Abstract
Carpels are leaf-like structures that bear ovules, and thus play a crucial role in the plant life cycle. In angiosperms, carpels are the last organs produced by the floral meristem and they differentiate a specialized meristematic tissue from which ovules develop. Members of the three-amino-acid-loop-extension (TALE) class of homeoproteins constitute major regulators of meristematic activity. This family contains KNOTTED-like (KNOX) and BEL1-like (BLH or BELL) homeodomain proteins, which function as heterodimers. KNOX proteins can have different BELL partners, leading to multiple combinations with distinct activities, and thus regulate many aspects of plant morphogenesis, including gynoecium development. TALE proteins act primarily through direct regulation of hormonal pathways and key transcriptional regulators. This review focuses on the contribution of TALE proteins to gynoecium development and connects TALE transcription factors to carpel gene regulatory networks.
Keywords: carpel, TALE, transcription factors, development, Arabidopsis
INTRODUCTION
In Arabidopsis, the female reproductive organ, or gynoecium, consists of an apical stigma, a style, and a basal ovary (Figure 1 and for reviews, Ferrándiz et al., 1999; Roeder and Yanofsky, 2006; Girin et al., 2009; Ferrándiz et al., 2010). The ovary is composed of two fused carpels (termed valves after fertilization) whose margins are joined by the replum. The inner (adaxial) side of the replum has a typical meristematic layered structure. This meristem gives rise to ovules and to two septum primordia, which grow and fuse to create the septum that divides the ovary into two locules. Two rows of ovules arise along the septum inside each locule. The septum differentiates a central transmitting tract tissue, which guides pollen tubes from the style to the ovule. Upon fertilization, ovules develop into seeds, and gynoecium structure changes dramatically: the fruit enlarges both longitudinally and laterally to accommodate seed growth and the valve margins undergo cell wall changes required for silique dehiscence and seed dispersal.
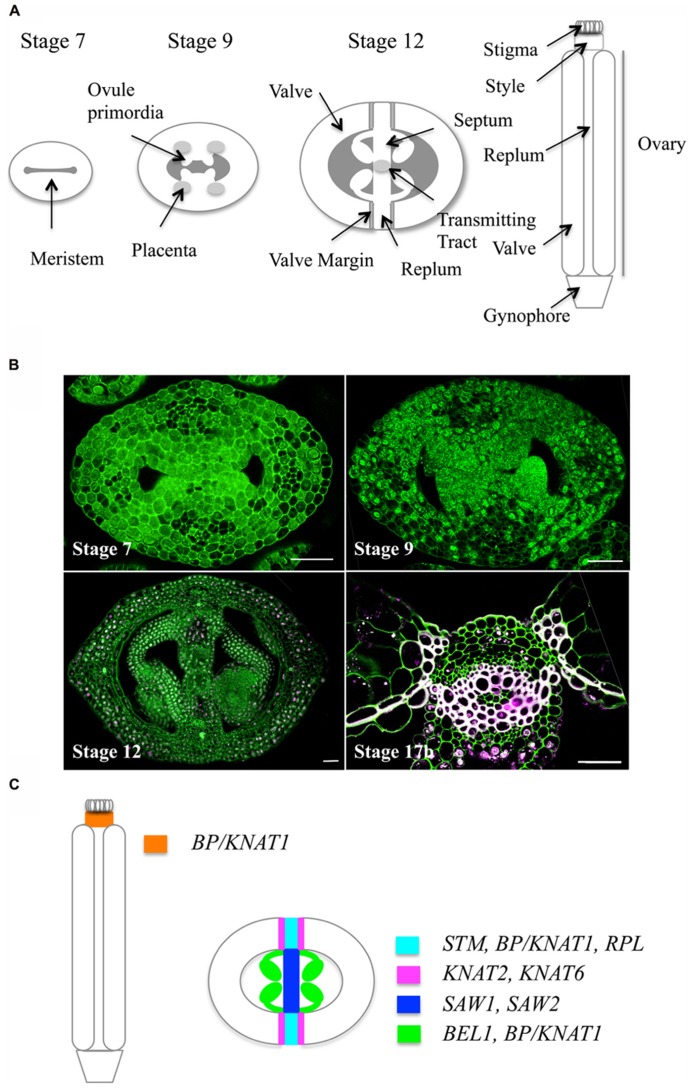
FIGURE 1.
Arabidopsis gynoecium development. (A) Schematic cross sections showing the different tissues of the gynoecium at three developmental stages according to Smyth et al. (1990). (B) Optical cross sections through the Arabidopsis gynoecium at four developmental stages stained with iodine green and carmine alum: upper left, stage 7, showing the layered structure of the meristem; upper right, stage 9, showing ovule primordia initiating from the placenta; lower left, stage 12, lower left, close-up of the medial tissue (stage 17b) showing the replum and lignin deposition at the valve margins and at the endocarp b layer. Scale bars represent 25 μm. (C) Schematic representation of expression patterns of TALE genes in the Arabidopsis gynoecium (stage 12).
