Abstract
Background:
RAD21 is a component of the cohesion complex and is integral to chromosome segregation and error-free DNA repair. RAD21 is functionally important in tumour progression but its role in colorectal carcinoma (CRC) is unclear. We therefore assessed its clinicopathological and prognostic significance in CRC, as well as its effect on chemosensitivity.
Methods:
A retrospective observation study examined RAD21 expression in 652 CRCs using a tissue microarray approach. Correlation with clinicopathological factors including gender, tumour grade, mucinous subtype, TNM stage, disease-specific survival (DSS), BRAF and KRAS mutation status, tumour p53 immunostaining, tumour microsatellite instability and tumour CpG island methylator phenotype was performed. Colorectal cancer cell clones with stable RAD21 knockdown were generated and tested for cellular sensitivity to conventional chemotherapeutic drugs.
Results:
RAD21 expression was significantly correlated with male gender (56.7% vs 43.3%, P=0.02), well-differentiated histology (14.4% vs 4.0%, P=0.0001), higher T-stage (36.1% vs 27.0%, P=0.01), presence of metastasis (18.8% vs 12.6%, P=0.03), and shorter DSS (hazard ratio (HR) 1.4, 95% CI 1.1 to 1.9, P=0.01) in both univariate and multivariate analysis. RAD21 expression was associated with shorter DSS in patients with KRAS mutant tumours (HR:2.6, 95% CI:1.4–4.3, P=0.001) and in patients receiving adjuvant chemoradiotherapy (HR:1.9, 95% CI:1.2–3.0, P=0.008). Colorectal cancer cells with RAD21 knockdown exhibited enhanced sensitivity to 5-fluorouracil, either alone or in combination with oxaliplatin.
Conclusions:
RAD21 expression in CRC is associated with aggressive disease especially in KRAS mutant tumours and resistance to chemoradiotherapy. RAD21 may be an important novel therapeutic target.
Keywords: RAD21, colorectal cancer, prognostic, predictive, KRAS
RAD21 is a key central component within the multi-protein cohesin complex (Xu et al, 2011a), the main function of which is to ensure balanced chromosome separation during the metaphase–anaphase transition of mitosis (Michaelis et al, 1997; Guacci, 2007). The complex is highly conserved through evolution between species such as yeast to humans (McKay et al, 1996; Sumara et al, 2000; Nasmyth, 2001). During the G1 phases of the cell cycle, the cohesin complex is thought to form a ring and entrap DNA helices and, with the aid of several accessory and regulatory proteins, facilitates binding of newly formed sister chromosomes and appropriate chromosome segregation during anaphase (Haering et al, 2008). The cohesin complex is also implicated in several other important processes including homologous recombinational repair or DNA damage (Birkenbihl and Subramani, 1992; Nasmyth et al, 2001; Sonoda et al, 2001; Watrin and Peters, 2006), in particular double-stranded breaks (Birkenbihl and Subramani, 1992; Bauerschmidt et al, 2010; Xu et al, 2010), cell cycle checkpoint control (Watrin and Peters, 2009), chromatin remodelling (Hakimi et al, 2002), gene regulation (Supernat et al, 2012) and prevention of read-through transcription. Presence of this versatile complex in subcellular locations outside the nucleus and within extracellular matrix has also been observed; however, the exact purpose of cohesin in these instances is unknown.
Several recent studies suggest that the cohesin complex is important in cancer development. Somatic mutations in genes encoding the core cohesin components SMC1A, SMC3 and STAG3, as well as cohesin loading protein NIBPL, were first reported in human colorectal cancers (CRC), leading to the suggestion that cohesion defects underpin chromosome instability in CRC (Barber et al, 2008). Unlike other cohesin subunits, mutation of RAD21 appears to be relatively rare (Barber et al, 2008; Xu et al, 2011a), and de-regulation of RAD21 expression primarily through increased gene copy number in CRC may be responsible (Xu et al, 2011a, 2011b).
However, to date, there are no studies evaluating RAD21 expression in CRC and its effect on patient survival and chemosensitivity. We, therefore, characterised RAD21 expression in CRC, correlated RAD21 expression with conventional clinicopathological features, molecular and prognostic data, and have assessed the role of RAD21 expression in treatment response.
Materials and methods
Patients were recruited from St Vincent's Hospital, Sydney (n=444) and the John Radcliffe Hospital, Oxford, UK (n=208) from between March 1993 to April 2007 (ethics approved by St Vincent's Campus Human Research Committee (approval numbers H02/022 and H07/002) and John Radcliff (approval number CO2.216)). These represent consecutive cases of CRC obtained from individuals undergoing resection with curative intent. Both cohorts include only cases with robust clinical, pathological and treatment data, as well as outcomes, whereas molecular data were available only on the Sydney cohort. The flow of patients is as per REMARK criteria as outlined in Supplementary Table 1. All cases were adenocarcinomas with a patient median age of 70.4 years (range 17.1–94.6 years). Patients were followed for up to 5 years with a median follow-up of 58.6 months (range 0.07–60.0 months). Two hundred and fifteen (33.0%) CRC-related deaths were seen during this period. Cases of synchronous tumours were excluded from the study.
Immunohistochemistry
Tissue microarrays were constructed using duplicate 1 mm cores from formalin-fixed paraffin-embedded tumour tissues. Sections of 4 μM thickness were dewaxed and rehydrated on silane-coated slides. Immunohistochemistry was performed using a rabbit polyclonal anti-RAD21 antibody (Abcam, Cambridge, UK), which was previously tested and validated (Xu et al, 2011b). Sections were counterstained with haematoxylin. Nuclear RAD21 expression was assessed for intensity (0, no staining; 1, weak; 2, moderate; and 3, strong) and percentage of positive cells (0, 0% 1, 0–25% 2, >25–50% 3, >50–75% and 4, >75%). The scores for intensity and percentage were multiplied and a cutoff of 6 was used to approximate the median, as this gave about equal-sized RAD21-negative (0–6 histoscore) and RAD21-positive (7–12 histoscore) expressing groups. Immunohistochemical analysis of p53 expression was performed as described previously using the mouse anti-human p53 antibody D07 (Dako, Dakopatts, Denmark) (Ward et al, 2001). Tumours were considered to show accumulation of p53 protein when >20% of tumour cells showed nuclear staining of moderate to high intensity in the absence of staining in the stromal cells and normal epithelium (Ward et al, 2001).
