Abstract
Background:
To elucidate clinicopathological characteristics of non-small-cell lung carcinoma (NSCLC) cases carrying RET rearrangements causing oncogenic fusions to identify responders to therapy with RET tyrosine kinase inhibitors.
Methods:
We investigated 1874 patients with carcinomas, including 1620 adenocarcinomas (ADCs), 203 squamous cell carcinomas (SCCs), 8 large cell carcinomas, and 43 sarcomatoid carcinomas (SACs). Fluorescence in situ hybridisation (FISH) and/or reverse transcription–PCR (RT–PCR) were performed to detect RET gene rearrangement.
Results:
In all, 22 cases (1.2%) showed RET rearrangements; all cases were of ADC histology. Of the 22 patients, 19 possessed KIF5B–RET fusion genes, whereas 3 possessed CCDC6–RET fusion genes. The RET-rearranged tumours were significantly more common in younger patients (P=0.038) and tended to occur in patients with no history of smoking (P=0.051). In addition, RET rearrangements were not associated with gender, occupational history (particularly radioactive exposure), tumour size, lymph node status, tumour stage, or patient survival. The predominant growth pattern in RET-rearranged ADCs was lepidic in 6 cases, papillary in 9 cases, acinar in 2 cases, micropapillary in 1 case, and solid in 4 cases. Cells with cytoplasmic mucin production were at least focally present in 12 of the 22 (54.5%) RET-rearranged ADC cases. Among the 21 analysed RET-rearranged tumours, RET immunopositivity was observed in 15 cases (71.4%), and was significantly associated with RET rearrangement (P<0.001).
Conclusions:
The RET rearrangements were observed in 1.2% of NSCLCs. All cases of RET rearrangement were ADCs. The RET rearrangements were more likely to be observed in younger patients. Although cytoplasmic mucin production was at least focally present in 54.5% of RET-rearranged ADCs, specific histological features were not detected.
Keywords: RET gene rearrangement, lung carcinoma, adenocarcinoma, fluorescence in situ hybridisation, immunohistochemistry
After the discovery of crucial ‘driver' oncogenic mutations in the epidermal growth factor receptor (EGFR) gene, the EGFR tyrosine kinase inhibitor (TKI) was found to improve survival in non-small-cell lung carcinoma (NSCLC) patients possessing an EGFR mutation (Lynch et al, 2004; Paez et al, 2004; Pao et al, 2004). A genomic alteration involving the transforming fusion gene joining the echinoderm microtubule-associated protein-like 4 gene (EML4) and the anaplastic lymphoma kinase gene (ALK) was identified in 3–13% of patients with NSCLC (Soda et al, 2007; Koivunen et al, 2008; Yoshida et al, 2011). A dramatic response has been observed in patients with ALK rearrangements under treatment with an ALK TKI crizotinib (PF-02341066) during a recent clinical trial (Kwak et al, 2010).
The rearranged during transfection (RET) proto-oncogene encodes a receptor tyrosine kinase for members of the glial cell line-derived neurotrophic factor family of extracellular signalling molecules (Knowles et al, 2006). This proto-oncogene is involved in the growth and differentiation of neural crest-derived tissues (Pachnis et al, 1993). Chromosomal rearrangements that generate a fusion gene consisting of the juxtaposition of the C-terminal region of the RET protein with the N-terminal portion of another protein can also lead to constitutive activation of the RET kinase. The RET gene rearrangements, as represented by papillary thyroid carcinoma (PTC), were most often observed as coiled-coil domain containing 6 (CCDC6)–RET (PTC1) (Grieco et al, 1990) and nuclear receptor coactivator 4 (NCOA4)–RET (PTC3) fusion genes (Santoro et al, 1994).
Several investigators, including the authors of the present study, have simultaneously reported on a novel fusion gene comprising parts of the kinesin family member 5B gene (KIF5B) and the RET gene in lung carcinoma (Ju et al, 2012; Kohno et al, 2012; Lipson et al, 2012; Takeuchi et al, 2012). Subsequently, other fusion partners of the RET genes CCDC6, NCOA4, and tripartite motif-containing 33 (TRIM33) were identified in NSCLCs (Wang et al, 2012; Drilon et al, 2013). These fusion transcripts were detected in 0.6–10% of pulmonary adenocarcinomas (ADCs).