In multicellular organisms, development relies on stem cells, which are defined by their ability to renew themselves and to give rise to daughter cells that contribute to organ production. In plants, stem cells are maintained within structures called meristems, and new organs are produced at the meristem periphery (for review, Sablowski, 2011). The shoot apical meristem (SAM) produces leaves and axillary meristems. Following floral evocation, the SAM becomes an inflorescence meristem (IM), which produces flower meristems (FMs) that give rise to flowers containing gynoecia. Carpels are thought to be modified leaves with their margins representing a lateral organ boundary (Frohlich, 2003). As such, similar interactions occurring between SAM-boundary-leaf apply to fruit patterning.
Within meristems, cell proliferation and differentiation are tightly controlled by networks of transcription factors (TFs), which integrate developmental cues such as position, differentiation, and growth (Sablowski, 2011). The KNOTTED1 (KN1) gene in maize was the first regulator of meristem activity identified in plants (Hake and Vollbrecht, 1989). In Arabidopsis, SHOOT MERISTEMLESS (STM), which is functionally related to KN1, and WUSCHEL (WUS) control meristem activity (for review, Aichinger et al., 2012). WUS is required to maintain the stem-cell population, as wus mutants lack stem cells at the center of the shoot apices while STM is required for SAM initiation and its maintenance in an undifferentiated state, as strong stm mutants fail to develop a meristem during embryogenesis and fail to produce lateral organs (Endrizzi et al., 1996; Long et al., 1996). STM is expressed in SAM, IM, FM, and in the inner side of the replum (Endrizzi et al., 1996; Long et al., 1996; Ragni et al., 2008). STM is down-regulated when cells become specified as primordium founder cells (Long et al., 1996).
STM belongs to the “Three-Amino-acid-Loop-Extension” (TALE) homeodomain superclass of TFs, which in Arabidopsis comprises 9 KNOTTED-like (KNAT or KNOX) and 13 BEL1-like (BLH or BELL) members (Box 1). The TALE factors function as KNOX-BELL heterodimers (for reviews, Hay and Tsiantis, 2010; Hamant and Pautot, 2010; Di Giacomo et al., 2013). STM maintains the pool of indeterminate meristematic cells through repression of gibberellin (GA) biosynthesis, activation of GA catabolism, and activation of cytokinin (CK) biosynthesis (Sakamoto et al., 2001; Chen et al., 2004; Jasinski et al., 2005; Bolduc and Hake, 2009). In addition, in the SAM, STM represses the ASYMMETRIC LEAVES1 (AS1) gene, which encodes a MYB TF involved in leaf patterning. AS1 represses other TALE-family members such as KNAT1/BREVIPEDICELLUS (BP), KNAT2, and KNAT6 in leaves (Byrne et al., 2000; Phelps-Durr et al., 2005). Subsequent organ initiation requires high auxin and GA levels and down regulation of STM and related TFs (Hay and Tsiantis, 2010).
Box 1. Meet the TALE gene family.
The TALE family is a superclass of homeodomain TFs which comprises eight KNOTTED-like proteins in Arabidopsis thaliana (KNAT or KNOX) plus a mini KNAT lacking the homeodomain (KNATM) and 13 BEL1-like (BLH or BELL) members (for a detailed review of the structure of this gene family, see Mukherjee et al. (2009) and for a phylogenetic tree of the Arabidopsis TALE family, see Hamant and Pautot, 2010). This family controls development in all eukaryotic lineages (Hay and Tsiantis, 2010). KNOX and BELL families occur in single copy in Green algae and have diversified in land plant (Lee et al., 2008). The KNOX family is divided into three classes based on sequence similarity and gene expression pattern (Hake et al., 2004; Magnani and Hake, 2008): Class I includes STM, BP/KNAT1, KNAT2 and KNAT6; Class II includes KNAT3, 4, 5, and 7. Class III contains KNATM, which can interact with other TALE members to modulate their activity (Kimura et al., 2008; Magnani and Hake, 2008). The BELL family comprises RPL/BLH9, PNF/BLH8, ATH1, SAW1/BLH2, SAW2/BLH8, BEL1 – whose functions have been characterized- and BLH1, BLH3, BLH5, BLH6, BLH7, BLH10, and BLH11 – whose functions are not yet known (Hamant and Pautot, 2010). The interaction of KNOX and BELL proteins is critical for their nuclear localization and their binding affinity to DNA, thereby imparting their activity (Smith et al., 2002; Rutjens et al., 2009; Kim et al., 2013). TALEs can also form complexes with other TFs, such as MADS-Box family members, to control ovule development (Brambilla et al., 2007) and with OVATE proteins, which negatively regulate KNOX-BELL heterodimers by relocalizing them from the nucleus to the cytoplasm (Hackbusch et al., 2005). STM protein traffics selectively through plasmodesmata, and this cell-to-cell movement, which involves chaperonins belonging to a group of cytosolic chaperones, is critical to maintain of the SAM (Xu et al., 2011).
Multiple combinations of TALE heterodimers with distinct activities are produced throughout the plant life cycle, controlling diverse developmental processes, such as SAM and boundary maintenance, leaf development and flowering. This review discusses the contribution of TALE TFs to gynoecium development in Arabidopsis, and links these proteins to the other key molecular players of carpel development.
