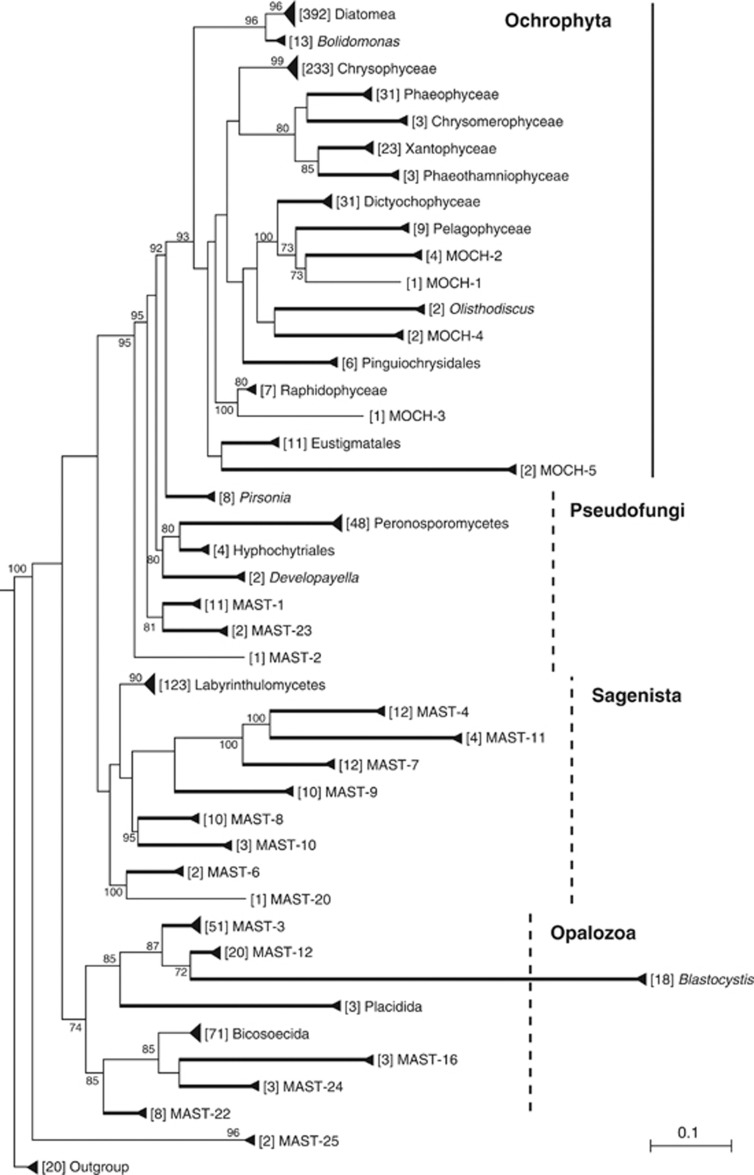
A phylogenetic tree with curated 18S rDNA sequences from GenBank and from the Tara-Oceans SAG collection was done with the aim of recovering all existing lineages within stramenopiles as well as minimizing artifacts like chimeras or unclear novel diversity (Figure 1). All groups were recovered with very high bootstrap values (except Bicosoecida, which had high support in other trees). MAST ribogroups, forming distinct lineages within basal heterotrophic stramenopiles, were reevaluated using this tree. Some previously defined ribogroups did not appear in the final tree because they affiliated to formal taxa or were based on chimeric sequences, and therefore its use should be abandoned. This applies to one of the 12 seminal groups (MAST-5), to most MAST groups later described (except MAST-16) and to many of the mystery heterokont (MH) clades (Table 1). Three MH clades were included in the MAST groups in agreement with the original authors (T Richards, personal communication): MH-IV (excluding ME1-24 that was MAST-6) renamed as MAST-20, MH-IX renamed as MAST-21 (not shown in the tree due to its long branch and ambiguous placement) and MH-XII renamed as MAST-22. A second aim of this phylogeny was identifying new ribogroups. Surprisingly, only three additional MAST groups were found (MAST-23 to -25). In addition, five new ribogroups emerged within Ochrophyta, the large division including all chlorophyll-containing stramenopiles, and were named as MOCH for Marine Ochrophyta (Figure 1). The number of sequences from independent molecular surveys populating the final set of MAST and MOCH ribogroups is shown in Table 2.
Figure 1.
High-rank phylogenetic tree of stramenopiles based on the 18S rDNA gene. The tree includes formal taxa described in Adl et al. (2012) and ribogroups within basal stramenopiles (MAST) and Ochrophyta (MOCH). Only MAST-21 is not shown. Numbers in brackets before group names indicate the sequences within the group (distinct OTU98). Groups displaying 100% bootstrap values after 1000 pseudoreplicates are shown as thickened lines. Bootstrap values above 70% for the remaining nodes are also shown. Some large clades are marked, as dashed lines if the clade requires validation. The scale bar indicates 0.1 substitutions per position.
Table 1. List of described MAST or MH ribogroups that are not supported phylogenetically and should be abandoned.
| Ribogroup | Reference | Accession # | Comment |
|---|---|---|---|
| MAST-5 | Massana et al. (2004) | AF290070 | Chimera (Labyrinthulomycetes and MALV-II) |
| MH I | Richards and Bass (2005) | Is MAST-5 | |
| MH II | Richards and Bass (2005) | Is MAST-4 | |
| MH III | Richards and Bass (2005) | Is MAST-7 | |
| MH V | Richards and Bass (2005) | AY180031 | Belongs to Peronosporomycetes |
| MH VI | Richards and Bass (2005) | AY046768 | Belongs to Peronosporomycetes |
| MH VII | Richards and Bass (2005) | Is MAST-1 | |
| MH VIII | Richards and Bass (2005) | Is MAST-8 | |
| MH X | Richards and Bass (2005) | AY046660 | Belongs to Peronosporomycetes |
| MH XI | Richards and Bass (2005) | Is MAST-3 | |
| MH XIII | Richards and Bass (2005) | Is MAST-2 | |
| MH XIV | Richards and Bass (2005) | AY919731 | Chimera (Bicosoecida and Cryptophyceae) |
| MH XV | Richards and Bass (2005) | Is MAST-12 | |
| MAST-13 | Zuendorf et al. (2006) | DQ103774 | Belongs to Bicosoecida |
| MAST-14 | Orsi et al. (2011) | GU823193 | Belongs to Bicosoecida |
| MAST-15 | Orsi et al. (2011) | GU823072 | Belongs to Diatomea |
| MAST-17 | Orsi et al. (2011) | GU823246 | Belongs to Labyrinthulomycetes |
| MAST-18 | Orsi et al. (2011) | GU823263 | Belongs to MAST-3 |
| MAST-19 | Orsi et al. (2011) | GU823065 | Belongs to Labyrinthulomycetes |
Abbreviations: MAST, MArine STramenopiles; MH, mystery heterokont; MALV, MArine ALveolates.