Notably, NSCLC cases that are positive for RET fusions have shown responses against existing RET TKIs, including cabozantinib and vandetanib (Drilon et al, 2013; Gautschi et al, 2013). Therefore, it is important to understand the clinicopathological characteristics of patients with RET fusion-positive NSCLCs for improved selection of patients who are likely to benefit from anti-RET therapy. In this study, we analysed RET fusions by fluorescence in situ hybridisation (FISH) combined with reverse transcription–PCR (RT–PCR) and RNA sequencing data from a large cohort (n=1874) and investigated distinct clinicopathological characteristics of RET fusion-positive cases.
Materials and methods
Case selection
The institutional review board of our hospital approved the study (2010-0077). The specimens used in this study were isolated from 1927 patients who underwent lung resection for ADC, squamous cell carcinoma (SCC), large cell carcinoma (LCC), or sarcomatoid carcinoma (SAC) at the National Cancer Center Hospital (Tokyo, Japan). We collected each patient's age, gender, smoking history, outcome, maximum tumour size (in cm), and pathologic stage (in p-stage). Staging was based on the tumour–necrosis–metastasis (TNM) classification (7th edition; Goldstraw, 2009). Among patients with RET rearrangements, we recorded the patients' occupational histories with particular reference to radioactive exposure.
Histological analysis
Histological diagnoses were based on the most recent World Health Organisation classification (Travis et al, 2004). Among all ADC cases, the predominant histological patterns were classified based on the International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society (IASLC/ATS/ERS) (Travis et al, 2011). In addition, in order to determine whether RET-rearranged lung cancer has any of the specific histological features described in prior reports (Wang et al, 2012), we performed a detailed histological analysis as follows: (1) the prevalent cytological feature (i.e., Clara/type II pneumocyte, columnar cell, or polygonal cell); (2) the presence of intracytoplasmic mucin production and mucinous cribriform pattern; and (3) the presence of intranuclear inclusion among 22 cases of RET-rearranged lung cancer.
Immunohistochemistry
For RET immunohistochemical staining, heat-induced epitope retrieval with Target Retrieval Solution (Dako Corporation, Carpinteria, CA, USA) was performed. The slides were subsequently incubated with primary antibodies against RET (EPR2871; 1 : 250 dilution; Epitomics, Burlingame, CA, USA). Immunoreactions were detected using EnVision-FLEX and LINKER (Dako). Immunopositive cases were defined as those showing cytoplasmic and/or membranous staining in ⩾10% of cells. We then divided the immunopositive cases into membrane staining-only and cytoplasmic-staining cases (irrespective of any membranous staining).
For the RET-rearranged NSCLCs, we performed immunohistochemical analysis to exclude metastatic thyroid carcinoma (see Supplementary Table 1).
FISH analysis for RET rearrangements
First, we performed using a dual-colour break-apart probe for the RET gene (Supplementary Table 2; Chromosome Science Labo, Inc., Sapporo, Japan). Among RET gene break-apart probe-positive cases, we next performed using break-apart probes for both KIF5B and CCDC6.
A total of 50 non-overlapping tumour cells with hybridisation signals examined for each case were captured using the Metafer Slide Scanning Platform (MetaSystems, Altlussheim, Germany). The signal in each cell was categorised into one of the following seven patterns: (1) fused 3′/5′ only; (2) fused 3′/5′ and both isolated 3′ and 5′ (split); (3) both isolated 3′ and 5′ (split) only; (4) fused 3′/5′ and isolated 5′ (5) fused 3′/5′ and isolated 3′ (6) isolated 5′ only; and (7) isolated 3′ only. A split signal was defined by 5′ and 3′ probes observed at a distance of greater than one-fold the signal size. A FISH-positive case was defined as ⩾20% of tumour cells having any split signals or any isolated 3′ (red) signals. The threshold for the RET gene was determined in 27 cases, yielding both FISH and previously reported RNA sequence data (Kohno et al, 2012) (Supplementary Figure 1).
RT–PCR analysis
Total RNA (500 ng) was reverse-transcribed onto cDNA using Superscript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA). Complementary DNA (corresponding to 10 ng total RNA) was subjected to multiplex PCR amplification using KAPA Taq DNA Polymerase (KAPA Biosystems, Woburn, MA, USA) and four primers (detection set in Supplementary Table 3). This PCR enabled the detection of all KIF5B/CCDC6-RET fusion variants identified to date. The reactions were conducted in a thermal cycler under the following conditions: 40 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 2 min, with a final extension cycle for 10 min at 72 °C. The housekeeping gene encoding glyceraldehyde-3-phosphate dehydrogenase was amplified to estimate the efficiency of cDNA synthesis. The PCR products were subjected to agarose gel electrophoresis. When visible bands were detected, the cDNA samples were further subjected to validation PCR (validation set in Supplementary Table 3). When visible bands were detected, the PCR products were subjected to Sanger sequencing in both directions by using the BigDye Terminator kit (Invitrogen) and an ABI 3130xl DNA Sequencer (Applied Biosystems, Foster City, CA, USA). The PCR primers used in the present study are shown in Supplementary Table 3.
