Abstract
Background:
Proteomics-based approaches for biomarker discovery are promising strategies used in cancer research. We present state-of-art label-free quantitative proteomics method to assess proteome of renal cell carcinoma (RCC) compared with noncancer renal tissues.
Methods:
Fresh frozen tissue samples from eight primary RCC lesions and autologous adjacent normal renal tissues were obtained from surgically resected tumour-bearing kidneys. Proteins were extracted by complete solubilisation of tissues using filter-aided sample preparation (FASP) method. Trypsin digested proteins were analysed using quantitative label-free proteomics approach followed by data interpretation and pathways analysis.
Results:
A total of 1761 proteins were identified and quantified with high confidence (MASCOT ion score threshold of 35 and P-value <0.05). Of these, 596 proteins were identified as differentially expressed between cancer and noncancer tissues. Two upregulated proteins in tumour samples (adipose differentiation-related protein and Coronin 1A) were further validated by immunohistochemistry. Pathway analysis using IPA, KOBAS 2.0, DAVID functional annotation and FLink tools showed enrichment of many cancer-related biological processes and pathways such as oxidative phosphorylation, glycolysis and amino acid synthetic pathways.
Conclusions:
Our study identified a number of differentially expressed proteins and pathways using label-free proteomics approach in RCC compared with normal tissue samples. Two proteins validated in this study are the focus of on-going research in a large cohort of patients.
Keywords: kidney cancer, proteomics, urinary biomarkers, Coronin 1A
Renal cell carcinoma (RCC) is a heterogeneous disease and its incidence has been increasing over the past 20 years (Drucker, 2005). It is the most common (>90%) kidney cancer and accounts for 2–3% of all adult malignancies (Rini and Atkins, 2009). Worldwide, there were ∼209 000 new cases of RCC diagnosed with corresponding 102 000 RCC-specific deaths in 2006 (Gupta et al, 2008). Every year, ∼4000 people die in the United Kingdom because of RCC. Renal cell carcinoma is often asymptomatic; the onset of symptoms such as haematuria and abdominal pain usually, but not always, indicate advanced disease. A third of cases, however, are metastatic at the time of their initial diagnosis (Weiss and Lin, 2006) and 30% of the patients develop metastatic disease following surgical extirpation of clinically localised disease (Campbell et al, 2003; Janzen et al, 2003).
Management of metastatic disease (>50% of RCC disease) is a real clinical challenge. Previous systemic treatment for metastatic disease with interleukin-2 and interferon-α was associated with modest survival benefit at best (Pyrhonen et al, 1999). Significant research, however, aimed at identification of abnormal signal transduction in RCC has provided insights into therapeutic molecular targets (Najjar and Rini, 2012) and pathways – the mTOR signalling pathway and the hypoxia-inducible pathway being the two main pathways involved in clear cell RCC. The clear cell histology is the most common subtype of RCC (Escudier et al, 2012). Targeted therapies developed in the recent years for metastatic disease inhibit tyrosine kinase (sunitinib, axitinib, pazopanib and sorafenib), vascular endothelial growth factor (VEGF) (bevacizumab) and mTOR (temsirolimus and everolimus) (Singer et al, 2013). Nevertheless, for the locally advanced RCC, there is no approved adjuvant therapy in place. Even with the recent advances in chemotherapy, durable and complete recovery remains elusive.
Given the enormity of the unmet clinical needs, research is needed to uncover the molecular basis of RCC. Even with the speedy progress in understanding cancer biology in general, the primary events leading to RCC remain unclear. Recent advances in high-throughput technologies have enabled us to analyse system-wide changes simultaneously and these advancements have been successfully used to understand molecular mechanisms and to identify biochemical markers of diseases. Although microarrays allow quantification of gene expression, studying at protein level remains a complementary method (Lichtenfels et al, 2009). Moreover, studying at protein level is desirable as mRNA levels do not always correlate well with the protein abundances. Proteomics-based approaches allow analyses not only at translational levels (Lichtenfels et al, 2009), but also at complex post-translational levels (Walther and Mann, 2010). Early in 2001, Sarto et al (2001) reviewed the application of two-dimensional electrophoresis-based proteomics in RCC and discussed the role of mitochondrial enzyme manganese superoxide dismutase in the regulatory functions of cells. In 2003, Seliger et al (2003) reviewed the progress in identifying RCC-associated biomarkers using proteomics and transcriptomics approaches and compared the complementarity between these two ‘omics' technologies. Their review showed a considerable number of proteins differentially expressed in RCC compared with healthy tissue: overexpression of manganese superoxide dismutase, heat shock protein 27, cytokeratin 8, stathmin and vimentin, and underexpression of ubiquitinol cytochrome C reductase, NADH-ubiquinone oxidoreductase complex 1 and isoforms of the plasma glutathione peroxidase in RCC. Recently, Masui et al (2013) used isobaric tags for relative and absolute quantitation (iTRAQ) proteomics method to compare protein expression profiles of metastatic and localised RCC and identified 29 proteins differentially expressed (12 overexpressed and 17 underexpressed in metastatic RCC) between them. Higher expressions of profilin-1, 14-3-3 ζ/δ and galectin-1 proteins were found in metastatic RCC in their study and correlated with poor prognosis. Perroud et al (2009) carried out liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based proteomics study on 50 FFPE samples (normal kidney and clear cell renal cancer). This study identified and quantified 777 proteins, of which 105 were differentially, expressed between Fuhrman grades 1–4 clear cell kidney cancer and normal kidney tissues. Further analysis showed grade-dependent alteration in glycolytic and amino acid synthetic pathways, in addition to proteins in acute phase and xenobiotic metabolism signalling.
Quantitative proteomics has been used to identify and quantify proteins in complex biological samples (Wang et al, 2008). The classical method for quantitative analysis of protein mixtures is by protein separation and comparison by two-dimensional polyacrylamide gel electrophoresis (2D-PAGE), followed by MS or MS/MS (Balabanov et al, 2001; Shi et al, 2004). However, 2D-PAGE technique suffers from its inability to analyse hydrophobic, very high or low molecular weight proteins (Wang et al, 2008). Moreover, this technique remains a labour-intensive approach, requiring several different experiments for high-throughput studies.
To address this, non-gel-based quantitative proteomics methods have been developed to widen the protein dynamic range and profile (Wang et al, 2008; Zhu et al, 2010). These approaches utilise isotope-labelled compounds that are identical to the properties of their natural compounds except in mass that allows for their identification in mass spectrometry. Stable labelling approaches deployed include isotope-coded affinity tag (ICAT), stable isotope labelling by amino acids in cell culture (SILAC), iTRAQ, 15N/14N metabolic labelling and other chemical labelling (Chen and Yates, 2007; Veenstra, 2007). These approaches are coupled to LC for separation before MS or MS/MS identification. Some limitations of the above approaches include increased sample preparation time, more complex methodology and higher costs attributed to labelling reagents (Wang et al, 2008). Furthermore, simultaneous quantification using labelling methods is only possible between few samples (Wang et al, 2008; Zhu et al, 2010). Label-free quantification is an alternate to often costly labelling strategies in complex samples like cancer tissues. The use of this approach has increased enormously in the past 10 years and has shown potential for identification and quantification of differentially expressed proteins in normal and diseased samples. The strategy of using label-free method in complex samples has the potential as a screening tool in biomarker discovery.
Here, we describe use of label-free MS to compare protein expression in RCC vs noncancer renal tissue from the same tumour-bearing kidneys. The major objectives were to discover differentially expressed proteins between RCC and noncancer renal tissues in order to infer altered signalling and metabolic pathways in RCC.
Materials and methods
Tayside Urological Cancer Network (TUCAN), Dundee, Scotland in collaboration with Tayside Tissue Bank, Dundee, Scotland has established a large bio-repository of resected renal cancer tissues with prior ethical approval (approval number 12/ES/0083). Using a validated protocol, renal tissue samples were prospectively collected from patients undergoing nephron-sparing or radical nephrectomy. From the same kidney specimen, two samples were collected: one from healthy renal tissue (noncancer tissue) and another from renal cancer (cancer tissue). In total, the study had eight pairs of tissues, providing 16 samples for further processing. Label-free quantitative proteomics approach of the present study included four basic steps: (1) sample preparation – protein extraction, reduction alkylation and digestion; (2) sample separation by LC and analysis by MS/MS; (3) data analyses – peak picking, ion abundance quantification, peptide and protein identification, quantification and statistical analyses; and (4) data interpretation and pathway analysis.
Protein extraction, reduction, alkylation and digestion
None of the participants received neoadjuvant chemotherapy, immunotherapy or radiotherapy. The tissue samples were washed with normal saline and stored at −70 °C following surgery. Before processing, samples were cut on dry ice to give approximate weights between 15 and 25 mg. Individual samples were soaked in 300 μl of butanol and heated at 95 °C for 5 min to remove lipids (modification of protocol for this study). During standardisation, filter-aided sample preparation (FASP) method without prior butanol treatment using fresh frozen tissues did not yield enough proteins. This was thought to be because of the presence of lipids known to significantly reduce protein extraction. Further development of the method included soaking and heating the samples in butanol and centrifugation to recover proteins for further processing. This standardised approach was then adopted in our study to overcome the interference from lipids in nanoLC-MS analysis. Proteins were recovered as pellets after centrifugation at 13 000 g, and were completely solubilised in sodium dodecyl sulphate and digested in solution with trypsin using the FASP method (Wisniewski et al, 2009).