Table 2. Number of sequences of MAST and MOCH ribogroups in GenBank (longer than ∼800 bp), in the SAG collection (derived from aplastidic or plastidic sorts), and in the BioMarKs data set.
| Ribogroup |
GenBank sequences |
SAG collection |
BioMarKs pyrotags |
||||
|---|---|---|---|---|---|---|---|
| n | OTU97 | % Clade | n (aplastidic) | n (plastidic) | n | OTU97 | |
| • Described MAST | |||||||
| MAST-1 | 421 | 10 | 88 | 14 | — | 7220 | 17 |
| MAST-2 | 55 | 1 | 97 | — | — | 1662 | 7 |
| MAST-3 | 339 | 42 | 83 | 80 | 2 | 11 553 | 103 |
| MAST-4 | 317 | 5 | 93 | 167 | 6 | 2941 | 8 |
| MAST-6 | 7 | 5 | 94 | — | — | 1112 | 17 |
| MAST-7 | 193 | 7 | 90 | 47 | 1 | 3601 | 11 |
| MAST-8 | 59 | 8 | 92 | 7 | — | 1010 | 16 |
| MAST-9 | 27 | 8 | 87 | 10 | — | 408 | 15 |
| MAST-10 | 13 | 1 | 97 | 2 | — | 511 | 2 |
| MAST-11 | 3 | 2 | 95 | 17 | — | 317 | 3 |
| MAST-12 | 109 | 20 | 85 | 3 | — | 1096 | 37 |
| MAST-16 | 4 | 2 | 96 | — | — | — | — |
| • Renamed MAST | |||||||
| MAST-20 (MH-IV) | 40 | 1 | 99 | — | — | 53 | 3 |
| MAST-21 (MH-IX) | 22 | 1 | 97 | — | — | — | — |
| MAST-22 (MH-XII) | 141 | 7 | 88 | — | — | 64 | 4 |
| • New MAST | |||||||
| MAST-23 | 7 | 2 | 96 | — | — | — | — |
| MAST-24 | 3 | 2 | 88 | — | — | 26 | 1 |
| MAST-25 | 2 | 2 | 93 | 6 | — | 223 | 2 |
| • New MOCH | |||||||
| MOCH-1 | 5 | 1 | 97 | — | 2 | 422 | 11 |
| MOCH-2 | 9 | 5 | 94 | — | 6 | 4358 | 44 |
| MOCH-3 | 4 | 1 | 98 | 1 | — | 653 | 4 |
| MOCH-4 | 7 | 2 | 93 | 4 | — | 115 | 2 |
| MOCH-5 | 11 | 2 | 95 | — | — | 366 | 7 |
Abbreviations: MAST, MArine STramenopiles; MH, mystery heterokont; MOCH, Marine OCHrophyta; OTU, operational taxonomic unit; SAG, single amplified genome.
For GenBank sequences, the number of OTUs at 97% similarity (OTU97) is shown, as well as the similarity that collapses the whole group in a single OTU (% clade). For pyrotags, OTU97 need to be found in at least two different samples.
While high bootstrap values characterized the groups shown in Figure 1, the relative order among them was less clear. One of the well-supported large clades was the Ochrophyta (93% bootstrap), and within them some relationships were recurrent: Diatomea and Bolidomonas (96%); Phaeophyceae, Chrysomerophyceae, Xantophyceae and Phaeothamniophyceae (80%); Dictyochophyceae, Pelagophyceae, MOCH-1 and -2 (100%). The placement of the remaining groups shifted in different trees. Non-ochrophyta stramenopiles were separated into three phylogenetic regions, following Cavalier-Smith and Scoble (2013). First, the Pseudofungi formed a set of separate lineages basal to Ochrophyta that included Pirsonia, Peronosporomyctes, Hyphochytriales, Developayella, and several MASTs. They were never monophyletic and their relative order shifted in different trees. Second, the Sagenista formed a clade binding Labyrinthulomycetes with many MAST groups. Although in the tree shown here this large clade did not have bootstrap values, it was generally supported in previous trees. Third, the Opalozoa formed another large clade uniting the remaining groups (74% bootstrap). Blastocystis, Placidida, MAST-3 and -12 were always related, whereas Bicosoecida related with other MAST groups. In the tree shown here, MAST-25 formed the most basal branch (but in other trees affiliated to Sagenista or Opalozoa).
New described ribogroups
Only three new MAST ribogroups were defined here, and they were formed by a limited number of sequences (Table 2). MAST-23 was retrieved in two sites with deep anoxic waters, the Cariaco Basin (Atlantic) and the Saanlich Inlet (Pacific), whereas was undetected in the pyrotag survey of European coastal waters. MAST-24 included GenBank sequences from anoxic waters (Cariaco Basin and Black Sea) together with pyrotags from anoxic waters. MAST-25 included GenBank sequences from surface Pacific picoplankton as well as SAGs from the Indian Ocean and coastal pyrotags. Whereas MAST-23 and MAST-24 seem to be anaerobic protists, MAST-25 likely is an aerobic heterotrophic picoplankter.
Five MOCH groups were described here at different positions within Ochrophyta (Figure 1). Each group included GenBank sequences from different marine regions, and all were well represented by BioMarKs pyrotags and, except MOCH-5, also by SAG sequences (Table 2). The BioMarKs data set indicated that MOCH cells formed part of the picoplankton and nanoplankton (Supplementary Table S3), and pointed MOCH-2 as particularly abundant (∼0.8% of pyrotags in both size fractions). MOCH-1 and -2 contained SAGs sorted as plastidic cells, and MOCH-5 included two cultures of phototrophs (now lost). So these three groups most likely represent new algal lineages. Conversely, MOCH-3 and MOCH-4 probably represent new heterotrophic lineages, since SAGs within them were sorted as aplastidic and some of their sequences derived from deep and dark samples.