CARPEL INITIATION: KNOX AND BELL INTERACTIONS WITH AGAMOUS
Carpels are the last organs to be produced by floral meristems. Weak STM alleles or weak STM RNAi lines show no carpel formation due to premature differentiation of meristematic cells (Endrizzi et al., 1996; Scofield et al., 2007). Consistent with this, CLAVATA (CLV) receptors control the proliferation and number of organs in developing gynoecia through STM activity. Mutations in CLV1, CLV2, and CORYNE (CRN) receptors lead to increased meristem size correlated with an enlarged STM expression pattern (Durbak and Tax, 2011). Unlike the SAM, which is indeterminate, the FM terminates after carpel initiation. This determinacy depends on a negative feedback loop involving the C-function homeotic MADS domain TF, AGAMOUS (AG) which acts in part via activation of the zinc finger protein KNUCKLES (KNU) to repress WUS expression (Lenhard et al., 2001; Lohmann et al., 2001; Sun et al., 2009).AG controls carpel identity in combination with another MADS BOX TF, SEPALLATA3 (SEP3)(Bowman et al., 1989; Honma and Goto, 2001; Pelaz et al., 2001). AG expression is first detectable in developing flowers at early stage 3, flower stages defined by Smyth et al. (1990), where it is initially localized in the center of the FM, and is later restricted to stamen and carpel primordia (Bowman et al., 1991; Drews et al., 1991). At late stage 5, the floral meristem forms a flattened oval where the gynoecium initiates (Smyth et al., 1990). This stage coincides presumably with the generation of auxin maxima similar to those observed at the initiation of other organs, although no expression of auxin-signaling reporters at stages 5–7 has been described (for review, Larsson et al., 2013). The BELL member, REPLUMLESS (RPL), also known as PENNYWISE (PNY), BELLRINGER (BLR), VAAMANA (VAN), or LARSON (LSN) and its close relative POUNDFOOLISH (PNF) together with STM, function in parallel with LEAFY (LFY) and WUS to promote carpel formation through positive regulation of AG (Byrne et al., 2003; Roeder et al., 2003; Smith and Hake, 2003; Bao et al., 2004; Bhatt et al., 2004; Yu et al., 2009). Interestingly, a previous report showed that RPL represses AG together with LEUNIG and SEUSS, two transcriptional co-regulators of AG (Bao et al., 2004). This study was based on analysis of two recessive rpl alleles (blr-4 and blr-5) whose flowers exhibit homeotic conversion of sepals to carpels at high temperature during late-stage flower development. This suggests that RPL could have two antagonistic activities depending presumably on its partner. However, no ectopic AG expression has been reported so far in null rpl mutants. An alternative hypothesis is that the point mutations within the homeobox region in blr-4 and blr-5 mutants cause the production of abnormal protein with regulatory defects.
GYNOECIUM PATTERNING
Once initiated, the gynoecium developmental program promotes correct patterning of the future fruit. Several specific tissues are formed (see above and Figure 1), some of which require the activity of TALE TFs. At stage 6, the gynoecium forms as a ridge of raised cells around a central cleft and starts to acquire its medio-lateral symmetry, comprising replum, valve margins and valves. In the transverse plane, the adaxial inner side of the replum has a typical meristematic layered structure (Figure 1B), and accordingly expresses the meristematic genes STM, CLV1/2,and CRN (Long et al., 1996; Durbak and Tax, 2011; Romera-Branchat et al., 2012).However, WUS is not expressed in the replum(Groß–Hardt et al.,2002). Recently, a role for WUS-LIKE HOMEOBOX13 (WOX13) in replum was reported (Romera-Branchat et al., 2012). Unlike WUS, which marks a few cells in the SAM, defining its organizing center, WOX13 has a broad expression pattern in replum, suggesting that the medial region of the gynoecium does not show typical SAM organization.
Consistent with a role for STM in initiating and maintaining meristems, weak alleles of STM or weak STM RNAi lines produce fewer ovules than the wild type (Endrizzi et al., 1996; Scofield et al., 2007). Two other TALE genes, RPL and BP, are also expressed in the replum. The rpl mutant shows defects in replum differentiation and in septum fusion (Roeder et al., 2003). RPL promotes replum identity through restriction of expression of the MADS-BOX genes SHATTERPROOF1/2 (SHP1/2) and the basic helix-loop-helix (bHLH) gene INDEHISCENT (IND), to the valve margins(Roeder et al., 2003; Liljegren et al., 2004; Dinneny et al., 2005), but RPL is not required per se for replum specification, since double or triple mutant combinations including rpl alleles develop a normal replum. In addition, RPL represses several valves-associated genes in the replum: JAGGED (JAG), and genes conferring abaxial fate FILAMENTOUS FLOWER (FIL)andYABBY3 (YAB3), which promote FRUITFULL (FUL), SHP1/2 and IND expression in the presumptive valve and valve margin tissues, respectively (Dinneny et al., 2005). BP, which interacts with RPL and activates its expression, contributes redundantly with RPL to replum development (Alonso-Cantabrana et al., 2007). Similarly to their role at the leaf/SAM interface, AS1 and the lateral organ boundary (LOB)-domain protein asymmetric leaves2 (AS2) restrict the expression of BP to the replum, exemplifying the co-option of this regulatory module in the SAM and carpel (Alonso-Cantabrana et al., 2007; González-Reig et al., 2012; Luo et al., 2012; Lodha et al., 2013). Together these studies led to proposition of a model in which antagonism between the lateral factors (JAG/FIL and AS1/2) and the medial factors (BP and RPL) determines the medio-lateral fruit pattern by regulating the formation and size of three domains: valve, valve margin and replum. Furthermore, APETALA2, a member of the AP2/Ethylene-responsive element binding protein (EREBP) TF family, limits growth of both replum and valve margins by repressing BP and RPL in the replum and SHP1/2 and IND in valve margins (Ripoll et al., 2011). Although BP, together with RPL, contributes to replum development, single bp loss-of-function mutants have wild-type repla (Alonso-Cantabrana et al., 2007; Ripoll et al., 2011). BP is also expressed in the style where it is required for radial growth (Venglat et al., 2002).
