Abstract
EpCAM is a 40-kD type I transmembrane protein that is overexpressed in human epithelial cancers, and is currently the target of molecular therapy based on its overexpression at the cell surface. Recently, we and others have demonstrated a role for EpCAM in cell signaling and carcinogenesis, and EpCAM expression appears to promote breast cancer invasion. Interleukin-8 (IL-8/CXCL-8) is an inflammatory cytokine that has recently been shown to modulate breast cancer invasion and angiogenesis. In preliminary experiments we identified a correlation between EpCAM and IL-8 expression in primary human breast cancers. Specific ablation of EpCAM in breast cancer cell lines results in decreased IL-8 expression, and IL-8 contributes to EpCAM-dependent breast cancer invasion. Specific ablation of EpCAM is also associated with decreased nuclear factor-κB (NF-κB) transcription factor activity, decreased phosphorylation of the NF-κB family member RELA, and increased IκBα protein expression. EpCAM modulates IL-8 expression at baseline, and following IL-1β stimulation, which is known to be a potent inducer of NF-κB in breast cancer. In functional rescue experiments, specific ablation of RELA, or forced expression of the NF-κB inhibitor protein IκBα prevented EpCAM-dependent rescue of IL-8 promoter activity. These studies demonstrate for the first time that EpCAM can modulate NF-κB transcription factor activity and IL-8 expression in breast cancer, and confirm the role of EpCAM signaling in modulating breast cancer invasion. Further study is required to define the molecular mechanism(s) of EpCAM signaling in breast cancer, and to direct the rational development of molecular therapies targeting EpCAM.
Keywords: EpCAM, IL-8, NF-κB, breast cancer
INTRODUCTION
The epithelial cell adhesion molecule (EpCAM, TACSTD1/CD326) is a type I transmembrane protein that is expressed at the basolateral membrane of the majority of normal epithelial tissues. EpCAM is perhaps best known for the fact that it is overexpressed in the majority of human epithelial carcinomas including colorectal, breast, gastric, prostate, ovarian and lung cancer (1, 2). EpCAM was the first human tumor-associated antigen to be identified with monoclonal antibodies (3), and was the first target of monoclonal antibody therapy in humans (4). Although initial attempts at clinical translation have been disappointing, there are a number of second-generation molecular therapies targeting EpCAM currently under investigation (5) .
Recent studies have demonstrated a role for EpCAM in cell signaling, carcinogenesis, and stem cell biology (6-9). We have focused on the impact of EpCAM signaling in breast cancer biology. In initial studies, we demonstrated that specific ablation of EpCAM is associated with decreased invasion in breast cancer (8, 10), and that EpCAM is directly regulated by the tumor suppressor protein p53 (11). In subsequent studies we demonstrated that the transcription factor activator protein 1 (AP-1) is an important downstream mediator of EpCAM signaling in breast cancer biology through the MEKK1-MKK7-JNK cascade, contributing to EpCAM-dependent breast cancer invasion (12). AP-1 is overexpressed in breast cancer, and is considered to be a central transcription factor in the regulation of invasion (13).
IL-8 is a member of the CXC chemokine family, and is a major mediator of inflammatory responses. In inflammatory responses, IL-8 is secreted by macrophages and other cell types and functions as a chemoattractant, and potent angiogenic factor (14). Recent studies have also highlighted the role of IL-8 in cancer biology (15). Increased expression of IL-8 and/or its receptors has been demonstrated on cancer cells, endothelial cells, and tumor-associated macrophages within the tumor microenvironment. IL-8 signaling has been shown to promote proliferation, survival and invasion of cancer cells, and angiogenesis in endothelial cells (16). In breast cancer, IL-8 is most commonly overexpressed in estrogen receptor-negative breast cancers (17). IL-8 expression is associated with increased breast cancer invasion in vitro (18), and serum levels of IL-8 in breast cancer patients appear to correlate with advanced disease (19). In this study we explore the relationship between EpCAM and IL-8 expression in breast cancer, demonstrating for the first time that EpCAM can modulate NF-κB transcription factor activity and IL-8 expression. These studies provide valuable insights into the role of EpCAM in the regulation of breast cancer invasion and angiogenesis.
MATERIALS AND METHODS
Cell lines and reagents
The MDA-231, MCF-7,and HUVEC cell lines were obtained from the American Type Culture Collection (ATCC, Rockville, MD). The CA1a breast cancer cell line was described previously (20). Breast cancer cells were maintained in DMEM supplemented with 10%FBS and antibiotics (GIBCO BRL). Recombinant IL-1β, and recombinant EpCAM-Fc were purchased from R&D Systems (Minneapolis, MN).
RNA interference
The pSicoR and related lentiviral shRNA constructs were obtained from Dr. Tyler Jacks (21). shRNA sequences targeting EpCAM in the 3’ UTR region, at nucleotide 271, and a scrambled control sequence were created as previously described (10).
Plasmid constructs
The full length open reading frame of EpCAM and IL-8 was amplified from the MCF-7 and MDA-231 breast cancer cell lines respectively and sub-cloned into both pcDNA3 and the retroviral vector pBABE. Mutant and wildtype IL-8 promoter reporter constructs are described elsewhere (22). shRELA (p65 shRNA) plasmid was obtained from Dr Steven Grant of the Medical College of Richmond, Va. pBabe-IkbαSS (super suppressor, S32A,S36A) was purchased form Addgene, Cambridge MA (Plasmid # 15291).
Retroviral transduction
To create stable EpCAM expression rescue (RES) cells, Phoenix-ECO packaging cells were transfected with 2.5 μg of pBABE-EpCAM using FuGENE HD (Promega, Madison, WI). After 24 hours the medium was replaced with 10% FBS. After 24 hours viral supernatants were collected, filtered through 0.45 micron filters and then added to growing cells with 8 μg/mL protamine sulfate. After two successive infections, cells were grown for 48 hours and selected for 2 weeks.
Primary breast cancer RNA samples
RNA from primary breast cancer and matched normal breast samples was obtained from Siteman Cancer Center Tissue Procurement Core (Washington University School of Medicine, Saint Louis, MO).
qRT-PCR
RNA was purified from cell lines using RNAeasy (Qiagen, Valencia, CA). Three micrograms of RNA were reverse transcribed using a cDNA synthesis kit (Ambion, Austin, TX). Quantitative mRNA expression was measured using SYBR green chemistry and an ABI Prism 7700 Sequence Detector (Life Technologies, Carlsbad, CA). Primer sequences are available upon request. Each reaction was performed in triplicate, and the data is representative of two independent RNA preparations.