DNA isolation, analysis of microsatellite instability, KRAS and BRAF mutation status, and DNA methylation
DNA was extracted from fresh and frozen tumour tissues received at St Vincent's Hospital, Sydney. Extraction and analysis of microsatellite instability, KRAS and BRAF mutation status, and tumour methylation status have been described previously for this cohort (Ward et al, 2001, 2004; Packham et al, 2009) and are included in greater detail in Supplementary Material File 1.
Cell culture and the generation of cell clones with stable RAD21 knockdown
Human CRC cell line LIM1215 (TP53, APC, BRAF and KRAS wild type) was obtained from Ludwig Institute (Whitehead et al, 1985; Zhang et al, 2009) and HCT116 (KRAS mutant) was obtained from ATCC. Cells were maintained in RPMI1640 containing 10% fetal calf serum, insulin 0.6 μg ml−1, hydrocortisone 1 μg ml−1 and 10 μM 1-thioglycerole. To generate cell clones with stable RAD21 knockdown, a single-cell clone LIM1215sc5 was established from the LIM1215 cell line and transfected with three small hairpin RNA constructs, sh57233, sh57224 and sh57226, which target different regions of the human RAD21 gene (Open Biosystems, Huntsville, AL, USA). A control shmir vector was introduced into cells in parallel. Cells were cultured in the presence of 2 μg ml−1 puromycin for 7–10 days, and under this condition only cells carrying the constructs survive. We previously successfully used these small hairpin RNA constructs to generate breast cancer cell clones with effectively silencing of RAD21 gene (Xu et al, 2011b).
Forced RAD21 expression was also attempted in the HCT116 cell line (harbouring KRAS mutation), which expresses only modest levels of RAD21. Western blot analysis of multiple cell clones carrying the RAD21 construct showed no apparent increase in the level of RAD21 protein. Furthermore, no significant difference in cell survival was observed following the treatment with 5-fluorouracil (5-FU) and oxaliplatin when compared with the parental cell clone (data not shown).
Western blot analysis and immunofluorescence
Western blot analysis was performed using a rabbit polyclonal anti-Rad21 antibody (Abcam) at a dilution of 1:1000 as described previously (Xu et al, 2004). For loading controls, the membranes were subsequently incubated in a mouse monoclonal anti-pan actin antibody (Cell Signalling Technology, Danvers, MA, USA) at a dilution of 1:5000 followed by an IRDye800-conjugated anti-mouse antibody (Rockland, Gilbertsville, PA, USA).
Immunofluorescence detection of RAD21 protein was performed essentially as described previously (Xu et al, 2004) using a primary rabbit polyclonal antibody (Abcam) (Supplementary Material File 1).
Drug treatments and cell survival assay
Exponentially growing cells were seeded at appropriate density in 96-well clear-bottom plates (Corning, Tewksbury, MA, USA). Six hours post seeding, cells were treated with graded concentrations of either 5-FU (Hospira Australia, Melbourne, VIC, Australia) for 18 h or oxaliplatin (Winthrop Pharmaceuticals, Macquarie Park, NSW, Australia) for 2 h. For the combination of 5-FU and oxaliplatin, a treatment regimen similar to FOLFOX was used. Briefly, cells were treated with oxaliplatin for 2 h, washed three times with culture medium and treated with 5-FU for 18 h. Following drug treatments, cells were washed three times with culture medium and cultured for 5 days. Cell survival was determined using the CellTiter 96 Aqueous One Solution Cell Proliferation Assay kit following the manufacturer's instructions (Promega, Madison, WI, USA). Absorbance was measured 4 h following the addition of the substrate at 450 nM using VersaMax microplate reader (Molecular Devices, Sunnyvale, CA, USA). Each data point represents the mean of three independent experiments and four replicate wells per experiment. Cell survival was calculated as the relative percentage of untreated cells. Survival curve fit was performed using the nonlinear regression linear-quadratic model, GraphPad Prism version 5.01 (GraphPad Software, San Diego, CA, USA).
RAD21 and c-Myc expression data mining
To ascertain mechanism of RAD21 overexpression in CRCs, we assessed the correlation of gene expression between RAD21 and c-Myc (found on the same amplicon). We obtained the normalised DNA microarray data from an independent Gene Expression Omnibus (GEO) data set: GSE14333 that included 293 primary CRCs (Jorissen et al, 2009). Probe annotation was downloaded from GEO for the corresponding microarray platforms and was used to identify the probes targeting RAD21 and c-Myc. Pearson correlation across the samples of interest was computed (using R) between RAD21 and c-Myc log-intensities as an indication of expression correlation. Data were plotted using R for manual assessment.
Statistical analysis
Correlations were examined using Students t-test or χ2-test where appropriate. Kaplan–Meier survival curves were calculated using CRC-related death (DSS) as end points and compared using a log-rank rest. Binary logistic regression was used for multivariate analyses and the Cox proportional hazard regression model was used to identify independent prognostic factors for disease-free and DSS. Analyses were performed in the R package for statistical analysis. A two-tailed P-value test was used in all analyses and a P-value <0.05 was considered as statistically significant.
