Abstract
Chemolithoautotrophic Epsilonproteobacteria are ubiquitous in sulfidic, oxygen-poor habitats, including hydrothermal vents, marine oxygen minimum zones, marine sediments and sulfidic caves and have a significant role in cycling carbon, hydrogen, nitrogen and sulfur in these environments. The isolation of diverse strains of Epsilonproteobacteria and the sequencing of their genomes have revealed that this group has the metabolic potential to occupy a wide range of niches, particularly at dynamic deep-sea hydrothermal vents. We expand on this body of work by examining the population genomics of six strains of Lebetimonas, a vent-endemic, thermophilic, hydrogen-oxidizing Epsilonproteobacterium, from a single seamount in the Mariana Arc. Using Lebetimonas as a model for anaerobic, moderately thermophilic organisms in the warm, anoxic subseafloor environment, we show that genomic content is highly conserved and that recombination is limited between closely related strains. The Lebetimonas genomes are shaped by mobile genetic elements and gene loss as well as the acquisition of novel functional genes by horizontal gene transfer, which provide the potential for adaptation and microbial speciation in the deep sea. In addition, these Lebetimonas genomes contain two operons of nitrogenase genes with different evolutionary origins. Lebetimonas expressed nifH during growth with nitrogen gas as the sole nitrogen source, thus providing the first evidence of nitrogen fixation in any Epsilonproteobacteria from deep-sea hydrothermal vents. In this study, we provide a comparative overview of the genomic potential within the Nautiliaceae as well as among more distantly related hydrothermal vent Epsilonproteobacteria to broaden our understanding of microbial adaptation and diversity in the deep sea.
Keywords: Epsilonproteobacteria, hydrothermal vent, population genomics, seamount
Introduction
Epsilonproteobacteria occupy a variety of niches in hydrothermal vent systems, including microbial mats, diffuse fluids, sulfide chimneys and animal-associated niches (Reysenbach et al., 2000; Corre et al., 2001; Huber et al., 2003; Higashi et al., 2004; Nakagawa et al., 2005a; Takai et al., 2005a; Campbell et al., 2006; Moussard et al., 2006; Huber et al., 2007). Since the isolation of the first Epsilonproteobacteria from hydrothermal vents (Campbell et al., 2001), representatives of several genera have been cultivated, revealing that most are chemolithoautotrophic and fall into two general groups: moderately thermophilic hydrogen oxidizers and mesophilic hydrogen and sulfur oxidizers (Takai et al., 2005a). Many are capable of ammonifying nitrate reduction and denitrification (Campbell et al., 2006). Given the dominance of Epsilonproteobacteria in molecular surveys of deep-sea communities worldwide, this group potentially has a large role in global carbon, hydrogen, sulfur and nitrogen cycling.
The genomes of seven hydrothermal vent Epsilonproteobacteria isolates, spanning six genera (Caminibacter, Nautilia, Nitratifractor, Nitratiruptor, Sulfurimonas and Sulfurovum), have previously been sequenced. These genomes revealed an abundance of hydrogenases (Nakagawa et al., 2007; Campbell et al., 2009), unique operon structures of sox sulfur oxidation genes (Nakagawa et al., 2007), the presence of pathogen-associated genes (Nakagawa et al., 2007), a novel nitrate assimilation pathway (Campbell et al., 2009) and the horizontally acquired reverse gyrase (rgy) gene in moderate thermophiles (Campbell et al., 2009). Within the vent-endemic Nautiliaceae, the genomes of Nautilia profundicola from the East Pacific Rise and Caminibacter mediatlanticus from the Mid-Atlantic Ridge have been sequenced (Campbell et al., 2009; Giovanelli et al., 2011). The third described genus of this family is Lebetimonas, of which there is only one named isolate, Lebetimonas acidiphila, from the TOTO Caldera in the Mariana Arc (Takai et al., 2005b).
Deep sequencing of hydrothermal vent fluid samples has shown that Lebetimonas is among the dominant Epsilonproteobacteria from Mariana Arc seamounts and that geographical isolation may have a role in structuring these populations (Huber et al., 2010). Lebetimonas has also been identified as dominant in hydrothermal vent deposits from the Eastern Lau Spreading Center, constituting more than half of all reads in some samples (Flores et al., 2012). Finally, Lebetimonas was detected as the only phylotype in the 16S rRNA gene clone library of white microbial mat at the Iceberg vent at NW Rota-1 seamount in one sample year and showed a decrease in abundance during a subsequent sample year, when diffuse fluid temperatures decreased and representatives of the mesophilic genera Sulfurovum and Sulfurimonas were detected (Davis and Moyer, 2008). While it is clear that biogeographical patterns exist in the structuring of hydrothermal vent Epsilonproteobacteria communities, less well constrained are the relative effects of biogeochemical conditions and geographical isolation in these systems. At NW Rota-1 seamount, the predominant magmatic sulfur gas is SO2, which readily dissolves in seawater to produce acidic diffuse fluids and elemental sulfur (Butterfield et al., 2011). The abundance of elemental sulfur rather than hydrogen sulfide is indicative of a highly reduced environment that may favor members of the strictly hydrogen-oxidizing, sulfur-reducing groups like Lebetimonas over sulfide-oxidizing Epsilonproteobacteria like Sulfurovum and Sulfurimonas, thus our enrichment strategy for this study targeted Nautiliaceae to gain insight into these vent-endemic organisms.
The use of model organisms for genomic and physiological studies from both terrestrial hot springs (Whitaker et al., 2003; Reno et al., 2009) and the surface ocean (Rocap et al., 2003) have dramatically increased our understanding of what drives microbial evolution and adaptation in natural environments. Population genomic studies of thermophilic archaea in terrestrial hot springs demonstrated a lack of gene flow between geographically isolated populations (Reno et al., 2009) and evidence for species divergence in two co-existing groups of strains within a single hot spring (Cadillo-Quiroz et al., 2012). Geographical isolation was also seen in the comparative genomics study of four bacterial isolates of Hydrogenobaculum sp. from a single hot spring that had highly conserved genomes that were distinct from a strain isolated at another site within Yellowstone National Park (Romano et al., 2013). To date, no similar comparative genomic studies have been carried out in the deep sea, where the isolated and often ephemeral nature of hydrothermal vents provides the opportunity to observe biogeographical patterns and evolutionary processes in marine microbial populations.
We isolated six strains of Lebetimonas from two sample years at the dynamic NW Rota-1 seamount where the first human observation of a submarine volcanic eruption occurred in 2004 (Embley et al., 2006). The seamount has been erupting ever since and between the 2009 and 2010 research expeditions, a landslide dramatically changed the topography of the seamount (Chadwick et al., 2012). Here, we use Lebetimonas as a model organism to examine the genomic diversity within one species of Epsilonproteobacteria from a single dynamic seamount, using low temperature diffuse vent fluids as a ‘window' into the subseafloor habitat (Deming and Baross, 1993). Our analysis provides insight into the genomic potential within the Nautiliaceae and other vent Epsilonproteobacteria, thus expanding our understanding of microbial adaptation and diversity in the deep sea.
Materials and methods
Strain enrichment and isolation
Diffuse hydrothermal vent fluids were collected at several vent sites on NW Rota-1 seamount in 2009 and 2010 using the ROV Jason 2 and the hydrothermal fluid and particle sampler (Butterfield et al., 2004; Supplementary Figure S1). Anaerobic enrichment media previously used for the isolation of Caminibacter profundus (Miroshnichenko et al., 2004) was inoculated with 1 ml of unfiltered diffuse flow fluids and incubated at 55 °C. Enrichments with positive microbial growth were isolated by three sets of dilution-to-extinction. The growth of Lebetimonas under varying conditions including alternative electron donor/acceptor pairs and with N2 gas as the sole nitrogen source was evaluated as described in the Supplementary Material. Growth of Lebetimonas strain JH369 with N2 gas as the sole nitrogen source was evaluated using anaerobic seawater media without yeast extract or ammonia and containing formate and elemental sulfur with an 80% N2 and 20% CO2 headspace. Strain JH369 was first weaned off the enrichment culture by three successive passes to media without yeast extract, then transferred by two passes to the formate/N2 media. RNA was extracted from the mid-log phase culture of the second pass of strain JH369 grown with N2 as the sole nitrogen source using a Norgen total RNA purification kit (Norgen, Thorold, ON, Canada), treated with Turbo DNase (Ambion Turbo DNA-free kit, Life Technologies, Carlsbad, CA, USA), and converted into cDNA with an Applied Biosystems (ABI) High Capacity RNA to cDNA kit (Life Technologies). The nitrogenase gene nifH was amplified from strain JH369 genomic DNA grown in the original enrichment media and from the cDNA of strain JH369 grown with N2 as the sole nitrogen source using previously described primers and thermal cycling conditions (Alain et al., 2004). The nifH PCR product from JH369 cDNA was cleaned with a MinElute PCR Purification kit (Qiagen, Hilden, Germany) and cloned with a PCR4 TOPO TA cloning kit (Life Technologies). Clones were screened for inserts by PCR amplification with M13F/R primers (Life Technologies). Positive PCR products from 14 clones were cleaned with a MinElute kit (Qiagen) and sequenced with an ABI3730XL (Applied Biosystems, Life Technologies).