Ecological analysis of MAST ribogroups
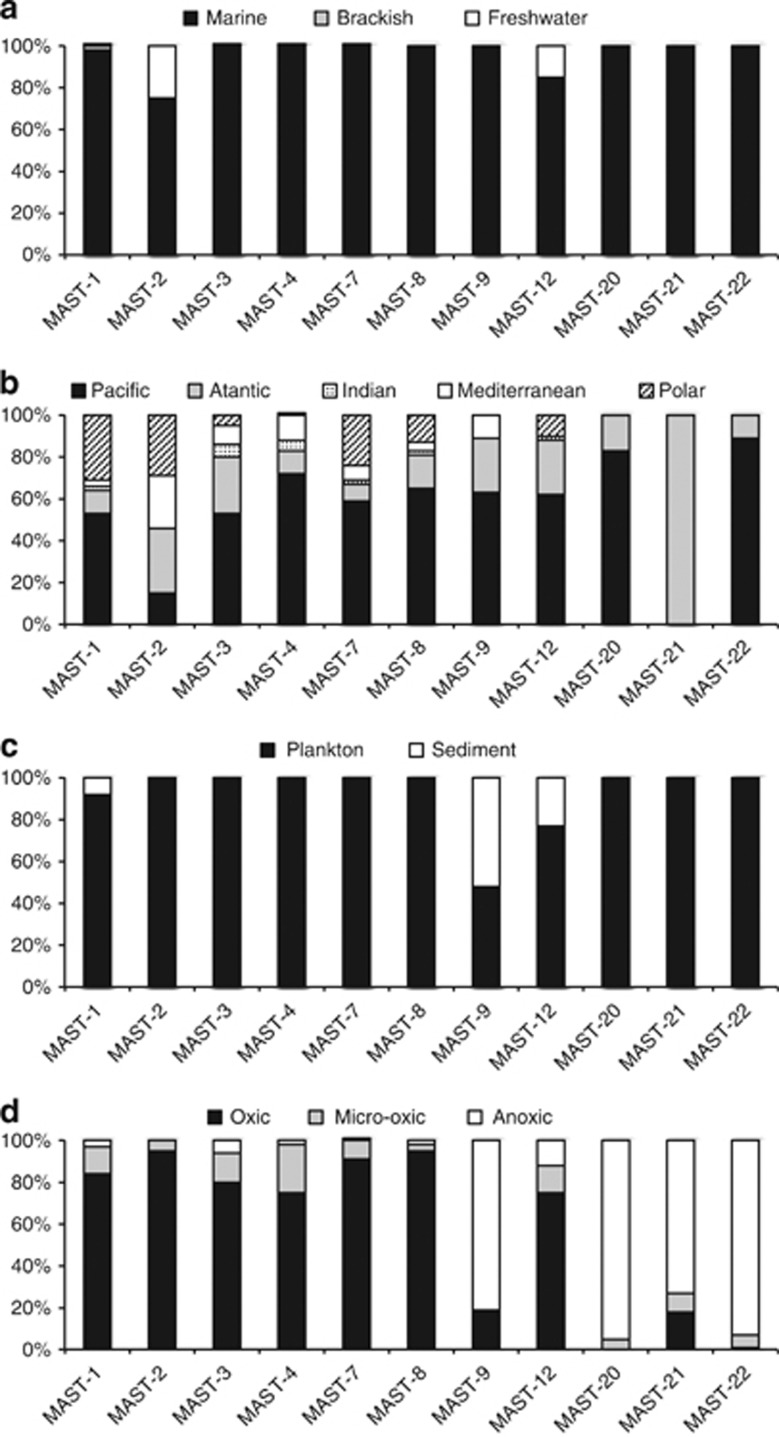
The ecological settings for each of the 18 MAST ribogroups were investigated by analyzing three independent data sets: sample properties originating GenBank sequences (Figure 2), the presence and sorting criteria of SAGs prepared from planktonic marine cells (Table 2), and the contribution of pyrotags in an European coastal survey (Table 3). Some clear patterns were inferred. First, the exhaustive GenBank screening did not reveal any ribogroup typical of freshwater systems. In addition, MAST groups were exclusively composed by marine sequences, with notable exceptions in MAST-2 and -12 (Figure 2a). Second, there were no particular geographic restrictions for any of the groups, which were consistently retrieved from samples in all oceans (Figure 2b). Third, four groups, MAST-1, -3, -4 and -7, dominated the data sets, contributing to 72% of GenBank sequences, 88% of SAGs and 80% of pyrotags (Table 2). Fourth, most groups derived from planktonic samples and were not detected in sediments, with the exception of MAST-1, -9 and -12 (Figure 2c). The BioMarKs survey demonstrated that MAST-6 and -20 were also important in sediments. Fifth, some groups were exclusive of anoxic habitats: MAST-16, -20, -21, -22, -23 and -24. These contributed little to the BioMarKs data set (which had a lower sequencing effort in anoxic habitats) and included no SAG (which derive from oxic waters). MAST-9 and -12 appeared both in oxic and in anoxic systems, whereas the rest were exclusive of oxic samples (Figure 2d). Virtually, all these oxic ribogroups included SAG representatives and were well represented in the BioMarKs data set, which also allowed inferring the cell size of these particular lineages. Thus, some were clearly picoplanktonic (MAST-4, -7, -8, -10, -11 and -25), whereas others were also nanoplanktonic (MAST-1, -2 and -3). Finally, the contribution of ribogroups was always larger than that of known taxa of basal heterotrophic stramenopiles (Table 2). Ribogroups accounted for a significant share of the picoplankton signal (5.6% on average) and had a lower contribution to nanoplankton (2.1%) and microplankton (0.2%).
Figure 2.
Ecological preferences of 11 MAST ribogroups inferred from sample attributes extracted from GenBank entries. Only ribogroups with a significant number of sequences (>20) are shown. Each panel shows the distribution of sequences within each group according to salinity (a), geographic origin (b), substrate (c) and oxygen (d).