From the maternal side, optimal seed production relies on adequate generation of ovules. Ovule primordia formation depends on auxin maxima (Bencivenga et al., 2012). Auxin levels are modulated by the combined activity of CUP-SHAPED COTYLEDON1 (CUC1) and CUC2 TFs, which are redundantly required to regulate the polar auxin transporter pin-formed1 (PIN1) expression (Galbiati et al., 2013). CK also regulates PIN1 expression during early stages of ovule development (Bencivenga et al., 2012). The interplay between hormones and TFs forms an integrative framework enabling ovule primordia initiation. Once initiated, an ovule differentiates a central nucellus containing the embryo sac, two integuments that envelop the nucellus, and a funiculus that connects the ovule to the placenta (for reviews, Colombo et al., 2008; Shi and Yang, 2011). Correct ovule development requires the activity of the BEL1 gene, the founding member of the BELL family. BEL1 is expressed in ovule integument primordia, and controls ovule integument identity (Robinson-Beers et al., 1992; Reiser et al., 1995). The bel1 mutant exhibits bell-shaped ovules – hence its name – caused by the abnormal development of integuments (Reiser et al., 1995). Occasionally, the bel1 mutant shows homeotic conversion of ovules into carpeloid structures due to prolonged AG expression during ovule development (Modrusan et al., 1994; Ray et al., 1994; Brambilla et al., 2007). BEL1 is required for auxin and CK signaling pathways during ovule development; the level and localization of PIN1 expression are controlled by CK in part viaBEL1 activity (Bencivenga et al., 2012).
Inside the future fruit, tissues required for successful fertilization and fruit compartmentalization are formed concomitantly.Two placenta ridges develop in the medial plane to give rise to a specialized structure compartmentalizing the fruit, the septum, which divides the fruit into two halves. In its center, the transmitting tract differentiates in the apical-basal axis to guide pollen tube growth. To date, little is known about the role of TALE genes in septum development. SAWTOOTH1 (SAW1)/BLH2 and SAW2/BLH4, members of the BELL family, are expressed in the transmitting tract, and interact with STM and BP, but their exact role in medial tissue development remains to be determined (Kumar et al., 2007).
POST-FERTILIZATION EVENTS
Upon fertilization, the gynoecium will develop into a fruit that contains the seeds. Gynoecium enlargement to accommodate the developing seeds relies on the coordinated growth of the entire organ, which strongly depends on hormonal balances (for review, Reyes-olalde et al., 2013). For instance, GA-deficient mutants show reduced fruit size, indicating that fruit development involves extensive GA-activated cell elongation (Koornneef and van der Veen, 1980; Chiang et al., 1995). While the fruit enlarges, differentiation processes take place to ensure efficient release of the seeds (Reyes-olalde et al., 2013). At the cellular level, this includes the differentiation of the dehiscence zone at the valve margins. This process depends on the activity of IND, which is responsible of the formation of a local auxin minimum at the valve margins through the regulation of PINOID and WAG2 kinases (Sorefan et al., 2009). The dehiscence zone consists of two cell layers: the lignified and the separation layers. The lignified layer, located at the boundary with the valve, is continuous with the lignified internal layer (endocarp b) and contributes to tension that builds up in the silique until dehiscence. The layer located on the replum side, which constitutes the separation layer, is composed of isodiametric cells that undergo middle lamella breakdown. This separation process involves the activity of specialized cell wall enzymes such as polygalacturonases (PGs) and pectin methylesterases (PMEs) that increase the ability of PGs to break down pectin (Ogawa et al., 2009 and for review, Wolf et al., 2009). A link between TALE proteins and cell wall modifications has been shown in studies of internode patterning in rpl and bp mutants (Mele et al., 2003; Smith and Hake, 2003; Peaucelle et al., 2011). BP prevents premature deposition of lignin during internode growth by direct repression of genes involved in lignin biosynthesis, and regulates other cell-wall-specific genes such as ones encoding PMEs or cellulose synthetase (Mele et al., 2003; Wang et al., 2006). RPL is involved in maintaining normal phyllotaxy via the regulation of PMEs, which are involved in the cell wall loosening necessary to allow growth (Peaucelle et al., 2011). Interestingly, KNAT6 and KNAT2, which act antagonistically to BP and RPL in stems, are expressed in valve margins (Ragni et al., 2008). This is consistent with KNAT6 expression in SAM and its role in maintaining boundaries between SAM and lateral organs (Belles-Boix et al., 2006). Inactivation of KNAT6 rescues replum formation in rpl mutants, showing that the antagonistic interaction between KNAT6 and RPL also controls fruit architecture. Consistent with their expression in valve margins, KNAT6, and KNAT2 positively regulate lignin deposition (Khan et al., 2012a, b). These factors also act antagonistically to BP during floral organ abscission, a process that also requires cell wall remodeling. BP regulates the timing of floral abscission by controlling abscission zone cell size. Upon activation of a signaling pathway including inflorescence deficient in abscission (IDA) and two receptor-like kinases, HAESA and HAESA-LIKE2 (HAE-HSL2), BP is inactivated, leading to an increase of KNAT2 and KNAT6 expression, which act as positive regulators of floral organ separation (Shi et al., 2011). The link between TALEs and cell wall remodeling enzymes was further confirmed with the identification of STM, KN1, and RPL targets, which include several genes involved in cell wall modifications (Spinelli et al., 2011; Bolduc et al., 2012; Etchells et al., 2012).
