Abstract
Background:
Transforming growth factor β-induced protein (TGFBI) is a secreted protein that mediates cell anchoring to the extracellular matrix. This protein is downregulated in lung cancer, and when overexpressed, contributes to apoptotic cell death. Using a small series of stage IV non-small cell lung cancer (NSCLC) patients, we previously suggested the usefulness of TGFBI as a prognostic and predictive factor in chemotherapy-treated late-stage NSCLC. In order to validate and extend these results, we broaden the analysis and studied TGFBI expression in a large series of samples obtained from stage I–IV NSCLC patients.
Methods:
TGFBI expression was assessed by immunohistochemistry in 364 completely resected primary NSCLC samples: 242 adenocarcinomas (ADCs) and 122 squamous cell carcinomas (SCCs). Kaplan–Meier curves, log-rank tests and the Cox proportional hazards model were used to analyse the association between TGFBI expression and survival.
Results:
High TGFBI levels were associated with longer overall survival (OS, P<0.001) and progression-free survival (PFS, P<0.001) in SCC patients who received adjuvant platinium-based chemotherapy. Moreover, multivariate analysis demonstrated that high TGFBI expression is an independent predictor of better survival in patients (OS: P=0.030 and PFS: P=0.026).
Conclusions:
TGFBI may be useful for the identification of a subset of NSCLC who may benefit from adjuvant therapy.
Keywords: lung carcinoma, prognosis, survival, TGFBI, adjuvant therapy
Lung cancer is the leading cause of cancer mortality worldwide (Siegel et al, 2011). Non-small cell lung cancer (NSCLC) constitutes the largest subgroup of lung cancers, accounting for ∼80% of all cases. Most patients with NSCLC are diagnosed with locally advanced or metastatic disease and have very low survival expectancy (Wang et al, 2010). Surgical resection is the most favourable treatment for early-stage NSCLC, although relapse is still high, especially in stage II and III (Win et al, 2008). Adjuvant cisplatin-based chemotherapy after complete resection is recommended in stage II and stage IIIA lung cancer patients to improve survival, although it is associated with adverse side effects and the proportion of patients who benefit from this therapy is still low (Robinson et al, 2007; Scott et al, 2007). To improve clinical outcomes, and to select the best performing adjuvant treatment for each patient, new tumour biomarkers that help to identify patients who are at the highest risk for recurrence are urgently needed. These biomarkers might enable clinicians to guide their treatment decisions.
Transforming growth factor β-induced protein (TGFBI) is a soluble extracellular matrix adaptor protein that mediates cell adhesion to extracellular proteins such as collagen, fibronectin and laminins through integrin binding (Kim et al, 2000). Transforming growth factor β-induced protein is differentially expressed in transformed tissues. Loss of TGFBI expression has been described in several cancers such as lung (Zhao et al, 2004; Shao et al, 2006), neuroblastoma (Becker et al, 2008), breast (Wen et al, 2011) and ovary carcinoma (Ahmed et al, 2007). Transforming growth factor β-induced protein is also overexpressed in several solid tumours such as colon (Kitahara et al, 2001), pancreas (Schneider et al, 2002) and kidney (Ivanov et al, 2008). This suggests that, depending on the tissue, TGFBI can exert either pro- or anti-tumoral functions, most likely depending on the integrins to which it binds to on the cell surface (Thapa et al, 2007). Interestingly, Zhao et al. demonstrated that TGFBI overexpression in lung adenocarcinoma (ADC) cells increases their sensitivity to etoposide-induced cell death (Zhao et al, 2006).
Recently, we suggested that TGFBI expression might be a predictive factor of response to chemotherapy in a series of stage IV NSCLC samples, as patients with high levels of this protein respond to chemotherapy, meanwhile patients who did not respond to the treatment showed low levels of TGFBI. Moreover, we have demonstrated that TGFBI-derived proteolytic fragments induce apoptotic cell death in NSCLC (Irigoyen et al, 2010). These significant results need to be validated in a greater number of samples in order to confirm TGFBI as a relevant biomarker in NSCLC. In recent years, a great number of prognostic biomarkers have been described in NSCLC (Coate et al, 2009; Aggarwal et al, 2010) and some are intensively evaluated for predictive value such as ERCC1 (Olaussen et al, 2006; Friboulet et al, 2013) or RRM1 (Rosell et al, 2004). The aim of this study was to analyse the prognostic and predictive impact of this protein on the survival of NSCLC patients.