Table 3. Relative abundance (% of pyrotags) of MAST ribogroups, basal taxa within stramenopiles, and unassigned stramenopiles in the different sample types of the BioMarKs data set.
| Ribogroup | Picoplankton | Nanoplankton | Microplankton | Anoxic planktona | Sediments |
|---|---|---|---|---|---|
| MAST-1 | 1.37 | 0.92 | 0.01 | 0.13 | 0.09 |
| MAST-2 | 0.26 | 0.22 | 0.01 | 0.05 | — |
| MAST-3 | 1.36 | 0.56 | 0.10 | 0.09 | 0.12 |
| MAST-4 | 0.75 | 0.04 | — | — | — |
| MAST-6 | 0.03 | 0.02 | — | — | 1.44 |
| MAST-7 | 1.14 | 0.12 | — | — | — |
| MAST-8 | 0.28 | 0.09 | — | — | — |
| MAST-9 | 0.08 | 0.02 | — | 0.59 | 0.01 |
| MAST-10 | 0.11 | 0.01 | — | — | — |
| MAST-11 | 0.04 | — | — | — | — |
| MAST-12 | 0.13 | 0.04 | 0.03 | 0.06 | 0.13 |
| MAST-20 | — | — | — | — | 0.04 |
| MAST-22 | — | — | — | 0.10 | 0.01 |
| MAST-24 | — | — | — | 0.06 | — |
| MAST-25 | 0.03 | — | — | — | — |
| All ribogroups | 5.59 | 2.05 | 0.17 | 1.09 | 1.83 |
| Bicosoecida | 0.14 | 0.03 | 0.06 | 0.30 | 0.30 |
| Developayella | — | — | — | — | 0.07 |
| Hyphochytriales | — | — | — | — | 0.04 |
| Peronosporomycetes | 0.08 | 0.06 | 0.14 | 0.29 | 0.96 |
| Pirsonia | 0.05 | 0.04 | — | — | 1.01 |
| Labyrinthulomycetes | 0.70 | 0.20 | 0.12 | 0.64 | 1.97 |
| All known taxa | 0.97 | 0.32 | 0.32 | 1.22 | 4.37 |
| Incertae sedis | 0.40 | 0.07 | 0.03 | 0.09 | 0.15 |
Abbreviation: MAST, MArine STramenopiles.
Picoplankton and nanoplankton combined.
Phylogenetic structure of MAST ribogroups
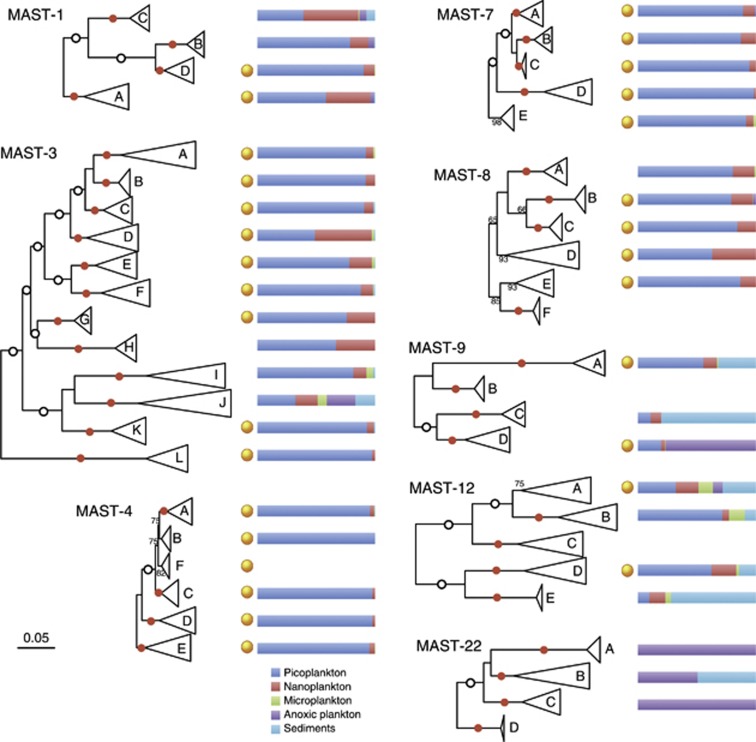
The diversity displayed by each of the 18 ribogroups was not equivalent, as shown by the number of OTUs detected at 97% similarity (OTU97) and by the similarity threshold that collapses the group into a single OTU (Table 2). Some groups, such as MAST-2, -10, -20, and -21, were quite homogenous and exhibited only one OTU97. Among these, MAST-2 was particularly interesting, since its low genetic diversity was structured into several clades, one of which included sequences from freshwater bodies only (Supplementary Figure S1). These sequences were highly similar among them, and differed by only ∼1% with marine MAST-2 sequences. The remaining groups exhibited a substantial genetic structure. In some cases (MAST-6, -11, -16, -23, -24 and -25), this structure was not characterized due to the limited number of GenBank sequences (less than 8, Table 2). For the rest, we did phylogenetic trees to define specific subclades and detect their putative ecological distinctiveness. The eight ribogroups analyzed here (Figure 3) were subdivided into 4–12 subclades and all except one (MAST-4B) were well supported by very high bootstrap values (75–100%). Most subclades (30 out of 46) included SAG representatives as well as pyrotags (41 out of 46), often derived from oxic picoplankton. Subclades generally collapsed at around 96% similarity (threshold that forms a single OTU), although some were more (90%) or less (99%) diverse (Table 4).
Figure 3.
Specific phylogenetic trees of eight MAST ribogroups (those with a large genetic structure and >25 GenBank sequences). Each group is divided into phylogenetic subclades that have generally the highest bootstrap support after 100 pseudoreplicates (red dots when 100%). White dots show 100% bootstrap values for internal nodes. The scale bar indicates 0.05 substitutions per position and applies to all trees. The presence of SAGs from planktonic protists within each subclade is noted by orange dots. The distribution of pyrotags in three planktonic size fractions, anoxic waters (pico- and nanoplankton together) and sediments for each subclade is noted in the horizontal bars.