Results
Association between RAD21 expression and clinicopathological and molecular characteristics in invasive cancer
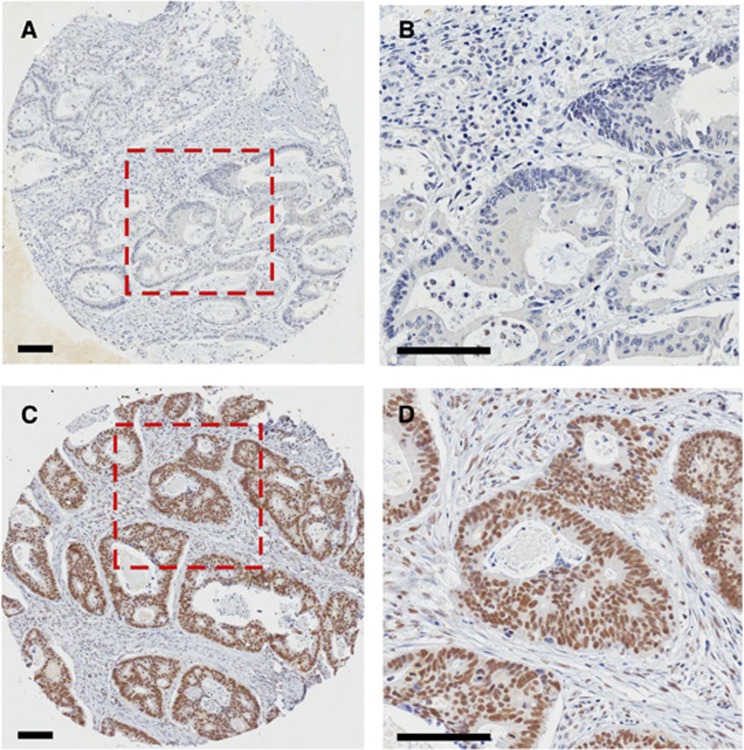
Three hundred and nineteen (48.9%) cases of 652 cases were positive for RAD21 nuclear staining (Figure 1C and D), and 333 cases (51.1%) were negative for RAD21 nuclear expression (Figure 1A and B, Table 1). RAD21 expression significantly correlated with male sex (56.7% vs 47.7%, P=0.02), histologically well-differentiated tumours (14.4% vs 4.0%, P=0.0001), advanced tumour stage (36.1% vs 27.0%, P=0.01) and the presence of metastases (18.8% vs 12.6%, P=0.03). There was no association between RAD21 expression and age (P=0.22), presence of nodal disease (P=0.75), mucinous histology phenotype (P=0.55), microsatellite instability status (P=0.89), methylation (CpG island methylator phenotype) (P=0.62), KRAS mutation status (P=0.83), BRAF mutation status (P=0.86) and p53 accumulation (P=0.91) (Table 1).
Figure 1.
RAD21 expression in CRC by immunohistochemistry. (A and B) Representative image showing RAD21-negative staining. (B) An enlarged view of the boxed region in A. (C) Strong RAD21 nuclear staining. (D) An enlarged view of the box region in C. Scale bars, 100 μM.
Table 1. Clinicopathological correlation of RAD21 expression.
| Total n (%) | RAD21 negative | RAD21 positive | P-value (t-test*/χ2-test) | |||
| Median age (range) – years | |
70.3 (29.2–91.2) | 70.4 (17.1–94.6) | 0.22* | ||
|
Gender | ||||||
| Male | 340 (52%) | 159 | 47.70% | 181 | 56.70% | 0.02 |
| Female |
312 (48%) |
174 |
52.30% |
138 |
43.30% |
|
|
Differentiation (grade) | ||||||
| Well | 56 (9%) | 13 | 4.00% | 43 | 14.40% | 0.0001 |
| Moderate | 496 (79%) | 274 | 84.00% | 222 | 74.20% | |
| Poorly |
73 (12%) |
39 |
12.00% |
34 |
11.40% |
|
|
TNM_T | ||||||
| 1 | 47 (7%) | 28 | 8.40% | 19 | 6.00% | 0.01 |
| 2 | 85 (13%) | 50 | 15.00% | 35 | 11.00% | |
| 3 | 313 (48%) | 165 | 49.50% | 148 | 46.40% | |
| 4 |
205 (32%) |
90 |
27.00% |
115 |
36.10% |
|
|
TNM_N | ||||||
| 0 | 373 (57%) | 193 | 58.00% | 180 | 56.40% | 0.75 |
| 1 | 151 (23%) | 80 | 24.00% | 71 | 22.30% | |
| 2 | 100 (15%) | 48 | 14.40% | 52 | 16.30% | |
| NA |
28 (4%) |
12 |
3.60% |
16 |
5.00% |
|
|
TNM_M | ||||||
| 0 | 550 (84%) | 291 | 87.40% | 259 | 81.20% | 0.03 |
| 1 |
102 (16%) |
42 |
12.60% |
60 |
18.80% |
|
|
Histological subtype (mucin producing) | ||||||
| Non-mucinous | 527 (81%) | 265 | 80.10% | 262 | 82.10% | 0.55 |
| Mucinous |
123 (19%) |
66 |
19.90% |
57 |
17.90% |
|
|
KRAS | ||||||
| Wild type | 290 (65%) | 182 | 64.80% | 108 | 66.30% | 0.83 |
| Mutation |
154 (35%) |
99 |
35.20% |
55 |
33.70% |
|
|
BRAF | ||||||
| Wild type | 385 (87%) | 245 | 87.50% | 140 | 86.40% | 0.86 |
| Mutation |
59 (13%) |
35 |
12.50% |
22 |
13.60% |
|
|
CIMP | ||||||
| Low | 362 (85%) | 230 | 85.80% | 132 | 83.50% | 0.62 |
| High |
64 (15%) |
38 |
14.20% |
26 |
16.50% |
|
|
MSI | ||||||
| Low | 382 (86%) | 241 | 86.10% | 141 | 87.00% | 0.89 |
| High |
60 (14%) |
39 |
13.90% |
21 |
13.00% |
|
|
p53 | ||||||
| Baseline | 154 (51%) | 98 | 48.50% | 56 | 49.10% | 0.91 |
| Accumulation |
162 (49%) |
104 |
51.50% |
58 |
50.90% |
|
|
Additional therapy | ||||||
| No adjuvant | 450 (69%) | 235 | 70.60% | 215 | 67.40% | 0.43 |
| Adjuvant | 202 (31%) | 98 | 29.40% | 104 | 32.60% | |
Abbreviations: CIMP=CpG island methylator phenotype; MSI=microsatellite instability.