Materials and Methods
Patients and tissue samples
A series of 364 patients diagnosed with NSCLC who underwent surgical resection in the M.D. Anderson Cancer Center (Houston, TX, USA) from 2003 to 2005 were included in this study. Histological diagnosis was carried out using the 2004 WHO classification system (Travis et al, 2004). Pathologic staging of the tumours was performed according to the International System for Staging Lung Cancer (Mountain, 1997). A tissue microarray containing representative areas of 511 NSCLC specimens was constructed with triplicated 1 mm tissue cores from each tumour as previously described (Kim et al, 2012). Only patients that complied the following inclusion criteria were included in this study: complete resection of the primary lung tumour, ADC or squamous cell carcinoma (SCC) histology, no malignancy in the last 5 years, no neoadjuvant therapy and available clinicopathological information. Adjuvant therapy was administered in 133 NSCLC patients after resection while 217 individuals were treated exclusively with surgery. Progression-free survival (PFS) was estimated as the time from surgery to recurrence or death from the disease. Overall survival (OS) was defined as the time from surgery to the date of death.
Detailed clinical and pathological information, including age, gender and stage, is summarised in Table 1.
Table 1. Clinical and pathological characteristics of patients.
| Patients (n=364) | n (%) |
|---|---|
| Age-year (median±SD) |
67±10 |
|
Gender | |
| Male | 176 (48.4) |
| Female |
188 (51.6) |
|
Stage | |
| I | 228 (62.6) |
| II | 60 (16.5) |
| III | 58 (15.9) |
| IV |
18 (5.0) |
|
pT | |
| T1 | 137 (37.6) |
| T2 | 187 (51.4) |
| T3 | 17 (4.7) |
| T4 |
23 (6.3) |
|
pN | |
| N0 | 263 (72.2) |
| N1 | 66 (18.1) |
| N2 | 34 (9.4) |
| N3 |
1 (0.3) |
|
Histology | |
| ADC | 242 (66.5) |
| SCC | 122 (33.5) |
Abbreviations: ADC=adenocarcinoma; pN=pathological N stage; pT=pathological T stage; SCC=squamous cell carcinoma; SD=standard deviation.
Tissue banking and the study protocol were approved by the institutional medical ethical committee. Written informed consent was obtained from each patient.
Reported recommendations for tumour marker prognostic studies (REMARK) criteria were followed throughout the study (Altman et al, 2012).
Immunohistochemical analysis
The expression of TGFBI was assessed using an indirect immunohistochemical staining, as previously described (Irigoyen et al, 2010). Briefly, tissue microarray sections were deparaffined in xylene and rehydrated with graded ethanol. Antigen retrieval was carried out in a pressure cooker in citrate buffer (10 mM, pH 6). Endogenous peroxidase activity was quenched with 3% hydrogen peroxide for 10 min. Non-specific binding sites were blocked with 5% normal goat serum in TBS-Tween (Wash buffer, Dako, Glostrup, Denmark) for 30 min. Sections were incubated with anti-TGFBI rabbit-polyclonal antibody (1 : 25; Proteintech group, Chicago, IL, USA) overnight at 4 °C. Detection was performed with ENVISION HRP system (Dako). The peroxidase activity was visualised with diaminobenzidine. Finally, sections were washed, lightly counterstained with hematoxylin, dehydrated and mounted. Negative controls were performed by omission of the primary antibody or incubation with an isotype control antibody.
As previously described (Irigoyen et al, 2010), the specificity of the antibody was assessed by western blot analysis in lung cancer cells in which TGFBI was overexpressed or inhibited.
Quantification of immunohistochemical staining
Two observers (MJP and ES) evaluated the samples independently and unaware of the outcomes of patients. The extension of the staining was scored as the percentage of positive cells (0–100%), and the intensity of the staining was assessed using a 4-value scoring system (0, below the level of detection; 1, weak; 2, moderate; and 3, strong). A final H score was calculated by adding the product of the percentage cells stained at a given staining intensity (0–100) and the staining intensity (0–3), as previously described (Pajares et al, 2012). The median value of all H scores was chosen as the cutoff point to separate low from high TGFBI expressing tumours. Discordant independent readings were resolved by simultaneous review by the two observers.
Statistical analysis
The association between TGFBI expression and clinicopathological parameters was analysed by Pearson's chi-square test. Cumulative survival of patients was estimated using Kaplan–Meier curves, and significant differences between groups were tested using the log-rank test. Follow-up time was calculated from the date of surgery to the date of progression, death, lost to follow-up or last contact with the patient. Univariate and multivariate Cox proportional hazards analyses were used to assess the prognostic role of TGFBI. Only those variables with P-value <0.1 in the univariate analysis were included in the multivariate analysis. Statistical analyses were performed using SPSS 15.0 software (Chicago, IL, USA). A P-value <0.05 was considered statistically significant.