Sample separation by LC and analysis by MS/MS
Following FASP digestion, peptides from the samples were quantified using NanoVue spectrophotometer (GE Healthcare, Hertfordshire, UK) and normalised before LC/MS analysis. Analysis of peptides was performed on a Velos orbitrap (Thermo Scientific, Bremen, Germany) mass spectrometer coupled with a Dionex Ultimate 3000 RS (Thermo Scientific). The following LC buffers were used: buffer A (2% acetonitrile and 0.1% formic acid in Milli-Q water (Merck, Darmstadt, Germany) (v/v)) and buffer B (80% acetonitrile and 0.08% formic acid in Milli-Q water (v/v)). Aliquots of 2 μl of each sample were loaded at 5 μl min−1 onto a trap column (100 μm × 2 cm, PepMap nanoViper C18 column, 5 μm, 100 Å, Thermo Scientific) and equilibrated in 98% buffer A. The trap column was washed for 3 min at the same flow rate and then the trap column was switched in-line with a Thermo Scientific, resolving C18 column (75 μm × 15 cm, PepMap RSLC C18 column, 2 μm, 100 Å). The peptides were eluted from the column at a constant flow rate of 300 nl min−1 with a linear gradient from 98% buffer A to 40% buffer B in 90 min and then to 90% buffer B by 92 min. The column was then washed with 98% buffer B for 10 min and reequilibrated in 98% buffer A for 24 min. LTQ-Orbitap Velos (Thermo Scientific) was used in data-dependent mode. A scan cycle comprised MS1 scan (m/z range from 335 to 1800) in the velos orbitrap followed by 10 sequential-dependant MS2 scans (the threshold value was set at 5000 and the minimum injection time was set at 200 ms) in LTQ with collision-induced dissociation. The resolution of the Orbitrap Velos was set at to 60 000. To ensure mass accuracy, the mass spectrometer was calibrated on the first day that the runs were performed. To monitor MS performance throughout the analysis, a QC sample consisting of 100 fmole of 6 bovine proteins digest (ARC Sciences, Hampshire, UK) was run between every 10 samples. The samples were randomised and ran in triplicate.
Abundance quantification
In total, 48 samples (8 specimens × 2; cancer and noncancer; × 3 technical replicates) were analysed in nanoLC-MS. The raw LC-MS/MS data were imported into Progenesis LC-MS (version 4.0, Nonlinear Dynamics Limited, Newcastle upon Tyne, UK) software and the ion intensity maps for all the 48 sample runs were examined visually for defects. There were no detectable defects in any of the runs and all the 48 samples were included in the analysis. Based on visual examination, total number of peaks and total intensity, one of the samples was selected as reference and all other ion intensity maps from rest of the samples were aligned to it using the ‘Automatic Alignment' function in the software. After all the runs were aligned with the reference, peptide ions (features) with charge state over 5 were considered unreliable and were filtered out and removed. Ion abundance quantifications were computed from the peak volumes of the ion chromatograms using protein correlation profiling method (Schulze and Usadel, 2010). Protein abundances were calculated from the sum of all normalised unique peptide ion abundances for a specific protein on each run.
Peptide/protein identification
We used MASCOT protein search engine (www.matrixscience.com – local installation on the server) to identify the peptides in the MS/MS spectra. To improve the quality of the spectral data being used in the search, MS/MS spectra of the top three ‘ranks' for each feature were exported to MASCOT search engine. Using the peptide score distribution from the MASCOT search results, ion score threshold of 35 (corresponding to P-value <0.05) was arrived and the identified peptides with ion scores of <35 were discarded. The filtered MASCOT search results were imported back into the Progenesis LC-MS and conflicts for peptide assignments at protein level were examined case by case and resolved appropriately. Only the proteins with at least two uniquely identified peptides were retained for further analysis.
Statistical analysis
To explore the patterns in the protein quantification data and to highlight the similarities and differences between the cancer and noncancer groups, we performed principal component analysis (in R language) and plotted the first three principal components using RGL package. We also performed hierarchical clustering using correlation as a distance metric on the protein quantification data in Progenesis LC-MS. In addition, we performed hierarchical clustering analysis using pvclust package in R (http://cran.r-project.org). To examine the differences in protein abundances between cancer and noncancer tissues, we performed analysis of variance (ANOVA) test between the cancer and noncancer tissue groups in Progenesis LC-MS. In addition, the protein abundance data were analysed in R–Bioconductor using ‘limma' package to identify differentially expressed proteins between these two groups.
Pathways analysis
As we were interested in the signalling networks and metabolic pathways enriched in the differentially expressed proteins, we analysed these using three different publically available pathways enrichment tools that use well-established databases. As these tools would not recognise some of the protein IDs, the protein identifiers (UniProtKB AC/ID, UniRef100 and IPI) were mapped to Entrez Gene IDs posting the data to UniProt (EMBL-EBI, Cambridge, UK; SIB, Geneva, Switzerland; PIR, Washington, DC, USA) Knowledgebase using a Perl script. The mapped Entrez Gene IDs from the differential expression analyses were used to identify enriched pathways and diseases in KOBAS 2.0 online tool (Xie et al, 2011), DAVID functional annotation tool (Huang da et al, 2009) and FLink (frequency-weighted links tool; http://www.ncbi.nlm.nih.gov/Structure/flink/flink.cgi). In addition, the differentially expressed proteins were analysed using IPA (Build version: 212183, Release date: 05-02-2013, Ingenuity Systems, www.ingenuity.com).
Immunohistochemical analysis
Among the identified proteins, we chose to validate two proteins that have been reported to be of prognostic significance in renal or any other cancers and have commercially available antibodies. However, validation of all other remaining proteins using immunohistochemistry is in progress. Immunohistochemical staining on tissue sections of the eight patients with renal cancer and noncancer tissue was performed as described previously (King et al, 2012). Antigen retrieval and deparaffinisation was performed using DAKO (Cambridge, UK) EnVision FLEX Target Retrieval solution (high pH) buffer in a DAKO PT Link. Immunostaining using DAKO EnVision FLEX system on a DAKO Autostainer Link 48 was carried out according to the manufacturer's protocol. Sections were incubated with primary antibodies specific for Coronin 1A (US Biological, Salem, MA, USA) and adipose differentiation-related protein (ADFP; Abcam, Cambridge, UK) for 30 min. The DAKO substrate working solution was used as a chromogenic agent for 2 × 5 min and sections were counterstained in EnVision FLEX haematoxylin. Sections known to stain positively were included in each batch and negative controls were prepared by replacing the primary antibody with DAKO antibody diluents. Results were scored by an experienced renal pathologist as diffuse or focal staining.
Results
The demographic details of the participants are shown in Table 1. Peptide extracts were randomised and analysed in triplicate by nanoLC-MS on a velos orbitrap. To account for technical variations and to strengthen the quality of the data, we aliquoted 3 technical replicates from each specimen that gave us a total of 48 nanoLC-MS runs. Using the workflow described in the methods, 65 753 spectra were used in the MASCOT search that identified 17 804 peptides (P<0.05, minimum ion 2). From the identified peptides, 1761 proteins showing at least two unique peptides per protein were identified and quantified.
Table 1. Demographic characteristics of the study cohort and tumour features.
| Sr. no. | Age (years) | Sex | Tumour size (mm) | Histopathology type | Surgery | Stage | Furhman grade |
|---|---|---|---|---|---|---|---|
| 1 |
78 |
Male |
55 |
Clear cell carcinoma |
Nephrectomy |
T3bN0MO |
2 |
| 2 |
66 |
Female |
65 |
Clear cell carcinoma |
Nephrectomy |
T3bN0MO |
3 |
| 3 |
68 |
Female |
55 |
Clear cell carcinoma |
Nephrectomy |
T1bN0MO |
2 |
| 4 |
67 |
Female |
3 |
Clear cell carcinoma |
Nephrectomy |
T1aN0MO |
3 |
| 5 |
50 |
Male |
115 |
Clear cell carcinoma |
Nephrectomy |
T3aN0MO |
3 |
| 6 |
60 |
Female |
46 |
Clear cell carcinoma |
Nephrectomy |
T1bN0MO |
2 |
| 7 |
82 |
Female |
82 |
Clear cell carcinoma |
Nephrectomy |
T3bN0MO |
3 |
| 8 | 74 | Male | 54 | Clear cell carcinoma | Nephrectomy | T3bN0MO | 4 |
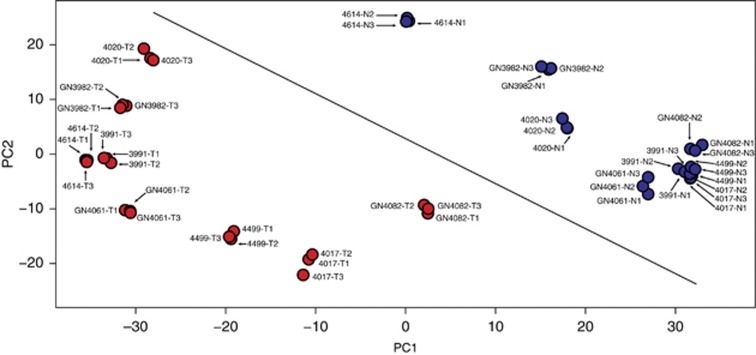
To explore the global protein expression patterns in RCC and noncancerous renal tissues, principal component analysis was performed. The first three principal components (PCs) captured 57.31, 11.5 and 6.6 percentages, respectively, of the total variance in the data set. Figure 1 shows complete separation of cancer and noncancer tissue groups by both PC1 and PC2, indicating a clear difference in protein expression between the two groups. Convincingly, all the three technical replicates from the same specimens are tightly grouped, giving us confidence on our study. The plots show possible existence of well-defined differences between the protein abundances of cancer and noncancer samples at the global level. To show the general pattern of proteome in cancer and noncancer samples in the data set, we have drawn an arbitrary diagonal line in the PCA plot (Figure 1). In this figure all three replicates of samples GN4082-tumour and 4614-normal have almost similar PC1 values, although they are very well separated by PC2. Furthermore, when we compare the individual specimens with their matching cancer and noncancer tissues, GN4082 tumour vs GN4082 normal and 4614 tumour vs 4614 normal, it is convincing that the cancer and noncancer samples are very well separated chiefly on the basis of PC1, giving confidence in the overall pattern. Hierarchical clustering of the protein quantification data also showed separation of the samples belonging to cancer and noncancer groups (Supplementary Data).