P-values <0.05 are in bold.
RAD21 expression and its correlation with DSS
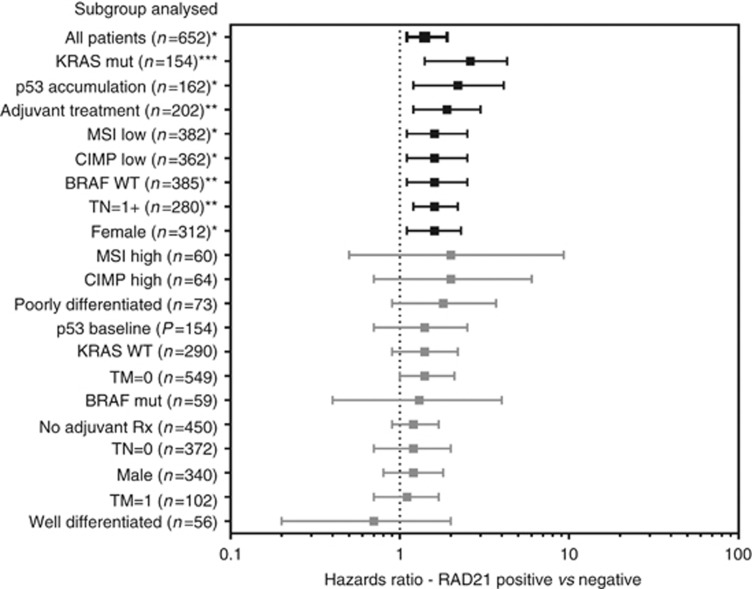
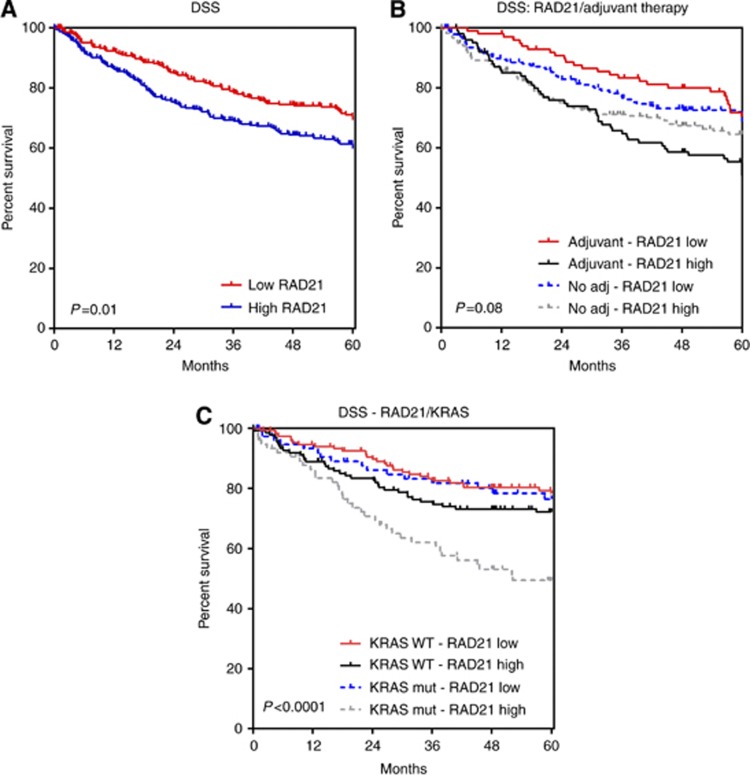
There was a significant correlation between RAD21 expression and shorter DSS on univariate analysis (HR:1.4 95% CI:1.1–1.9, P=0.01) (Figures 2 and 3A, Supplementary Table 2) and multivariate analysis (HR:2.56, P=0.01) (Table 2).
Figure 2.
Analysis of the prognostic effect (HR for disease-specific survival) of positive RAD21 expression in subgroups (as listed on the y-axis). HR represented by line point with end bars extending to 95% confidence intervals. *P-value <0.05, **P=<0.01, ***P=<0.001.
Figure 3.
Kaplan–Meier curves – disease-specific survival. (A) Overall; (B) stratified by adjuvant therapy; (C) stratified by KRAS mutation status.
Table 2. Multivariate analysis.
| Variable | Coefficient | Exp (coefficient) | s.e. (coef) | Z | P-value |
|---|---|---|---|---|---|
|
Low grade |
−0.49 |
0.61 |
0.19 |
−2.6 |
0.01 |
|
Stage 2 |
0.95 |
2.58 |
0.41 |
2.32 |
0.02 |
|
Stage 3 |
1.93 |
6.91 |
0.4 |
4.89 |
<0.01 |
|
Stage 4 |
3.31 |
27.46 |
0.4 |
8.36 |
<0.01 |
| RAD21 negative | −0.39 | 0.68 | 0.15 | −2.56 | 0.01 |
RAD21 expression on DSS was further assessed in patients stratified by treatment with conventional chemoradiotherapy (combinations of 5-FU, oxaliplatin+radiotherapy). An equal proportion of patients received chemoradiotherapy in both RAD21-negative and RAD21-positive cohorts (29.4% vs 32.6%, P=0.43). Here positive levels of RAD21 expression was a marker of shorter DSS (HR:1.9, 95% CI:1.2–3.0, P=0.008) (Figures 2 and 3B, Supplementary Table 2). In the cohort of patients not receiving adjuvant therapy, RAD21 expression showed no prognostic significance (Figure 2, Supplementary Table 2). Stratification of patients by KRAS mutational status also showed a striking effect with RAD21, showing a much more pronounced prognostic effect (increased HR and significance) in KRAS mutant cases when compared with wild type (Figures 2 and 3C, Supplementary Table 2).