Results
TGFBI expression in NSCLC
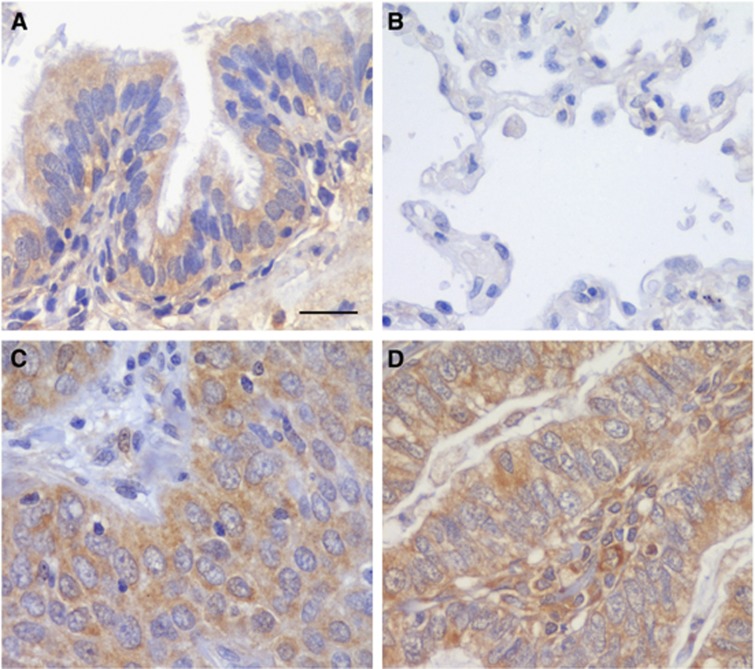
A series of 364 NSCLC patients was analysed by immunohistochemistry in this study. Transforming growth factor β-induced protein staining was found in the stroma of normal and tumour tissue (Figure 1). Specifically, in non-tumour tissues, lung bronchiolar epithelia showed immunoreactivity for TGFBI (Figure 1A) meanwhile the alveoli were negative for this protein (Figure 1B). In lung tumours, TGFBI was detected in the cytoplasm of the cells in all the samples (Figure 1C and D). Patients were dichotomised into high and low by the median TGFBI expression H score. No significant association was found between TGFBI expression and clinicopathological features of the patients (Table 2). However, when patients were stratified according to the histological subtype, a significant correlation was found between TGFBI expression and smoking status in ADC patients (P=0.021). More interestingly, in the SCC subgroup, TGFBI expression was higher in early-stage tumours (P=0.014) and in patients without lymph node involvement (P=0.019).
Figure 1.
Transforming growth factor β-induced protein expression in NSCLC samples. Immunohistochemical staining for TGFBI protein in non-tumour tissue (A and B) and lung carcinomas (C and D). Immunoreactivity was observed in the cytoplasm of bronchiole epithelial cells (A), ADC (C) and SCC (D). No imunostaining was detected in lung alveoli (B). Scale bar, 50 μm.
Table 2. Relationship between TGFBI expression and clinicopathological features of NSCLC patients.
|
NSCLC (n=364) |
ADC (n=242) |
SCC (n=122) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
n
(%) |
|
n
(%) |
|
n
(%) |
|
||||
| Low | High | P | Low | High | P | Low | High | P | |
|
Age | |||||||||
| ⩽70 | 107 (59.8) | 72 (40.2) | 0.284 | 71 (43.8) | 91 (56.2) | 0.146 | 37 (56.1) | 29 (43.9) | 0.692 |
| >70 |
121 (65.4) |
64 (34.6) |
|
43 (53.7) |
37 (46.3) |
|
29 (51.8) |
27 (48.2) |
|
|
Gender | |||||||||
| Male | 91 (51.7) | 85 (48.3) | 0.405 | 49 (48) | 53 (52) | 0.804 | 42 (56.8) | 32 (43.2) | 0.464 |
| Female |
89 (47.3) |
99 (52.7) |
|
65 (46.4) |
75 (53.6) |
|
24 (50) |
24 (50) |
|
|
Stage | |||||||||
| I–II | 136 (47.2) | 152 (52.8) | 0.098 | 89 (46.6) | 102 (53.4) | 0.758 | 47 (48.5) | 50 (51.5) | 0.014 |
| III–IV |
44 (57.9) |
32 (42.1) |
|
25 (49) |
26 (51) |
|
19 (76) |
6 (24) |
|
|
pT | |||||||||
| T1 | 63 (46) | 74 (54) | 0.304 | 41 (41.8) | 57 (58.2) | 0.175 | 22 (56.4) | 17 (43.6) | 0.725 |
| T2/T3/T4 |
117 (51.5) |
110 (48.5) |
|
73 (50.7) |
71 (49.3) |
|
44 (53) |
39 (47) |
|
|
pN | |||||||||
| N0 | 124 (47.1) | 139 (52.9) | 0.156 | 88 (47.6) | 97 (52.4) | 0.796 | 36 (46.2) | 42 (53.8) | 0.019 |
| N1–N2–N3 |
56 (55.4) |
45 (44.6) |
|
26 (45.6) |
31 (54.4) |
|
30 (68.2) |
14 (31.8) |
|
|
Histology | |||||||||
| ADC | 114 (80.3) | 128 (19.7) | 0.208 | ||||||
| SCC |
66 (54.1) |
56 (45.9) |
|
|
|
|
|
|
|
|
Smoking statusa | |||||||||
| Never | 22 (57.9) | 16 (42.1) | 0.204 | 22 (57.9) | 16 (42.1) | 0.021 | — | — | 0.496 |
| Former | 90 (52.6) | 81 (47.4) | 58 (52.7) | 52 (47.3) | 32 (52.5) | 29 (47.5) | |||
| Current | 68 (44.2) | 86 (55.8) | 34 (36.2) | 60 (63.8) | 34 (56.7) | 26 (43.3) | |||
Abbreviations: ADC=adenocarcinoma; NSCLC=non-small cell lung cancer; pN=pathological N stage; pT=pathological T stage; SCC=squamous cell carcinoma.