FUTURE DIRECTIONS
Gynoecium development is critical for Angiosperm reproductive success, and is therefore tightly controlled by interconnected networks of TFs. Here, we reviewed the role of TALE TFs in the control of carpel development, and present the state of knowledge of the molecular interactions within this gene regulatory network. To date, the studies concerning the contribution of TALE TFs to carpel development focused on a few members of this family. Despite the number of studies, several pieces of the puzzle that will be needed to decipher the entire carpel regulatory network are still missing. In particular, the role of the KNAT class II members in carpels has not been investigated. Although several TALE members are expressed in carpels, the detailed expression pattern remains to be characterized for most of them. A precise map of TALE expression and co-localization of KNOX and BELL in the gynoecium will provide clues about putative partners and redundancies. Despite evidence linking TALE TFs, CK and GA pathways, the exact role of this regulatory node and its precise contribution to carpel development are not yet well established. Recently, the direct targets of KN1 in maize inflorescences were identified, and these data confirm that TALE TFs function as major orchestrators of hormone synthesis or response (Bolduc et al., 2012). Importantly, a clear link between KN1 and the auxin pathway was demonstrated. Furthermore, key developmental regulators such as homeodomain TFs are highly represented among KN1 targets, suggesting that KN1 orchestrates upper levels of regulatory networks controlling development. New strategies based on next generation sequencing to identify targets of TFs have begun to shed light on the molecular interactions downstream of key TFs, providing crucial insight into the mechanisms controlling development and opening new perspectives regarding carpel development. The integration of these data into comprehensive models accounting for spatial and temporal information represents a challenge to fully understand how fruits develop. Developing mathematical models will be particularly useful for understanding how fruit morphology can vary and how their astonishing diversity of shape can be achieved among plant species.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Olivier Grandjean for technical assistance, Mark Tepfer, Thomas Girin, Patrick Laufs, and Shelley Hepworth for critical comments.
REFERENCES
- Aichinger E., Kornet N., Friedrich T., Laux T. (2012). Plant stem cell niches. Annu. Rev. Plant Biol. 63 615–636 10.1146/annurev-arplant-042811-105555 [DOI] [PubMed] [Google Scholar]
- Alonso-Cantabrana H., Ripoll J. J., Ochando I., Vera A., Ferrándiz C., Martínez-Laborda A. (2007). Common regulatory networks in leaf and fruit patterning revealed by mutations in the Arabidopsis ASYMMETRIC LEAVES1 gene. Development 134 2663–2671 10.1242/dev.02864 [DOI] [PubMed] [Google Scholar]
- Bao X., Franks R. G., Levin J. Z., Liu Z. (2004). Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. Plant Cell 16 1478–1489 10.1105/tpc.021147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belles-Boix E., Hamant O., Witiak S. M., Morin H., Traas J., Pautot V. (2006). KNAT6: an Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell 18 1900–1907 10.1105/tpc.106.041988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencivenga S., Simonini S., Benková E., Colombo L. (2012). The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis. Plant Cell 24 2886–2897 10.1105/tpc.112.10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt A. M., Etchells J. P., Canales C., Lagodienko A., Dickinson H. (2004). VAAMANA-a BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene 328 103–111 10.1016/j.gene.2003.12.033 [DOI] [PubMed] [Google Scholar]
- Bolduc N., Hake S. (2009). The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21 1647–1658 10.1105/tpc.109.068221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N., Yilmaz A., Mejia-Guerra M. K., Morohashi K., Connor D. O., Grotewold E., et al. (2012). Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 26 1685–1690 10.1101/gad.193433.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J. L., Drews G. N., Meyerowitz E. M. (1991). Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell 3 749–758 10.1105/tpc.3.8.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J. L., Smyth D. R., Meyerowitz E. M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1 37–52 10.1105/tpc.1.1.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla V., Battaglia R., Colombo M., Masiero S., Bencivenga S., Kater M. M., et al. (2007). Genetic and molecular interactions between BELL1 and MADS box factors support ovule development in Arabidopsis. Plant Cell 19 2544–2556 10.1105/tpc.107.051797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M. E., Barley R., Curtis M., Arroyo J. M., Dunham M., Hudson A., et al. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971 10.1038/35050091 [DOI] [PubMed] [Google Scholar]