Immunoblotting
For phosphoprotein immunoblots, stimulated cells were washed with ice-cold PBS and lysed in cell lysis buffer with protease inhibitor cocktail (Cell Signaling Technology, Danvers, MA). Protein concentrations were determined by BCA protein assay (Pierce, Rockford, IL). 20-30 μg protein was subjected to SDS-PAGE (NuPAGE, Life Technologies), and transferred by electrophoresis to a PVDF membrane. Antibodies were obtained from Santa Cruz Biotechnology Santa Cruz, CA (EpCAM, β-Actin, RELA), and Cell Signaling Technology Beverly, MA (phospho-RELA-ser-536, IκBα). Signal detection was performed using the SuperSignal West Pico chemiluminescent immunodetection system (Thermo Scientific, Rockford, IL). To quantify band density, immunoblots were developed on film, scanned and pixels in each band measured using Image J software. Plots of each lane were generated, and the area under the peak corresponding to the appropriate control sample was determined, and arbitrarily set at 1.0.
IL-8 ELISA
Conditioned media from MDA-231, CA1a and MCF-7 cells were collected after 24 hours and analyzed in triplicate using a human IL-8/CXCL-8-specific ELISA kit (R&D Systems).
Analysis of breast cancer gene expression data
To assess EpCAM and IL-8 expression in primary breast cancers, we used the GOBO datasets described (23). We used the Gene Set Analysis (GSA) tool to assess EpCAM and IL-8 expression in a precompiled data sets consisting of gene expression data and annotation data for 1881 primary breast cancer specimens.
Invasion assay
Transfected or stably transduced cells (4×104) were added to matrigel transwell invasion chambers or control transwell chambers (BD Biosciences, San Jose, CA) and incubated for 24-72 hours with chemoattractant media (Clonetics, Lonza, Walkersville, MD) supplemented with growth factors. Cells invading through the matrigel or control membranes were fixed using 70% ethanol, stained with 0.1% crystal violet and photographed in four fields to cover the entire area. Cells were counted from all fields by a scientist blinded to the experimental conditions.
Angiogenesis assay
The angiogenesis assay was carried out using the in vitro angiogenesis assay kit from Millipore. Briefly 5 × 103 HUVEC cells were seeded in a 96-well plate containing polymerized EC matrix. After 2 hours, conditioned media from MDA-231 cells was added for 8-12 hours and images were captured and branch points were counted as recommended by the manufacturer.
Reporter assays
pTA-Luc, pTA-AP-1-Luc, pTA-NF-κB-Luc and pRL-TKRen-Luc were obtained from Clontech Laboratories (Mountain View, CA). The pGL3-C/EBPb reporter construct was obtained from Dr. J. Cardinaux (University of Lausanne, Switzerland). 400 ng control pTA-Luc or the indicated reporter constructs and 20 ng of pRL-TK-Luc were transiently transfected using either Lipofectamine LTX (Life Technologies) FuGENE HD (Promega) for MCF-7 cells. After 24 hours, cells were washed and placed in serum-free media. 24 hours later, reporter activity was determined using the Dual-Luciferase kit (Promega).
Statistical analysis
Numerical data are presented as the mean values ± the standard deviation. Statistical significance was evaluated by the Student's t test. p-values < 0.05 were considered to be statistically significant. Significant results are indicated in the appropriate figures with an asterisk.
RESULTS
EpCAM and IL-8 expression are correlated in human breast cancer
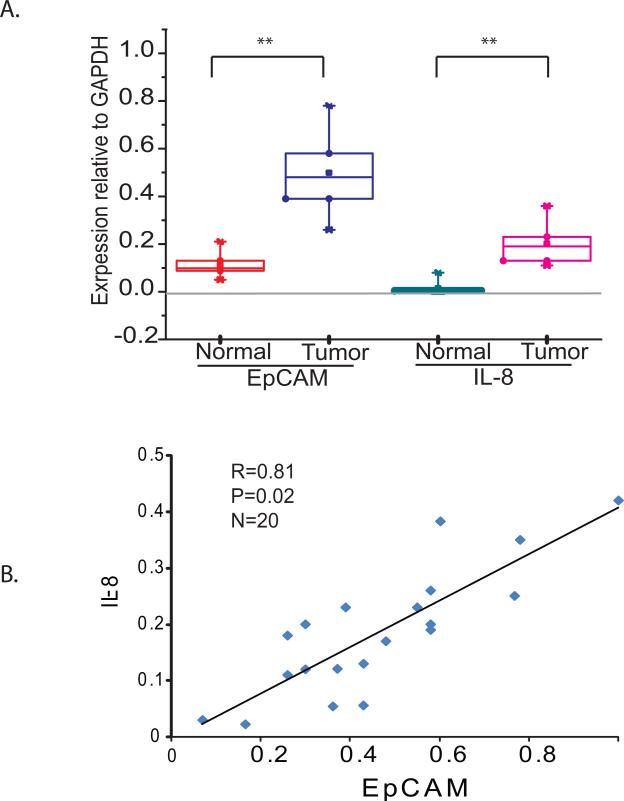
To investigate whether EpCAM and IL-8 expression are correlated in primary human breast cancers, we obtained normal breast tissue and matched primary breast cancer tissue samples from the Siteman Cancer Center Tissue Procurement Core. EpCAM and IL-8 mRNA expression levels were measured by quantitative RT-PCR. Both EpCAM and IL-8 expression are increased in primary breast cancers compared to normal breast tissue (Figure 1A). In addition, EpCAM and IL-8 expression appear to be correlated in primary breast cancers (Figure 1B, R = 0.81, p value = 0.02). To extend these analyses, we evaluated gene expression profiling data available online using “Gene expression-based Outcome for Breast cancer Online, GOBO” (23). In a precompiled and clinically annotated data set consisting of gene expression data from 1881 primary breast cancer specimens, we found that EpCAM and IL-8 expression are coordinately expressed in high-grade tumors, estrogen receptor-negative tumors, and in specific intrinsic subtypes of breast cancer (basal-like, HER2+) (Figure S1).
Figure 1. EpCAM and IL-8 expression is increased and correlated in human breast cancer.
(A) RNA from normal breast tissue and matched primary breast cancers (n = 10 each) was obtained from the Siteman Cancer Center Tissue Procurement Core. EpCAM and IL-8 expression in individual samples was measured by quantitative RT-PCR and the results are summarized by box plot. (B) EpCAM and IL-8 expression was measured by quantitative RT-PCR in twenty primary breast cancer specimens. The strength of the linear relationship between EpCAM and IL-8 expression was assessed by calculating the Pearson correlation coefficient (R) P=0.02.