Smoking status data were available for n=363 patients. The values highlighted in bold are statistically significant (P<0.05).
TGFBI and clinical outcome in NSCLC
To further extend our previous study about the potential value of TGFBI as a relevant biomarker in NSCLC (Irigoyen et al, 2010), we sought to investigate whether TGFBI may influence disease outcome in a series of 364 NSCLC patients.
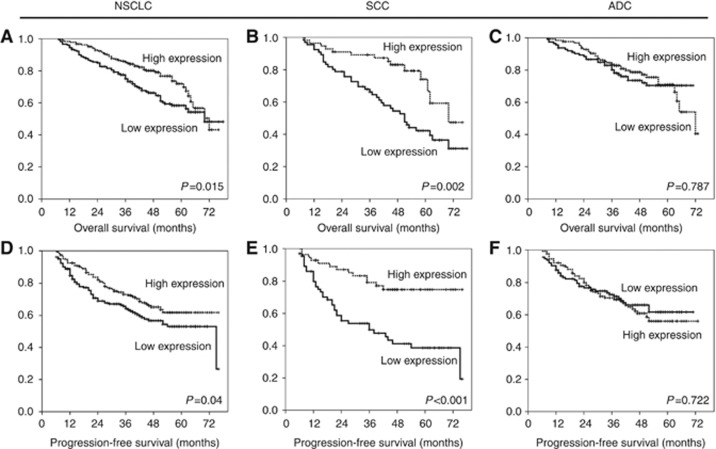
First, we analysed the expression of TGFBI in the entire cohort of NSCLC patients. Patients with high levels of TGFBI showed better OS (P=0.015) (Figure 2A) and longer PFS (P=0.04) (Figure 2D) than patients with low TGFBI expression. Interestingly, when we stratified these patients according to histology (242 ADC and 122 SCC), the same association between high TGFBI expression and better prognosis was restricted to lung SCC patients (OS: P=0.002 and PFS: P<0.001) (Figure 2B and E, respectively). In contrast to those findings, no significant association was found between TGFBI expression and OS or PFS in lung ADC patients (OS: P=0.787; PFS: P=0.722) (Figure 2C and F).
Figure 2.
High TGFBI levels were associated with longer survival in patients with SCC of the lung. Kaplan–Meier curves of OS (A–C) and PFS (D–F) for high and low expression of TGFBI. The median was used as the cutoff point.