Genomic library preparation and sequencing
Genomic DNA was extracted from pure cultures at log phase using a CTAB extraction (Dempster et al., 1999). Libraries were prepared using Nextera DNA sample prep kits (Illumina, San Diego, CA, USA) and sequenced by Roche 454 GS FLX Titanium (454 Life Sciences, Branford, CT, USA) and/or using Illumina HiSeq 2000 paired reads (Illumina). In the case of strains sequenced with multiple platforms, the same genomic DNA extraction was used for all library preparations, with the exception of strain JS085. Genomes were assembled using several tools as described in the Supplementary material.
Genome annotation and analysis
Six draft genomes were submitted to the Joint Genome Institute's Integrated Microbial Genomes (IMG) system for annotation and comparative analyses (Markowitz et al., 2012), including the Dot Plot synteny viewer generated from nucleotide alignments using the nucmer component of MUMmer (Delcher et al., 2002). Annotated genomes were also aligned with progressiveMauve for visualization and comparative analyses, including counting of single-nucleotide polymorphisms (Darling et al., 2010). A comparative genome map was created with the CGView comparative tool (Grant et al., 2012). Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) regions were identified in IMG and with CRISPRFinder (Grissa et al., 2007). Potential prophages were identified with Prophage Finder (Bose and Barber, 2006) and genomic islands (GIs) were identified with IslandViewer (Langille and Brinkman, 2009). A core genome alignment was created with progressiveMauve and highly variable regions were removed so that all core alignment blocks were longer than 500 nt. Rates of mutation and recombination were determined with the LDHat package (McVean et al., 2002) through the Recombination Detection Program v4.19 (RDP4; Martin et al., 2010). Rates of mutation and recombination were also determined for a subset of seven housekeeping genes: atpA, efp, aspA, glnA, pgm, glyA and trpC.
Results
Isolation and growth of Lebetimonas
Six Lebetimonas strains were enriched and isolated from four different diffuse flow vent sites at NW Rota-1 seamount in 2009 and 2010, with in situ temperatures ranging from 30 °C to 45 °C (Figure 1; Table 1). One site (Marker 110) was sampled in both 2009 and 2010 and one site (Arrowhead) was sampled twice in 2010. All six strains grew in anaerobic saltwater media with hydrogen gas or formate as the sole electron donor and elemental sulfur as the sole electron acceptor (additional culturing details are available in the Supplementary text).
Figure 1.
High-resolution bathymetric map of NW Rota-1 seamount generated by multibeam sonar with the locations of sampling sites of Lebetimonas strains in 2009 (left) and 2010 (right). Map courtesy of W. Chadwick and S. Merle, NOAA/PMEL.
Table 1. Sampling locations of Lebetimonas isolates.
| Isolate name | Sample year | Vent | Fluid sample | Depth (m) | In situ fluid sample temperature (°C) |
|---|---|---|---|---|---|
| JH292 | 2009 | Marker 110/Iceberg 09-2 | FS657 | 535 | 36 |
| JH369 | 2009 | Marker 103/Floc Rock | FS678 | 521 | 30 |
| JS032 | 2010 | Arrowhead | FS730 | 545 | 45 |
| JS085 | 2010 | Marker 110/Iceberg 2009 | FS739 | 533 | 33 |
| JS138 | 2010 | Arrowhead | FS749 | 546 | 33 |
| JS170 | 2010 | Marker 117 | FS755 | 537 | 41 |
General genome properties
The nearly complete draft genomes (GenBank accession numbers: ATHP00000000, ATHQ00000000, ATHR00000000, ATHS00000000, ATHT00000000 and ATHU00000000) of all six Lebetimonas strains were composed of five contigs, the largest of which was at least 1.45 Mbp and the smallest of which was at least 18 Kbp. The draft genome sizes ranged from 1.64 to 1.74 Mbp, with up to 1989 annotated genes per genome (Table 2). GC content in the six genomes was 31–32%.
Table 2. Summary of sequencing and assembly results for Lebetimonas genomes.
| Genome name | Sequencing status | Bases | Genes | GC content (%) | Sequencing coverage by 454 | Sequencing coverage by Illumina |
|---|---|---|---|---|---|---|
| Lebetimonas sp. JH292 | Draft | 1 641 000 | 1955 | 31 | 22 × | 1376 × |
| Lebetimonas sp. JH369 | Draft | 1 740 444 | 1989 | 31 | — | 3618 × |
| Lebetimonas sp. JS032 | Draft | 1 709 134 | 1913 | 31 | 27 × | 2588 × |
| Lebetimonas sp. JS085 | Draft | 1 740 329 | 1929 | 31 | 50 × | 2506 × |
| Lebetimonas sp. JS138 | Draft | 1 688 674 | 1882 | 32 | 30 × | 1765 × |
| Lebetimonas sp. JS170 | Draft | 1 740 475 | 1914 | 31 | — | 2206 × |
The similarity of 16S ribosomal RNA genes was 98.8–100% between the six Lebetimonas strains isolated in this study and 99–99.4% between these strains and the type strain Lebetimonas acidiphila (GenBank accession AB167820) (Takai et al., 2005b). All six strains had two complete ribosomal operons, with very similar (>99.7%) but not identical 16S rRNA genes within strains (Figure 3b), as well as a partial 23S rRNA gene located near the end of one of the smaller contigs. This minor intragenomic heterogeneity between ribosomal operons was consistent in Lebetimonas, as the first full ribosomal operon (located at around 200 kb in the aligned draft genomes, Figure 2), including the 16S–23S internal transcribed spacer region, was completely identical in four strains (JH369, JS085, JS138 and JS170) as was the second operon (located at around 700 kb), but the operons within one strain were only 99.3% similar. The two ribosomal operons within strains JH292 and JS032 were 98.6 and 99.0% similar, respectively, with most differences occurring in the internal transcribed spacer region. Examination of seven other genomes of Epsilonproteobacteria from hydrothermal vents, including Caminibacter mediatlanticus (Giovannelli et al., 2011), Nautilia profundicola (Campbell et al., 2009), Nitratifractor salsuginis (Anderson et al., 2011), Nitratiruptor strain SB155-2 (Nakagawa et al., 2007), Nitratiruptor tergarcus (Gi13756), Sulfurimonas autotrophica (Sikorski et al., 2010) and Sulfurovum strain NBC37-1 (Nakagawa et al., 2007), revealed that Sulfurimonas autotrophica also had heterogenous ribosomal operons. The S. autotrophica genome contained four ribosomal operons that are only 92.9% similar due to large differences in the internal transcribed spacer region, whereas the 16S sequences were 99.7% similar.
Figure 2.
Comparative genome map of largest contig (1.55 Mbp) from six Lebetimonas strains with highly conserved synteny. Contigs 2–5, totaling 200 kbp, are not shown. Coding sequences from the reference strain, Lebetimonas sp. JS085, are indicated in the outermost ring. Inner rings are colored by BLAST similarity to the reference strain for strains JS170, JH369, JS138, JS032 and JH292, in order from outside to inside. Ring 7 (black plot) displays the G+C content for the reference strain and ring 8 displays the GC skew from the average genomic G+C content where a positive skew is green and a negative skew is purple. The locations of four genomic islands (GI I–IV), two ribosomal operons (rRNA) and one CRISPR region are indicated.