Specific ablation of EpCAM decreases IL-8 expression
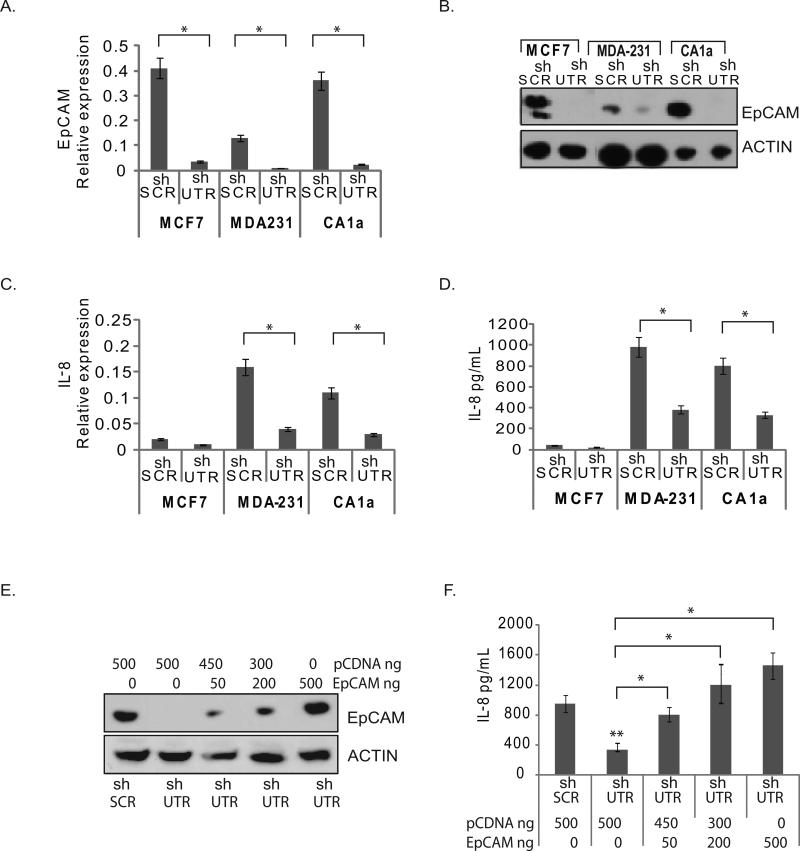
To determine if EpCAM modulates IL-8 expression in breast cancer, we manipulated EpCAM expression in three well-characterized breast cancer cell lines (MCF-7, MDA-231, and CA1a), using loss-of-function and gain-of-function experimental systems. In our initial studies, we transduced the MCF-7, MDA-231, and CA1a breast cancer cell lines with lentiviral shRNA vectors targeting EpCAM or an irrelevant nucleotide sequence. Stable transduction with shRNA vectors targeting EpCAM (shUTR) was associated with greater than 90% decrease in EpCAM mRNA and protein expression in all three cell lines (Figure 2A, 2B), and greater than 60% decrease in IL-8 mRNA and protein expression in the MDA-231 and CA1a breast cancer cell lines (Figure 2C, 2D). To control for the off-target effects of RNA interference, additional studies were performed in CA1a cells using shRNA constructs targeting EpCAM at a second nucleotide sequence, with similar results (Figure S2). Of note, MCF-7 has minimal IL-8 expression at baseline. However, EpCAM does modulate IL-8 expression in MCF-7 cells following stimulation with IL-1β (Figure S3). As an additional control, we rescued EpCAM expression using a cDNA construct resistant to RNA interference. Increasing doses of EpCAM cDNA are associated with increasing EpCAM protein expression as measured by immunoblot, and increasing IL-8 protein expression as measured by ELISA or qRT-PCR (Figure 2E, 2F, and Figure S2).
Figure 2. Specific ablation of EpCAM decreases IL-8 expression.
(A-D) MCF-7, MDA-231 and CA1a breast cancer cell lines were transduced with lentiviral shRNA constructs targeting EpCAM (shUTR) or an irrelevant nucleotide sequence (shSCR). Stably transduced cells were subsequently analyzed for EpCAM expression by qRT-PCR (A), and protein immunoblot (B). IL-8 expression was measured by RT-PCR (C) and ELISA (D). For the ELISA assay, transduced cells were seeded onto 6-well plates, and conditioned media was collected after 24 hrs. (E, F) Transduced breast cancer cells were rescued with empty vector (pcDNA) or an EpCAM construct resistant to RNA interference at the indicated doses. Increasing doses of the EpCAM construct resulted in a dose-dependent increase in EpCAM expression as measured by protein immunoblot (E), and IL-8 expression as measured by ELISA (F).Asterisks indicate significance at P value <0.05 when compared to control.