Figure 1.
Principal component analysis of global protein profiles in cancer and noncancer renal tissues. The plot shows principal component 1 (PC1) on X axis and principal component 2 (PC2) on Y axis, and they capture 57.31% and 11.5%, respectively, of the total variance. Blue data points denote noncancer renal tissues and red data points denote RCC tissues. The data points are marked with the sample identifiers (specimen number, noncancer/tumour tissue and technical replicate 1/2/3). An arbitrary diagonal line was drawn across the plot to show the separation of cancer and noncancer tissues. A full colour version of this figure is available at the British Journal of Cancer journal online.
To identify the proteins that were expressed differentially between the RCC and noncancer tissues, ANOVA test was performed in Progenesis LC-MS that identified 558 proteins (P<0.05 and fold change over as differentially expressed between the cancer and noncancer groups). In addition, to increase our confidence on differential expression analysis and to correct for multiple testing (not available in Progenesis LC-MS), we used linear modelling to identify differentially expressed proteins. Using FDR-adjusted P<0.05 and log2 fold change over 1.0 (linear scale fold change over 2) as cutoff, 596 proteins were identified from the linear modelling analysis as differentially expressed between the cancer and noncancer groups (Supplementary Table 1). Tables 2 and 3 show the top 25 (ranked by fold change with significant FDR-adjusted P-value) upregulated and downregulated proteins in RCC respectively. Some of the upregulated proteins in cancerous tissues include thymidine phosphorylase, annexin A4, periostin, aggrecan core protein, serpin H1, ADFP, 6-phosphofructokinase type C, Coronin 1A, von Willebrand factor, integrin-β, tyrosine-protein kinase receptor and sialic acid synthase. Proteins downregulated in cancerous tissue include synaptopodin 2, ubiquitin carboxyl-terminal hydrolase isozyme L1, L-xylulose reductase, cadherin-16, fructose-bisphosphate aldolase B, uromodulin and NADH dehydrogenase (ubiquinone).
Table 2. Top upregulated proteins in cancerous tissues compared with the normal renal parenchyma (ranked by fold change).
| Sr. no. | Protein name | Protein ID | Fold change (log2) cancer vs noncancer | P-value | FDR-adjusted P-value |
|---|---|---|---|---|---|
| 1 |
Integrin β |
tr|B4E0R1|B4E0R1_HUMAN |
5.918997073 |
8.05452E−10 |
2.86278E−09 |
| 2 |
Von Willebrand factor |
UniRef100_P04275 |
3.252970919 |
1.76848E−14 |
2.59377E−13 |
| 3 |
Ectonucleotide pyrophosphatase/phosphodiesterase family member 3 |
UniRef100_O14638 |
3.113004422 |
1.65797E−08 |
4.05426E−08 |
| 4 |
Full-length cDNA clone CS0DI085YI08 of placenta of Homo sapiens |
tr|Q86TV4|Q86TV4_HUMAN |
3.011741403 |
0.023659047 |
0.024837091 |
| 5 |
ADFP protein |
tr|Q6FHZ7|Q6FHZ7_HUMAN |
2.950019975 |
1.12143E−11 |
7.07877E−11 |
| 6 |
Histone H3.1 |
UniRef100_UPI0001D3410D |
2.898432243 |
1.31146E−09 |
4.36579E−09 |
| 7 |
Thymidine phosphorylase |
UniRef100_UPI0000E571B6 |
2.888733547 |
1.32789E−19 |
4.4794E−18 |
| 8 |
Aggrecan core protein |
tr|E7EX88|E7EX88_HUMAN |
2.745397619 |
6.5538E−06 |
9.50207E−06 |
| 9 |
Interferon-γ-inducible protein 16, isoform |
tr|D3DUZ3|D3DUZ3_HUMAN |
2.725425564 |
1.91243E−14 |
2.76483E−13 |
| 10 |
Coronin 1A |
IPI00010133 |
2.717496528 |
3.83009E−15 |
6.68285E−14 |
| 11 |
NNMT protein |
tr|Q6FH49|Q6FH49_HUMAN |
2.713371423 |
1.7417E−09 |
5.64935E−09 |
| 12 |
cDNA FLJ59379, highly similar to haematopoietic lineage cell-specific protein |
tr|B4DQ92|B4DQ92_HUMAN |
2.65230756 |
2.87995E−15 |
5.20448E−14 |
| 13 |
Fatty acid binding protein 7 |
tr|Q9H047|Q9H047_HUMAN |
2.617050396 |
5.83826E−06 |
8.56278E−06 |
| 14 |
Periostin |
tr|B1ALD8|B1ALD8_HUMAN |
2.60827884 |
3.85862E−09 |
1.15609E−08 |
| 15 |
cDNA FLJ52464, highly similar to GTPase, IMAP family member 4 |
tr|B4DWA5|B4DWA5_HUMAN |
2.527846248 |
1.22822E−15 |
2.3452E−14 |
| 16 |
ENO2 protein |
tr|Q6FHV6|Q6FHV6_HUMAN |
2.492047979 |
1.45322E−10 |
6.47865E−10 |
| 17 |
Histone H1.5 |
sp|P16401|H15_HUMAN |
2.436089334 |
3.03607E−16 |
6.98295E−15 |
| 18 |
Insulin-like growth factor binding protein 7 |
tr|B4E1N2|B4E1N2_HUMAN |
2.418191943 |
2.65323E−10 |
1.08269E−09 |
| 19 |
Apoptosis-associated speck-like protein containing a CARD |
sp|Q9ULZ3|ASC_HUMAN |
2.402662738 |
4.75155E−17 |
1.20214E−15 |
| 20 |
cDNA FLJ43948 fis, clone TESTI4014924, highly similar to Homo sapiens cytoplasmic FMR1 interacting protein 1 (CYFIP1), transcript variant 1, mRNA |
tr|B3KWV6|B3KWV6_HUMAN |
2.331192861 |
4.53312E−15 |
7.64586E−14 |
| 21 |
Sialic acid synthase |
sp|Q9NR45|SIAS_HUMAN |
2.23177376 |
1.29297E−07 |
2.51631E−07 |
| 22 |
Annexin A4 |
UniRef100_P09525 |
2.209750305 |
8.41075E−13 |
7.53246E−12 |
| 23 |
6-phosphofructokinase type C |
UniRef100_Q01813 |
2.14408679 |
5.29845E−12 |
3.69795E−11 |
| 24 |
ATP-dependent RNA helicase DDX1 |
tr|A3RJH1|A3RJH1_HUMAN |
2.137678406 |
0.000942011 |
0.001063967 |
| 25 | Adenosine deaminase | tr|F5GWI4|F5GWI4_HUMAN | 2.131392334 | 3.03952E−12 | 2.27852E−11 |
Abbreviations: ADFP=adipose differentiation-related protein; FDR=false discovery rate; NNMT=nicotinamide N-methyltransferase.
Table 3. Top down-regulated proteins in cancerous tissues compared with the normal renal parenchyma (ranked by fold change).