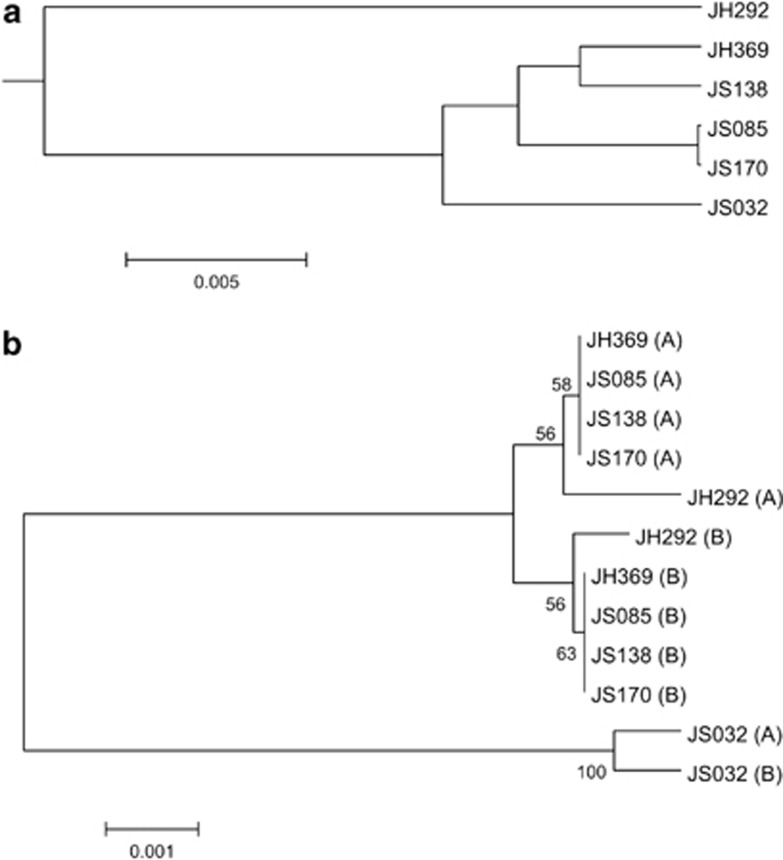
Overall, the six strains of Lebetimonas had very similar genomes with highly conserved synteny (Figure 2; Supplementary Figure S2). There were no full-length genes unique to any individual strain, rather all potentially functional genes were found in at least two genomes. Open reading frames that were unique to one strain consisted only of hypothetical proteins shorter than 300 bp, which were actually fragments of longer genes present in other Lebetimonas strains. The proportion of genes shared by any two genomes with greater than 60% similarity ranged from 89 to 100% (Supplementary Table S1). Within the Nautiliaceae, >55% of genes were shared by Lebetimonas and Nautilia or Caminibacter. The average nucleotide identity of shared genes between Lebetimonas strains ranged from 95.7 to 99.9% (Supplementary Table S2). In contrast, the average nucleotide identity of shared genes between Lebetimonas and Nautilia or Caminibacter was 77%. A total of 76 855 single-nucleotide polymorphisms were found in the core genome alignment, of which 4034 were due to insertions or deletions (indels). The major differences between these genomes are what is missing or degraded in one or more strains, especially strains JH292 and JS138 (Supplementary Table S2). In addition, strain JH292 had the most sequence differences, as reflected in the clustering of core genomes (Figure 3a). The relationship of aligned core genomes did not match the phylogeny based on 16S rRNA genes (Figure 3b).
Figure 3.
(a) UPGMA clustering of aligned core genomes and (b) neighbor-joining tree of full-length 16S rRNA genes with two heterogenous copies per genome. Bootstrap values from 500 replicate trees are shown next to the branches. Evolutionary distances were calculated with the Maximum Composite Likelihood method and are shown in units of the number of base substitutions per site.
Homologous recombination
Homologous recombination between these six Lebetimonas strains based on their core genome alignment was three orders of magnitude lower than the mutation rate. The average rate of recombination per site (rho) was 2.272 × 10−5, with a 95th percentile lower bound of 1.882 × 10−5 and an upper bound of 2.632 × 10−5. The rate of mutation per site (Watterson theta) was 2.043 × 10−2. The average recombination and mutation rates were similar when comparing just the four strains collected during the 2010 research cruise. The rate of recombination based on seven housekeeping genes was higher (average rho per site of 1.281 × 10−2) while the mutation rate per site was similar (1.342 × 10−2), resulting in a less drastic rho/theta of 0.95.
Non-homologous gene flow
Regions of dissimilarity in the Lebetimonas genomes were often a result of or response to viral infection. One clearly defined CRISPR region was found in all six strains, containing 27–30 spacers. The 30-bp repeats in the CRISPR region were identical in five strains, while strain JH292 had a repeat sequence with two nucleotide differences. The sequence identity of the spacer regions was variable among most strains; however, strains JS085 and JS170 had identical CRISPR regions. All six strains also had a suite of CRISPR-associated (cas) genes, although strain JH292 was missing up to four of the cas genes found in other strains and cas3 was degraded in strains JH369 and JS085. CRISPR regions are common in gut-associated genera such as Campylobacter and Helicobacter, but are more sporadically distributed in the free-living Epsilonproteobacteria. CRISPRs were detected in the genomes of several hydrothermal vent isolates, including Caminibacter mediatlanticus, Nitratifractor salsuginis and Nitratiruptor tergarcus. However, no CRISPRs were found in the genomes of Nautilia profundicola, Nitratiruptor strain SB155-2, Sulfurimonas autotrophica and Sulfurovum strain NBC37-1, all of which are closed genomes. All six Lebetimonas genomes also contained multiple sets of type I restriction modification system genes for protection against bacteriophages. One set was most closely related to homologs in Nitratiruptor sp. SB155-2, whereas the two other sets were distantly related to homologs in Bacteroidetes and Spirochaetes. No homologs of the Lebetimonas type I restriction modification system genes were detected in Nautilia and Caminibacter.
A total of four genomic islands (GIs) were identified by annotation of phage-related genes and/or distinct codon usage (Table 3; Figure 2). GIs made up 1–5% of the draft genomes. Three of the four GIs contained integrated phage genes that were present in all six Lebetimonas genomes. However, these prophages were in the process of being degraded in some strains. One large prophage (GI I) was identified in strain JS085 that spanned almost 30 Kbp and included genes for phage integrases, phage/plasmid primase, phage terminase and DNA competence among 33 coding sequences. A putative DNA helicase within this prophage is conserved in Nautiliaceae, with sequence similarities of 76 and 74% in Nautilia and Caminibacter, respectively. This large prophage was also found in strains JS032, JS170 and JH369 but was severely degraded in strains JH292 and JS138 (Figure 2, GI I).
Table 3. Location and description of genomic islands in Lebetimonas strain JS085.
| Genomic island | Start | End | Size (bp) | Description |
|---|---|---|---|---|
| GI I | 131 241 | 158 050 | 26 809 | Large prophage |
| GI II | 760 941 | 776 683 | 15 742 | Small prophage |
| GI III | 835 812 | 859 393 | 23 491 | Glycotransferases |
| GI IV | 1 483 161 | 1 495 179 | 12 018 | Transposon |
A second smaller prophage (GI II) was identified that spanned roughly 15 Kbp and 11 coding sequences, including phage integrase and restriction endonucleases. This smaller prophage was shared by three strains and almost completely degraded in strains JH292, JS138 and JS032 (Figure 2, GI II). The third prophage (GI IV) appeared to be a transposable prophage as it was flanked by mutator family transposases (COG3328). The 10 structural genes within this transposable element included phage integrase and transcriptional regulators. All six strains had two flanking transposases, however, all structural genes within the transposable element were degraded in strains JH292 and JS032 (Figure 2, GI IV). In strain JH292, this GI was followed by a region of 43 degraded or missing genes including an operon of nitrogenase genes. The region following GI IV was conserved in all other strains.
All six Lebetimonas genomes contained an average of 14 mutator-type transposases (COG3328) in addition to transposases belonging to COG1943 and COG0675. Transposases made up ∼1% of gene content of each Lebetimonas strain. Transposases were present near GI I and GI IV and at one or both ends of three of the five scaffolds making up each draft genome. In contrast, the genomes of Caminibacter and Nautilia each contain just one short 53 amino acids-long putative transposase (CTMB2_01059 and NAMH_0938). An examination of all 198 currently available Epsilonproteobacteria genomes revealed that, with the exception of two Helicobacter species, Lebetimonas had the highest number of transposases, with an average of 17 transposases per genome (s.d. 0.98). The average number of transposases in all other genomes from hydrothermal vent Epsilonproteobacteria was 5 (±5.9). Notably, Nautilia and Caminibacter had no annotated transposases.
The final GI (GI III) and adjacent variable region were identified by codon usage and included a large region of glycotransferases involved in cell wall biosynthesis and genes for protein N-glycosylation, a post-translational protein modification system common in pathogenic Epsilonproteobacteria (Nothaft and Szymanski, 2010). Strain JH292 had 37 degraded or missing genes in this region and strain JS138 had 27 degraded or missing genes (Figure 2, GI III). Within GI III, full-length orthologs of the pglB/stt3 oligosaccharyltransferase needed for protein N-glycosylation were present in all Lebetimonas strains except for JH292, in which this gene is interrupted by two stop codons. Oligosaccharyltransferase (pglB/stt3) was previously identified in Nitratiruptor and Sulfurovum genomes (Nakagawa et al., 2007) and the availability of additional non-pathogenic Epsilonproteobacteria genomes has revealed orthologs of pglB/stt3 in Caminibacter, Nautilia, Nitratifractor and Sulfurimonas. Lebetimonas strain JH292 was the only Epsilonproteobacterial genome from hydrothermal vents to lack a full-length copy of this gene.