IL-8 contributes to EpCAM-dependent breast cancer invasion
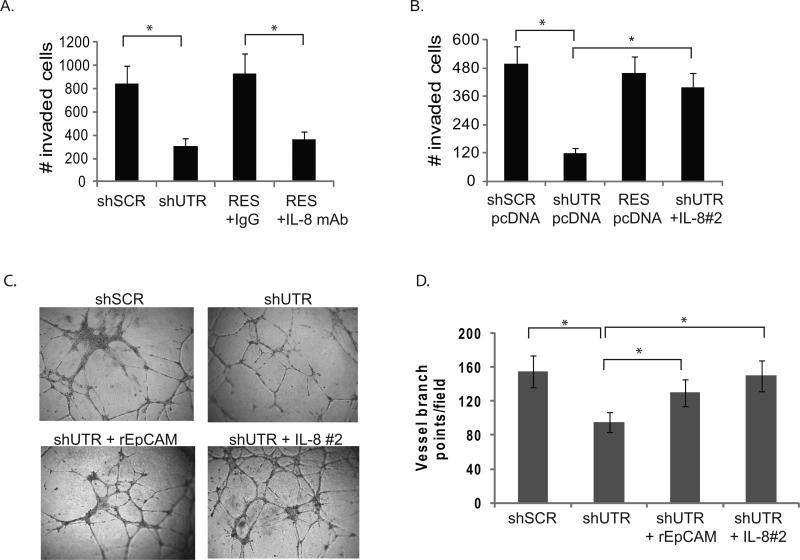
Both EpCAM and IL-8 have been reported to contribute to breast cancer invasion. Given that EpCAM and IL-8 expression is highly correlated in primary human breast cancers, and the evidence that EpCAM can modulate IL-8 expression in breast cancer cell lines in vitro, we hypothesized that IL-8 contributes to EpCAM-dependent breast cancer invasion and angiogenesis. To test this hypothesis, we specifically ablated EpCAM expression in MDA-231 cells, and then rescued IL-8 expression using genetic constructs (Figure 3A). Invasion was measured in transwell invasion assays. As observed previously (8, 12), specific ablation of EpCAM significantly decreases MDA-231 invasion (Figure 3A). Invasion can be rescued by forced expression of an EpCAM construct resistant to RNA interference, but addition of IL-8 neutralizing antibody abrogates the ability of EpCAM to rescue invasion (Figure 3A). Of note, forced expression of IL-8 using two independent clones is able to rescue invasion following specific ablation of EpCAM (Figure 3B and Figure S4). This data is in agreement with recent report showing IL-8 neutralizing antibody decreased invasion of breast cancer cells (24). IL-8 is a multifunctional cytokine, and IL-8 signaling has been implicated in the recruitment and activation of immune cells, proliferation, survival and invasion of cancer cells, and promotion of angiogenesis in endothelial cells (16). To assess the impact of EpCAM expression on angiogenesis, we specifically ablated EpCAM expression in MDA-231 breast cancer cells. Additional experimental conditions included rescue of EpCAM signaling with soluble recombinant EpCAM, or IL-8 rescue with a genetic construct. Conditioned media was harvested and then added to human umbilical vein endothelial cells (HUVEC) plated on matrigel, a standard angiogenesis assay (16). Conditioned media from MDA-231 cells transduced with an irrelevant control shRNA induced HUVEC cells to form numerous elongated tube-like structures within 24 hr (Figure 3C, 3D). Conditioned media from MDA-231 cells transduced with shRNA targeting EpCAM, induced HUVEC cells to form partially broken tube-like structures, but no thick vessels or extensions. This decrease in vessel formation was rescued with either recombinant EpCAM or forced expression of IL-8. Taken together, these studies confirm that modulation of IL-8 expression contributes to EpCAM-dependent breast cancer invasion and angiogenesis.
Figure 3. EpCAM-dependent modulation of IL-8 expression contributes to breast cancer invasion and angiogenesis.
(A-B) MDA-231 cells were stably transduced with lentiviral shRNA constructs targeting EpCAM (shUTR), or an irrelevant nucleotide sequence (shSCR). Transduced MDA-231 cells were subsequently rescued with vector (shUTR, see Figure S2). Trypsinized cells were washed with 10% FBS and Opti-MEM (Invitrogen). Mouse IgG or IL-8 neutralizing antibody (R&D systems, clone 6217.111) was added to cells, and the cells were transferred to matrigel chambers. Triplicate sets were harvested after 24h stained and counted. B. In independent experiments, tumor cell invasion was assessed using MDA-231 cells containing, shSCR, shUTR, shIL-8, EpCAM resistant to RNA interference (RES), or with a genetic construct encoding IL-8 (two clones were analyzed independently, designated IL-8#1, IL-8#2 see Figure S4). Invasion was evaluated in a matrigel transwell invasion assay. (C, D) MDA-231 cells were stably transduced with the lentiviral vectors as detailed above. EpCAM signaling was rescued by addition of 100 ng/mL recombinant EpCAM, or IL-8 expression was rescued with a genetic construct. 100 μL of conditioned media was added to HUVEC cells growing in matrigel in a 96 well plate. After 14 hours the cells were washed and photographed, and the number of vessel branch points/high-power field was determined by a scientist blinded to the experimental conditions. Asterisks indicate significance at P value <0.05 when compared to control.
Specific ablation of EpCAM is associated with decreased IL-8 and NF-κB transcription factor activity
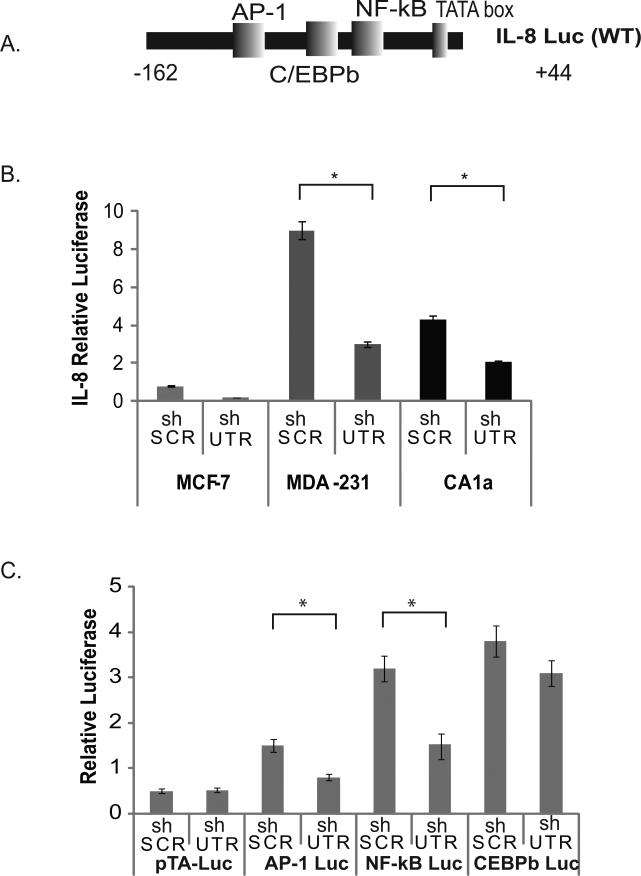
We used a reporter construct containing the core IL-8 promoter fused to luciferase (Figure 4A) to demonstrate that specific ablation of EpCAM is associated with decreased IL-8 promoter activity in the MCF-7, MDA-231 and CA1a breast cancer cell lines (Figure 4B). In order to determine which transcription factors are most likely to contribute to EpCAM-dependent modulation of IL-8 expression in breast cancer, we used luciferase reporters containing response elements from the AP-1, NF-κB and C/EBPβ transcription factors (Figure 4C). Transfection of these reporter constructs into stably transduced MDA-231 breast cancer cell lines demonstrates that specific ablation of EpCAM is associated with decreased AP-1, and NF-κB transcription factor activity but minimal change in C/EBPβ transcription factor activity (Figure 4C). Similar results were observed in CA1a cells (data not shown). To determine the role of AP-1 and NF-κB in EpCAM-dependent modulation of IL-8 expression, we used IL-8 reporter constructs with mutated AP-1 and NF-κB binding sites (Figure S5). In these studies, deletion of the AP-1 binding site results in a 50% reduction in IL-8 promoter activity, and mutation of the NF-κB binding site results in an 80% reduction in IL-8 promoter activity (Figure S5), suggesting an important contribution from NF-κB.