| Sr. no. | Protein name | Protein ID | Fold change (log2) cancer vs noncancer | P-value | FDR-adjusted P-value |
|---|---|---|---|---|---|
| 1 |
Metallothionein |
tr|Q8WVB5|Q8WVB5_HUMAN |
−7.070631065 |
1.06E−14 |
1.68E−13 |
| 2 |
Synaptopodin 2 |
tr|B9EG60|B9EG60_HUMAN |
−5.063445471 |
9.29E−12 |
6.1E−11 |
| 3 |
NADH dehydrogenase (ubiquinone) 1α subcomplex, 8, 19 kDa |
tr|B1AM93|B1AM93_HUMAN |
−3.81124236 |
2.51E−36 |
2.54E−33 |
| 4 |
Ubiquitin carboxyl-terminal hydrolase isozyme L1 |
sp|P09936|UCHL1_HUMAN |
−3.744218547 |
1.11E−21 |
4.86E−20 |
| 5 |
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13 kDa (NADH-coenzyme Q reductase), isoform CRA_a |
tr|Q6IBC4|Q6IBC4_HUMAN |
−3.686259992 |
3.59E−24 |
2.02E−22 |
| 6 |
L-xylulose reductase |
sp|Q7Z4W1|DCXR_HUMAN |
−3.583275422 |
4.51E−20 |
1.57E−18 |
| 7 |
Non-secretory ribonuclease |
UniRef100_P10153 |
−3.548814593 |
8.25E−10 |
2.92E−09 |
| 8 |
NADH dehydrogenase (ubiquinone) iron–sulphur protein 8, mitochondrial (fragment) |
tr|E9PPW7|E9PPW7_HUMAN |
−3.432935132 |
2.5E−27 |
3.17E−25 |
| 9 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24 kDa |
tr|Q6IPW4|Q6IPW4_HUMAN |
−3.413973508 |
8.26E−35 |
4.18E−32 |
| 10 |
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
tr|Q5VU50|Q5VU50_HUMAN |
−3.35833422 |
3.88E−22 |
1.78E−20 |
| 11 |
Cadherin-16 |
UniRef100_O75309 |
−3.349171259 |
3.13E−27 |
3.52E−25 |
| 12 |
Calbindin 1, 28 kDa, isoform CRA_b |
tr|B2R696|B2R696_HUMAN |
−3.345341765 |
4.53E−11 |
2.33E−10 |
| 13 |
Probable N-acetyltransferase 8 |
sp|Q9UHE5|NAT8_HUMAN |
−3.334487761 |
2.4E−11 |
1.36E−10 |
| 14 |
ATPase, Na+/K+ transporting, β1 polypeptide, isoform CRA_a |
tr|A3KLL5|A3KLL5_HUMAN |
−3.332531661 |
4.34E−26 |
4E−24 |
| 15 |
Sodium/glucose cotransporter |
tr|Q8WY15|Q8WY15_HUMAN |
−3.263975139 |
9.13E−10 |
3.18E−09 |
| 16 |
Fructose-bisphosphate aldolase B |
UniRef100_P05062 |
−3.222229924 |
2.6E−15 |
4.78E−14 |
| 17 |
NADH dehydrogenase (ubiquinone) 1α subcomplex subunit 2 |
UniRef100_O43678 |
−3.21030869 |
7.56E−31 |
2.55E−28 |
| 18 |
D-β-hydroxybutyrate dehydrogenase, mitochondrial |
UniRef100_Q02338 |
−3.116941388 |
7.68E−15 |
1.23E−13 |
| 19 |
Ubiquinone biosynthesis protein COQ9, mitochondria |
UniRef100_O75208 |
−3.110815803 |
1.26E−11 |
7.81E−11 |
| 20 |
Uromodulin, secreted form |
tr|E9PEA4|E9PEA4_HUMAN |
−3.088130282 |
1.78E−17 |
4.87E−16 |
| 21 |
cDNA FLJ60317, highly similar to Aminoacylase-1 (EC 3.5.1.14) |
tr|B4DNW0|B4DNW0_HUMAN |
−3.066759162 |
2.07E−18 |
6.17E−17 |
| 22 |
PDZK1-interacting protein 1 |
IPI00011858 |
−3.053007695 |
8.63E−16 |
1.78E−14 |
| 23 |
NADH dehydrogenase (ubiquinone) iron–sulphur protein 7, mitochondrial |
tr|F5GXJ1|F5GXJ1_HUMAN |
−3.048863326 |
2.46E−30 |
4.98E−28 |
| 24 |
UDP-glucuronosyltransferase 2B7 |
UniRef100_P16662 |
−3.046009696 |
6.41E−09 |
1.78E−08 |
| 25 | Glutathione peroxidase 3 | UniRef100_P22352 | −3.037043306 | 1.02E−13 | 1.08E−12 |
Abbreviation: FDR=false discovery rate.
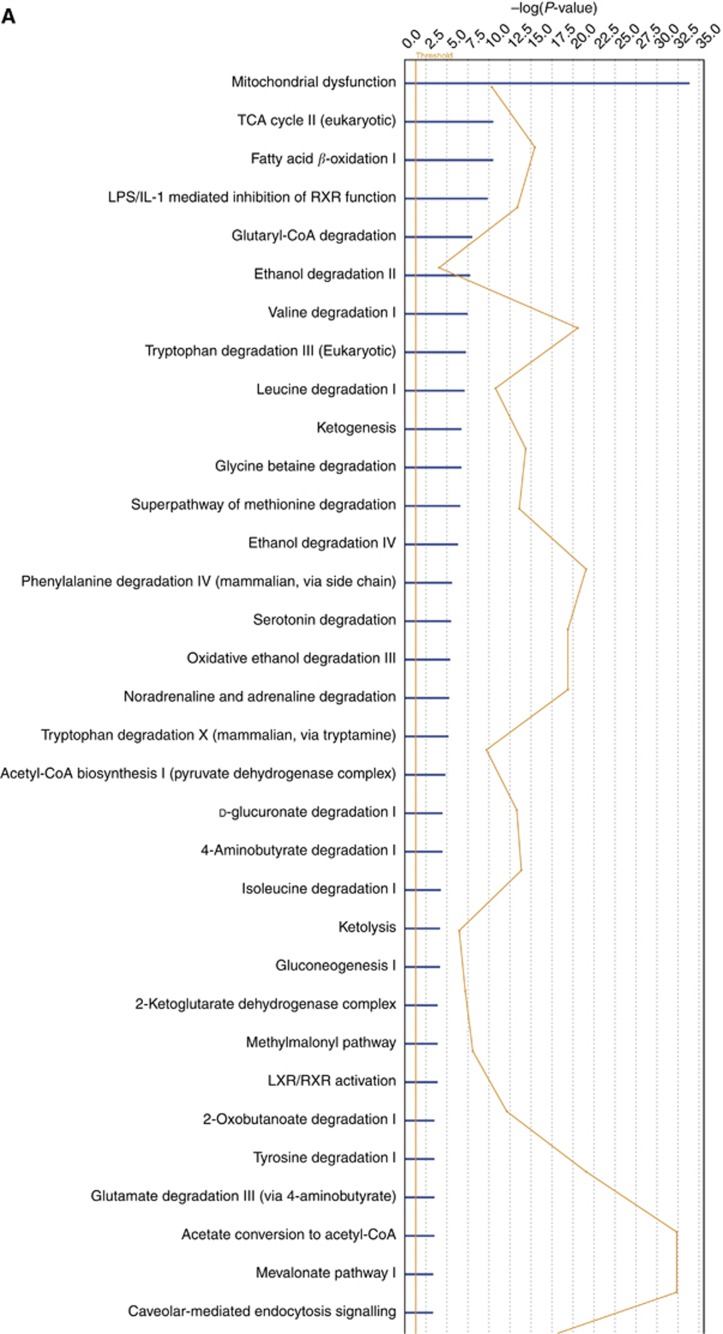
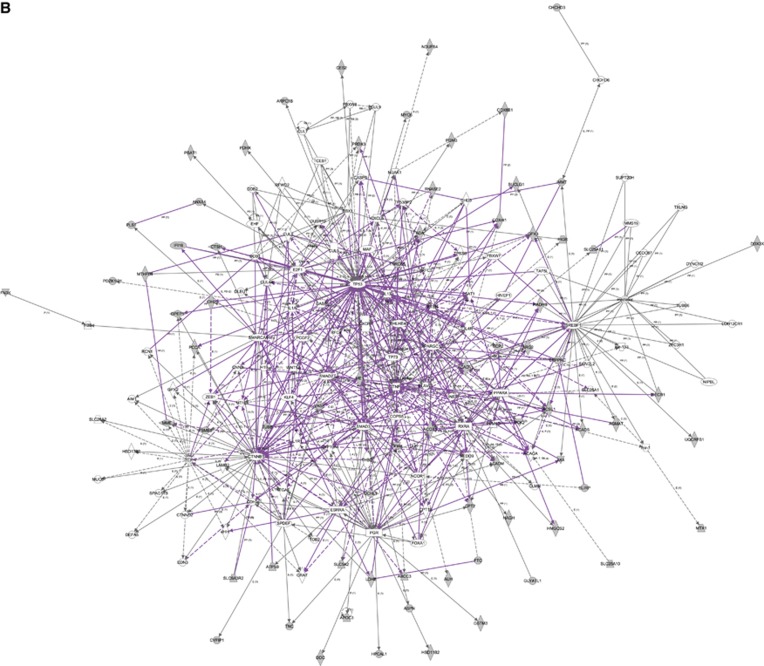
We used three publically available pathways analysis tools (webservers), namely, KOBAS 2.0, DAVID and FLink, for finding enriched pathways in the 596 differentially expressed proteins. The KOBAS 2.0 webserver uses KEGG Pathway, BioCyc, Reactome, Pathway Interaction Database and Panther databases to identify statistically enriched pathways in the differentially expressed proteins (Entrez Gene IDs), against the background of all the genes in the human genome. The important enriched pathways include oxidative phosphorylation, TCA cycle, peroxisome proliferator-activated receptor (PPAR) signalling pathway and integrin signalling pathway (Table 4). The DAVID functional annotation tool uses over 40 annotation categories incorporating many databases including Gene Ontology, protein–protein interactions, protein functional domains and pathway databases and finds enriched terms similar to KOBAS 2.0. The results from the DAVID functional annotation chart are given in Supplementary Table 1. The FLink uses a subset of Entrez databases to rank enriched terms using ‘frequency-weight' method, and the top enriched terms ranked by percentage coverage are given in Supplementary Table 2. We also used IPA for analysing the differentially expressed proteins. Mitochondrial dysfunction, LPS/IL-1-modulated inhibition of RXR and LXR/RXR are the top enriched pathways in IPA analysis (Figure 2A). The IPA network analysis showed important upstream regulators and downstream effectors involved in carcinogenesis such as PGR, PPARA, TNF, RXRA, p53, PPARGC-1-α, SMAD3 and CTNNB1 at the centre of the merged networks (Figure 2B). However, these data and confirmation of their involvement in carcinogenesis need further studies by western blot and/or immunohistochemistry.