We defined the cases with RET rearrangement that were RT–PCR-positive or RET break-apart probe-positive as well as cases that were KIF5B break-apart probe-positive or CCDC6 break-apart probe-positive in the absence of RT–PCR data.
Analysis of EGFR mutational status and ALK rearrangement
We detected two common EGFR mutations (deletions in exon 19 (DEL) and a point mutation at codon 858 in exon 21 (L858R)) by using high-resolution melting analysis (Fukui et al, 2008). The ALK rearrangement was analysed by immunohistochemistry, RT–PCR, and/or FISH assay (Yoshida et al, 2011).
Statistical analysis
Statistical analysis was performed using SPSS Statistics 21 software (IBM Corporation, Somers, NY, USA). Student's t-test was used to analyse continuous variables and χ2 tests were used to analyse categorical variables. Overall survival (OS) curves were calculated using the Kaplan–Meier method. Curves were compared using the log-rank test. Univariate survival analysis was performed using a log-rank test. Statistical significance was set at P<0.05.
Results
Clinicopathological background
Among the 1927 cases investigated, 53 cases were excluded because they lacked RT–PCR data and the FISH analysis failed. Therefore, the final cohort included 1874 cases: 1620 ADCs (56 in situ, 41 microinvasive, 366 lepidic-predominant, 179 acinar-predominant, 577 papillary-predominant, 101 micropapillary-predominant, 236 solid-predominant, and 64 invasive mucinous), 203 SCCs, 8 LCCs, and 43 SACs. Among the 1620 ADC cases, 830 cases consisted of consecutively resected cases from 1998 to 2002 (Supplementary Figure 2).
Background clinicopathological data are displayed in Table 1. Because lymph node status was recorded in 1860 cases (99.3%), pathological staging was performed in 1860 cases. The mean follow-up time for all 1874 patients was 62.3 months (range, 0.1–163 months), with 1292 patients still alive at follow-up.
Table 1. Clinicopathologic factors of RET-rearranged lung carcinomas.
|
Total |
RET
rearrangement |
|||
|---|---|---|---|---|
| 1874 | Negative (%) | Positive (%) | P-value | |
|
Age (year) | ||||
| Median | 63.1 | 63.2 | 57.5 | 0.038 |
| Range |
23–89 |
23–89 |
28–78 |
|
|
Gender | ||||
| Female | 809 | 798 (43.1) | 11 (50) | 0.524 |
| Male |
1065 |
1054 (56.9) |
11 (50) |
|
|
Smoking | ||||
| Never | 867 | 852 (46.1) | 15 (68.2) | 0.051 |
| Former/current |
1007 |
1000 (53.9) |
7 (31.8) |
|
|
Tumour size (cm) | ||||
| Median | 3.0 | 3.0 | 2.8 | 0.598 |
| Range |
0.4–17.5 |
0.4–17.5 |
1.4–8.0 |
|
|
N status | ||||
| Negative | 1377 | 1362 (74.1) | 15 (68.2) | 0.624 |
| Positive |
483 |
475 (25.9) |
7 (31.8) |
|
|
p-Stage | ||||
| I+II | 1496 | 1480 (80.5) | 16 (72.7) | 0.189 |
| III+IV |
364 |
358 (19.5) |
6 (27.3) |
|
|
Histology | ||||
| ADC | 1620 | 1598 (86.3) | 22 | 0.322 |
| SQC | 203 | 203 (11.0) | 0 | |
| LCC | 8 | 8 (0.4) | 0 | |
| SAC |
43 |
43 (2.3) |
0 |
|
|
RET immunostaining | ||||
| Negative | 1527 (86.1) | 7 (33.3) | <0.001 | |
| Positive | 247 (13.9) | 14 (66.7) | ||
Abbreviations: ADC=adenocarcinoma; LCC=large cell carcinoma; RET=rearranged during transfection; SAC=sarcomatoid carcinoma; SQC = squamous cell carcinoma.