- Byrne M. E., Groover A. T., Fontana J. R., Martienssen R. A. (2003). Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development 130 3941–3950 10.1242/dev.00620 [DOI] [PubMed] [Google Scholar]
- Chen H., Banerjee A. K., Hannapel D. J. (2004). The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1. Plant J. 38 276–284 10.1111/j.1365-313X.2004.02048.x [DOI] [PubMed] [Google Scholar]
- Chiang H. H., Hwang I., Goodman H. M. (1995). Isolation of the Arabidopsis GA4 locus. Plant Cell 7 195–201 10.1105/tpc.7.2.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo L., Battaglia R., Kater M. M. (2008). Arabidopsis ovule development and its evolutionary conservation. Trends Plant Sci. 13 444–450 10.1016/j.tplants.2008.04.011 [DOI] [PubMed] [Google Scholar]
- Dinneny J. R., Weigel D., Yanofsky M. F. (2005). A genetic framework for fruit patterning in Arabidopsis thaliana. Development 132 4687–4696 10.1242/dev.02062 [DOI] [PubMed] [Google Scholar]
- Drews G. N., Bowman J. L., Meyerowitz E. M. (1991). Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65 991–1002 10.1016/0092-8674(91)90551-9 [DOI] [PubMed] [Google Scholar]
- Durbak A. R., Tax F. E. (2011). CLAVATA signaling pathway receptors of Arabidopsis regulate cell proliferation in fruit organ formation as well as in meristems. Genetics 189 177–194 10.1534/genetics.111.130930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endrizzi K., Moussian B., Haecker A., Levin J. Z., Laux T. (1996). The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 10 967–979 10.1046/j.1365-313X.1996.10060967.x [DOI] [PubMed] [Google Scholar]
- Etchells J., Moore L., Jiang W. (2012). A role for BELLRINGER in cell wall development is supported by loss-of-function phenotypes. BMC Plant Biol. 12:212 10.1186/1471-2229-12-212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz C., Fourquin C., Nathanael P., Scutt C., Sundberg E., Christophe T., et al. (2010). Carpel development. Adv. Bot. Res. 1–7320703330 [Google Scholar]
- Ferrándiz C., Pelaz S., Yanofsky M. F. (1999). Control of carpel and fruit development in Arabidopsis. Annu. Rev. Biochem. 68 321–354 10.1146/annurev.biochem.68.1.321 [DOI] [PubMed] [Google Scholar]
- Frohlich M. W. (2003). An evolutionary scenario for the origin of flowers. Nat. Rev. Genet. 4 559–566 10.1038/nrg1114 [DOI] [PubMed] [Google Scholar]
- Di Giacomo E., Iannelli M., Frugis G. (2013). TALE and Shape: how to Make a Leaf Different. Plants 2 317–342 10.3390/plants2020317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati F., Sinha Roy D., Simonini S., Cucinotta M., Ceccato L., Cuesta C., et al. (2013). An integrative model of the control of ovule primordia formation. Plant J. 76 446–455 10.1111/tpj.12309 [DOI] [PubMed] [Google Scholar]
- Girin T., Sorefan K, østergaard L. (2009). Meristematic sculpting in fruit development. J. Exp. Bot. 60 1493–1502 10.1093/jxb/erp031 [DOI] [PubMed] [Google Scholar]
- González-Reig S., Ripoll J. J., Vera A., Yanofsky M. F, Martínez-Laborda A. (2012). Antagonistic gene activities determine the formation of pattern elements along the mediolateral axis of the Arabidopsis fruit. PLoS Genet. 8:e1003020 10.1371/journal.pgen.1003020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groß-Hardt R., Lenhard M., Laux T. (2002). WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev. 1129–1138 10.1101/gad.225202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackbusch J., Richter K., Müller J., Salamini F., Uhrig J. F. (2005). A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc. Natl. Acad. Sci. U.S.A. 102 4908–4912 10.1073/pnas.0501181102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S., Smith H. M. S., Holtan H., Magnani E., Mele G., Ramirez J. (2004). The Role of KNOX Genes in Plant Development. Annu. Rev. Cell Dev. Biol. 20 126–151 10.1146/annurev.cellbio.20.031803.093824 [DOI] [PubMed] [Google Scholar]
- Hake S., Vollbrecht E. (1989). Cloning Knotted, the dominant morphological mutant in maize using Ds2 as atransposon tag. EMBO 8 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O., Pautot V. (2010). Plant development: a TALE story. C. R. Biol. 333 371–381 10.1016/j.crvi.2010.01.015 [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2010). KNOX genes: versatile regulators of plant development and diversity. Development 3165 3153–3165 10.1242/dev.030049 [DOI] [PubMed] [Google Scholar]
- Honma T., Goto K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409 525–529 [DOI] [PubMed] [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., et al. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15 1560–1565 10.1016/j.cub.2005.07.023 [DOI] [PubMed] [Google Scholar]
- Khan M., Xu M., Murmu J., Tabb P., Liu Y., Storey K., et al. (2012a). Antagonistic interaction of BLADE-ON-PETIOLE1 and 2 with BREVIPEDICELLUS and PENNYWISE regulates Arabidopsis inflorescence architecture. Plant Physiol. 158 946–960 10.1104/pp.111.188573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Tabb P., Hepworth S. R. (2012b). BLADE-ON-PETIOLE1 and 2 regulate Arabidopsis inflorescence architecture in conjunction with homeobox genes KNAT6 and ATH1. Plant Signal. Behav. 7 1–5 10.4161/psb.20599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Cho Y. H., Ryu H., Kim Y., Kim T. H., Hwang I. (2013). BLH1 and KNAT3 modulate ABA responses during germination and early seedling development in Arabidopsis. Plant J. 75 755–766 10.1111/tpj.12236 [DOI] [PubMed] [Google Scholar]