Table 4. Top pathways enriched in the differentially expressed proteins – results from KOBAS 2.0.
| Sr. no. | Pathways | Database | ID | P-value | FDR (Benjamini and Hochberg)-corrected P-value |
|---|---|---|---|---|---|
| 1 |
Oxidative phosphorylation |
KEGG PATHWAY |
hsa00190 |
0 |
0 |
| 2 |
Valine, leucine and isoleucine degradation |
KEGG PATHWAY |
hsa00280 |
2.54241E−14 |
3.69497E−12 |
| 3 |
Pyruvate metabolism |
KEGG PATHWAY |
hsa00620 |
2.3728E−10 |
2.29897E−08 |
| 4 |
Citrate cycle (TCA cycle) |
KEGG PATHWAY |
hsa00020 |
3.09356E−10 |
2.69758E−08 |
| 5 |
Propanoate metabolism |
KEGG PATHWAY |
hsa00640 |
8.215E−10 |
5.96957E−08 |
| 6 |
Branched-chain amino acid catabolism |
PID Reactome |
500780 |
8.18667E−09 |
5.49137E−07 |
| 7 |
Glycolysis/Gluconeogenesis |
KEGG PATHWAY |
hsa00010 |
4.03624E−08 |
2.514E−06 |
| 8 |
Fatty acid metabolism |
KEGG PATHWAY |
hsa00071 |
7.23503E−08 |
3.94309E−06 |
| 9 |
ATP synthesis |
PANTHER |
P02721 |
4.77073E−07 |
2.4471E−05 |
| 10 |
Respiratory electron transport |
PID Reactome |
500282 |
2.98826E−06 |
0.000137145 |
| 11 |
Butanoate metabolism |
KEGG PATHWAY |
hsa00650 |
4.85495E−06 |
0.000211676 |
| 12 |
Arginine and proline metabolism |
KEGG PATHWAY |
hsa00330 |
1.15415E−05 |
0.000437574 |
| 13 |
Tryptophan metabolism |
KEGG PATHWAY |
hsa00380 |
1.24063E−05 |
0.00045076 |
| 14 |
Alanine, aspartate and glutamate metabolism |
KEGG PATHWAY |
hsa00250 |
9.2847E−05 |
0.003113944 |
| 15 |
PPAR signalling pathway |
KEGG PATHWAY |
hsa03320 |
0.000113691 |
0.003540661 |
| 16 |
Glyoxylate and dicarboxylate metabolism |
KEGG PATHWAY |
hsa00630 |
0.000125436 |
0.003771744 |
| 17 |
Lysine degradation |
KEGG PATHWAY |
hsa00310 |
0.000425581 |
0.010644352 |
| 18 |
Integrin signalling pathway |
PANTHER |
P00034 |
0.000441775 |
0.01070076 |
| 19 |
ECM–receptor interaction |
KEGG PATHWAY |
hsa04512 |
0.000637655 |
0.015027987 |
| 20 |
Pentose and glucuronate interconversions |
KEGG PATHWAY |
hsa00040 |
0.000908772 |
0.020161889 |
| 21 |
Peroxisome |
KEGG PATHWAY |
hsa04146 |
0.001069004 |
0.02273588 |
| 22 |
Cytoskeletal regulation by Rho GTPase |
PANTHER |
P00016 |
0.001258049 |
0.026119495 |
| 23 |
Ascorbate and aldarate metabolism |
KEGG PATHWAY |
hsa00053 |
0.001388723 |
0.026910372 |
| 24 |
Collecting duct acid secretion |
KEGG PATHWAY |
hsa04966 |
0.001388723 |
0.026910372 |
| 25 | Role of mitochondria in apoptotic signalling | PID BioCarta | 100106 | 0.002004916 | 0.038006226 |
Abbreviations: ATP=adenosine triphosphate; ECM=extracellular matrix; FDR=false discovery rate; PPAR=peroxisome proliferator-activated receptor; TCA=tricarboxylic acid.
Figure 2.
(A) Pathway analysis showing the top enriched canonical pathways. The enrichment of canonical pathways in the differentially expressed proteins identified by IPA software is shown. The enrichment shows many important pathways that have vital role in tumourigenesis. (B) Network showing important upstream regulators and downstream effectors in the differentially expressed proteins. Ingenuity Pathway Analysis showing the interrelationship among some of the differentially expressed proteins including upstream regulators and downstream effectors.
Validation of proteins that are overexpressed in renal cancer but not in normal renal tissue
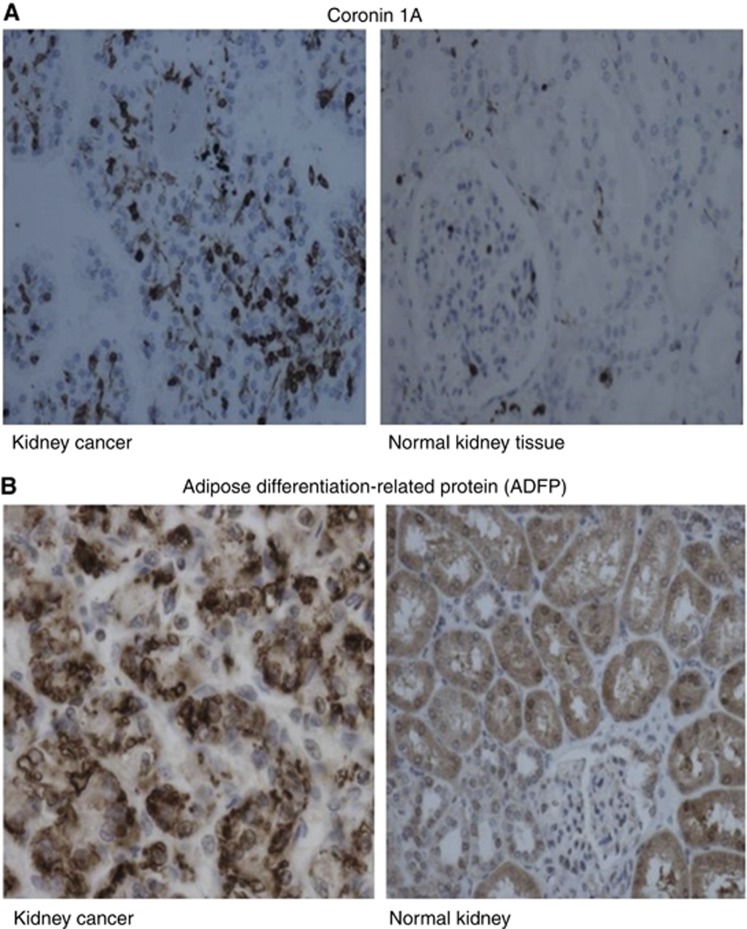
Given the large number of proteins differentially expressed in tumour and normal tissues as demonstrated by the results, we chose to use immunohistochemistry to validate two of the proteins that were upregulated in tumour samples. These include Coronin 1A and ADFP. The choice was based on the reports of these two proteins involved in key pathways of kidney or any other cancers in the literature as suggested in the Materials and Methods. Coronin 1A is an actin remodelling protein that has been reported to be overexpressed in breast cancer (Kim et al, 2009a; Klopfleisch et al, 2010). Our immunohistochemical results revealed that Coronin 1A is not expressed by the cells of the primary renal cancer (Figure 3A), but was highly expressed in the infiltrating lymphocytes in all the eight renal cancer specimens. The ADFP is a major lipid droplet protein present in all cells that accumulate lipids either normally or abnormally. It was expressed in the cytoplasm-surrounding lipid droplets within the renal cancer cells (Figure 3B). The staining was seen in the majority of the tumour cells in all the cases.
Figure 3.
(A) Expression of Coronin 1A in cancerous tissues compared with the corresponding normal kidney tissues. (B) Expression of adipose differentiation-related protein in cancerous tissues compared with the corresponding normal kidney tissues.
Discussion
Renal cell carcinoma is the third most common urological cancer; however, there are limited curative treatment options for both localised and metastatic diseases. For localised RCC tumours measuring up to 7 cm, the recommended treatment is partial nephrectomy (wherever technically possible), with radio frequency or cryoablative treatments being other alternatives in small cortical or bilateral tumours. For tumours over 7 cm, radical nephrectomy is the commonly offered option. Currently, there is no recommended adjuvant therapy available to treat locally advanced RCC, although nonspecific immunotherapy with cytokines IL-2 and/or IFN-α were tried with questionable benefits in the past. Lack of progress is mainly because of nonavailability of biomarkers to stratify patients into those who may or may not benefit from the treatment. Specific immunotherapy with tumour vaccines is still under development. For metastatic disease, targeted therapies with single or combination of inhibitors of tyrosine kinase, VEGF and mTOR are recommended. Both specific immunotherapy and targeted therapies require biomarkers to predict whether such therapy would be suitable to the patient and actually provide desired benefits. This underlines the importance of biomarker discovery in RCC and there is urgent need to discover the following categories of biomarkers:
Early diagnostic biomarkers that could also be used for routine screening.
Therapeutic biomarkers for choosing the right course of treatment and
Prognostic biomarkers for predicting metastatic possibility, tumour recurrence and treatment outcomes.
High-throughput omics technologies, including proteomics, have remarkable ability to capture changes in thousands of variables simultaneously and are found to be useful in biomarker research. In the present study we used label-free quantitative proteomics approach for profiling the proteome of resected cancer and autologous normal kidney tissues. This approach provided a snapshot of proteins expressed in kidney and also differences in protein expression between the normal and the diseased tissues. A total of 1761 proteins showing at least two unique peptides per protein were identified and quantified. Of these, 596 were found to be differentially expressed between the cancer and noncancer groups.