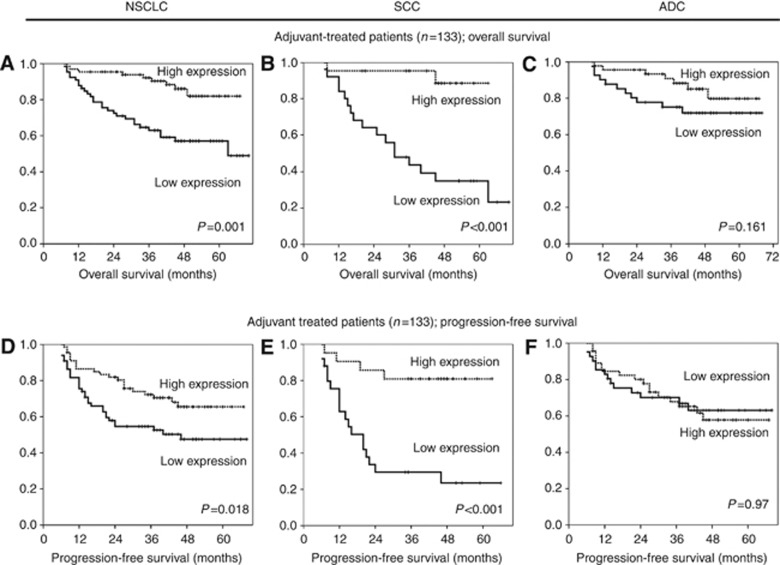
To get further insights into the potential value of TGFBI as a predictive biomarker in NSCLC, we analysed the clinical outcome after patient stratification based on adjuvant therapy. In patients who received platinum-based chemotherapy after surgery, an association between higher TGFBI levels and longer OS and PFS was found (P=0.001 and P=0.018, respectively) (Figure 3A and D). We further sought to determine whether this behaviour occurred in both histological subtypes. Among patients treated with adjuvant therapy, the SCC subgroup showed similar results, that is, high TGFBI expression was associated with better OS (P<0.001) (Figure 3B) and PFS (P<0.001) (Figure 3E). In contrast, TGFBI expression in adjuvant-treated ADC patients showed no statistical differences in OS or PFS (P=0.161 and P=0.97, respectively) (Figure 3C and F). Moreover, patients treated only with surgery did not show any correlation between TGFBI levels and survival in SCC (OS: P=0.263 and PFS: P=0.088) or ADC subtypes (OS: P=0.362 and PFS: P=0.386).
Figure 3.
High TGFBI is a good prognostic factor in lung SCC patients who received adjuvant treatment. Kaplan–Meier curves of OS (A–C) and PFS (D–F) for high and low expression of TGFBI. The median was used as the cutoff point.
Multivariate analysis using the Cox regression model was performed on the complete series to determine independent risk factor for OS and PFS in NSCLC. Transforming growth factor β-induced protein expression (P=0.044), age (P=0.009) and stage (P=0.001) were found to be independent predictor factors for OS (Table 3 and Supplementary Table 1). In regards to PFS, only tumour stage reached the level of statistical significance (P=0.001), although we observed a trend towards an association between high TGFBI expression and better outcome (P=0.085). Interestingly, when we evaluated adjuvant-treated SCC, TGFBI and stage were found to be independent predictor factors for OS (TGFBI, P=0.012; stage, P=0.001) and PFS (TGFBI, P=0.003; stage, P=0.001) (Table 3).
Table 3. Multivariate analysis for the effect of TGFBI expression on OS or PFS in adjuvant-treated NSCLC patients.
|
OS |
PFS |
|||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
|
NSCLC | ||||
|
TGFBI H-score | ||||
| ⩽173 | ||||
| >173 |
0.680 (0.467–0.990) |
P=0.044 |
0.739 (0.524–1.043) |
P=0.085 |
|
Age | ||||
| <70 | ||||
| ⩾70 |
1.628 (1.1287–2.351) |
P=0.009 |
|
|
|
Stage | ||||
| I–II | ||||
| III–IV |
1.940 (1.295–2.906) |
P=0.001 |
2.536 (1.762–3.649) |
P<0.001 |
|
SCC | ||||
|
TGFBI H-score | ||||
| ⩽173 | ||||
| >173 |
0.450 (0.241–0.838) |
P=0.012 |
0.377 (0.197–0.720) |
P=0.003 |
|
Stage | ||||
| I–II | ||||
| III–IV | 2.557 (1.411–4.633) | P=0.001 | 2.719 (1.483–4.985) | P=0.001 |
Abbreviations: NSCLC=non-small cell lung cancer; OS=overall survival; PFS=progression-free survival; SCC=squamous cell carcinoma.
Together, these findings indicate that TGFBI expression may be useful for the identification of a subset of NSCLC who may benefit from adjuvant platinum-based chemotherapy after resection.
Discussion
TGFBI was originally isolated from a cDNA library performed in A549 lung ADC cells treated with TGFβ. Mostly studied in dystrophy of the cornea, TGFBI is linked to cancer due to its function as adapter between the integrins expressed on the cell surface and some extracellular matrix proteins. Early reports from Zhao et al, 2002 demonstrated that TGFBI/Bigh3 expression was downregulated in asbestos induced lung cancer models in mice, as well as in several NSCLC-derived human cell lines compared with primary bronchial epithelial cells (Zhao et al, 2002; Shao et al, 2006). Reports from the same group showed TGFBI promoter hypermethylation in lung cancer cell lines as one possible mechanistic explanation for its decreased expression in transformed cells (Shao et al, 2006; Shah et al, 2008).