Table 4. Number of sequences in the three data sets, similarity value that forms a single OTU and the accession number of a representative sequence for subclades shown in Figure 3.
| Ribogroup | Subclade |
Number of sequences |
% Subclade | Representative sequence | ||
|---|---|---|---|---|---|---|
| GenBank | SAGs | Pyrotags | ||||
| MAST-1 | MAST-1A | 177 | 1 | 1167 | 95 | AF363190 |
| MAST-1B | 70 | — | 778 | 97 | JQ782001 | |
| MAST-1C | 155 | — | 3610 | 94 | JQ782036 | |
| MAST-1D | 19 | 13 | 1484 | 97 | JQ781940 | |
| MAST-3 | MAST-3A | 71 | 48 | 176 | 96 | AJ402357 |
| MAST-3B | 6 | 1 | 55 | 98 | AY129068 | |
| MAST-3C | 55 | 3 | 232 | 94 | JQ781904 | |
| MAST-3D | 25 | 2 | 470 | 95 | JN832731 | |
| MAST-3E | 45 | 1 | 1146 | 94 | JQ782025 | |
| MAST-3F | 35 | 17 | 559 | 94 | AY381157 | |
| MAST-3G | 4 | 1 | 317 | 98 | JQ223005 | |
| MAST-3H | 4 | — | 58 | 97 | GU825495 | |
| MAST-3I | 41 | — | 6141 | 92 | JQ781998 | |
| MAST-3J | 29 | — | 1661 | 92 | AY116221 | |
| MAST-3K | 6 | 4 | 378 | 95 | JQ782000 | |
| MAST-3L | 15 | 3 | 263 | 95 | EU371189 | |
| MAST-4 | MAST-4A | 130 | 31 | 1599 | 96 | JQ782049 |
| MAST-4B | 19 | 23 | 6 | 98 | AY129066 | |
| MAST-4C | 13 | 77 | 203 | 96 | EF172962 | |
| MAST-4D | 53 | 3 | 304 | 97 | JQ782028 | |
| MAST-4E | 95 | 33 | 778 | 97 | AF363189 | |
| MAST-4F | 6 | 6 | — | 98 | JQ782011 | |
| MAST-7 | MAST-7A | 149 | 4 | 1071 | 95 | JQ781903 |
| MAST-7B | 24 | 34 | 2103 | 98 | AY129061 | |
| MAST-7C | 9 | 1 | 119 | 98 | JQ781968 | |
| MAST-7D | 5 | 8 | 137 | 95 | JQ781935 | |
| MAST-7E | 5 | 1 | 138 | 98 | AY381191 | |
| MAST-8 | MAST-8A | 42 | — | 233 | 96 | JQ955868 |
| MAST-8B | 5 | 3 | 577 | 98 | JQ782091 | |
| MAST-8C | 4 | 1 | 103 | 98 | GU823321 | |
| MAST-8D | 4 | 1 | 43 | 94 | AY116220 | |
| MAST-8E | 2 | 2 | 33 | 97 | GU823095 | |
| MAST-8F | 2 | — | — | 99 | GU823363 | |
| MAST-9 | MAST-9A | 17 | 7 | 196 | 95 | AY381217 |
| MAST-9B | 6 | — | — | 99 | GU823160 | |
| MAST-9C | 3 | — | 16 | 96 | AB275041 | |
| MAST-9D | 1 | 3 | 195 | 95 | DQ504337 | |
| MAST-12 | MAST-12A | 69 | 1 | 432 | 92 | FR874441 |
| MAST-12B | 15 | — | 177 | 94 | FR874479 | |
| MAST-12C | 15 | — | — | 95 | EU162644 | |
| MAST-12D | 3 | 1 | 100 | 94 | AF167414 | |
| MAST-12E | 3 | — | 52 | 99 | AB275039 | |
| MAST-22 | MAST-22A | 128 | — | 4 | 97 | AY256330 |
| MAST-22B | 3 | — | 46 | 90 | AB505560 | |
| MAST-22C | 4 | — | 15 | 95 | EF526979 | |
| MAST-22D | 6 | — | — | 99 | GU824307 | |
Abbreviations: MAST, MArine STramenopiles; OTU, operational taxonomic unit; SAGs, single amplified genomes.
MAST-1 was subdivided into four subclades. Two of them, MAST-1A and -1C, appeared in both the pico- and nanoplankton, and one MAST-1C lineage (not shown) concentrated GenBank sequences and pyrotags from sediments and anoxic waters. MAST-3 was the most diverse group (42 OTU97 and collapsing at 83%) and was subdivided into 12 subclades. Most were picoplanktonic, except some that were also present in the nanoplankton (MAST-3D and -3H) and MAST-3J that was also important in anoxic waters and sediments. Curiously, this clade contained the cultured Incisomonas uniciliatida. The other described MAST species from the genus Solenicola affiliated within MAST-3I. These two clades with described species, together with MAST-3H, were the only subclades without SAGs. The ribogroups MAST-4, -7 and -8 displayed five to six subclades each with little ecological distinctiveness, since almost all included SAGs and pyrotags from the oxic picoplankton. MAST-4F was one exception, since it did not contain pyrotags and included some GenBank sequences from anoxic waters (3 out of 6). Within MAST-8, the exceptions were MAST-8A (no SAGs), MAST-8D (also in the nanoplankton) and MAST-8F (no SAGs or pyrotags). Again, one of the two GenBank sequences from MAST-8F derived from anoxic plankton.
The remaining three groups were clearly distinct with respect to their ecological preferences (Figure 3), having a strong signal in anoxic plankton and sediments (as noted in Figure 2). Within MAST-9, two clades (MAST-9A and -9C) appeared to be more important in sediments (often anoxic), whereas MAST-9B and -9D appeared to be more important in anoxic plankton. Only MAST-9A and -9D had a significant contribution of pyrotags from oxic picoplankton and SAGs. MAST-12 was the group exhibiting the highest ecological differentiation among subclades. Thus, two subclades appeared as oxic picoplankton (MAST-12B and -12D, the later with one SAG), one was important in sediments (MAST-12E), one seemed to be very versatile, with one SAG and sequences from plankton, sediments and anoxic sites (MAST-12A) and one subclade was typical of freshwater and marine plankton, oxic non-marine sediments, and did not contain any pyrotag or SAG (MAST-12C). Finally, the last ribogroup, MAST-22 was divided into four subclades, being all of them retrieved from anoxic plankton, with the exception of MAST-22B that also included sequences retrieved from sediments.
Discussion
Novel diversity within stramenopiles
Identifying novel high-rank diversity was one of the primary goals of environmental molecular surveys (Dawson and Pace, 2002; Epstein and López-García, 2008) and indeed some high-rank groups were detected and characterized (Not et al., 2007). Nevertheless, often the putative novel groups were based on chimeric sequences or were divergent lineages within well-known taxa (Berney et al., 2004), highlighting the importance of robust phylogenies and careful sequence curation. Here, we did not try to find novel diversity at the highest level but within one of the most important eukaryotic supergroups, the stramenopiles, which include the ribogroups collectively named as MASTs (Massana et al., 2004). By compiling 18S rDNA sequences from different sources we aimed at updating the MAST groups. This was prone to similar artifacts, as many sequences that initially appeared as novel lineages were chimeras or formed long branches within described taxa (generally Labyrinthulomycetes or Bicosoecida). So, a number of the defined MAST ribogroups did not stand our phylogenetic evaluation and were discarded (listed in Table 1). Nevertheless, many of the described MAST groups (Massana et al., 2004; Richards and Bass, 2005; Orsi et al., 2011) remained valid in our phylogenies and were further supported by sequences from single planktonic cells (that is, SAGs) and pyrotags (Table 2).