There are several proteomics studies in kidney cancer using 2D-PAGE/MS (Balabanov et al, 2001; Shi et al, 2004; Lichtenfels et al, 2009; Valera et al, 2010; Giribaldi et al, 2013) and shot gun proteomics (Perroud et al, 2009). Our method consisting of complete solubilisation of kidney proteins using FASP followed by nanoLC-MS has yielded a higher number of proteins compared with previous studies (Hwa et al, 2005; Okamura et al, 2008; Perroud et al, 2009; Valera et al, 2010; Zhu et al, 2010; Junker et al, 2011). The method in the present study is known to solubilise membrane and lipid-associated proteins that may explain the large number of proteins identified. In addition, the number of proteins differentially expressed between the two groups is higher in our study. Many of the proteins discovered by other groups (Perroud et al, 2009; Valera et al, 2010; Giribaldi et al, 2013) are consistent with the finding reported here. In general, we found our label-free approach was relatively more comprehensive, rapid and ideal for analysing large number of clinical samples.
The differences in the abundance of proteins between the RCC and noncancer renal tissues highlighted important cancer-related proteins including von Willebrand factor, Ectonucleotide pyrophosphatase/phosphodiesterase family member 3, adipose differentiation-related protein, Coronin 1A, thymidine phosphorylase, nicotinamide N-methyltransferase, fatty-acid binding protein 5, annexin A4, laminin, vimentin, NADH dehydrogenase, metallothionein, ubiquitin carboxyl-terminal hydrolase isozyme L1 and L-xylulose reductase. These proteins are of particular importance for a number of reasons:
Ectonucleotide pyrophosphatase/phosphodiesterase family member 3 (E-NPP3) is overexpressed in RCC (log2 fold change=3.11, FDR-adjusted P=4.05E−08). E-NPP3 has an alkaline phosphodiesterase domain and a nucleotide pyrophosphatase domain. It cleaves a number of phosphodiester and phosphosulphate bonds including deoxynucleotides, nucleotide sugars and NAD. The E-NPP3 has been shown to be upregulated in invasive bile duct cancers. It has been shown to be a marker for cell activation in allergic responses (Buhring et al, 2004). Von Willebrand factor (von Willebrand antigen II) is overexpressed in RCC (log2 fold change=3.25, FDR-adjusted P=2.59E−13). The vWF is localised in the extracellular matrix and is involved in cell–substrate adhesion. It is found to be upregulated in many cancers of other tissues and linked to tumour angiogenesis (Plate et al, 1993; Zanetta et al, 2000; Franchini et al, 2013), Thymidine phosphorylase (TP) is overexpressed in RCC (log2 fold change=2.89, FDR-adjusted P=4.48E−18). It promotes angiogenesis in vivo and activates growth of endothelial cells in vitro. It has very high target cell specificity acting only on endothelial cells. It is implicated in many tumours for its role in tumour aggressiveness and angiogenesis. The TP targeting has been suggested for antiangiogenic therapy in tumours (Bronckaers et al, 2009; Bijnsdorp et al, 2011). Nicotinamide N-methyltransferase (NNMT) is overexpressed in RCC (log2 fold change=2.71, FDR-adjusted P=5.65E−09). It is a key enzyme in drug and xenobiotic compounds metabolism as it catalyses the N-methylation of nicotinamide and other pyridines. Overexpression of NNMT in RCC has been widely reported. It has been suggested as a potential prognostic biomarker in RCC and hepatocellular carcinoma where its higher expression is linked to poor prognosis (Kim et al, 2009b; Zhang et al, 2010; D'Andrea et al, 2011). Fatty-acid binding protein 5 (FABP5) is upregulated (log2 fold change=1.7, FDR-adjusted P=1.24E−06) in kidney cancer tissues. It binds to PPAR-β/δ and its overexpression in cancer tissues, particularly in prostate cancer, has been reported (Morgan et al, 2010; Tolle et al, 2011).
Coronin 1A, a 57 kDa protein, has been shown to play an important role in signalling of T lymphocytes (Mugnier et al, 2008) and is an important member of the coronin family of proteins (seven members are coded by the mammalian genome). This may play a crucial role in a number of cytoskeleton-dependant processes such as cell migration, morphogenesis, cell trafficking and cytokinesis. A number of human cancers show tumour-infiltrating lymphocytes (TILs) with known diversity in their phenotypic expression and function depending on the type of tumour. The TILs of renal cancers phenotypically differ from the peripheral blood of the same patients, suggesting an active interaction between the lymphocytes and tumour cells at the local level (Kowalczyk et al, 1997). Coronin 1A could be a potential biomarker to assess the degree of TILs in the renal cancer, but this needs to be further studied. Coronin 1A was shown to be significantly correlated with development and migration of breast cancer cells (Kim et al, 2009a), compared with normal cell lines. Upregulation of Coronin 1A has been reported by others in RCC samples (Perroud et al, 2009). Immunostaining showed predominant expression of this protein in infiltrating lymphocytes rather than in renal cancer cells. This is an interesting observation, as RCC is considered as immunogenic malignancy and responds to immunotherapy. Furthermore, RCCs express common antigenic determinants that can be recognised by MHC-restricted T lymphocytes and create a local environment (Schendel and Gansbacher, 1993; Wang et al, 2008). Bromwich et al (2003) reported poor correlation between intratumour CD4+ T-lymphocyte infiltrates and outcomes of renal cancer following surgical treatment, and this was independent of the grade of tumour. Coronin 1A identified in the present study could be used as a biomarker for quantification of TILs in renal cancer. However, further work needs to be done to explore the possibility of measuring this protein in urine and blood as a biomarker for lymphocytic activation in RCC.
In contrast to Coronin 1A, ADFP has been shown to be expressed by clear cell carcinoma cells and its expression level has been reported to correlate with differentiation of cells (Yao et al, 2005). The ADFP, also known as adipophilin, is a member of PAT family of proteins that have the ability to bind intracellular lipid droplets. The presence of a large number of lipid droplets and glycogen granules is a characteristic feature of clear cell RCC (Yao et al, 2005, 2007; Tickoo and Gopalan, 2008; Urahama et al, 2008). Expression of ADFP has been reported in previous studies and its expression has shown correlation with the survival of renal cancer. Moreover, it has been reported that the upregulation of ADFP is caused by the disruption of VHL/HIF pathway in clear cell RCC (Yao et al, 2007), and the VHL/HIF pathway is a key target for the tyrosine-kinase inhibitor therapy in advanced renal cell carcinoma. The ADFP is overexpressed and secreted in the urine of patients with RCC and in those with benign kidney disease, compared with normal healthy adults (Morrissey et al, 2010). There are emerging reports exploring the role of ADFP measurement in urine as prognostic marker or marker of response to systemic therapy (Grebe and Erickson, 2010).
Our findings confirm previous reports that ADFP could be a biomarker for diagnosis, predicting prognosis or response to treatment (Morrissey et al, 2010). Pathway analysis allowed us to identify enrichment of various metabolic and signalling pathways, which are known to play key role in oncogenesis or cancer progression, from the differentially expressed proteins in this study. Various pathways such as glycolysis reported previously (Mathupala et al, 1997; Unwin et al, 2003; Perroud et al, 2006, 2009) were confirmed by our data. The large number of proteins identified and demonstrated to be differentially expressed in RCC could be validated by using a larger number of samples and this may become a part of our future research. The PPAR signalling pathway, enriched in our study, could be a new potential therapeutic target in RCC. The transcription factors in PPAR pathways strongly influence molecular events in cancer cells (Michalik et al, 2004). The PPARs are nuclear hormone receptors and exist in three isotopes: α, γ and δ. They heterodimerise with another nuclear hormone receptor, retinoid X receptor (RXR), and exert their effects via regulation of gene transcription upon binding of ligands (Chinetti et al, 2001). The PPAR/RXR heterodimer exists in active and inactive forms. In the presence of a ligand, the PPAR disassociates itself from co-suppressor and becomes active (Chinetti et al, 2001). The transcription factors in PPAR pathways strongly influence molecular events in cancer cells (Jackson et al, 2003; Michalik et al, 2004). There are reports that PPAR pathway activity promotes colonic cell carcinogenesis and has been shown to be suppressed by nonsteroidal anti-inflammatory drugs. Similarly, Rho GTPases pathway is dysregulated during hypoxia in renal cell carcinoma as shown by mRNA levels (Turcotte et al, 2004); Rho A expression is required for HIF-1-α accumulation during hypoxia, and it could also become a therapeutic target. Our study also showed a strong evidence of altered mitochondrial function as demonstrated by downregulation of a number of mitochondrial enzymes: fumarate hydratase, succinate semialdehyde dehydrogenase, methyl-CoA mutase, agmatinase, adenylate kinase isoenzyme 4, mitochondrial peptide methionine sulphoxide reductase, sulphite oxydase, D-β-hydroxybutyrate dehydronenase, cytochrome C oxydase subunit 4, pyruvate carboxylase, methylcrotonoyl-CoA carboxylase β-chain and propionyl-CoA carboxylase α-chain. Indeed, metabolic and molecular alterations in mitochondria are well documented in cancers (Modica-Napolitano et al, 2007; Gasparre et al, 2013) and these alterations are linked to shift in tumour metabolism from mitochondrial oxidative phosphorylation to aerobic glycolysis (the Warburg effect), and possible other unknown metabolic pathways (Vander Heiden et al, 2009) allowing tumour cells to satisfy their bioenergetic and biosynthetic requirements that are higher than those of normal cells (Vander Heiden et al, 2009; Zhao et al, 2013).
Conclusions
Our findings using quantitative label-free proteomics analysis in RCC revealed a number of differentially expressed proteins with potential diagnostic and prognostic role as biomarkers in RCC. The dysregulated pathways identified have a significant potential for therapeutic targets. The two proteins (Coronin 1A and ADFP) that were followed up in-depth using immunohistochemistry show differential expression between cancer and normal renal parenchymal tissue.