Hydrogen metabolism
Like other genomes from hydrothermal vent Epsilonproteobacteria, the Lebetimonas genomes contain multiple hydrogenases (Table 4). The Lebetimonas genomes had Group 1 [Ni-Fe]-hydrogenases (hyd) for hydrogen uptake and Group 2 [Ni-Fe]-hydrogenases for sensing and regulating hydrogen uptake. Group 1 and 2 [Ni-Fe]-hydrogenases are found in an operon that is generally conserved in all 13 hydrothermal vent Epsilonproteobacteria. Multiple forms of energy-conserving Group 4 [Ni-Fe]-hydrogenases were identified in Lebetimonas, each of which was part of an operon including formate dehydrogenase (fdh). In Nautilia and Caminibacter, only the Group 4 hyc hydrogenase was associated with formate dehydrogenase. Where multiple hydrogenases of the same type are present, each has a unique sequence (Figure 4). For example, the Group 4 hyc hydrogenases in Nautilia and Caminibacter were unique and Lebetimonas had one Nautilia-like hyc and one Caminibacter-like hyc. In general, all of the genomes within the strictly hydrogen-oxidizing Nautiliaceae have a higher number and greater diversity of hydrogenases when compared with other hydrothermal vent Epsilonproteobacteria (Table 4). The Lebetimonas genomes are the only vent Epsilons to also contain an [Fe-Fe]-hydrogenase most closely related to Thermodesulfobium, which is part of an operon containing Firmicutes-like nitrogenase genes, described below.
Table 4. Comparison of genes involved in hydrogen, nitrogen and sulfur metabolism in Epsilonproteobacteria genomes from hydrothermal vents.
|
Lebetimonas strains |
Nautilia profundicola |
Caminibacter mediatlanticus |
Nitratifractor salsuginis |
Nitratiruptor tergarcus |
Nitratiruptor sp. SB155-2 |
Sulfurovum sp. NBC37-1 |
Sulfurimonas autotrophica |
|
|---|---|---|---|---|---|---|---|---|
| H2-oxidizing | H2-oxidizing | H2-oxidizing | H2-oxidizing | H2-oxidizing | H2- and SX- oxidizing | H2- and SX- oxidizing | SX- oxidizing | |
| Hydrogen oxidation and reduction | ||||||||
| [Ni-Fe]-hydrogenases | ||||||||
| Group 1, hyd | 1 | 2 | 2 | 1 | 1 | 1 | 2 | 1 |
| Group 2, hup | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Group 4, hyc | 2 | 1 | 1 | 1 | — | 1 | 1 | 1 |
| Group 4, coo | 1 | 1 | 1 | — | — | — | — | — |
| Group 4, ech | 1 | 1 | 1 | — | — | — | — | — |
| [Fe-Fe]-hydrogenase | 1 | — | — | — | — | — | — | — |
| Nitrate reduction | ||||||||
| Periplasmic nitrate reductase, napAGHBFLD | 1a | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Nitrate reductase, narB | 1 | — | 2 | — | — | 1 | 1 | 1 |
| Nitrite ammonification | ||||||||
| Nitrite and sulfite reductase/Ferredoxin-nitrite reductase, nirA | 1 | — | 1 | 1 | 1 | 2 | 2 | 2 |
| Novel nitrite reductase (reverse hydroxylamine oxidoreductase) | 1a | 1 | 1 | — | — | — | — | — |
| Cytochrome c protein in the NapC/NrfH/ cM552 protein superfamily | — | 1 | 1 | — | — | — | — | — |
| Hydroxylamine reductase | 1 | 1 | 1 | — | — | — | — | — |
| Denitrification | ||||||||
| Cytochrome cd1 nitrite reductase (NO-forming), nirS | — | — | — | 1 | 1 | 1 | 1 | 1 |
| Nitric oxide reductase, norC | — | — | — | 1 | 1 | 1 | 1 | 1 |
| Nitrous oxide reductase, nosZ | — | — | — | 1 | 1 | 1 | 1 | 1 |
| Nitrogen fixation | ||||||||
| Nitrogenase molybdenum-iron protein, nifDK | 2a | — | — | — | — | — | — | — |
| Nitrogenase iron protein, nifH | 2a | — | — | — | — | — | — | — |
| Sulfur assimilation | ||||||||
| Sulfate permease | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Sulfate adenylyltransferase, cysD/cysN | 1a | — | — | — | — | — | 1 | 1 |
| Sulfate adenylyltransferase, met3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Adenylylsulfate kinase, apsK/cysC | 1a | — | — | — | — | — | 1 | 1 |
| Phosphoadenosine phosphosulfate reductase/PAPS reductase | 1a | — | — | — | — | 1 | — | 1 |
| NAD(P)H:polysulfide oxidoreductase | — | 1 | — | — | — | — | — | — |
| Serine O-acetyltransferase, cysE | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Cysteine synthase, cysK | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Sulfur reduction | ||||||||
| Polysulfide reductase | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Sulfur oxidation | ||||||||
| Sulfur oxidation system, sox | — | — | — | 1 | 1 | 1 | 1 | 1 |
| Sulfide:quinone oxidoreductase, sqr | 1 | — | 2 | 3 | 3 | 3 | 6 | 4 |
| Sulfite:cytochrome c oxidoreductase, sorAB | — | — | — | — | — | — | 2 | 1 |
Some strains contain split genes.
Figure 4.
Neighbor-joining tree of full-length hydrogenase genes from free-living Epsilonproteobacteria and Lebetimonas strain JS085. Bootstrap values from 500 replicate trees are shown next to the branches. Evolutionary distances were calculated with the Maximum Composite Likelihood method and are shown in units of the number of base substitutions per site.
Sulfur metabolism
Sulfur in hydrothermal vent Epsilonproteobacteria may be assimilated from sulfide or potentially from sulfate (Table 4). Partial sulfate assimilation pathways were present in all 13 vent Epsilonproteobacteria genomes. All 13 genomes had sulfate permease and sulfate adenylyltransferase/ATP sulfurylase (cysDN and/or met3) to convert sulfate into adenosine 5′-phosphatase (APS), but none contain adenylylsulfate reductase to convert APS into sulfite. Only Lebetimonas strains JH369, JS085 and JS170 and Sulfurimonas autotrophica had homologs of adenylylsulfate kinase (cysC/apsK) and 3′-phosphoadenosine-5′-phosphosulfate (PAPS) reductase to complete conversion of APS into PAPS and PAPS into sulfite. In Lebetimonas, adenylylsulfate kinase was located in GI III where strains JH292 and JS138 were severely degraded and the gene was either missing or degraded and PAPS reductase was located within GI IV where strains JH292, JS138 and JS032 were severely degraded. Sulfite reductase is needed for the final step of converting sulfite into sulfide for assimilation. All 13 genomes had a nitrite and sulfite reductase/ferredoxin-nitrite reductase (nirA) from the protein family Pfam NIR_SIR_Ferr of closely related nitrite reductases and sulfite reductases (Table 4). This gene may only be used for nitrite reduction, given its location near either a periplasmic or a cytoplasmic nitrate reductase in most of the Epsilonproteobacteria genomes. While the sulfate assimilation pathway appears to be incomplete in hydrothermal vent Epsilonproteobacteria, all 13 genomes, including the six Lebetimonas strains, contained homologs of serine O-acetyltransferase (cysE) and cysteine synthase (cysK) for the biosynthesis of cysteine from sulfide.
All 13 vent Epsilonproteobacteria genomes contained homologs of polysulfide reductase (psr) for the reduction of polysulfides for energy conservation. Sulfur oxidation genes in the sox sulfur oxidation system are present only outside the Nautiliaceae. All of the vent Epsilonproteobacteria genomes, except for Nautilia profundicola, contained at least one copy of sulfide:quinone oxidoreductase (sqr) for the oxidation of hydrogen sulfide to elemental sulfur. Sulfite:cytochrome c oxidoreductase (sorAB) was present only in the Sulfurovum and Sulfurimonas genomes. None of the 13 vent Epsilonproteobacteria genomes contain homologs of genes for dissimilatory sulfate and sulfite reduction.