Figure 4. EpCAM modulates IL-8 promoter activity and NF-κB transcription factor activity.
(A) A schematic diagram of the IL-8 promoter is indicated, demonstrating AP-1, C/EBPβ, and NF-κB binding sites. (B) MCF-7, MDA-231, and CA1a breast cancer cells were stably transduced with lentiviral shRNA constructs targeting EpCAM (shUTR) or an irrelevant nucleotide sequence (shSCR). To measure IL-8 promoter activity, transduced cells were transfected with constructs containing the IL-8 promoter fused to luciferase, and a transfection control construct, TK-renilla. Relative luciferase activity was analyzed using a dual-luciferase kit. (C) MDA-231 cells transduced with the indicated lentiviral constructs were transfected with non-promoter luciferase reporter constructs for AP-1, C/EBPβ, NF-κB, and transfection control TK-renilla. Relative luciferase activity was analyzed using a dual-luciferase kit (Promega). Asterisks indicate significance at P value <0.05 when compared to control.
EpCAM modulates RELA phosphorylation and NF-κB contributes to EpCAM-dependent IL-8 expression
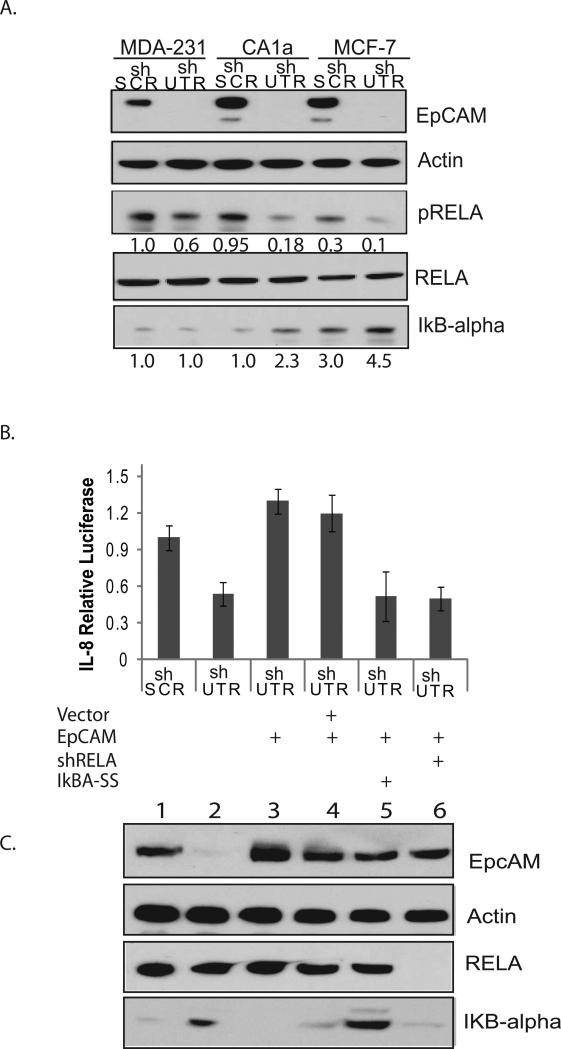
The NF-κB family is composed of homo-dimers and heterodimers of REL family proteins including p65 (RELA), c-REL, RELB, p52 and p50 (25). The predominant form of NF-κB is a heterodimer composed of p50 and RELA. At baseline, NF-κB is sequestered in the cytoplasm by IκB proteins. Upon activation, IκB proteins are degraded and NF-κB translocates to the nucleus. Phosphorylation of RELA is required for maximal NF-κB transcriptional activity, and the p50/RELA heterodimer is activated in the majority of ER-negative breast cancers (25). To evaluate the impact of EpCAM expression on NF-κB transcriptional activity, we specifically ablated EpCAM in MCF-7, MDA-231, and CA1a breast cancer cells and measured RELA phosphorylation (ser-536), and IκBα protein expression. Specific ablation of EpCAM results in a > 90% decrease in EpCAM expression, an approximate 40-70% decrease in RELA phosphorylation, and increased IκBα protein expression (Figure 5A). These studies suggest that modulation of NF-κB may contribute to EpCAM-dependent regulation of IL-8 in breast cancer.
Figure 5. NF-κB contributes to EpCAM-dependent IL-8 expression.
(A) MCF-7, MDA-231, and CA1a breast cancer cells stably transduced with the indicated lentiviral shRNA constructs and were grown to 70% confluence. EpCAM, phosphorylated RELA, IκBα, and actin expression were measured by immunoblot. Band density was analyzed by densitometric analysis using ImageJ software, arbitrarily assigning the band density of shSCR at 1.0. (B, C) CA1a breast cancer cells were stably transduced with the indicated lentiviral constructs, and then transiently transfected with an EpCAM cDNA construct resistant to RNA interference, and genetic constructs targeting NF-κB. Expression of EpCAM, RELA and IκBα was confirmed by immunoblot, and the impact on IL-8 promoter activity was measured using a dual-luciferase kit.
To evaluate EpCAM-dependent regulation of IL-8 transcription in more detail, we specifically ablated EpCAM in CA1a breast cancer cells, and then rescued EpCAM with a cDNA construct resistant to RNA interference. Specific ablation and rescue of EpCAM expression were confirmed by immunoblot (Figure 5C). Rescue of EpCAM expression increases IL-8 promoter activity, establishing a functional rescue model system (Figure 5B). CA1a rescue cells were then transiently transfected with genetic constructs targeting NF-κB (RELA shRNA and dominant-negative IκBαSS). IL-8 promoter activity was then measured using luciferase reporters. Specific ablation of RELA or forced expression of the dominant-negative construct IκBα-SS prevented EpCAM-dependent rescue of IL-8 promoter activity. These results confirm that NF-κB is a key downstream mediator of EpCAM-dependent regulation of IL-8 in breast cancer.