RET FISH and RT–PCR
Among the 1874 cases investigated, 1823 cases yielded break-apart FISH data, 477 cases yielded RT–PCR data, and 456 cases yielded both FISH and RT–PCR data. Fifty (2.7%) cases were RET break-apart probe positive cases (Figure 1). The RT–PCR analysis of 29 of the 50 FISH-positive cases for which RNAs were available verified that 14 (44.8%) cases possessed the KIF5B–RET fusion and 2 possessed the CCDC6–RET fusion. The most prevalent variant of KIF5B–RET was variant K15;R12 (10/14; 71.4%), whereas the other variants were observed in 1 case each (7.1% each; Supplementary Table 4). On the other hand, RT–PCR results were negative for all 406 FISH-negative cases. Based on these RT–PCR data, the split signal sensitivity and specificity was 100% and 44.8%, respectively. The average FISH split signal for RT–PCR-positive and -negative cases was 40.9% (range, 22–72) and 7.4% (range, 0–40), respectively (P<0.001). Among the 50 RET break-apart probe-positive cases, 13 out of the 40 analysed cases were confirmed to be KIF5B break-apart probe-positive cases and 3 out of the 46 analysed cases were confirmed to be CCDC6 break-apart probe-positive cases. In conjunction with RT–PCR data, 19 cases with KIF5B–RET rearrangement and 3 cases with CCDC6–RET rearrangement were detected (Supplementary Table 4).
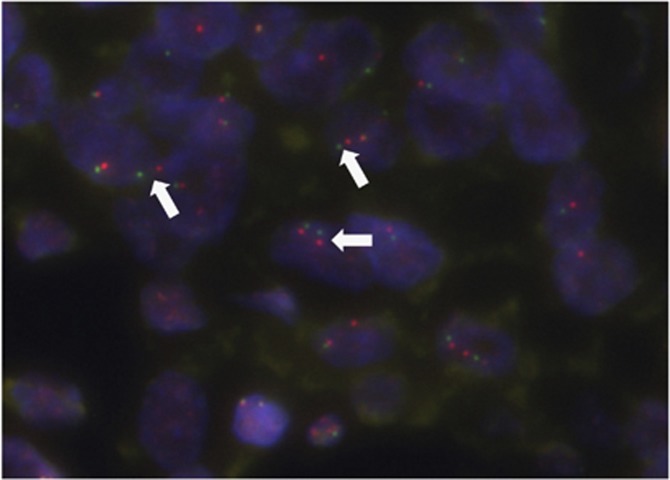
Figure 1.
Representative image of fluorescence in situ hybridisation using a break-apart probe for RET-rearranged carcinoma. White arrows indicate split red–green signals. A full colour version of this figure is available at the British Journal of Cancer journal online.
Clinical characteristics of patients with RET-rearranged NSCLCs
Based on the aforementioned FISH and RT–PCR data, 22 of the 1874 cases (1.2%) were considered to be RET rearrangements. The clinical characteristics of patients with RET-rearranged NSCLCs are displayed in Table 1 and Supplementary Table 4. Among 22 cases with RET rearrangements, 6 cases were reported previously (case nos. 2, 4, 7, 12, 14, and 15) (Kohno et al, 2012). All RET-rearranged cases were ADCs. When analysing consecutively resected ADC cases alone, RET rearrangements were observed in 7 of 830 ADC cases (0.8%). Of the 22 RET-rearranged cases, 19 (86%) possessed KIF5B–RET and 3 (14%) had CCDC6–RET fusions.
The RET-rearranged tumours were significantly more common in younger patients (P=0.026) and tended to occur in patients with no history of smoking (P=0.051). The RET rearrangements were not associated with gender, smoking status, tumour size, tumour stage, or lymph node status. Clinical records revealed that there were no patient histories of occupational exposure to radioactivity (Supplementary Table 5).
Among the 1874 cases examined, EGFR mutation was observed in 663 of the 1585 analysed cases (42.7%), and ALK rearrangement was observed in 55 of the 1860 analysed cases (3.0% Table 1). All cases were detected exclusively with additional driver genetic changes (i.e., RET, EGFR, and ALK).
Histological and immunohistochemical characteristics of patients with RET-rearranged NSCLCs
Histological findings are summarised in Table 2. The predominant growth pattern was lepidic in 6 cases, papillary in 9 cases, acinar in 2 cases, micropapillary in 1 case, and solid in 4 cases (Figures 2A and B). Focal lepidic, papillary, acinar, micropapillary, and solid patterns were observed in 16, 20, 16, 10, and 7 cases, respectively.