Nitrogen metabolism
Based on the genome content, these Lebetimonas strains are capable of nitrate reduction with either a periplasmic (napA) or a respiratory nitrate reductase (narB) (Table 4). The periplasmic nitrate reductase in strains JH292 and JS138 was split into two coding sequences. All six Lebetimonas strains had a complete ferredoxin-nitrite reductase (nirA), which was found in six other hydrothermal vent Epsilonproteobacteria genomes (Table 4). The exception is Nautilia profundicola, which is proposed to have a novel nitrite reductase pathway, using a reverse hydroxylamine oxidoreductase, a cytochrome C protein in the NapC/NrfH/cM552 superfamily and hydroxylamine reductase (Campbell et al., 2009). Caminibacter mediatlanticus had homologs for both nitrite reduction pathways, while Lebetimonas lacked the cytochrome C protein in the novel pathway and reverse hydroxylamine oxidoredutase was split into two coding sequences in strains JH369 and JS138 (Table 4). The hydrothermal vent Epsilonproteobacteria genomes outside the Nautiliaceae, including Nitratifractor, Nitratiruptor, Sulfurimonas and Sulfurovum, each had a ferredoxin-nitrite reductase (nirA) as well as a cytochrome cd1 nitrite reductase (nirS), nitric oxide reductase (norC) and nitrous oxide reductase (nosZ) for denitrification to N2.
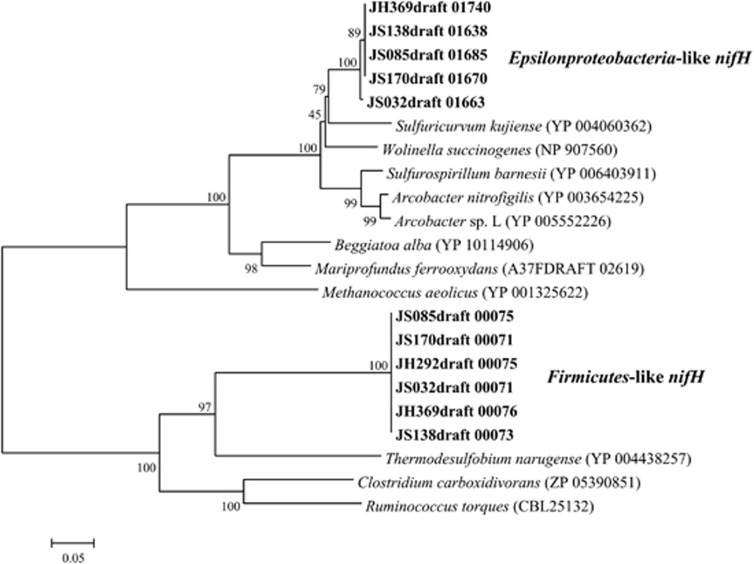
Among hydrothermal vent Epsilonproteobacteria, only Lebetimonas contains genes for nitrogen fixation. All six strains of Lebetimonas had two nitrogenase operons, including the alpha and beta chains of nitrogenase molybdenum-iron protein (COG2710) and the nitrogenase subunit nifH (COG1348). One set of nitrogenase genes, including nifHDKENB, as well as nifA and sigma factor σ54 that are used for the regulation of nitrogenase in Proteobacteria (Dixon and Kahn, 2004), was most closely related to nitrogenase genes in terrestrial isolates of Epsilonproteobacteria, such as Sulfuricurvum and Arcobacter (Figure 5). Five Lebetimonas strains had nearly identical genetic sequences for the Epsilonproteobacteria-like nifH. However, the Epsilonproteobacteria-like nifH in JH292 was degraded into three coding sequences and was part of the region of gene degradation downstream of GI IV.
Figure 5.
Neighbor-joining tree of amino-acid sequences of full-length nifH genes in Lebetimonas with comparisons to nifH in other sequenced genomes. Bootstrap values from 1000 replicate trees are shown next to the branches. Evolutionary distances were calculated with the Poisson correction method and are shown in units of the number of amino-acid substitutions per site.
The second set of nitrogenase genes (nifHDK) was most closely related to nitrogenase genes in Firmicutes. The closest BLAST hit was 65% sequence similarity to the nitrogenase in Thermodesulfobium narugense, an isolate from terrestrial hot springs in Japan (Mori et al., 2003). All six strains shared identical Firmicutes-like nifH genes (Figure 5), which are part of a region of nine Firmicutes-like genes.
Lebetimonas strain JH369 grew weakly in media without ammonia and with N2 gas as a sole nitrogen source compared with the enrichment media (additional culturing details are available in the Supplementary text). While cultures grown in the enrichment media containing yeast extract became turbid in 24 h, cultures grown with N2 gas as a sole nitrogen source did not become turbid when incubated for 2 weeks, but cell growth was verified by phase contrast microscopy. The nitrogenase gene nifH was successfully amplified from strain JH369 in both genomic DNA and RNA extracted from cultures grown without organic nitrogen, indicating that Lebetimonas may be fixing nitrogen during growth with only N2 gas rather than just scavenging nitrogen from dead cells in the media. Sequencing of nifH from the RNA fraction of JH369 grown under N2 gas revealed that only the Firmicutes-like gene was expressed.
Reverse gyrase
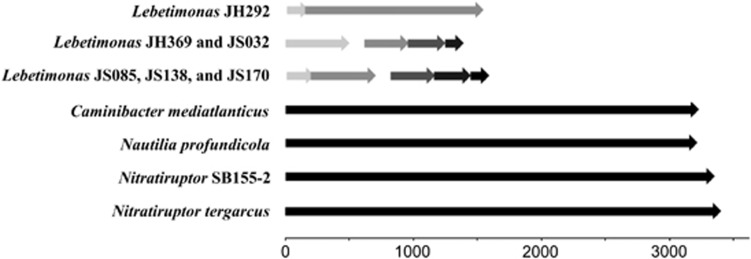
Like other members of the Nautiliaceae, Lebetimonas had homologs of the gene for reverse gyrase (rgy); however, in all six strains, reverse gyrase was degraded. Reverse gyrase is typically made up of two domains: a helicase domain and a type IA topoisomerase domain (Rodríguez and Stock, 2002). In Lebetimonas, reverse gyrase was interrupted by one to four stop codons and lacked the helicase domain (Figure 6). No homologs of the reverse gyrase-associated helicase domain were found elsewhere in the genome. Genes neighboring rgy in Lebetimonas were not degraded and bear the highest sequence similarity to genes in Caminibacter and Nautilia. In contrast, full-length reverse gyrase genes, totaling over 3000 bp in length, were found in Caminibacter, Nautilia, Nitratiruptor sp. SB155-2 and Nitratiruptor tergarcus (Figure 6).
Figure 6.
Comparison of open reading frames annotated as reverse gyrase (rgy) in hydrothermal vent Epsilonproteobacteria. All six Lebetimonas strains have only the topoisomerase domain of rgy, while Caminibacter, Nautilia and Nitratiruptor strains have both the topoisomerase and helicase domains.
Discussion
Overall, the six Lebetimonas genomes show highly conserved synteny, which was also seen in comparative genome analyses of co-occurring strains of Aquificales in Yellowstone (Romano et al., 2013), co-occurring strains of Salinibacter ruber in a Spanish saltern (Peña et al., 2010), and in widespread strains of surface ocean-dwelling SAR11 (Grote et al., 2012). Despite the conservation of both gene content and order, the genomes analysed here contained abundant single-nucleotide differences and showed little evidence of recombination between strains, suggesting that these populations are not co-occurring within niches in the subseafloor. While we lack the ability to clearly visualize the mixing and transportation of hydrothermal fluids beneath the seafloor, it is likely that the continuous eruption of the NW Rota-1 seamount creates an unstable and rapidly changing subseafloor environment. In addition, the reservoir of subseafloor habitat that is accessible by diffuse flow at the seafloor is very large, limiting our ability to capture co-occurring strains, even from fluids collected from the same vent. In this study, Lebetimonas strains JS032 and JS138, which were isolated from Arrowhead vent fluids collected 6 days apart, had less similar genomes than strains JH369 and JS085, which were isolated from different vents a year apart.