EpCAM expression modulates IL-8 expression and NF-κB transcription factor activity following IL-1β stimulation
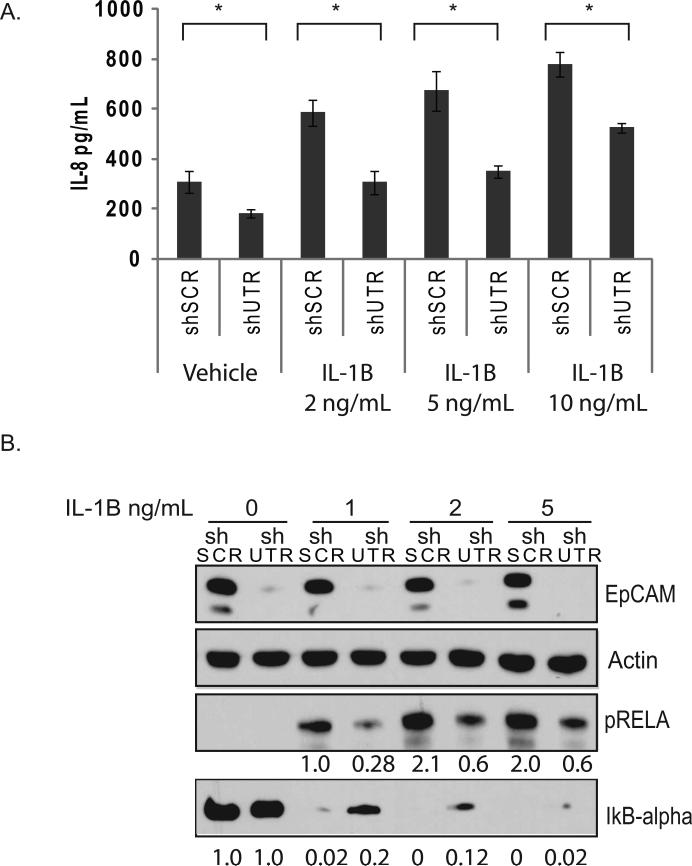
To confirm and extend the observation that EpCAM modulates NF-κB transcription factor activity and IL-8 expression, we performed additional experiments in the presence of IL-1β, an inflammatory cytokine that is known to activate NF-κB in breast cancers (26). CA1a breast cancer cells were stably transduced with lentiviral vectors targeting EpCAM (shUTR) or control nucleotide sequences, serum-starved for 24 hours, and then stimulated with increasing doses of IL-1β. IL-8 protein expression (Figure 6A) increases when stimulated with IL-1β, but specific ablation of EpCAM decreases IL-8 expression at all concentrations of IL-1β tested. Similar results were also observed in the MCF-7 breast cancer cell line (Figure S3). To evaluate NF-κB activity following IL-1β stimulation, we stimulated CA1a cells with IL-1β, and then measured RELA phosphorylation and IκBα protein expression. Specific ablation of EpCAM results in a > 90% decrease in EpCAM expression, and an approximate 70% decrease in RELA phosphorylation at all concentrations of IL-1β tested (Figure 6B). These studies provide additional evidence that EpCAM-dependent modulation of NF-κB contributes to the regulation of IL-8 in breast cancer.
Figure 6. EpCAM modulates IL-1β-induced IL-8 expression, and RELA phosphorylation.
(A) CA1a cells were stably transduced with the indicated shRNA constructs and stimulated for 18 hours with the indicated doses of IL-1β (ng/mL). IL-8 protein expression was measured by ELISA. (B) Stably transduced CA1a cells were serum-starved for 24 hours and then stimulated for 15 minutes with the indicated doses of IL-1β (ng/mL). EpCAM, phosphorylated RELA, IκBα, and actin expression were measured by immunoblot. Band density was analyzed by densitometric analysis using ImageJ. Asterisks indicate significance at P value <0.05 when compared to shSCR.
DISCUSSION
EpCAM is a cell surface molecule that is overexpressed in the majority of human epithelial cancers, including breast cancer. We have previously shown that EpCAM expression modulates breast cancer invasion, through the JNK/AP-1 signal transduction pathway (8, 12). The present study builds on these findings, demonstrating for the first time that EpCAM expression can also modulate IL-8 expression, and the NF-κB signal transduction pathway in Ras positive cells. EpCAM and IL-8 expression is increased in human breast cancer specimens, and manipulation of EpCAM expression in vitro modulates IL-8 expression and NF-κB signaling at baseline, and following IL-1β stimulation.
NF-κB is a pleiotropic sequence-specific transcription factor that contributes to the regulation of such diverse processes as inflammation, innate immunity, apoptosis, and cell proliferation (25). NF-κB is often constitutively activated in cancers, including breast cancer (27, 28). NF-κB is activated in cancers by a range of stimuli, including various pro-inflammatory cytokines (IL-1β, and tumor necrosis factor α), growth factors (epidermal growth factor), DNA-damaging agents (radiation) and oncogenes (Ras). Of note, NF-κB and IL-1β are known to be overexpressed in estrogen receptor-negative breast cancers (25), and IL-1β stimulation has been associated with breast cancer invasion and IL-8 expression (29). Our studies indicate that EpCAM expression modulates IL-1β-induced NF-κB activation, and concomitant IL-8 production.
IL-8 is known to be regulated primarily by the AP-1, NF-κB and C/EBPβ transcription factors, depending on the cell type, and/or signaling pathway involved (29). We used luciferase reporters containing response elements from the AP-1, NF-κB and C/EBPβ transcription factors to determine how EpCAM modulates IL-8 in breast cancers. Specific ablation of EpCAM results in a marginal decrease in C/EBPβ reporter activity, a 50% decrease in AP-1 reporter activity, and an 80% decrease in NF-κB reporter activity. Deletion of the NF-κB binding site results in a greater than 80% reduction in IL-8 promoter activity. Our data are consistent with the hypothesis that EpCAM regulates IL-8 expression by modulating NF-κB transcription factor activity. However, as EpCAM is also known to modulate AP-1 transcription factor activity, it is likely that EpCAM-dependent modulation of AP-1transcription factor activity also contributes to the regulation of IL-8.
Recently Maetzel et al. confirmed the role of EpCAM in cell signaling, defining a potential pathway based on regulated intramembrane proteolysis (RIP) (6). In these studies, activation of EpCAM is dependent on RIP, with shedding of the ectodomain of EpCAM, and nuclear translocation of the intracellular domain of EpCAM (EpCAMICD). EpCAMICD is able to form a nuclear complex with FHL2, β-catenin, and Lef-1 to induce gene transcription and Wnt signaling. It is not clear if EpCAM modulates IL-8 expression and NF-κB transcription factor activity through a similar pathway that is dependent upon RIP. Although many of the changes observed are occurring in the cytoplasm (IκBα protein expression, RELA phosphorylation), these could be secondary to changes induced following EpCAMICD nuclear localization. It has been previously reported (30) that EpCAM expression in squamous cell carcinoma is negatively regulated by TNF-alpha through the activation of NF-κB. This raises the possibility that some type of feedback loop may contribute to the regulation of EpCAM expression and NF-κB activity. Additional study is required to assess this possibility.