Surprisingly enough, we identified very little novel diversity within basal heterotrophic stramenopiles, only three additional MAST clades (Table 2). These are rare in environmental surveys (0.7% of GenBank sequences, 1.7% of SAGs and 0.8% of pyrotags), which could explain why they had not been described before; most likely they are minor components among commonly sampled environments. In addition, our extensive phylogenetic survey unveiled novel diversity within Ochrophyta. These novel groups were sometimes pointed out in the original publication but remained unnamed (Le Gall et al., 2008; Bachy et al., 2011; Stock et al., 2012). Here, we defined five novel ochrophyta ribogroups, and one of them (MOCH-2) seemed to be a quantitatively important member of oxic pico- and nanoplankton. On the basis of the provenance of SAGs and GenBank sequences, it appeared that some MOCH groups were novel algal lineages while others were most likely heterotrophs.
One may wonder how much space is left for the discovery of novel diversity within basal heterotrophic stramenopiles. In our phylogenies, we removed single sequences that formed independent lineages, and some may represent low abundant novel taxa, but their validation requires the support of related sequences. Within the BioMarKs data set, pyrotags assigned to stramenopiles without a clear group affiliation (incertae sedis in Table 3) were always a small fraction. For instance, they only accounted for 0.4% of pyrotags in the picoplankton. These unassigned stramenopiles were dominated by a single lineage related to GenBank entry HQ867108, but the phylogenetic placement of these partial sequences could not be resolved. Unassigned pyrotags were also scarce in the other samples suggesting that new MAST groups, if they existed, would be minor members of natural assemblages. Nevertheless, particularly isolated systems like anoxic deep-sea basins or hydrothermal vents are generally less connected and overall underinvestigated (Dawson and Pace, 2002; López-García et al., 2003; Wylezich and Jürgens, 2011), so some could still potentially harbor a significant amount of undescribed diversity.
High-rank phylogeny of stramenopiles
The tree shown in Figure 1 captured the essence of stramenopile diversity sampled using 18S rDNA gene sequencing. Our extensive approach maximized the genetic diversity detected while excluded chimeras and artifacts by manual sequence curation. Stramenopiles appeared formed by a set of formally described taxa, following the recent protist classification (Adl et al., 2012), together with a number of phylogenetically dispersed ribogroups. Overall, 18 MAST and 5 MOCH ribogroups were defined. Interestingly, since sequences were obtained at random from GenBank and SAGs and then clustered at 98% similarity, the number of sequences in the tree provided a rough estimate of the diversity contained in each group. Thus, of a total of 1206 OTU98 displayed in this particular tree, Diatomea was the most diverse taxa (392 OTU98), followed by Chrysophyceae (233), Labyrinthulomycetes (123) and Bicosoecida (71).
The main division of stramenopiles is between Ochrophyta, which unites all photosynthetic taxa, and basal heterotrophic taxa (Cavalier-Smith and Chao, 2006). The dichotomy between photosynthetic ochrophyta and basal heterotrophs was evident when analyzing the SAG collection. Thus, most SAGs from MOCH clades were sorted as plastidic cells (8 of 13), whereas virtually all SAGs from MAST lineages were sorted as aplastidic cells (353 out of 362). This strong dichotomy gives support to name differently ribogroups in these two parts of the tree (MAST versus MOCH). There has been a substantial effort in organizing the stramenopile tree in smaller categories using phylogeny (Cavalier-Smith and Chao, 2006; Cavalier-Smith and Scoble, 2013). Thus, Ochrophyta were subdivided into two large clades, the Khakista (Diatomea plus Bolidomonas) and the Phaeista (the remaining taxa), and this was supported in our tree (Figure 1). Regarding the higher rank taxonomic names for basal stramenopiles, they were generally unstable. Thus, Pseudofungi did not form a monophyletic clade, Sagenista showed little bootstrap support, and only Opalozoa appeared to be robust in Figure 1. Validating these large clades would require multigene phylogenies, which may yield slightly different topologies than when using the single 18S rDNA gene (Riisberg et al., 2009).
The main barriers for MAST colonization
The analysis of GenBank sequences showed that most ribogroups contained sequences from all over the world (Figure 2b), suggesting the absence of marked geographic barriers for dispersal of these minute protists in the marine environment. There were cases when one particular group was absent from a given ocean, such as MAST-4 from polar systems, but this seemed to be an environmental (temperature) barrier (Rodríguez-Martínez et al., 2013). Our results confirmed the perception that small microeukaryotes exhibited the capacity for global dispersal, and that their establishment was determined by environmental conditions.
One striking finding was the absence of any typical freshwater ribogroup, despite freshwater sequences being retrieved in our database mining and also the fact that stramenopiles are considered to be important members of freshwater microbial assemblages (Boenigk and Arndt, 2002). Sampling effort in marine systems is certainly larger, but still there is a number of protist surveys in freshwater systems, and in these surveys the stramenopile sequences most often affiliate within Chrysophyceae and Bicosoecida (Richards et al., 2005). Therefore, MASTs (defined initially as Marine Stramenopiles) remain as typical marine groups. Our analysis, however, identified two ribogroups with a number of sequences deriving from freshwaters (Figure 2a), forming a separate subclade within each group. This points to two examples of marine freshwater transitions, a relatively infrequent phenomenon in the microbial world (Logares et al., 2009). Within MAST-2, almost identical sequences were retrieved from lake plankton in Europe, Asia and America (Supplementary Figure S1). This suggests a recent freshwater adaptation and subsequent global colonization. On the other hand, subclade MAST-12C contained more diverse sequences (clustering at 95%) deriving from more habitats (lake plankton, soils and marine plankton).