Alternatively, the lack of recombination between these strains may be the result of undetermined barriers to recombination that exist between strains, even if they are sympatric, as seen in Sulfolobus strains from terrestrial hot springs (Cadillo-Quiroz et al., 2012). However, in the large and intricately structured subseafloor habitat, it is likely that these Lebetimonas strains are allopatric. Finally, comparative studies of recombination in bacteria and archaea have demonstrated that the relative rate of recombination versus mutation is quite variable, from low to intermediate rates in extremophiles to very high rates in marine and aquatic species (Vos and Didelot, 2009). Relative rates of homologous recombination are not necessarily conserved phylogenetically (Vos and Didelot, 2009), and this may be the case for Epsilonproteobacteria, which exhibit a wide range of lifestyles from pathogenic and gut-associated groups to free-living and biofilm-forming groups (Campbell et al., 2006). The rate of recombination in hydrothermal vent Epsilonproteobacteria such as Lebetimonas may be naturally low given their status as extremophiles, in contrast to the high rates of recombination seen in opportunistically pathogenic Campylobacter (Yu et al., 2012) and Helicobacter (Falush et al., 2001) strains.
In Lebetimonas, the largest differences between genomes were related to gene loss associated with mobile genetic elements. While mobile genetic elements are a source of innovation in microbial genomes (Boucher et al., 2003), acquired genes that do not confer an advantage are quickly degraded (Mira et al., 2001), sometimes en masse (Nilsson et al., 2005). Several large regions of gene loss were apparent in Lebetimonas and these were either within or adjacent to GIs, including both prophages and transposons. Comparative genomics of co-occurring strains have previously shown that genomic differences are often associated with such mobile genetic elements (Cuadros-Orellana et al., 2007; Reno et al., 2009; Peña et al., 2010). In Lebetimonas, we identified a GI (GI III) that is both a region which may have been acquired through horizontal gene transfer and a source of variation between strains as two of the strains are in the process of concurrently losing dozens of genes. This region contained glycotransferases and other proteins related to cell surface changes, which may impact susceptibility to bacteriophages. Horizontally transferred genes tend to be biased toward specific functional groups, including those involved in cell surface manipulation (Nakamura et al., 2004) and this bias has been noted in the population genomics of Haloquadratum walsbyi (Cuadros-Orellana et al., 2007), Salinibacter ruber (Peña et al., 2010) and Pelagibacter ubique (Grote et al., 2012).
Other genes being degraded in Lebetimonas included reverse gyrase (rgy), a DNA chaperonin believed to confer resistance to the denaturation of DNA at high temperatures (Kampmann and Stock, 2004). Reverse gyrase was once thought to be the hallmark protein for hyperthermophilic organisms (Forterre, 2002) and has been transferred from archaea to bacteria multiple times (Brochier-Armanet and Forterre, 2007). The gene has since been identified as common among moderately thermophilic Epsilonproteobacteria at deep-sea hydrothermal vents and is expressed by Nautilia profundicola during heat stress (Campbell et al., 2009). While the genomes of other moderately thermophilic hydrogen-oxidizing groups such as Caminibacter, Nautilia and Nitratiruptor have full-length reverse gyrase genes, this gene does not appear to confer an advantage to Lebetimonas where reverse gyrase is highly fragmented and lacks the helicase domain. In the parasitic archaeon Nanoarchaeum equitans, the two domains of reverse gyrase are split within the genome, yet they still function together (Capp et al., 2010). A similar scenario may be possible for Lebetimonas; however, no helicase domains homologous to the reverse gyrase-associated helicase in other Nautiliaceae were detected. This may have important implications for the distribution of functional reverse gyrase genes in moderately thermophilic Epsilonproteobacteria as the primers designed by Campbell et al. (2009) amplify only the topoisomerase domain of reverse gyrase. Future laboratory experiments with Lebetimonas isolates will help elucidate alternative strategies that hydrothermal vent Epsilonproteobacteria may have to survive heat stress.
In contrast to the degradation of rgy and other genes, the acquisition of nitrogenase genes is clearly advantageous to Lebetimonas. Fixed nitrogen can be limiting in warm (>30 °C) hydrothermal fluids (Lilley et al., 1983), especially ammonia, which is not detectable in vent fluids from the Juan de Fuca Ridge, the East Pacific Rise, and the Mid-Atlantic Ridge (Kelley et al., 2002). However, dissolved N2 gas is abundant in hydrothermal vent fluids (Charlou et al., 2000), providing a source of inorganic nitrogen for organisms that can perform nitrogen fixation. Nitrogenase (nifH) genes have been identified in hydrothermal vent samples from the Juan de Fuca Ridge (Mehta et al., 2003, 2005) and from Lost City (Brazelton et al., 2011), but neither of these studies identified nifH from Epsilonproteobacteria, despite the ability of the primers used to amplify nitrogenase from widespread groups of both archaea and bacteria. To date, nitrogen fixation in isolates from hydrothermal vents has only been demonstrated in hyperthermophilic methanogens (Mehta and Baross, 2006). Lebetimonas strain JH369 expressed the Firmicutes-like nifH in cultures grown with N2 gas as the sole nitrogen source, indicating that is also capable of nitrogen fixation. The current study provides the first indication of nitrogen fixation in any Epsilonproteobacteria from deep-sea hydrothermal vents and may represent an important adaptation for Lebetimonas. Future laboratory experiments will examine whether these nitrogenase genes are expressed by all six Lebetimonas strains during conditions of limited organic nitrogen and if both versions of the nifH gene are functional. These future studies will also include assays of nitrogenase activity to provide conclusive evidence that Lebetimonas can fix inorganic nitrogen.
The population genomics of Lebetimonas reveals that mobile genetic elements shape the genomes of closely related Epsilonproteobacteria in hydrothermal vents through both gene loss and gain. While only a few potentially functional differences were detected between strains, a stable microdiversity may be promoted by the presence of bacteriophages. In the surface ocean, it is thought that heavy phage predation leads to stable populations containing many co-occurring bacterial strains whose genomes differ mainly in their sensitivity to phages (Rodriguez-Valera et al., 2009) and a similar scenario may also play out in the deep sea. A biogeographical study of thermophilic Persephonella from hydrothermal vents in the Okinawa Trough and the South Mariana Trough showed a significant isolation between strains from different troughs (Mino et al., 2013). Using multi-locus sequence analysis, 35 unique sequence types were detected in 36 strains with >98.7% similar 16S rRNA gene sequences, supporting the idea of stable microdiversity in hydrothermal vent microbial populations. Four of the six Lebetimonas strains examined in this study contained identical 16S rRNA genes, yet no two genomes were identical. No geographical or temporal patterns were identified in the distribution of Lebetimonas genomes at NW Rota-1 seamount. However, these six genomes offer a framework for future metagenomic analyses to assess the distribution of Lebetimonas across space and time.
Comparison of the functional repertoire in hydrothermal vent Epsilonproteobacteria genomes shows a gradient of potential phenotypes with Nautiliaceae at one end and Nitratifractor, Sulfurovum and Sulfurimonas at the other, reflecting their tolerance to oxygen and heat. The Nautiliaceae are the only Epsilonproteobacteria with sequenced genomes that lack cytochrome c oxidase for aerobic respiration and they are the only group to contain three different types of energy-conserving Group 4 hydrogenases. In contrast, all vent Epsilonproteobacteria genomes outside Nautiliaceae have genes not only for aerobic respiration, but also for denitrification and for sulfur oxidation through the sox pathway. The major metabolic gene content of Nitratiruptor is most similar to mesophilic vent Epsilonproteobacteria, although both Nitratiruptor tergarcus and Nitratiruptor strain SB155-2 have an optimal growth temperature of 55 °C (Nakagawa et al., 2005b, 2007), similar to the Nautiliaceae, and Nitratiruptor also has the gene for reverse gyrase. Collectively, the genomes of deep-sea Epsilonproteobacteria highlight the capacity of this group to colonize a wide range of niches at hydrothermal vents. The addition of six Lebetimonas genomes to this body of knowledge demonstrates that while core metabolic genes are conserved, a stable microdiversity created by mobile genetic elements exists between strains. Functional genes acquired through horizontal gene transfer, such as nitrogenase in Lebetimonas, may lead to adaptation and speciation at dynamic deep-sea hydrothermal vents.