RAD21 expression correlates with cellular sensitivity to 5-FU but not with oxaliplatin
RAD21 knockdown was therefore performed in the LIM1215 cell line that expresses high levels of RAD21 (data not shown) and is wild type for APC, KRAS, BRAF and p53 (Zhang et al, 2009). Of the three LIM1215 cell clones carrying RAD21 small hairpin RNAs, sh57224 showed a reduction in RAD21 protein level on western blot analysis compared with the parental and shmir (vector) cell clones (Supplementary Figure 1A). There was no difference in RAD21 protein levels between the parental LIM1215sc5 and shmir clones (Supplementary Figure 1A). The level of SMC1, another core cohesin subunit and RAD21's binding partner, was not affected (Supplementary Figure 1A). Immunofluorescence staining for RAD21 also showed a reduction in nuclear RAD21 expression in sh57224 clone when compared with the parental clone (Supplementary Figure 1B) with a 2.3-fold reduction in nuclear RAD21 protein level in sh57224 clones compared with the parental clone (Supplementary Figure 1C). This clone was used for subsequent analysis.
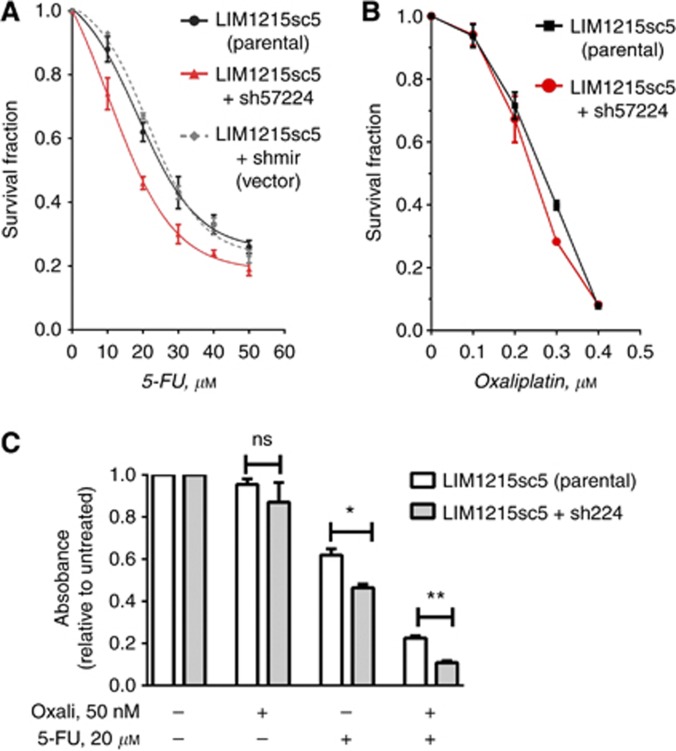
Treatment of all three LIM1215 clones, the parental LIM1215sc5, shmir and sh57224 cells, with graded doses (10–50 μM) of 5-FU resulted a reduction in cell survival in a dose-dependent manner (Figure 4A). Importantly, the sh57224 RAD21 knockdown clone showed an enhanced cellular sensitivity to 5-FU compared with both the parental and shmir clones (Figure 4A). There was no difference in cell survival between the parental LIM1215 single-cell clone (LIM1215sc5) and shmir clones. The IC50 of sh57224 was 16.0 μM, representing a significant reduction compared with the IC50 of the parental (22.0 μM) and the shmir (23.83 μM).
Figure 4.
The effect of RAD21 knockdown on 5-FU and oxaliplatin treatment in LIM1215 cells. (A) Survival fraction following treatment with 5-FU at concentrations are indicated. (B) Survival fraction following treatment with oxaliplatin at concentrations are indicated; (C) Survival fraction of a combined oxaliplatin and 5-FU treatment in relation to the singular treatment. Error bars=s.e.m. *P<0.01; **P<0.001.
Similar to 5-FU, treatment of cells with graded doses (100–400 nm) of oxaliplatin led to a decrease in cell survival in a dose-dependent manner (Figure 4B). However, there was no significant difference between the survival of sh57224 and parental cells (Figure 4B), suggesting that RAD21 expression has a negligible effect on cellular sensitivity to oxaliplatin alone in vitro.
As oxaliplatin and 5-FU is commonly given in combination with oxaliplatin, for example, FOLFOX, we further evaluated the effect of RAD21 expression on cell survival following the combined treatment of oxaliplatin and 5-FU. To better recapitulate the FOLFOX treatment schedule used in patients, we first treated cells with a single dose (50 μM) of oxaliplatin followed by a single dose (20 μM) of 5-FU. Cell survival fraction, relative to untreated cells, following the treatment of oxaliplatin alone showed no statistical difference between the two cell clones (Figure 4C). However, the treatment with 5-FU alone reduced cellular survival fraction, and there was a statistically significant difference between the two clones (P=0.011) (Figure 4C). As expected, the combination of the two drugs resulted in a significant reduction in cell survival for both the parental and sh57224 clones (Figure 4C). There was a 2.75-fold reduction in cell survival fraction of the parental clone, from 0.61 for 5-FU alone to 0.22 for the combined treatment. Strikingly, cell survival of the sh57224 clone was reduced from 0.48 for 5-FU alone to 0.1 for oxaliplatin/5-FU combination, thus representing a reduction of 4.76-fold (Figure 4C).