Besides the marine-freshwater boundary, there are other strong barriers for protist colonization, being perhaps the most important the oxygen status, strongly conditioning cell metabolism. In our survey, seven MAST groups were typical of anoxic systems, and these affiliated with Pseudofungi, Sagenista or Opalozoa. Additional anoxic sequences were detected in three MAST groups, always restricted to specific subclades: MAST-1C, MAST-3J and MAST-12A. The eight remaining ribogroups were retrieved almost exclusively from oxic sites. These data further support the view of oxygen as a fundamental driver of protist community structure, which creates a marked phylogenetic dichotomy between oxic and anoxic assemblages (Dawson and Pace, 2002; Wylezich and Jürgens, 2011).
Small heterotrophic flagellates in planktonic microbial food webs
A diverse assemblage of MAST cells from 11 separate evolutionary lineages (MAST-1 to -4, -7 to -12 and -25) thrives in surface oceans, arguably one of the largest biomes on earth. They include SAGs sorted as aplastidic planktonic protists, confirming their relevance in marine ecosystems and their heterotrophic nature. The groups studied in more detail, that is, MAST-1, -2, -3 and -4 (Massana et al., 2006, 2009; Lin et al., 2012; Piwosz et al., 2013) have been shown to be bacterial grazers, and it is likely that the other groups are also grazers. Thus, MAST cells form a heterogenous collection of small bacterial grazers that probably exhibit physiological differentiation and adaptation. Just by looking at the OTU number, which provides a conservative estimate of diversity contained in these uncultured protists, 98 OTU97 can be identified. Each of these phylogenetic units deserves a careful inspection, which can be based on FISH probes or targeted sequencing as has been recently done for MAST-4 to evaluate its genetic structure (Rodríguez-Martínez et al., 2012) and biogeography (Rodríguez-Martínez et al., 2013). Also, culturing attempts should be continued (Cavalier-Smith and Scoble, 2013; del Campo et al., 2013), and the potential of genomes obtained from single cells should be exploited by phylogenomics and genome reconstructions (Yoon et al., 2011). The ecological attributes and putative specialization of the different clades is an intriguing aspect to be analyzed. The key open question is what drives and maintains the diversity of these phylogenetically diverse but apparently functionally redundant small bacterial grazers.
Acknowledgments
Financial support has been provided by projects FLAME (CGL2010-16304, MICINN, Spain) and BioMarKs (2008-6530, ERA-net Biodiversa, EU) to RM and by the Marie Curie Fellowship (PIEF-GA-2009-235365) and Juan de la Cierva program (JCI-2010-06594) to RL. Large-scale computing resources were provided by the Barcelona Supercomputer Center and the Spanish Network of Supercomputing (grants BCV-2010-3-0003 and 2011-2-0003/3-0005 to RL and RM). We thank Colomban de Vargas for coordinating the BioMarKs project and the protist team at the Tara-Oceans expedition.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol. 2008;6:431–440. doi: 10.1038/nrmicro1872. [DOI] [PubMed] [Google Scholar]
- Adl SM, Simpson AGB, Lane CE, Lukes J, Bass D, Bowser SS, et al. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bachy C, López-García P, Vereshchaka A, Moreira D. Diversity and vertical distribution of microbial eukaryotes in the snow, sea ice and seawater near the North Pole at the end of the polar night. Front Microbiol. 2011;2:106. doi: 10.3389/fmicb.2011.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berney C, Fahrni J, Pawlowski J. How many novel eukaryotic "kingdoms"? Pitfalls and limitations of environmental DNA surveys. BMC Biol. 2004;2:1–13. doi: 10.1186/1741-7007-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boenigk J, Arndt H. Bacterivory by heterotrophic flagellates: community structure and feeding strategies. Antonie Van Leeuwenhoek. 2002;81:465–480. doi: 10.1023/a:1020509305868. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EEY. Phylogeny and megasystematics of phagotrophic heterokonts (Kingdom Chromista) J Mol Evol. 2006;62:388–420. doi: 10.1007/s00239-004-0353-8. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Scoble JM. Phylogeny of Heterokonta: Incisomonas marina, a uniciliate gliding opalozoan related to Solenicola (Nanomonadea), and evidence that Actinophryida evolved from raphidophytes. Eur J Protistol. 2013;49:328–353. doi: 10.1016/j.ejop.2012.09.002. [DOI] [PubMed] [Google Scholar]
- Dawson SC, Pace NR. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc Natl Acad Sci USA. 2002;99:8324–8329. doi: 10.1073/pnas.062169599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campo J, Not F, Forn I, Sieracki ME, Massana R. Taming the smallest predators of the oceans. ISME J. 2013;7:351–358. doi: 10.1038/ismej.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Epstein S, López-García P. ‘Missing' protists: a molecular prospective. Biodivers Conserv. 2008;17:261–276. [Google Scholar]
- Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive earth's biogeochemical cycles. Science. 2008;320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- Guillou L, Viprey M, Chambouvet A, Welsh RM, Kirkham AR, Massana R, et al. Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata) Environ Microbiol. 2008;10:397–408. doi: 10.1111/j.1462-2920.2008.01731.x. [DOI] [PubMed] [Google Scholar]
- Guillou L, Bachar D, Audic S, Bass D, Berney C, Bittner L, et al. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote Small SubUnit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2013;41:D597–D604. doi: 10.1093/nar/gks1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MV, Zmasek CM. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics. 2009;10:356. doi: 10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heywood JL, Sieracki ME, Bellows W, Poulton NJ, Stepanauskas R. Capturing diversity of marine heterotrophic protists: one cell at a time. ISME J. 2011;5:674–684. doi: 10.1038/ismej.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti E, Acinas SG, Bork P, Bowler C, de Vargas C, Raes J, et al. A holistic approach to marine eco-systems biology. PLoS Biol. 2011;9:e1001177. doi: 10.1371/journal.pbio.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Harrison JW, Sudek S, Jones MDM, Wilcox HM, Richards TA, et al. Newly identified and diverse plastid-bearing branch on the eukaryotic tree of life. Proc Natl Acad Sci USA. 2011;108:1496–1500. doi: 10.1073/pnas.1013337108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall F, Rigaut-Jalabert F, Marie D, Garczarek L, Viprey M, Gobet A, et al. Picoplankton diversity in the South-East Pacific Ocean from cultures. Biogeosci Discuss. 2008;4:2699–2732. [Google Scholar]