In this work, we have analysed TGFBI expression by immunohistochemistry in 364 samples of NSCLC (242 ADC and 122 SCC). Data obtained from lung SCC samples showed that decreased TGFBI expression significantly correlated with advanced tumour stages and the presence of metastasis in lymph nodes. As already mentioned, TGFBI sustains cell adhesion to substrates (Kim et al, 2000), therefore, losses in the structures that provide anchorage to the substrates might increase cell motility and the chance to metastasise. In fact, two recent reports showed that TGFBI overexpression in breast and lung cancer cell lines suppressed their in vitro and in vivo ability to metastasise (Ween et al, 2011; Wen et al, 2011). Our results are in line with those reports, and provide further clinical relevance to support them.
The use of genes related to cell metastasis and survival as markers to predict outcome is based on their physiological role and offers them as potential targets for therapy. Building on the findings from our group (Irigoyen et al, 2010) and others (Zhao et al, 2006), we hypothesised that TGFBI could serve as good predictor for survival in NSCLC patients treated with adjuvant platinum-based chemotherapy. As showed herein, a strong correlation exists between high TGFBI expression and both PFS and OS in NSCLC patients. When samples were stratified according to tumour histology, statistical significance upheld only for SCC patients. Multivariate analysis confirmed the value of TGFBI as an independent predictor for survival in adjuvant-treated SCC.
Besides its contribution to cell adhesion, there is another feature of TGFBI that might be relevant for tumour progression: TGFBI-derived fragments are pro-apoptotic in several cell types (Kim et al, 2003; Morand et al, 2003). In this respect, we have previously demonstrated that TGFBI-derived proteolytic peptides provoke cell death through binding to αvβ3 integrins and induction of caspase 3 activation (Irigoyen et al, 2010). The reason why TGFBI induces cell death in some cell types and promotes metastasis in others is still unknown. One possible explanation may come from the activation of cell-specific proteases capable of degrading TGFBI (Wen et al, 2011), such as plasmin (Ween et al, 2011), or from its binding to cell type-restricted integrins, leading to activation different cellular responses (Thapa et al, 2007). In fact, the observed dichotomised behaviour of this marker between ADC and SCC samples is probably a consequence of their phenotypical heterogeneity. The finding of tissue-specific biomarkers is not trivial, as results of recent clinical trials advocate for histology-based decision-making for therapy of NSCLC (Okamoto et al, 2006; Chang, 2011; Maus et al, 2013). To improve cancer treatment efficacy, it is important to have reliable predictive markers that help clinicians to distinguish those patients who need more aggressive therapies from those who do not. This is especially important in order to carefully select treatment options while keeping in mind the principles of maximum efficacy with minimal toxicity.
In summary, the results presented herein propose TGFBI as a new predictive marker of survival in lung SCC patients treated with adjuvant platinum-based chemotherapy, and emphasise the differences in the biology of the two main types of NSCLC (Langer et al, 2010).
Acknowledgments
The authors thank Patricia Martín and Saray Garasa for technical support. This work has been supported by ‘UTE project CIMA' European Union (grants no. QLG1-CT-2002-01735 and HEALTH-F2-2010-258677); Spanish Government (ISCIII-RTICC RD12/0036/0040), European Regional Development Fund (ERDF) ‘Una manera de hacer Europa' and Spanish Ministry of Science and Innovation (FIS) (PI10/00166 and PI10/02131).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Aggarwal C, Somaiah N, Simon G. Biomarkers with predictive and prognostic function in non-small cell lung cancer: ready for prime time. J Natl Compr Canc Netw. 2010;8:822–832. doi: 10.6004/jnccn.2010.0059. [DOI] [PubMed] [Google Scholar]