- Kimura S., Koenig D., Kang J. (2008). Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr. Biol. 18 1–6 10.1016/j.cub.2008.04.008 [DOI] [PubMed] [Google Scholar]
- Koornneef M, van der Veen J. H. (1980). Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh. Theor. Appl. Genet. 58 257–263 10.1007/BF00265176 [DOI] [PubMed] [Google Scholar]
- Kumar R., Kushalappa K., Godt D., Pidkowich M. S., Pastorelli S., Hepworth S. R., et al. (2007). The Arabidopsis BEL1-LIKE HOMEODOMAIN proteins SAW1 and SAW2 act redundantly to regulate KNOX expression spatially in leaf margins. Plant Cell 19 2719–2735 10.1105/tpc.106.048769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson E., Franks R. G., Sundberg E. (2013). Auxin and the Arabidopsis thaliana gynoecium. J. Exp. Bot. 64 2619–2627 10.1093/jxb/ert099 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Lin H., Joo S., Goodenough U. (2008). Early sexual origins of homeoprotein heterodimerization and evolution of the plant KNOX/BELL family. Cell 133 829–840 10.1016/j.cell.2008.04.028 [DOI] [PubMed] [Google Scholar]
- Lenhard M., Bohnert A., Jürgens G., Laux T. (2001). Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105 805–814 10.1016/S0092-8674(01)00390-7 [DOI] [PubMed] [Google Scholar]
- Liljegren S. J., Roeder A. H., Kempin S. A., Gremski K., østergaard L., Guimil S., et al. (2004). Control of fruit patterning in Arabidopsis by indehiscent. Cell 116 843–853 10.1016/S0092-8674(04)00217-X [DOI] [PubMed] [Google Scholar]
- Lodha M., Marco C. F, Timmermans M. C. P. (2013). The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of Polycomb-repressive complex2. Genes Dev. 27 596–601 10.1101/gad.211425.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann J. U., Hong R. L., Hobe M., Busch M., Parcy F., Simon R., et al. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105 793–803 10.1016/S0092-8674(01)00384-1 [DOI] [PubMed] [Google Scholar]
- Long J. A., Moan E. I., Medford J. I., Barton M. K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69 10.1038/379066a0 [DOI] [PubMed] [Google Scholar]
- Luo M., Yu C.-W., Chen F.-F., Zhao L., Tian G., Liu X., et al. (2012). Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in Arabidopsis. PLoS Genet. 8:e1003114 10.1371/journal.pgen.1003114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani E., Hake S. (2008). KNOX Lost the OX: the Arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain. Plant Cell 20 875–887 10.1105/tpc.108.058495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele G., Ori N., Sato Y., Hake S. (2003). The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev. 17 2088–2093 10.1101/gad.1120003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrusan Z., Reiser L., Feldmann K. A., Fischer R. L., Haughn G. W. (1994). Homeotic transformation of ovules into carpel-like structures in Arabidopsis. Plant Cell 6 333–349 10.1105/tpc.6.3.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K., Brocchieri L, Bürglin T. R. (2009). A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 26 2775–94 10.1093/molbev/msp201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Kay P., Wilson S., Swain S. M. (2009). Arabidopsis DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are Polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21 216–233 10.1105/tpc.108.063768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A., Louvet R., Johansen J. N., Salsac F., Morin H., Fournet F., et al. (2011). The transcription factor BELLRINGER modulates phyllotaxis by regulating the expression of a pectin methylesterase in Arabidopsis. Development 138 4733–4741 10.1242/dev.072496 [DOI] [PubMed] [Google Scholar]
- Pelaz S., Tapia-López R., Alvarez-Buylla E. R., Yanofsky M. F. (2001). Conversion of leaves into petals in Arabidopsis. Curr. Biol. 11 182–184 10.1016/S0960-9822(01)00024-0 [DOI] [PubMed] [Google Scholar]
- Phelps-Durr T. L., Thomas J., Vahab P, Timmermans M. C. P. (2005). Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17 2886–2898 10.1105/tpc.105.035477.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragni L., Belles-Boix E., Gunl M., Pautot V. (2008). Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis Inflorescences. Plant Cell 20 888–900 10.1105/tpc.108.058230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., Robinson-beerst K. A. Y., Ray S., Bakert S. C., Lang J. D., Preusst D., et al. (1994). Arabidopsis floral homeotic gene BELL (BELI) controls ovule development through negative regulation of AGAMOUS gene (AG). Proc. Natl. Acad. Sci. U.S.A. 91 5761–5765 10.1073/pnas.91.13.5761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser L., Modrusan Z., Margossian L., Samach A., Ohad N., Haughn G. W., et al. (1995). The BELL1 gene encodes a homeodomain involved in pattern formation in the Arabidopsis ovule primordium. Cell 83 735–742 10.1016/0092-8674(95)90186-8 [DOI] [PubMed] [Google Scholar]