There is evidence to suggest that the impact of EpCAM signaling is tissue-dependent. For example, EpCAM expression in primary cancer specimens has been studied extensively, and a number of studies in the surgical pathology literature have evaluated the association between EpCAM expression and prognosis. One inconsistency in the literature is that EpCAM expression in primary cancer specimens appears to be associated with a favorable prognosis in some cancer types, and an unfavorable prognosis in other cancer types. EpCAM expression in primary breast cancer appears to be associated with decreased patient survival (31-33), but EpCAM expression in colorectal cancer appears to be associated with improved patient survival (34). This inconsistency is paralleled in functional studies of EpCAM biology performed in vitro. Loss-of-function analyses using RNA interference suggest that EpCAM expression is associated with increased invasion in breast cancer (8), and gain-of-function analyses in colorectal and lung cancer suggest that EpCAM expression is associated with decreased cancer invasion in these cancer types (35, 36).
The context-dependent impact of EpCAM signaling has important implications in breast cancer. Recent gene expression studies using DNA microarrays demonstrate that breast cancer can be subdivided into different intrinsic subtypes (37). These intrinsic subtypes differ markedly in prognosis (38-40), and represent a developing paradigm shift in the clinical management of breast cancer and the application of targeted therapies. The studies reported here highlight the role of EpCAM expression in modulating signaling pathways that are predominant in ER-negative breast cancers, as discussed above. A better understanding of the relationship between EpCAM and breast cancer invasion in specific breast cancer intrinsic subtypes will clearly facilitate the rational design, and successful application of molecular therapies targeting EpCAM in epithelial carcinomas.
In conclusion, this study confirms the role of EpCAM in regulating breast cancer invasion, angiogenesis, and demonstrates for the first time that EpCAM can also modulate IL-8 expression, and an autocrine NF-κB signal transduction pathway. These findings have important implications for the rational development of molecular therapies targeting EpCAM, and suggest that further study is required to define the molecular mechanism(s) of EpCAM-dependent modulation of breast cancer invasion.
Supplementary Material
Acknowledgements and Grant support
Support for this study was provided by Susan G. Komen for the Cure. The authors thank Jason Weber for helpful discussions. We are grateful to Dr Steven Grant of the Medical College of Richmond, Va, for providing NF-κB p65 shRNA plasmid.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interests were disclosed.
Authors' Contributions
Conception and design: N Sankpal, WE Gillanders.
Development of methodology: N Sankpal, T Fleming.
Acquisition of data: N Sankpal, T Fleming.
Analysis and interpretation of data: N Sankpal, T Fleming, WE Gillanders.
Writing of the manuscript: WE Gillanders, N Sankpal, T Fleming.
REFERENCES
- 1.Baeuerle PA, Gires O. EpCAM (CD326) finding its role in cancer. Br J Cancer. 2007;96:417–23. doi: 10.1038/sj.bjc.6603494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Went PT, Lugli A, Meier S, Bundi M, Mirlacher M, Sauter G, et al. Frequent EpCam protein expression in human carcinomas. Hum Pathol. 2004;35:122–8. doi: 10.1016/j.humpath.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 3.Herlyn M, Steplewski Z, Herlyn D, Koprowski H. Colorectal carcinoma-specific antigen: detection by means of monoclonal antibodies. Proc Natl Acad Sci U S A. 1979;76:1438–42. doi: 10.1073/pnas.76.3.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sears HF, Atkinson B, Mattis J, Ernst C, Herlyn D, Steplewski Z, et al. Phase-I clinical trial of monoclonal antibody in treatment of gastrointestinal tumours. Lancet. 1982;1:762–5. doi: 10.1016/s0140-6736(82)91811-6. [DOI] [PubMed] [Google Scholar]
- 5.Schmoll HJ, Arnold D. When wishful thinking leads to a misty-eyed appraisal: the story of the adjuvant colon cancer trials with edrecolomab. J Clin Oncol. 2009;27:1926–9. doi: 10.1200/JCO.2008.20.6284. [DOI] [PubMed] [Google Scholar]
- 6.Maetzel D, Denzel S, Mack B, Canis M, Went P, Benk M, et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol. 2009;11:162–71. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- 7.Munz M, Kieu C, Mack B, Schmitt B, Zeidler R, Gires O. The carcinoma-associated antigen EpCAM upregulates c-myc and induces cell proliferation. Oncogene. 2004;23:5748–58. doi: 10.1038/sj.onc.1207610. [DOI] [PubMed] [Google Scholar]
- 8.Osta WA, Chen Y, Mikhitarian K, Mitas M, Salem M, Hannun YA, et al. EpCAM is overexpressed in breast cancer and is a potential target for breast cancer gene therapy. Cancer Res. 2004;64:5818–24. doi: 10.1158/0008-5472.CAN-04-0754. [DOI] [PubMed] [Google Scholar]
- 9.Munz M, Baeuerle PA, Gires O. The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res. 2009;69:5627–9. doi: 10.1158/0008-5472.CAN-09-0654. [DOI] [PubMed] [Google Scholar]
- 10.Sankpal NV, Fleming TP, Gillanders WE. Dual expression lentiviral vectors for concurrent RNA interference and rescue. J Surg Res. 2009;156:50–6. doi: 10.1016/j.jss.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sankpal NV, Willman MW, Fleming TP, Mayfield JD, Gillanders WE. Transcriptional repression of epithelial cell adhesion molecule contributes to p53 control of breast cancer invasion. Cancer Res. 2009;69:753–7. doi: 10.1158/0008-5472.CAN-08-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sankpal NV, Mayfield JD, Willman MW, Fleming TP, Gillanders WE. Activator protein 1 (AP-1) contributes to EpCAM-dependent breast cancer invasion. Breast Cancer Res. 2011;13:R124. doi: 10.1186/bcr3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozanne BW, Spence HJ, McGarry LC, Hennigan RF. Transcription factors control invasion: AP-1 the first among equals. Oncogene. 2007;26:1–10. doi: 10.1038/sj.onc.1209759. [DOI] [PubMed] [Google Scholar]