- Lin YC, Campbell T, Chung CC, Gong GC, Chiang KP, Worden AZ. Distribution patterns and phylogeny of marine stramenopiles in the North Pacific Ocean. Appl Environ Microbiol. 2012;78:3387–3399. doi: 10.1128/AEM.06952-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logares R, Bråte J, Bertilsson S, Clasen JL, Shalchian-Tabrizi K, Rengefors K. Infrequent marine-freshwater transitions in the microbial world. Trends Microbiol. 2009;17:414–422. doi: 10.1016/j.tim.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Logares R, Audic S, Santini S, Pernice MC, de Vargas C, Massana R. Diversity patterns and activity of uncultured marine heterotrophic flagellates unveiled with pyrosequencing. ISME J. 2012;6:1823–1833. doi: 10.1038/ismej.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-García P, Philippe H, Gail F, Moreira D. Autochthonous eukaryotic diversity in hydrothermal sediment and experimental microcolonizers at the Mid-Atlantic Ridge. Proc Natl Acad Sci USA. 2003;100:697–702. doi: 10.1073/pnas.0235779100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia M, Brazel D, Poulton NJ, Swan BK, Lluesma Gomez M, Mashland D, et al. Unveiling in situ interactions between marine protists and bacteria through single cell sequencing. ISME J. 2012;6:703–707. doi: 10.1038/ismej.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana R, Castresana J, Balagué V, Guillou L, Romari K, Groisillier A, et al. Phylogenetic and ecological analysis of novel marine stramenopiles. Appl Environ Microbiol. 2004;70:3528–3534. doi: 10.1128/AEM.70.6.3528-3534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana R, Pedrós-Alió C. Unveiling new microbial eukaryotes in the surface ocean. Curr Opin Microbiol. 2008;11:213–218. doi: 10.1016/j.mib.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Massana R, Terrado R, Forn I, Lovejoy C, Pedrós-Alió C. Distribution and abundance of uncultured heterotrophic flagellates in the world oceans. Environ Microbiol. 2006;8:1515–1522. doi: 10.1111/j.1462-2920.2006.01042.x. [DOI] [PubMed] [Google Scholar]
- Massana R, Unrein F, Rodríguez-Martínez R, Forn I, Lefort T, Pinhassi J, et al. Grazing rates and functional diversity of uncultured heterotrophic flagellates. ISME J. 2009;3:588–596. doi: 10.1038/ismej.2008.130. [DOI] [PubMed] [Google Scholar]
- Not F, Valentin K, Romari K, Lovejoy C, Massana R, Töbe K, et al. Picobiliphytes: a marine picoplanktonic algal group with unknown affinities to other eukaryotes. Science. 2007;315:252–254. doi: 10.1126/science.1136264. [DOI] [PubMed] [Google Scholar]
- Orsi W, Edgcomb V, Jeon S, Leslin C, Bunge J, Taylor GT, et al. Protistan microbial observatory in the Cariaco Basin, Caribbean. II. Habitat specialization. ISME J. 2011;5:1357–1373. doi: 10.1038/ismej.2011.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwosz K, Pernthaler J. Seasonal population dynamics and trophic role of planktonic nanoflagellates in coastal surface waters of the Southern Baltic Sea. Environ Microbiol. 2010;12:364–377. doi: 10.1111/j.1462-2920.2009.02074.x. [DOI] [PubMed] [Google Scholar]
- Piwosz K, Wiktor JM, Niemi A, Tatarek A, Michel C. Mesoscale distribution and functional diversity of picoeukaryotes in the first-year sea ice of the Canadian Arctic. ISME J. 2013;7:1461–1471. doi: 10.1038/ismej.2013.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards TA, Bass D. Molecular screening of free-living microbial eukaryotes: diversity and distribution using a meta-analysis. Curr Opin Microbiol. 2005;8:240–252. doi: 10.1016/j.mib.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Richards TA, Vepritskiy AA, Gouliamova DE, Nierzwicki-Bauer SA. The molecular diversity of freshwater picoeukaryotes from an oligotrophic lake reveals diverse, distinctive and globally dispersed lineages. Environ Microb. 2005;7:1413–1425. doi: 10.1111/j.1462-2920.2005.00828.x. [DOI] [PubMed] [Google Scholar]
- Riisberg I, Orr RJS, Kluge R, Schalchian-Tabrizi K, Bowers HA, Patil V, et al. Seven gene phylogeny of heterokonts. Protist. 2009;160:191–204. doi: 10.1016/j.protis.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Robertson CE, Harris JK, Spear JR, Pace NR. Phylogenetic diversity and ecology of environmental Archaea. Curr Opin Microbiol. 2005;8:638–642. doi: 10.1016/j.mib.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Martínez R, Rocap G, Logares R, Romac S, Massana R. Low evolutionary diversification in a widespread and abundant uncultured protist MAST-4. Mol Biol Evol. 2012;29:1393–1406. doi: 10.1093/molbev/msr303. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Martínez R, Rocap G, Salazar G, Massana R. Biogeography of the uncultured marine picoeukaryote MAST-4: temperature driven distribution patterns. ISME J. 2013;7:1531–1543. doi: 10.1038/ismej.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seenivasan R, Sausen N, Medlin LK, Melkonian M. Picomonas judraskeda Gen. et Sp. Nov.: The first identified member of the Picozoa phylum Nov., a widespread group of picoeukaryotes, formerly known as ‘Picobiliphytes'. PLoS One. 2013;8:e59565. doi: 10.1371/journal.pone.0059565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stock A, Breiner HW, Pachiadaki M, Edgcomb V, Filker S, La Cono V, et al. Microbial eukaryote life in the new hypersaline deep-sea basin Thetis. Extremophiles. 2012;16:21–34. doi: 10.1007/s00792-011-0401-4. [DOI] [PubMed] [Google Scholar]
- Stoeck T, Bass D, Nebel M, Christen R, Jones MD, Breiner HW, et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol. 2010;19(Suppl 1):21–31. doi: 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- Wylezich C, Jürgens K. Protist diversity in suboxic and sulfidic waters of the Black Sea. Environ Microbiol. 2011;13:2939–2956. doi: 10.1111/j.1462-2920.2011.02569.x. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Price DC, Stepanauskas R, Rajah VD, Sieracki ME, Wilson WH, et al. Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science. 2011;322:714–717. doi: 10.1126/science.1203163. [DOI] [PubMed] [Google Scholar]
- Zuendorf A, Bunge J, Behnke A, Barger KJA, Stoeck T. Diversity estimates of microeukaryotes below the chemocline of the anoxic Mariager Fjord, Denmark. FEMS Microbiol Ecol. 2006;58:476–491. doi: 10.1111/j.1574-6941.2006.00171.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.