Acknowledgments
This study was supported by Tayside Oncology Funds; Tayside Tissue Bank; FingerPrints Proteomics Facility, College of Life Sciences, University of Dundee; and The Barton Group, College of Life Sciences, University of Dundee.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Balabanov S, Zimmermann U, Protzel C, Scharf C, Klebingat KJ, Walther R. Tumour-related enzyme alterations in the clear cell type of human renal cell carcinoma identified by two-dimensional gel electrophoresis. Eur J Biochem. 2001;268:5977–5980. doi: 10.1046/j.0014-2956.2001.02546.x. [DOI] [PubMed] [Google Scholar]
- Bijnsdorp IV, Capriotti F, Kruyt FA, Losekoot N, Fukushima M, Griffioen AW, Thijssen VL, Peters GJ. Thymidine phosphorylase in cancer cells stimulates human endothelial cell migration and invasion by the secretion of angiogenic factors. Br J Cancer. 2011;104:1185–1192. doi: 10.1038/bjc.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromwich EJ, McArdle PA, Canna K, McMillan DC, McNicol AM, Brown M, Aitchison M. The relationship between T-lymphocyte infiltration, stage, tumour grade and survival in patients undergoing curative surgery for renal cell cancer. Br J Cancer. 2003;89:1906–1908. doi: 10.1038/sj.bjc.6601400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronckaers A, Gago F, Balzarini J, Liekens S. The dual role of thymidine phosphorylase in cancer development and chemotherapy. Med Res Rev. 2009;29:903–953. doi: 10.1002/med.20159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhring HJ, Streble A, Valent P. The basophil-specific ectoenzyme E-NPP3 (CD203c) as a marker for cell activation and allergy diagnosis. Int Arch Allergy Immunol. 2004;133:317–329. doi: 10.1159/000077351. [DOI] [PubMed] [Google Scholar]
- Campbell SC, Flanigan RC, Clark JI. Nephrectomy in metastatic renal cell carcinoma. Curr Treat Options Oncol. 2003;4:363–372. doi: 10.1007/s11864-003-0037-4. [DOI] [PubMed] [Google Scholar]
- Chen EI, Yates JR., 3rd Cancer proteomics by quantitative shotgun proteomics. Mol Oncol. 2007;1:144–159. doi: 10.1016/j.molonc.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinetti G, Lestavel S, Bocher V, Remaley AT, Neve B, Torra IP, Teissier E, Minnich A, Jaye M, Duverger N, Brewer HB, Fruchart JC, Clavey V, Staels B. PPAR-alpha and PPAR-gamma activators induce cholesterol removal from human macrophage foam cells through stimulation of the ABCA1 pathway. Nat Med. 2001;7:53–58. doi: 10.1038/83348. [DOI] [PubMed] [Google Scholar]
- D'Andrea FP, Safwat A, Kassem M, Gautier L, Overgaard J, Horsman MR. Cancer stem cell overexpression of nicotinamide N-methyltransferase enhances cellular radiation resistance. Radiother Oncol. 2011;99:373–378. doi: 10.1016/j.radonc.2011.05.086. [DOI] [PubMed] [Google Scholar]
- Drucker BJ. Renal cell carcinoma: current status and future prospects. Cancer Treat Rev. 2005;31:536–545. doi: 10.1016/j.ctrv.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Escudier B, Eisen T, Porta C, Patard JJ, Khoo V, Algaba F, Mulders P, Kataja V, ESMO Guidelines Working Group Renal cell carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2012;23 (Suppl 7:vii65–vii71. doi: 10.1093/annonc/mds227. [DOI] [PubMed] [Google Scholar]
- FLink: Frequency weighted links [Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information. 2010—[cited 2012/06/12]. Available from http://ncbi.nlm.nih.gov/Structure/flink/flink.cgi .
- Franchini M, Frattini F, Crestani S, Bonfati C, Lippi G. Von Willebrand factor and cancer: a renewed interest. Thromb Res. 2013;131 (4:290–292. doi: 10.1016/j.thromres.2013.01.015. [DOI] [PubMed] [Google Scholar]
- Gaspare G, Porcelli AM, Lenaz G, Romeo G. Relevance of mitochondrial genetics and metabolism in cancer development. Cold spring Harb Perspect Biol. 2013;5 (2:1–17. doi: 10.1101/cshperspect.a011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giribaldi G, Barbero G, Mandili G, Daniele L, Khadjavi A, Notarpietro A, Ulliers D, Prato M, Minero VG, Battaglia A, Allasia M, Bosio A, Sapino A, Gontero P, Frea B, Fontana D, Destefanis P. Proteomic identification of reticulocalbin 1 as potential tumor marker in renal cell carcinoma. J Proteomics. 2013;91C:385–392. doi: 10.1016/j.jprot.2013.07.018. [DOI] [PubMed] [Google Scholar]
- Gupta K, Miller JD, Li JZ, Russell MW, Charbonneau C. Epidemiologic and socioeconomic burden of metastatic renal cell carcinoma (mRCC): a literature review. Cancer Treat Rev. 2008;34:193–205. doi: 10.1016/j.ctrv.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Grebe SK, Erickson LA. Screening for kidney cancer: is there a role for aquaporin-1 and adipophilin. Mayo Clin Proc. 2010;85 (5:410–412. doi: 10.4065/mcp.2010.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hwa JS, Park HJ, Jung JH, Kam SC, Park HC, Kim CW, Kang KR, Hyun JS, Chung KH. Identification of proteins differentially expressed in the conventional renal cell carcinoma by proteomic analysis. J Korean Med Sci. 2005;20:450–455. doi: 10.3346/jkms.2005.20.3.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L, Wahli W, Michalik L, Watson SA, Morris T, Anderton K, Bell DR, Smith JA, Hawkey CJ, Bennett AJ. Potential role for peroxisome proliferator activated receptor (PPAR) in preventing colon cancer. Gut. 2003;52:1317–1322. doi: 10.1136/gut.52.9.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen NK, Kim HL, Figlin RA, Belldegrun AS. Surveillance after radical or partial nephrectomy for localized renal cell carcinoma and management of recurrent disease. Urol Clin North Am. 2003;30:843–852. doi: 10.1016/s0094-0143(03)00056-9. [DOI] [PubMed] [Google Scholar]
- Junker H, Venz S, Zimmermann U, Thiele A, Scharf C, Walther R. Stage-related alterations in renal cell carcinoma—comprehensive quantitative analysis by 2D-DIGE and protein network analysis. PLoS One. 2011;6:e21867. doi: 10.1371/journal.pone.0021867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Bae J, Lee JW, Kim SY, Kim YH, Bae JY, Yi JK, Yu MH, Noh DY, Lee C. Proteomic analysis of breast cancer tissue reveals upregulation of actin-remodeling proteins and its relevance to cancer invasiveness. Proteomics Clin Appl. 2009;3:30–40. doi: 10.1002/prca.200800167. [DOI] [PubMed] [Google Scholar]
- Kim J, Hong SJ, Lim EK, Yu YS, Kim SW, Roh JH, Do IG, Joh JW, Kim DS. Expression of nicotinamide N-methyltransferase in hepatocellular carcinoma is associated with poor prognosis. J Exp Clin Cancer Res. 2009;28:20. doi: 10.1186/1756-9966-28-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SI, Purdie CA, Bray SE, Quinlan PR, Jordan LB, Thompson AM, Meek DW. Immunohistochemical detection of Polo-like kinase-1 (PLK1) in primary breast cancer is associated with TP53 mutation and poor clinical outcom. Breast Cancer Res. 2012;14:R40. doi: 10.1186/bcr3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfleisch R, Klose P, Weise C, Bondzio A, Multhaup G, Einspanier R, Gruber AD. Proteome of metastatic canine mammary carcinomas: similarities to and differences from human breast cancer. J Proteome Res. 2010;9:6380–6391. doi: 10.1021/pr100671c. [DOI] [PubMed] [Google Scholar]
- Kowalczyk D, Skorupski W, Kwias Z, Nowak J. Flow cytometric analysis of tumour-infiltrating lymophocytes in patients with renal cell carcinoma. Br J Urol. 1997;4:543–547. doi: 10.1046/j.1464-410x.1997.00408.x. [DOI] [PubMed] [Google Scholar]
- Lichtenfels R, Dressler SP, Zobawa M, Recktenwald CV, Ackermann A, Atkins D, Kersten M, Hesse A, Puttkammer M, Lottspeich F, Seliger B. Systematic comparative protein expression profiling of clear cell renal cell carcinoma: a pilot study based on the separation of tissue specimens by two-dimensional gel electrophoresis. Mol Cell Proteomics. 2009;8:2827–2842. doi: 10.1074/mcp.M900168-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui O, White NM, DeSouza LV, Krakovska O, Matta A, Metias S, Khalil B, Romaschin AD, Honey RJ, Stewart R, Pace K, Bjarnason GA, Siu KW, Yousef GM. Quantitative proteomic analysis in metastatic renal cell carcinoma reveals a unique set of proteins with potential prognostic significance. Mol Cell Proteomics. 2013;12:132–144. doi: 10.1074/mcp.M112.020701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathupala SP, Rempel A, Pedersen PL. Aberrant glycolytic metabolism of cancer cells: a remarkable coordination of genetic, transcriptional, post-translational, and mutational events that lead to a critical role for type II hexokinase. J Bioenerg Biomembr. 1997;29:339–343. doi: 10.1023/a:1022494613613. [DOI] [PubMed] [Google Scholar]
- Michalik L, Desvergne B, Wahli W. Peroxisome-proliferator-activated receptors and cancers: complex stories. Nat Rev Cancer. 2004;4:61–70. doi: 10.1038/nrc1254. [DOI] [PubMed] [Google Scholar]