- 14.Moser B, Wolf M, Walz A, Loetscher P. Chemokines: multiple levels of leukocyte migration control. Trends Immunol. 2004;25:75–84. doi: 10.1016/j.it.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Waugh DJ, Wilson C. The interleukin-8 pathway in cancer. Clin Cancer Res. 2008;14:6735–41. doi: 10.1158/1078-0432.CCR-07-4843. [DOI] [PubMed] [Google Scholar]
- 16.Li A, Dubey S, Varney ML, Dave BJ, Singh RK. IL-8 directly enhanced endothelial cell survival, proliferation, and matrix metalloproteinases production and regulated angiogenesis. J Immunol. 2003;170:3369–76. doi: 10.4049/jimmunol.170.6.3369. [DOI] [PubMed] [Google Scholar]
- 17.Yao C, Lin Y, Chua MS, Ye CS, Bi J, Li W, et al. Interleukin-8 modulates growth and invasiveness of estrogen receptor-negative breast cancer cells. Int J Cancer. 2007;121:1949–57. doi: 10.1002/ijc.22930. [DOI] [PubMed] [Google Scholar]
- 18.Freund A, Chauveau C, Brouillet JP, Lucas A, Lacroix M, Licznar A, et al. IL-8 expression and its possible relationship with estrogen-receptor-negative status of breast cancer cells. Oncogene. 2003;22:256–65. doi: 10.1038/sj.onc.1206113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benoy IH, Salgado R, Van Dam P, Geboers K, Van Marck E, Scharpe S, et al. Increased serum interleukin-8 in patients with early and metastatic breast cancer correlates with early dissemination and survival. Clin Cancer Res. 2004;10:7157–62. doi: 10.1158/1078-0432.CCR-04-0812. [DOI] [PubMed] [Google Scholar]
- 20.Santner SJ, Dawson PJ, Tait L, Soule HD, Eliason J, Mohamed AN, et al. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res Treat. 2001;65:101–10. doi: 10.1023/a:1006461422273. [DOI] [PubMed] [Google Scholar]
- 21.Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, et al. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci U S A. 2004;101:10380–5. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Hara AM, Bhattacharyya A, Mifflin RC, Smith MF, Ryan KA, Scott KG, et al. Interleukin-8 induction by Helicobacter pylori in gastric epithelial cells is dependent on apurinic/apyrimidinic endonuclease-1/redox factor-1. J Immunol. 2006;177:7990–9. doi: 10.4049/jimmunol.177.11.7990. [DOI] [PubMed] [Google Scholar]
- 23.Ringner M, Fredlund E, Hakkinen J, Borg A, Staaf J. GOBO: gene expression-based outcome for breast cancer online. PLoS One. 2011;6:e17911. doi: 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aceto N, Duss S, Macdonald G, Meyer DS, Roloff TC, Hynes NE, et al. Co-expression of HER2 and HER3 receptor tyrosine kinases enhances invasion of breast cells via stimulation of interleukin-8 autocrine secretion. Breast Cancer Res. 2012;14:R131. doi: 10.1186/bcr3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basseres DS, Baldwin AS. Nuclear factor-kappaB and inhibitor of kappaB kinase pathways in oncogenic initiation and progression. Oncogene. 2006;25:6817–30. doi: 10.1038/sj.onc.1209942. [DOI] [PubMed] [Google Scholar]
- 26.Buss H, Dorrie A, Schmitz ML, Hoffmann E, Resch K, Kracht M. Constitutive and interleukin-1-inducible phosphorylation of p65 NF-{kappa}B at serine 536 is mediated by multiple protein kinases including I{kappa}B kinase (IKK)-{alpha}, IKK{beta}, IKK{epsilon}, TRAF family member-associated (TANK)-binding kinase 1 (TBK1), and an unknown kinase and couples p65 to TATA-binding protein-associated factor II31-mediated interleukin-8 transcription. J Biol Chem. 2004;279:55633–43. doi: 10.1074/jbc.M409825200. [DOI] [PubMed] [Google Scholar]
- 27.Biswas DK, Shi Q, Baily S, Strickland I, Ghosh S, Pardee AB, et al. NF-kappa B activation in human breast cancer specimens and its role in cell proliferation and apoptosis. Proc Natl Acad Sci U S A. 2004;101:10137–42. doi: 10.1073/pnas.0403621101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao Y, Karin M. NF-kappaB in mammary gland development and breast cancer. J Mammary Gland Biol Neoplasia. 2003;8:215–23. doi: 10.1023/a:1025905008934. [DOI] [PubMed] [Google Scholar]
- 29.Freund A, Jolivel V, Durand S, Kersual N, Chalbos D, Chavey C, et al. Mechanisms underlying differential expression of interleukin-8 in breast cancer cells. Oncogene. 2004;23:6105–14. doi: 10.1038/sj.onc.1207815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gires O, Kieu C, Fix P, Schmitt B, Munz M, Wollenberg B, et al. Tumor necrosis factor alpha negatively regulates the expression of the carcinoma-associated antigen epithelial cell adhesion molecule. Cancer. 2001;92:620–8. doi: 10.1002/1097-0142(20010801)92:3<620::aid-cncr1362>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 31.Spizzo G, Obrist P, Ensinger C, Theurl I, Dunser M, Ramoni A, et al. Prognostic significance of Ep-CAM AND Her-2/neu overexpression in invasive breast cancer. Int J Cancer. 2002;98:883–8. doi: 10.1002/ijc.10270. [DOI] [PubMed] [Google Scholar]
- 32.Spizzo G, Went P, Dirnhofer S, Obrist P, Simon R, Spichtin H, et al. High Ep-CAM expression is associated with poor prognosis in node-positive breast cancer. Breast Cancer Res Treat. 2004;86:207–13. doi: 10.1023/B:BREA.0000036787.59816.01. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt M, Hasenclever D, Schaeffer M, Boehm D, Cotarelo C, Steiner E, et al. Prognostic effect of epithelial cell adhesion molecule overexpression in untreated node-negative breast cancer. Clin Cancer Res. 2008;14:5849–55. doi: 10.1158/1078-0432.CCR-08-0669. [DOI] [PubMed] [Google Scholar]
- 34.Went P, Vasei M, Bubendorf L, Terracciano L, Tornillo L, Riede U, et al. Frequent high-level expression of the immunotherapeutic target Ep-CAM in colon, stomach, prostate and lung cancers. Br J Cancer. 2006;94:128–35. doi: 10.1038/sj.bjc.6602924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basak S, Speicher D, Eck S, Wunner W, Maul G, Simmons MS, et al. Colorectal carcinoma invasion inhibition by CO17-1A/GA733 antigen and its murine homologue. J Natl Cancer Inst. 1998;90:691–7. doi: 10.1093/jnci/90.9.691. [DOI] [PubMed] [Google Scholar]
- 36.Tai KY, Shiah SG, Shieh YS, Kao YR, Chi CY, Huang E, et al. DNA methylation and histone modification regulate silencing of epithelial cell adhesion molecule for tumor invasion and progression. Oncogene. 2007;26:3989–97. doi: 10.1038/sj.onc.1210176. [DOI] [PubMed] [Google Scholar]
- 37.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 38.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A. 2003;100:10393–8. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.