- Ahmed A, Mills A, Ibrahim AEK, Temple J, Blenkiron C, Vias M, Massie C, Iyer NG, McGeoch A, Crawford R, Nicke B, Downward J, Swanton C, Bell S, Earl H, Laskey R, Caldas C, Brenton J. The extracellular matrix protein TGFBI induces microtubule stabilization and sensitizes ovarian cancers to paclitaxel. Cancer Cell. 2007;12:514–527. doi: 10.1016/j.ccr.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman D, McShane L, Sauerbrei W, Taube S. Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. BMC Med. 2012;10:51–51. doi: 10.1186/1741-7015-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Volland S, Noskova I, Schramm A, Schweigerer L, Wilting J. Keratoepithelin reverts the suppression of tissue factor pathway inhibitor 2 by MYCN in human neuroblastoma: a mechanism to inhibit invasion. Int J Oncol. 2008;32:235–240. [PubMed] [Google Scholar]
- Chang A. Chemotherapy, chemoresistance and the changing treatment landscape for NSCLC. Lung Cancer. 2011;71:3–10. doi: 10.1016/j.lungcan.2010.08.022. [DOI] [PubMed] [Google Scholar]
- Coate L, John T, Tsao M, Shepherd F. Molecular predictive and prognostic markers in non-small-cell lung cancer. Lancet Oncol. 2009;10:1001–1010. doi: 10.1016/S1470-2045(09)70155-X. [DOI] [PubMed] [Google Scholar]
- Friboulet L, Olaussen KA, Pignon JP, Shepherd FA, Tsao MS, Graziano S, Kratzke R, Douillard JY, Seymour L, Pirker R, Filipits M, André F, Solary E, Ponsonnailles F, Robin A, Stoclin A, Dorvault N, Commo F, Adam J, Vanhecke E, Saulnier P, Thomale J, Le Che. ERCC1 isoform expression and DNA repair in non-small-cell lung cancer. N Engl J Med. 2013;368:1101–1110. doi: 10.1056/NEJMoa1214271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irigoyen M, Pajares MJ, Agorreta J, Ponz-Sarvise M, Salvo E, Lozano MD, Pio R, Gil-Bazo I, Rouzaut A. TGFBI expression is associated with a better response to chemotherapy in NSCLC. Mol Cancer. 2010;9:130. doi: 10.1186/1476-4598-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S, Ivanova A, Salnikow K, Timofeeva O, Subramaniam M, Lerman M. Two novel VHL targets, TGFBI (BIGH3) and its transactivator KLF10, are up-regulated in renal clear cell carcinoma and other tumors. Biochem Biophys Res Commun. 2008;370:536–540. doi: 10.1016/j.bbrc.2008.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Kim SJ, Lee BH, Park RW, Kim KS, Kim IS. Identification of motifs for cell adhesion within the repeated domains of transforming growth factor-beta-induced gene, betaig-h3. J Biol Chem. 2000;275:30907–30915. doi: 10.1074/jbc.M002752200. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim E, Liu D, Lee JJ, Solis L, Behrens C, Lippman S, Hong W, Wistuba I, Lee H. Prognostic impact of insulin receptor expression on survival of patients with nonsmall cell lung cancer. Cancer. 2012;118:2454–2465. doi: 10.1002/cncr.26492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim S, Jeong H, Lee B, Choi J, Park R, Park J, Kim I. RGD peptides released from beta ig-h3, a TGF-beta-induced cell-adhesive molecule, mediate apoptosis. Oncogene. 2003;22:2045–2053. doi: 10.1038/sj.onc.1206269. [DOI] [PubMed] [Google Scholar]
- Kitahara O, Furukawa Y, Tanaka T, Kihara C, Ono K, Yanagawa R, Nita ME, Takagi T, Nakamura Y, Tsunoda T. Alterations of gene expression during colorectal carcinogenesis revealed by cDNA microarrays after laser-capture microdissection of tumor tissues and normal epithelia. Cancer Res. 2001;61:3544–3549. [PubMed] [Google Scholar]
- Langer C, Besse B, Gualberto A, Brambilla E, Soria J. The evolving role of histology in the management of advanced non-small-cell lung cancer. J Clin Oncol. 2010;28:5311–5320. doi: 10.1200/JCO.2010.28.8126. [DOI] [PubMed] [Google Scholar]
- Maus MKH, Mack P, Astrow S, Stephens C, Zeger G, Grimminger P, Hsiang J, Huang E, Li T, Lara P, Danenberg K, Gandara D. Histology-related associations of ERCC1, RRM1, and TS biomarkers in patients with non-small-cell lung cancer: implications for therapy. J Thorac Oncol. 2013;8:582–586. doi: 10.1097/JTO.0b013e318287c3c5. [DOI] [PubMed] [Google Scholar]
- Morand S, Buchillier V, Maurer F, Bonny C, Arsenijevic Y, Munier F, Schorderet D. Induction of apoptosis in human corneal and HeLa cells by mutated BIGH3. Invest Ophthalmol Vis Sci. 2003;44:2973–2979. doi: 10.1167/iovs.02-0661. [DOI] [PubMed] [Google Scholar]
- Mountain CF. Revisions in the International System for Staging Lung Cancer. Chest. 1997;111:1710–1717. doi: 10.1378/chest.111.6.1710. [DOI] [PubMed] [Google Scholar]
- Okamoto T, Maruyama R, Suemitsu R, Aoki Y, Wataya H, Kojo M, Ichinose Y. Prognostic value of the histological subtype in completely resected non-small cell lung cancer. Interact Cardiovasc Thorac Surg. 2006;5:362–366. doi: 10.1510/icvts.2005.125989. [DOI] [PubMed] [Google Scholar]