- Modica-Napolitano JS, Kulawiec M, Singh KK. Mitochondria and human cancer. Curr Mol Med. 2007;7:121–131. doi: 10.2174/156652407779940495. [DOI] [PubMed] [Google Scholar]
- Morgan E, Kannan-Thulasiraman P, Noy N. Involvement of fatty acid binding protein 5 and PPARbeta/delta in prostate cancer cell growth. PPAR Res. 2010;2010:1–9. doi: 10.1155/2010/234629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey JJ, London AN, Luo J, Kharasch ED. Urinary biomarkers for the early diagnosis of kidney cancer. Mayo Clin Proc. 2010;85:413–421. doi: 10.4065/mcp.2009.0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugnier B, Nal B, Verthuy C, Boyer C, Lam D, Chasson L, Nieoullon V, Chazal G, Guo XJ, He HT, Rueff-Juy D, Alcover A, Ferrier P. Coronin-1A links cytoskeleton dynamics to TCRαβ-induced cell signaling. PLoS One. 2008;3 (10:e3467. doi: 10.1371/journal.pone.0003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najjar YG, Rini BI. Novel agents in renal carcinoma: a reality check. Ther Adv Med Oncol. 2012;4:183–194. doi: 10.1177/1758834012443725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura N, Masuda T, Gotoh A, Shirakawa T, Terao S, Kaneko N, Suganuma K, Watanabe M, Matsubara T, Seto R, Matsumoto J, Kawakami M, Yamamori M, Nakamura T, Yagami T, Sakaeda T, Fujisawa M, Nishimura O, Okumura K. Quantitative proteomic analysis to discover potential diagnostic markers and therapeutic targets in human renal cell carcinoma. Proteomics. 2008;8:3194–3203. doi: 10.1002/pmic.200700619. [DOI] [PubMed] [Google Scholar]
- Perroud B, Lee J, Valkova N, Dhirapong A, Lin PY, Fiehn O, Kultz D, Weiss RH. Pathway analysis of kidney cancer using proteomics and metabolic profiling. Mol Cancer. 2006;5:64. doi: 10.1186/1476-4598-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud B, Ishimaru T, Borowsky AD, Weiss RH. Grade-dependent proteomics characterization of kidney cancer. Mol Cell Proteomics. 2009;8 (5:971–985. doi: 10.1074/mcp.M800252-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plate KH, Breier G, Millauer B, Ullrich A, Risau W. Up-regulation of vascular endothelial growth factor and its cognate receptors in a rat glioma model of tumor angiogenesis. Cancer Res. 1993;53:5822–5827. [PubMed] [Google Scholar]
- Pyrhonen S, Salminen E, Ruutu M, Lehtonen T, Nurmi M, Tammela T, Juusela H, Rintala E, Hietanen P, Kellokumpu-Lehtinen PL. Prospective randomized trial of interferon alfa-2a plus vinblastine versus vinblastine alone in patients with advanced renal cell cancer. J Clin Oncol. 1999;17:2859–2867. doi: 10.1200/JCO.1999.17.9.2859. [DOI] [PubMed] [Google Scholar]
- Rini BI, Atkins MB. Resistance to targeted therapy in renal-cell carcinoma. Lancet Oncol. 2009;10:992–1000. doi: 10.1016/S1470-2045(09)70240-2. [DOI] [PubMed] [Google Scholar]
- Sarto C, Deon C, Doro G, Hochstrasser DF, Mocarelli P, Sanchez JC. Contribution of proteomics to the molecular analysis of renal cell carcinoma with an emphasis on manganese superoxide dismutase. Proteomics. 2001;1:1288–1294. doi: 10.1002/1615-9861(200110)1:10<1288::AID-PROT1288>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Schendel DJ, Gansbacher B. Tumor-specific lysis of human renal cell carcinomas by tumor-infiltrating lymphocytes: modulation of recognition through retroviral transduction of tumor cells with interleukin 2 complementary DNA and exogenous alpha interferon treatment. Cancer Res. 1993;53:4020–4025. [PubMed] [Google Scholar]
- Schulze WX, Usadel B. Quantitation in mass-spectrometry-based proteomics. Annu Rev Plant Biol. 2010;61:491–516. doi: 10.1146/annurev-arplant-042809-112132. [DOI] [PubMed] [Google Scholar]
- Seliger B, Lichtenfels R, Kellner R. Detection of renal cell carcinoma-associated markers via proteome- and other ‘ome'-based analyses. Brief Funct Genomic Proteomic. 2003;2:194–212. doi: 10.1093/bfgp/2.3.194. [DOI] [PubMed] [Google Scholar]
- Shi T, Dong F, Liou LS, Duan ZH, Novick AC, DiDonato JA. Differential protein profiling in renal-cell carcinoma. Mol Carcinog. 2004;40:47–61. doi: 10.1002/mc.20015. [DOI] [PubMed] [Google Scholar]
- Singer EA, Gupta GN, Marchalik D, Srinivasan R. Evolving therapeutic targets in renal cell carcinoma. Curr Opin Oncol. 2013;25 (3:273–280. doi: 10.1097/CCO.0b013e32835fc857. [DOI] [PubMed] [Google Scholar]
- Tickoo SK, Gopalan A. Pathologic features of renal cortical tumors. Urol Clin North Am. 2008;35:551–561. doi: 10.1016/j.ucl.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Tolle A, Suhail S, Jung M, Jung K, Stephan C. Fatty acid binding proteins (FABPs) in prostate, bladder and kidney cancer cell lines and the use of IL-FABP as survival predictor in patients with renal cell carcinoma. BMC Cancer. 2011;11:302. doi: 10.1186/1471-2407-11-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte S, Desrosiers RR, Beliveau R. Hypoxia upregulates von Hippel-Lindau tumor-suppressor protein through RhoA-dependent activity in renal cell carcinoma. Am J Physiol Renal Physiol. 2004;286:F338–F348. doi: 10.1152/ajprenal.00254.2003. [DOI] [PubMed] [Google Scholar]
- Unwin RD, Craven RA, Harnden P, Hanrahan S, Totty N, Knowles M, Eardley I, Selby PJ, Banks RE. Proteomic changes in renal cancer and co-ordinate demonstration of both the glycolytic and mitochondrial aspects of the Warburg effect. Proteomics. 2003;3:1620–1632. doi: 10.1002/pmic.200300464. [DOI] [PubMed] [Google Scholar]
- Urahama Y, Ohsaki Y, Fujita Y, Maruyama S, Yuzawa Y, Matsuo S, Fujimoto T. Lipid droplet-associated proteins protect renal tubular cells from fatty acid-induced apoptosis. Am J Pathol. 2008;173:1286–1294. doi: 10.2353/ajpath.2008.080137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324 (5930:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valera VA, Li-Ning-T E, Walter BA, Roberts DD, Linehan WM, Merino MG. Protein expression profiling in the spectrum of renal cell carcinomas. Journal of Cancer. 2010;1:184–196. doi: 10.7150/jca.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra TD. Global and targeted quantitative proteomics for biomarker discovery. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;847:3–11. doi: 10.1016/j.jchromb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Walther TC, Mann M. Mass spectrometry-based proteomics in cell biology. J Cell Biol. 2010;190:491–500. doi: 10.1083/jcb.201004052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, You J, Bemis KG, Tegeler TJ, Brown DP. Label-free mass spectrometry-based protein quantification technologies in proteomic analysis. Brief Funct Genomic Proteomic. 2008;7:329–339. doi: 10.1093/bfgp/eln031. [DOI] [PubMed] [Google Scholar]
- Weiss RH, Lin PY. Kidney cancer: identification of novel targets for therapy. Kidney Int. 2006;69:224–232. doi: 10.1038/sj.ki.5000065. [DOI] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Mann M. Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J Proteome Res. 2009;8:5674–5678. doi: 10.1021/pr900748n. [DOI] [PubMed] [Google Scholar]
- Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Huang Y, Shioi K, Hattori K, Murakami T, Nakaigawa N, Kishida T, Nagashima Y, Kubota Y. Expression of adipose differentiation-related protein: a predictor of cancer-specific survival in clear cell renal carcinoma. Clin Cancer Res. 2007;13:152–160. doi: 10.1158/1078-0432.CCR-06-1877. [DOI] [PubMed] [Google Scholar]
- Yao M, Tabuchi H, Nagashima Y, Baba M, Nakaigawa N, Ishiguro H, Hamada K, Inayama Y, Kishida T, Hattori K, Yamada-Okabe H, Kubota Y. Gene expression analysis of renal carcinoma: adipose differentiation-related protein as a potential diagnostic and prognostic biomarker for clear-cell renal carcinoma. J Pathol. 2005;205:377–387. doi: 10.1002/path.1693. [DOI] [PubMed] [Google Scholar]
- Zanetta L, Marcus SG, Vasile J, Dobryansky M, Cohen H, Eng K, Shamamian P, Mignatti P. Expression of Von Willebrand factor, an endothelial cell marker, is up-regulated by angiogenesis factors: a potential method for objective assessment of tumor angiogenesis. Int J Cancer. 2000;85:281–288. doi: 10.1002/(sici)1097-0215(20000115)85:2<281::aid-ijc21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Butler EB, Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Disease. 2013;4:e532. doi: 10.1038/cddis.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Xie XY, Yang SW, Wang J, He C. Nicotinamide N-methyltransferase protein expression in renal cell cancer. J Zhejiang Univ Sci B. 2010;11:136–143. doi: 10.1631/jzus.B0900249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Smith JW, Huang CM. Mass spectrometry-based label-free quantitative proteomics. J Biomed Biotechnol. 2010;2010:840518. doi: 10.1155/2010/840518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.