Acknowledgments
Cruise participation and sample collection were made possible through NSF grants OCE-0751776 to William Chadwick and OCE-0751699 to David Butterfield. Sheryl Murdock, David Butterfield and the ROV Jason II provided critical support during expeditions in 2009 and 2010. Laboratory work and analysis was supported through a National Aeronautics and Space Administration (NASA) Astrobiology Science and Technology for Exploring Planets grant (NNX09AB756), the Neal Cornell Endowed Research Fund to JAH and through a Center for Dark Energy Biosphere Investigations Postdoctoral Fellowship to JLM. This is C-DEBI contribution #180.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alain K, Zbinden M, Le Bris N, Lesongeur F, Quérellou J, Gaill F, et al. Early steps in microbial colonization processes at deep-sea hydrothermal vents. Environ Microbiol. 2004;6:227–241. doi: 10.1111/j.1462-2920.2003.00557.x. [DOI] [PubMed] [Google Scholar]
- Anderson I, Sikorski J, Zeytun A, Nolan M, Lapidus A, Lucas S, et al. Complete genome sequence of Nitratifractor salsuginis type strain (E9I37-1) Stand Genomic Sci. 2011;4:322–330. doi: 10.4056/sigs.1844518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose M, Barber RD. Prophage Finder: a prophage loci prediction tool for prokaryotic genome sequences. In Silico Biol. 2006;6:223–227. [PubMed] [Google Scholar]
- Boucher Y, Douady CJ, Papke RT, Walsh DA, Boudreau MER, Nesbø CL, et al. Lateral gene transfer and the origins of prokaryotic groups. Annu Rev Genet. 2003;37:283–328. doi: 10.1146/annurev.genet.37.050503.084247. [DOI] [PubMed] [Google Scholar]
- Brazelton WJ, Mehta MP, Kelley DS, Baross JA. Physiological differentiation within a single-species biofilm fueled by serpentinization. mBio. 2011;2:e00127–11. doi: 10.1128/mBio.00127-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier-Armanet C, Forterre P. Widespread distribution of archaeal reverse gyrase in thermophilic bacteria suggests a complex history of vertical inheritance and lateral gene transfers. Archaea. 2007;2:83–93. doi: 10.1155/2006/582916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield DA, Nakamura K, Takano B, Lilley MD, Lupton JE, Resing JA, et al. High SO2 flux, sulfur accumulation, and gas fractionation at an erupting submarine volcano. Geology. 2011;39:803–806. [Google Scholar]
- Butterfield DA, Roe KK, Lilley MD, Huber JA, Baross JA, Embley RW, et al. 2004Mixing, reaction, and microbial activity in the sub-seafloor revealed by temporal and spatial variation in diffuse flow vents at Axial VolcanoIn Wilcock WSD, DeLong EF, Kelley DS, Baross JA, Cary SC (eds)The Subseafloor Biosphere at Mid-Ocean Ridges American Geophysical Union: Washington, DC; 269–289. [Google Scholar]
- Cadillo-Quiroz H, Didelot X, Held NL, Herrera A, Darling A, Reno ML, et al. Patterns of gene flow define species of thermophilic archaea. PLoS Biol. 2012;10:e1001265. doi: 10.1371/journal.pbio.1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ, Engel AS, Porter ML, Takai K. The versatile epsilon-proteobacteria: key players in sulphidic habitats. Nat Rev Microbiol. 2006;4:458–468. doi: 10.1038/nrmicro1414. [DOI] [PubMed] [Google Scholar]
- Campbell BJ, Jeanthon C, Kostka JE, Luther GW, III, Cary SC. Growth and phylogenetic properties of novel bacteria belonging to the epsilon subdivision of the Proteobacteria enriched from Alvinella pompejana and deep-sea hydrothermal vents. Appl Environ Microbiol. 2001;67:4566–4572. doi: 10.1128/AEM.67.10.4566-4572.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ, Smith JL, Hanson TE, Klotz MG, Stein LY, Lee CK, et al. Adaptations to submarine hydrothermal environments exemplified by the genome of Nautilia profundicola. PLoS Genet. 2009;5:e1000362. doi: 10.1371/journal.pgen.1000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capp C, Qian Y, Sage H, Huber H, Hsieh TS. Separate and combined biochemical activities of the subunits of a naturally split reverse gyrase. J Biol Chem. 2010;285:39637–39645. doi: 10.1074/jbc.M110.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick WW, Dziak RP, Haxel JH, Embley RW, Matsumoto H. Submarine landslide triggered by volcanic eruption recorded by in situ hydrophone. Geology. 2012;40:51–54. [Google Scholar]
- Charlou JL, Donval JP, Douville E, Jean-Baptiste P, Radford-Knoery J. Compared geochemical signatures and the evolution of Menez Gwen (37°50′N) and Lucky Strike (37°17′N) hydrothermal fluids, south of the Azores Triple Junction on the Mid-Atlantic Ridge. Chem Geol. 2000;171:49–75. [Google Scholar]
- Corre E, Reysenbach A, Prieur D. ɛ-Proteobacterial diversity from a deep-sea hydrothermal vent on the Mid-Atlantic Ridge. FEMS Microbiol Lett. 2001;205:329–335. doi: 10.1111/j.1574-6968.2001.tb10968.x. [DOI] [PubMed] [Google Scholar]
- Cuadros-Orellana S, Martin-Cuadrado A-B, Legault B, D'Auria G, Zhaxybayeva O, Papke RT, et al. Genomic plasticity in prokaryotes: the case of the square haloarchaeon. ISME J. 2007;1:235–245. doi: 10.1038/ismej.2007.35. [DOI] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RE, Moyer CL. Extreme spatial and temporal variability of hydrothermal microbial mat communities along the Mariana Island Arc and southern Mariana back-arc system. J Geophys Res. 2008;113:1–17. [Google Scholar]
- Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deming JW, Baross JA. Deep-sea smokers: windows to a subsurface biosphere. Geochim Cosmochim Acta. 1993;57:3219–3230. doi: 10.1016/0016-7037(93)90535-5. [DOI] [PubMed] [Google Scholar]
- Dempster EL, Pryor KV, Francis D, Young JE, Rogers HJ. Rapid DNA extraction from ferns for PCR-based analyses. Biotechniques. 1999;27:66–68. doi: 10.2144/99271bm13. [DOI] [PubMed] [Google Scholar]
- Dixon R, Kahn D. Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol. 2004;2:621–631. doi: 10.1038/nrmicro954. [DOI] [PubMed] [Google Scholar]
- Embley RW, Chadwick WW, Baker ET, Butterfield DA, Resing JA, De Ronde CEJ, et al. Long-term eruptive activity at a submarine arc volcano. Nature. 2006;441:494–497. doi: 10.1038/nature04762. [DOI] [PubMed] [Google Scholar]
- Falush D, Kraft C, Taylor NS, Correa P, Fox JG, Achtman M, et al. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc Natl Acad Sci USA. 2001;98:15056–15061. doi: 10.1073/pnas.251396098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores GE, Shakya M, Meneghin J, Yang ZK, Seewald JS, Wheat CG, et al. Inter-field variability in the microbial communities of hydrothermal vent deposits from a back-arc basin. Geobiology. 2012;10:333–346. doi: 10.1111/j.1472-4669.2012.00325.x. [DOI] [PubMed] [Google Scholar]
- Forterre P. A hot story from comparative genomics: reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 2002;18:236–237. doi: 10.1016/s0168-9525(02)02650-1. [DOI] [PubMed] [Google Scholar]
- Giovannelli D, Ferriera S, Johnson J, Kravitz S, Pérez-Rodríguez I, Ricci J, et al. Draft genome sequence of Caminibacter mediatlanticus strain TB-2T, an epsilonproteobacterium isolated from a deep-sea hydrothermal vent. Stand Genomic Sci. 2011;5:135–143. doi: 10.4056/sigs.2094859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant JR, Arantes AS, Stothard P. Comparing thousands of circular genomes using the CGView Comparison Tool. BMC Genomics. 2012;13:202. doi: 10.1186/1471-2164-13-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote J, Thrash JC, Huggett MJ. Streamlining and core genome conservation among highly divergent members of the SAR11 clade. mBio. 2012;3:e00252–12. doi: 10.1128/mBio.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi Y, Sunamura M, Kitamura K, Nakamura K, Kurusu Y, Ishibashi J, et al. Microbial diversity in hydrothermal surface to subsurface environments of Suiyo Seamount, Izu-Bonin Arc, using a catheter-type in situ growth chamber. FEMS Microbiol Ecol. 2004;47:327–336. doi: 10.1016/S0168-6496(04)00004-2. [DOI] [PubMed] [Google Scholar]
- Huber JA, Butterfield DA, Baross JA. Bacterial diversity in a subseafloor habitat following a deep-sea volcanic eruption. FEMS Microbiol Ecol. 2003;43:393–409. doi: 10.1111/j.1574-6941.2003.tb01080.x. [DOI] [PubMed] [Google Scholar]
- Huber JA, Cantin HV, Huse SM, Mark Welch DB, Sogin ML, Butterfield DA. Isolated communities of Epsilonproteobacteria in hydrothermal vent fluids of the Mariana Arc seamounts. FEMS Microbiol Ecol. 2010;73:538–549. doi: 10.1111/j.1574-6941.2010.00910.x. [DOI] [PubMed] [Google Scholar]
- Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA, et al. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- Kampmann M, Stock D. Reverse gyrase has heat-protective DNA chaperone activity independent of supercoiling. Nucleic Acids Res. 2004;32:3537–3545. doi: 10.1093/nar/gkh683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DS, Baross JA, Delaney JR. Volcanoes, fluids, and life at mid-ocean ridge spreading centers. Annu Rev Earth Planet Sci. 2002;30:385–491. [Google Scholar]
- Langille MGI, Brinkman FSL. IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics. 2009;25:664–665. doi: 10.1093/bioinformatics/btp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley MD, Baross JA, Gordon LI.1983Reduced gases and bacteria in hydrothermal fluids: the Galapagos Spreading Center and 21N East Pacific RiseIn Rona PA, Bostrom K, Laubier L, Smith KL (eds)Hydrothermal Processes at Seafloor Spreading Centers Plenum Publishing: New York, NY; 411–449. [Google Scholar]
- Lopez-Garcia P, Duperron S, Philippot P, Foriel J, Susini J, Moreira D. Bacterial diversity in hydrothermal sediment and epsilonproteobacterial dominance in experimental microcolonizers at the Mid-Atlantic Ridge. Environ Microbiol. 2003;5:961–976. doi: 10.1046/j.1462-2920.2003.00495.x. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, Chen IA, Palaniappan K, Chu K, Szeto E, Grechkin Y, et al. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G, Awadalla P, Fearnhead P. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics. 2002;160:1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta MP, Baross JA. Nitrogen fixation at 92 degrees C by a hydrothermal vent archaeon. Science. 2006;314:1783–1786. doi: 10.1126/science.1134772. [DOI] [PubMed] [Google Scholar]
- Mehta MP, Butterfield DA, Baross JA. Phylogenetic diversity of nitrogenase (nifh) genes in deep-sea and hydrothermal vent environments of the Juan de Fuca Ridge. Appl Environ Microbiol. 2003;69:960–970. doi: 10.1128/AEM.69.2.960-970.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta MP, Huber JA, Baross JA. Incidence of novel and potentially archaeal nitrogenase genes in the deep Northeast Pacific Ocean. Environ Microbiol. 2005;7:1525–1534. doi: 10.1111/j.1462-2920.2005.00836.x. [DOI] [PubMed] [Google Scholar]
- Mino S, Makita H, Toki T, Miyazaki J, Kato S, Watanabe H, et al. Biogeography of Persephonella in deep-sea hydrothermal vents of the Western Pacific. Front Microbiol. 2013;4:107. doi: 10.3389/fmicb.2013.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- Miroshnichenko ML, L'Haridon S, Schumann P, Spring S, Bonch-Osmolovskaya EA, Jeanthon C, et al. Caminibacter profundus sp. nov., a novel thermophile of Nautiliales ord. nov. within the class “Epsilonproteobacteria”, isolated from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol. 2004;54:41–45. doi: 10.1099/ijs.0.02753-0. [DOI] [PubMed] [Google Scholar]
- Mori K, Kim H, Kakegawa T, Hanada S. A novel lineage of sulfate-reducing microorganisms: Thermodesulfobiaceae fam. nov., Thermodesulfobium narugense, gen. nov., sp. nov., a new thermophilic isolate from a hot spring. Extremophiles. 2003;7:283–290. doi: 10.1007/s00792-003-0320-0. [DOI] [PubMed] [Google Scholar]
- Moussard H, Corre E, Cambon-Bonavita MA, Fouquet Y, Jeanthon C. Novel uncultured Epsilonproteobacteria dominate a filamentous sulphur mat from the 13 degrees N hydrothermal vent field, East Pacific Rise. FEMS Microbiol Ecol. 2006;58:449–463. doi: 10.1111/j.1574-6941.2006.00192.x. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Takai K, Inagaki F, Hirayama H, Nunoura T, Horikoshi K, et al. Distribution, phylogenetic diversity and physiological characteristics of epsilon-Proteobacteria in a deep-sea hydrothermal field. Environ Microbiol. 2005;7:1619–1632. doi: 10.1111/j.1462-2920.2005.00856.x. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Takai K, Inagaki F, Horikoshi K, Sako Y. Nitratiruptor tergarcus gen. nov., sp. nov. and Nitratifractor salsuginis gen. nov., sp. nov., nitratre-reducing chemolithoautotrophs of the ɛ-Proteobacteria isolated from a deep-sea hydrothermal system in the Mid-Okinawa Trough. Int J Syst Evol Microbiol. 2005;55:925–933. doi: 10.1099/ijs.0.63480-0. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Takaki Y, Shimamura S, Reysenbach AL, Takai K, Horikoshi K. Deep-sea vent ɛ-proteobacterial genomes provide insights into emergence of pathogens. Proc Natl Acad Sci USA. 2007;104:12146–12150. doi: 10.1073/pnas.0700687104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Itoh T, Matsuda H, Gojobori T. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat Genet. 2004;36:760–766. doi: 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- Nilsson AI, Koskiniemi S, Eriksson S, Kugelberg E, Hinton JCD, Andersson DI. Bacterial genome size reduction by experimental evolution. Proc Natl Acad Sci USA. 2005;102:12112–12116. doi: 10.1073/pnas.0503654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft H, Szymanski CM. Protein glycosylation in bacteria: sweeter than ever. Nat Rev Microbiol. 2010;8:765–778. doi: 10.1038/nrmicro2383. [DOI] [PubMed] [Google Scholar]
- Peña A, Teeling H, Huerta-Cepas J, Santos F, Yarza P, Brito-Echeverría J, et al. Fine-scale evolution: genomic, phenotypic and ecological differentiation in two coexisting Salinibacter ruber strains. ISME J. 2010;4:882–895. doi: 10.1038/ismej.2010.6. [DOI] [PubMed] [Google Scholar]
- Reno ML, Held NL, Fields CJ, Burke PV, Whitaker RJ. Biogeography of the Sulfolobus islandicus pan-genome. Proc Natl Acad Sci USA. 2009;106:8605–8610. doi: 10.1073/pnas.0808945106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reysenbach AL, Banta AB, Boone DR, Cary SC, Luther GW. Microbial essentials at hydrothermal vents. Nature. 2000;404:835. doi: 10.1038/35009029. [DOI] [PubMed] [Google Scholar]
- Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- Rodríguez AC, Stock D. Crystal structure of reverse gyrase: insights into the positive supercoiling of DNA. EMBO J. 2002;21:418–426. doi: 10.1093/emboj/21.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Valera F, Martin-Cuadrado AB, Rodríguez-Brito B, Pasić L, Thingstad TF, Rohwer F, et al. Explaining microbial population genomics through phage predation. Nat Rev Microbiol. 2009;7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- Romano C, D'Imperio S, Woyke T, Mavromatis K, Lasken R, Shock EL, et al. Comparative genomic analysis of phylogenetically closely-related Hydrogenobaculum sp. from Yellowstone National Park. Appl Environ Microbiol. 2013;79:2932–2943. doi: 10.1128/AEM.03591-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert SM, Vetriani C. Chemoautotrophy at deep-sea vents: past, present, and future. Oceanography. 2012;25:218–233. [Google Scholar]
- Sikorski J, Munk C, Lapidus A, Djao ODN, Lucas S, Del Rio TG, et al. Complete genome sequence of Sulfurimonas autotrophica type strain (OK10) Stand Genomic Sci. 2010;3:194–202. doi: 10.4056/sigs.1173118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K, Campbell BJ, SC Cary, Suzuki M, Oida H, Nunoura T, et al. Enzymatic and genetic characterization of carbon and energy metabolisms by deep-sea hydrothermal chemolithoautotrophic isolates of Epsilonproteobacteria. Appl Environ Microbiol. 2005;71:7310–7320. doi: 10.1128/AEM.71.11.7310-7320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K, Hirayama H, Nakagawa T, Suzuki Y, Nealson KH, Horikoshi K. Lebetimonas acidiphila gen. nov., sp. nov., a novel thermophilic, acidophilic, hydrogen-oxidizing chemolithoautotroph within the “Epsilonproteobacteria”, isolated from a deep-sea hydrothermal fumarole in the Mariana Arc. Int J Syst Evol Microbiol. 2005;55:183–189. doi: 10.1099/ijs.0.63330-0. [DOI] [PubMed] [Google Scholar]
- Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 2009;3:199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- Whitaker RJ, Grogan DW, Taylor JW. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003;301:976–978. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- Yu S, Fearnhead P, Holland BR, Biggs P, Maiden M, French N. Estimating the relative roles of recombination and point mutation in the generation of single locus variants in Campylobacter jejuni and Campylobacter coli. J Mol Evol. 2012;74:273–280. doi: 10.1007/s00239-012-9505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.