- Olaussen K, Dunant A, Fouret P, Brambilla E, Andr F, Haddad V, Taranchon E, Filipits M, Pirker R, Popper H, Stahel R, Sabatier L, Pignon J, Tursz T, Le Chevalier T, Soria J. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- Pajares M, Agorreta J, Larrayoz M, Vesin A, Ezponda T, Zudaire I, Torre W, Lozano M, Brambilla E, Brambilla C, Wistuba I, Behrens C, Timsit J, Pio R, Field J, Montuenga L. Expression of tumor-derived vascular endothelial growth factor and its receptors is associated with outcome in early squamous cell carcinoma of the lung. J Clin Oncol. 2012;30:1129–1136. doi: 10.1200/JCO.2011.37.4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson L, Ruckdeschel J, Wagner H, Stevens C. Treatment of non-small cell lung cancer-stage IIIA: ACCP evidence-based clinical practice guidelines (2nd edition) Chest. 2007;132:243S–265S. doi: 10.1378/chest.07-1379. [DOI] [PubMed] [Google Scholar]
- Rosell R, Danenberg K, Alberola V, Bepler G, Sanchez J, Camps C, Provencio M, Isla D, Taron M, Diz P, Artal A. Ribonucleotide reductase messenger RNA expression and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res. 2004;10:1318–1325. doi: 10.1158/1078-0432.ccr-03-0156. [DOI] [PubMed] [Google Scholar]
- Schneider D, Kleeff J, Berberat P, Zhu Z, Korc M, Friess H, Bchler M. Induction and expression of betaig-h3 in pancreatic cancer cells. Biochim Biophys Acta. 2002;1588:1–6. doi: 10.1016/s0925-4439(02)00052-2. [DOI] [PubMed] [Google Scholar]
- Scott W, Howington J, Feigenberg S, Movsas B, Pisters K. Treatment of non-small cell lung cancer stage I and stage II: ACCP evidence-based clinical practice guidelines (2nd edition) Chest. 2007;132:234S–242S. doi: 10.1378/chest.07-1378. [DOI] [PubMed] [Google Scholar]
- Shah J, Shao G, Hei T, Zhao Y. Methylation screening of the TGFBI promoter in human lung and prostate cancer by methylation-specific PCR. BMC Cancer. 2008;8:284–284. doi: 10.1186/1471-2407-8-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao G, Berenguer J, Borczuk A, Powell C, Hei T, Zhao Y. Epigenetic inactivation of Betaig-h3 gene in human cancer cells. Cancer Res. 2006;66:4566–4573. doi: 10.1158/0008-5472.CAN-05-2130. [DOI] [PubMed] [Google Scholar]
- Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- Thapa N, Lee BH, Kim IS. TGFBIp/betaig-h3 protein: a versatile matrix molecule induced by TGF-beta. Int J Biochem Cell Biol. 2007;39:2183–2194. doi: 10.1016/j.biocel.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Travis WD, Brambilla E, Müller-Hermelink H, Harris C. Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. IARC Press: Lyon; 2004. [Google Scholar]
- Wang T, Nelson R, Bogardus A, Grannis F. Five-year lung cancer survival: which advanced stage nonsmall cell lung cancer patients attain long-term survival. Cancer. 2010;116:1518–1525. doi: 10.1002/cncr.24871. [DOI] [PubMed] [Google Scholar]
- Ween MP, Lokman NA, Hoffmann P, Rodgers RJ, Ricciardelli C, Oehler MK. Transforming growth factor-beta-induced protein secreted by peritoneal cells increases the metastatic potential of ovarian cancer cells. Int J Cancer. 2011;128:1570–1584. doi: 10.1002/ijc.25494. [DOI] [PubMed] [Google Scholar]
- Wen G, Partridge M, Li B, Hong M, Liao W, Cheng S, Zhao Y, Calaf G, Liu T, Zhou J, Zhang Z, Hei T. TGFBI expression reduces in vitro and in vivo metastatic potential of lung and breast tumor cells. Cancer Lett. 2011;308:23–32. doi: 10.1016/j.canlet.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win T, Sharples L, Groves A, Ritchie A, Wells F, Laroche C. Predicting survival in potentially curable lung cancer patients. Lung. 2008;186:97–102. doi: 10.1007/s00408-007-9067-1. [DOI] [PubMed] [Google Scholar]
- Zhao Y, El Gabry M, Hei T. Loss of Betaig-h3 protein is frequent in primary lung carcinoma and related to tumorigenic phenotype in lung cancer cells. Mol Carcinog. 2006;45:84–92. doi: 10.1002/mc.20167. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Piao C, Hei T. Downregulation of Betaig-h3 gene is causally linked to tumorigenic phenotype in asbestos treated immortalized human bronchial epithelial cells. Oncogene. 2002;21:7471–7477. doi: 10.1038/sj.onc.1205891. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Shao G, Piao C, Berenguer J, Hei T. Down-regulation of Betaig-h3 gene is involved in the tumorigenesis in human bronchial epithelial cells induced by heavy-ion radiation. Radiat Res. 2004;162:655–659. doi: 10.1667/rr3270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.