RAD21 overexpression correlates with c-Myc amplification
Analysis of the GEO data set GSE14333 showed a correlation between RAD21 and c-Myc overexpression (correlation: 0.47, P-value <0.0001) (Supplementary Figure 2).
Discussion
This multi-institution translational study is the first to comprehensively analyse the novel chromosomal DNA repair protein, RAD21 cohesin, in a large cohort of CRC. Our study revealed that positive RAD21 expression is a novel prognostic biomarker in CRC, is associated with disease progression and is also highly predictive in treatment outcome when using conventional chemoradiotherapy.
As CIN is a feature of CRC evolution, participation of RAD21 in maintaining genomic stability through its roles in accurate chromosome segregation, homologous recombination and telomere preservation demonstrated in vitro appears important in early tumour development. RAD21, unlike other cohesin genes, is not typically mutated in CRC (Barber et al, 2008). Instead, it is upregulated as demonstrated by prominent constitutive expression in primary cancers. We suggest that upregulation of RAD21 expression may be required for promoting CIN, either directly by favouring hyper-recombination or indirectly, via dysregulated chromosome segregation or by promoting global gene de-regulation, or in combination. This notion is supported biologically by its association with unfavourable primary tumour characteristics such as advanced tumour stage, as shown in both our current study and previous work in breast (Xu et al, 2011b) and endometrial cancer (Supernat et al, 2012). Interestingly, RAD21 expression also lost its prognostic significance in microsatellite instability high and CpG island methylator phenotype high subsets of tumours, but maintained significance in the p53-positive group. Interestingly, a significantly greater proportion of well-differentiated tumours were present in the RAD21-positive group. This is different to the association with higher grade seen in breast cancer, and as numbers of well-differentiated tumours are low in our overall cohort (and more so in our negative RAD21 group) a larger study focused specifically on well-differentiated tumours, and highly differentiated colon cell lines may elucidate RAD21-associated mechanisms further in this subset.
The mechanism of RAD21 upregulation is yet to be determined. We and others found that β-catenin activation is capable of regulating the transcriptional activity of RAD21 or SMC3 gene promoters in vitro, suggesting that at least one route is by Wnt/β-catenin signalling activation (Ghiselli et al, 2003; Xu et al, unpublished). Alternatively or additionally, elevated RAD21 expression may be a consequence of RAD21 gene/8q24 amplicon amplification as shown in high-grade breast cancer (Xu et al, 2011b). This possibility is supported by results of our in silico analysis showing correlating RAD21 and c-Myc expression in an independent CRC data set (GEO data set: GSE14333).
To date, both in vitro and in vivo data demonstrate that RAD21 expression influences cellular sensitivity to DNA-damaging agents such as those used in chemotherapy or chemoradiotherapy (Atienza et al, 2005; Bauerschmidt et al, 2010; Xu et al, 2010; Xu et al, 2011a, 2011b). An increased susceptibility of cancer cell lines to conventional chemotherapeutics was demonstrated following knockdown of RAD21 or other cohesin gene (Atienza et al, 2005; Xu et al, 2011b; Supernat et al, 2012). Similarly, yeast mutants and mammalian cells with reduced functional RAD21 have also been shown to have increased sensitivity to ionising radiation secondary to deficiency in the repair of DNA double-stranded breaks (Birkenbihl and Subramani, 1992; Bauerschmidt et al, 2010; Xu et al, 2010). In vitro, we have further verified the effect of RAD21 depletion within a CRC cell line to the sensitivity of commonly used CRC chemotherapeutics. As we reported in breast cancer, stable knockdown of RAD21 in a CRC cell line significantly enhanced cellular sensitivity, in a dose-dependent manner, to 5-FU and combined 5-FU and oxaliplatin treatments. The 5-FU antimetabolite is incorporated as 5-fluoro-dUTP into DNA, inducing its fragmentation and the subsequent repair of DNA damage involving several DNA repair pathways including homologous recombination (Wyatt and Wilson, 2009). The enhanced cellular sensitivity to 5-FU is thus consistent with the known role of RAD21 in homologous recombination. Intriguingly, the reduced RAD21 expression has no apparent effect on cells treated by oxaliplatin, but potentiates the effect of a combined oxaliplatin and 5-FU treatment regimen similar to the commonly used FOLFOX regimen (Andre et al, 2004, 2009; Tournigand et al, 2012). Oxaliplatin is a platinum-based chemotherapeutic that elicits DNA damage by forming DNA–platinum monoadducts with guanines that are subsequently converted to diadducts. Subsequent repair is predominantly through the nucleotide excision repair pathway (Arnould et al, 2003). The absence of any effect with oxaliplatin-based therapy as a single agent may be explained by DNA repair mechanisms by nucleotide excision repair that are adequate for singular oxaliplatin therapy but inadequate in the face of combination therapy.
The subanalysis of patients who had received adjuvant chemoradiotherapy supports the above in vitro findings. While we were not able to logistically separate the various therapeutic regimens present within our patient population, our findings that the patients receiving adjuvant therapy who had tumours positive for RAD21 expression showed a significantly shorter overall survival than patients whose tumours were negative for RAD21 expression is similar to that demonstrated in breast cancer. This observation further strengthens the role of RAD21 for predicting therapeutic response and also as a potential target for increasing tumour susceptibility to conventional and novel targeted agents.