- Reyes-olalde J. I., Zuniga-Mayo V. M., Chavez Montes R. A., Marsch-Martιnez N, de Folter S. (2013). Inside the gynoecium: at the carpel margin. Trends Plant Sci. 18 644–655 10.1016/j.tplants.2013.08.002 [DOI] [PubMed] [Google Scholar]
- Ripoll J. J., Roeder A. H. K., Ditta G. S., Yanofsky M. F. (2011). A novel role for the floral homeotic gene APETALA2 during Arabidopsis fruit development. Development 138 5167–76 10.1242/dev.073031 [DOI] [PubMed] [Google Scholar]
- Robinson-Beers K., Pruitt R. E., Gasser C. S. (1992). Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell 4 1237–1249 10.1105/tpc.4.10.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder A. H. K., Ferra C., Yanofsky M. F. (2003). The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr. Biol. 13 1630–1635 10.1016/j.cub.2003.08.027 [DOI] [PubMed] [Google Scholar]
- Roeder A. H. K., Yanofsky M. F. (2006). Fruit development in Arabidopsis. Arabidopsis Book 4:e0075 10.1199/tab.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romera-Branchat M., Ripoll J. J., Yanofsky M. F., Pelaz S. (2012). The WOX13 homeobox gene promotes replum formation in the Arabidopsis thaliana fruit. Plant J. 37–49 10.1111/tpj.12010 [DOI] [PubMed] [Google Scholar]
- Rutjens B., Bao D., van Eck-Stouten E., Brand M., Smeekens S., Proveniers M. (2009). Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. Plant J. 58 641–654 10.1111/j.1365-313X.2009.03809.x [DOI] [PubMed] [Google Scholar]
- Sablowski R. (2011). Plant stem cell niches: from signalling to execution. Curr. Opin. Plant Biol. 14 4–9 10.1016/j.pbi.2010.08.001 [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Kamiya N., Ueguchi-tanaka M., Iwahori S., Matsuoka M. (2001). KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Gene 15 581–590 10.1101/gad.867901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield S., Dewitte W., Murray J. A. (2007). The KNOX gene SHOOT MERISTEMLESS is required for the development of reproductive meristematic tissues in Arabidopsis. Plant J. 50 767–781 10.1111/j.1365-313X.2007.03095.x [DOI] [PubMed] [Google Scholar]
- Shi C.-L., Stenvik G.-E., Vie A. K., Bones A. M., Pautot V., Proveniers M., et al. (2011). Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway. Plant Cell 23 2553–2567 10.1105/tpc.111.084608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D.-Q., Yang W.-C. (2011). Ovule development in Arabidopsis: progress and challenge. Curr. Opin. Plant Biol. 14 74–80 10.1016/j.pbi.2010.09.001 [DOI] [PubMed] [Google Scholar]
- Smith H. M., Boschke I., Hake S. (2002). Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc. Natl. Acad. Sci. U.S.A. 99 9579–9584 10.1073/pnas.092271599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. M., Hake S. (2003). The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 15 1717–1727 10.1105/tpc.012856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D. R., Bowman J. L., Meyerowitz E. M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767 10.1105/tpc.2.8.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K., Girin T., Liljegren S. J., Ljung K., Robles P., Galván-Ampudia C. S., et al. (2009). A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature 459 583–586 10.1038/nature07875 [DOI] [PubMed] [Google Scholar]
- Spinelli S. V, Martin A. P., Viola I. L., Gonzalez D. H., Palatnik J. F. (2011). A mechanistic link between STM and CUC1 during Arabidopsis development. Plant Physiol. 156 1894–1904 10.1104/pp.111.177709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Xu Y., Ng K., Ito T. (2009). A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 23 1791–1804 10.1101/gad.1800409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venglat S. P., Dumonceaux T., Rozwadowski K., Parnell L., Babic V., Keller W., et al. (2002). The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 99 4730–4735 10.1073/pnas.072626099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Q., Xu W. H., Ma L. G., Fu Z. M., Deng X. W., Li J. Y., et al. (2006). Requirement of KNAT1/BP for the development of abscission zones in Arabidopsis thaliana. J. Integr. Plant Biol. 48 15–26 10.1111/j.1365-313X.2007.03096.x [DOI] [Google Scholar]
- Wolf S., Mouille G., Pelloux J. (2009). Homogalacturonan methyl-esterification and plant development. Mol. Plant 2 851–860 10.1093/mp/ssp066 [DOI] [PubMed] [Google Scholar]
- Xu X. M., Wang J., Xuan Z., Goldshmidt A., Borrill P. G. M., Hariharan N., et al. (2011). Chaperonins facilitate KNOTTED1 cell-to-cell trafficking and stem cell function. Science 333 1141–1144 10.1126/science.1205727 [DOI] [PubMed] [Google Scholar]
- Yu L., Patibanda V, Smith H. M. S. (2009). A novel role of BELL1-like homeobox genes, PENNYWISE and POUND-FOOLISH, in floral patterning. Planta 229 693–707 10.1007/s00425-008-0867-861 [DOI] [PubMed] [Google Scholar]