Of particular interest is the finding that RAD21 expression showed specific prognostic significance in the context of KRAS mutational status. In both KRAS wild-type and mutant groups, elevated RAD21 expression associates with worse prognosis when compared with those with negative RAD21 expression. The data suggest that RAD21 or cohesin may be an important modifier of the KRAS signalling pathway. Indeed, forced expression in vitro of another cohesin component SMC3 has been shown to alter the expression of multiple genes modulating the Ras and Rho GTPase-dependent signalling pathways, in particular the upregulation of RhoB (Ghiselli and Liu, 2005). The elevated RAD21 expression may have a similar consequence in modulating RAS signalling through Rho. In addition to the prognostic relevance, the potential functional cross talk between RAD21 and KRAS demonstrated in our analysis may have therapeutic significance. KRAS mutational status is an established predictor of anti-EGFR monoclonal antibody therapy, as tumours with KRAS mutations respond poorly to cetuximab or panitumumab treatment (Allegra et al, 2009; Brand and Wheeler, 2012). Even within KRAS wild-type tumours, a subset fail to respond to anti-EGRF monoclonal antibodies (Linardou et al, 2009). Our data suggest that RAD21 and/or its network of cohesin regulatory proteins may be potential therapeutic targets for the treatment of resistant tumours such as those carrying KRAS mutations.
Conclusion
This study of RAD21 expression in CRC provides strong evidence that RAD21 expression is a novel prognostic marker, particularly in the context of KRAS mutations and most likely within cancers arising through the chromosomal instability pathway. Furthermore, both in vitro and in vivo data suggest that RAD21 expression affects the response to chemotherapy or chemoradiotherapy. Our findings in CRC are consistent with our previous studies in breast cancer (Xu et al, 2011b; Yan et al, 2012), suggesting that elevated RAD21 expression and its association with poor prognosis, as well as resistance to conventional chemoradiotherapy, may be a common feature of epithelial cancers.
Acknowledgments
This work is support by the National Health and Medical Research Council (NHMRC) project grant to RGR, HX and MJM. Tissue microarrays were provided by the Lowy Biorepository, University of NSW, Sydney, with support from the Translational Cancer Research Network, NSW. We thank the microscopy core for assistance with histology and image analysis.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, Mcallister PK, Morton RF, Schilsky RL. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- Andre T, Boni C, Mounedji-Boudiaf L, Navarro M, Tabernero J, Hickish T, Topham C, Zaninelli M, Clingan P, Bridgewater J, Tabah-Fisch I, de Gramont A. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med. 2004;350:2343–2351. doi: 10.1056/NEJMoa032709. [DOI] [PubMed] [Google Scholar]
- Andre T, Boni C, Navarro M, Tabernero J, Hickish T, Topham C, Bonetti A, Clingan P, Bridgewater J, Rivera F, de Gramont A. Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J Clin Oncol. 2009;27:3109–3116. doi: 10.1200/JCO.2008.20.6771. [DOI] [PubMed] [Google Scholar]
- Arnould S, Hennebelle I, Canal P, Bugat R, Guichard S. Cellular determinants of oxaliplatin sensitivity in colon cancer cell lines. Eur J Cancer. 2003;39:112–119. doi: 10.1016/s0959-8049(02)00411-2. [DOI] [PubMed] [Google Scholar]
- Atienza JM, Roth RB, Rosette C, Smylie KJ, Kammerer S, Rehbock J, Ekblom J, Denissenko MF. Suppression of RAD21 gene expression decreases cell growth and enhances cytotoxicity of etoposide and bleomycin in human breast cancer cells. Mol Cancer Ther. 2005;4:361–368. doi: 10.1158/1535-7163.MCT-04-0241. [DOI] [PubMed] [Google Scholar]
- Barber TD, Mcmanus K, Yuen KW, Reis M, Parmigiani G, Shen D, Barrett I, Nouhi Y, Spencer F, Markowitz S, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C, Hieter P. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci USA. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerschmidt C, Arrichiello C, BURDAK-Rothkamm S, Woodcock M, Hill MA, Stevens DL, Rothkamm K. Cohesin promotes the repair of ionizing radiation-induced DNA double-strand breaks in replicated chromatin. Nucleic Acids Res. 2010;38:477–487. doi: 10.1093/nar/gkp976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Subramani S. Cloning and characterization of rad21 an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res. 1992;20:6605–6611. doi: 10.1093/nar/20.24.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand TM, Wheeler DL. KRAS mutant colorectal tumors: past and present. Small GTPases. 2012;3:34–39. doi: 10.4161/sgtp.18751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiselli G, Coffee N, Munnery CE, Koratkar R, Siracusa LD. The cohesin SMC3 is a target the for beta-catenin/TCF4 transactivation pathway. J Biol Chem. 2003;278:20259–20267. doi: 10.1074/jbc.M209511200. [DOI] [PubMed] [Google Scholar]
- Ghiselli G, Liu CG. Global gene expression profiling of cells overexpressing SMC3. Mol Cancer. 2005;4:34. doi: 10.1186/1476-4598-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V. Sister chromatid cohesion: the cohesin cleavage model does not ring true. Genes Cells. 2007;12:693–708. doi: 10.1111/j.1365-2443.2007.01093.x. [DOI] [PubMed] [Google Scholar]
- Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- Hakimi MA, Bochar DA, Schmiesing JA, Dong Y, Barak OG, Speicher DW, Yokomori K, Shiekhattar R. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature. 2002;418:994–998. doi: 10.1038/nature01024. [DOI] [PubMed] [Google Scholar]
- Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J, Kerr D, Aaltonen LA, Arango D, Kruhoffer M, Orntoft TF, Andersen CL, Gruidl M, Kamath VP, Eschrich S, Yeatman TJ, Sieber OM. Metastasis-associated gene expression changes predict poor outcomes in patients with Dukes stage B and C colorectal cancer. Clin Cancer Res. 2009;15:7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardou H, Dahabreh IJ, Bafaloukos D, Kosmidis P, Murray S. Somatic EGFR mutations and efficacy of tyrosine kinase inhibitors in NSCLC. Nat Rev Clin Oncol. 2009;6:352–366. doi: 10.1038/nrclinonc.2009.62. [DOI] [PubMed] [Google Scholar]
- McKay MJ, Troelstra C, van der Spek P, Kanaar R, Smit B, Hagemeijer A, Bootsma D, Hoeijmakers JH. Sequence conservation of the rad21 Schizosaccharomyces pombe DNA double-strand break repair gene in human and mouse. Genomics. 1996;36:305–315. doi: 10.1006/geno.1996.0466. [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Peters JM, Uhlmann F.2001Splitting the chromosome: cutting the ties that bind sister chromatids Novartis Found Symp 237113–133.discussion 133–138, 158–163. [DOI] [PubMed] [Google Scholar]
- Packham D, Ward RL, Ap Lin V, Hawkins NJ, Hitchins MP. Implementation of novel pyrosequencing assays to screen for common mutations of BRAF and KRAS in a cohort of sporadic colorectal cancers. Diagn Mol Pathol. 2009;18:62–71. doi: 10.1097/PDM.0b013e318182af52. [DOI] [PubMed] [Google Scholar]
- Sonoda E, Matsusaka T, Morrison C, Vagnarelli P, Hoshi O, Ushiki T, Nojima K, Fukagawa T, Waizenegger IC, Peters JM, Earnshaw WC, Takeda S. Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev Cell. 2001;1:759–770. doi: 10.1016/s1534-5807(01)00088-0. [DOI] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supernat A, Lapinska-Szumczyk S, Sawicki S, Wydra D, Biernat W, Zaczek AJ. Deregulation of RAD21 and RUNX1 expression in endometrial cancer. Oncol Lett. 2012;4:727–732. doi: 10.3892/ol.2012.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournigand C, Andre T, Bonnetain F, Chibaudel B, Lledo G, Hickish T, Tabernero J, Boni C, Bachet JB, Teixeira L, de Gramont A. Adjuvant therapy with fluorouracil and oxaliplatin in stage II and elderly patients (between ages 70 and 75 years) with colon cancer: subgroup analyses of the Multicenter International Study of Oxaliplatin, Fluorouracil, and Leucovorin in the Adjuvant Treatment of Colon Cancer trial. J Clin Oncol. 2012;30:3353–3360. doi: 10.1200/JCO.2012.42.5645. [DOI] [PubMed] [Google Scholar]
- Ward R, Meagher A, Tomlinson I, O'connor T, Norrie M, Wu R, Hawkins N. Microsatellite instability and the clinicopathological features of sporadic colorectal cancer. Gut. 2001;48:821–829. doi: 10.1136/gut.48.6.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RL, Williams R, Law M, Hawkins NJ. The CpG island methylator phenotype is not associated with a personal or family history of cancer. Cancer Res. 2004;64:7618–7621. doi: 10.1158/0008-5472.CAN-03-3978. [DOI] [PubMed] [Google Scholar]
- Watrin E, Peters JM. Cohesin and DNA damage repair. Exp Cell Res. 2006;312:2687–2693. doi: 10.1016/j.yexcr.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Watrin E, Peters JM. The cohesin complex is required for the DNA damage-induced G2/M checkpoint in mammalian cells. EMBO J. 2009;28:2625–2635. doi: 10.1038/emboj.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead RH, Macrae FA, St John DJ, Ma J. A colon cancer cell line (LIM1215) derived from a patient with inherited nonpolyposis colorectal cancer. J Natl Cancer Inst. 1985;74:759–765. [PubMed] [Google Scholar]
- Wyatt MD, Wilson DM., 3RD Participation of DNA repair in the response to 5-fluorouracil. Cell Mol Life Sci. 2009;66:788–799. doi: 10.1007/s00018-008-8557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Balakrishnan K, Malaterre J, Beasley M, YAN Y, Essers J, Appeldoorn E, Thomaszewski JM, Vazquez M, Verschoor S, Lavin MF, Bertonchello I, Ramsay RG, Mckay MJ. Rad21-cohesin haploinsufficiency impedes DNA repair and enhances gastrointestinal radiosensitivity in mice. PLoS One. 2010;5:e12112. doi: 10.1371/journal.pone.0012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Beasley M, Verschoor S, Inselman A, Handel MA, Mckay MJ. A new role for the mitotic RAD21/SCC1 cohesin in meiotic chromosome cohesion and segregation in the mouse. EMBO Rep. 2004;5:378–384. doi: 10.1038/sj.embor.7400121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Tomaszewski JM, Mckay MJ. Can corruption of chromosome cohesion create a conduit to cancer. Nat Rev Cancer. 2011;11:199–210. doi: 10.1038/nrc3018. [DOI] [PubMed] [Google Scholar]
- Xu H, Yan M, Patra J, Natrajan R, Yan Y, Swagemakers S, Tomaszewski JM, Verschoor S, Millar EK, van der Spek P, Reis-Filho JS, Ramsay RG, O'Toole SA, McNeil CM, Sutherland RL, McKay MJ, Fox SB. Enhanced RAD21 cohesin expression confers poor prognosis and resistance to chemotherapy in high grade luminal, basal and HER2 breast cancers. Breast Cancer Res. 2011;13:R9. doi: 10.1186/bcr2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Xu H, Waddell N, Shield-Artin K, Haviv I, Mckay MJ, Fox SB. Enhanced RAD21 cohesin expression confers poor prognosis in BRCA2 and BRCAX, but not BRCA1 familial breast cancers. Breast Cancer Res. 2012;14:R69. doi: 10.1186/bcr3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HH, Walker F, Kiflemariam S, Whitehead RH, Williams D, Phillips WA, Mikeska T, Dobrovic A, Burgess AW. Selective inhibition of proliferation in colorectal carcinoma cell lines expressing mutant APC or activated B-Raf. Int J Cancer. 2009;125:297–307. doi: 10.1002/ijc.24289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.