Table 2. Pathological, cytological, and immunohistological features for RET-rearranged cases.
| Histologic pattern | Cytological features | Immunohistochemical results | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
No |
Fusion partner |
Predominant |
LEP |
PAP |
ACI |
MPC |
SOL |
Cell type |
Mucin production |
Nuclear inclusion |
SRC |
M-Crib |
RET |
Staining pattern |
TTF-1 |
Napsin A |
PAX8 |
Thyroglobulin |
| 1 |
KIF5B |
MPC |
2 |
3 |
1 |
4 |
0 |
Type II |
+ |
− |
− |
− |
Pos |
C |
Pos |
Neg |
Neg |
Neg |
|
2 |
KIF5B |
LEP |
8 |
2 |
0 |
0 |
0 |
Type II |
− |
+ |
− |
− |
Pos |
C |
Pos |
Pos |
Neg |
Neg |
| 3 |
KIF5B |
ACI |
0 |
1 |
6 |
0 |
3 |
Polygonal |
+ |
− |
− |
− |
Pos |
C |
Pos |
Pos |
Neg |
Neg |
|
4 |
KIF5B |
LEP |
5 |
3 |
1 |
1 |
0 |
Type II |
− |
+ |
− |
− |
Pos |
C |
Pos |
Pos |
Neg |
Neg |
| 5 |
KIF5B |
PAP |
3 |
6 |
0 |
1 |
0 |
Type II |
− |
+ |
− |
− |
Pos |
C |
Pos |
Pos |
Neg |
Neg |
| 6 |
KIF5B |
PAP |
0 |
4 |
2 |
4 |
0 |
Columnar |
+ |
− |
− |
− |
Pos |
C |
Pos |
Pos |
Neg |
Neg |
|
7 |
KIF5B |
ACI |
3 |
2 |
5 |
0 |
0 |
Type II |
+ |
− |
+ |
− |
Pos |
C |
Pos |
Pos |
Neg |
Neg |
| 8 |
KIF5B |
PAP |
1 |
4 |
3 |
0 |
2 |
Columnar |
− |
− |
− |
− |
Neg |
− |
Pos |
Pos |
Neg |
Neg |
| 9 |
KIF5B |
PAP |
1 |
4 |
2 |
3 |
0 |
Columnar |
+ |
− |
+ |
+ |
Neg |
− |
Pos |
Pos |
Neg |
Neg |
| 10 |
KIF5B |
LEP |
8 |
2 |
0 |
0 |
0 |
Type II |
− |
+ |
− |
− |
Neg |
− |
Pos |
Pos |
Neg |
Neg |
| 11 |
KIF5B |
PAP |
2 |
6 |
1 |
1 |
0 |
Type II |
− |
+ |
− |
− |
Pos |
C |
Pos |
Pos |
Neg |
Neg |
|
12 |
KIF5B |
PAP |
3 |
6 |
0 |
0 |
0 |
Type II |
− |
+ |
− |
− |
Pos |
C |
Pos |
Pos |
Neg |
Neg |
| 13 |
KIF5B |
PAP |
4 |
5 |
0 |
0 |
0 |
Type II |
− |
− |
− |
− |
Neg |
− |
Pos |
Pos |
Neg |
Neg |
|
14 |
KIF5B |
LEP |
5 |
4 |
0 |
1 |
0 |
Type II |
+ |
+ |
− |
− |
Pos |
C |
Pos |
Pos |
Neg |
Neg |
|
15 |
KIF5B |
PAP |
0 |
4 |
3 |
3 |
0 |
Type II |
+ |
− |
+ |
+ |
#N/A |
#N/A |
#N/A |
#N/A |
#N/A |
#N/A |
| 16 |
KIF5B |
SOL |
0 |
2 |
3 |
0 |
5 |
Polygonal |
+ |
− |
− |
− |
Pos |
C |
Pos |
Pos |
Neg |
Neg |
| 17 |
KIF5B |
SOL |
1 |
2 |
1 |
1 |
5 |
Polygonal |
+ |
− |
− |
− |
Pos |
C |
Pos |
Pos |
Neg |
Neg |
| 18 |
KIF5B |
PAP |
2 |
3 |
2 |
2 |
1 |
Polygonal |
+ |
− |
+ |
− |
Pos |
C |
Pos |
Neg |
Neg |
Neg |
| 19 |
KIF5B |
LEP |
8 |
0 |
2 |
0 |
0 |
Type II |
− |
+ |
− |
− |
Neg |
− |
Pos |
Pos |
Neg |
Neg |
| 20 |
CCDC6 |
LEP |
5 |
4 |
1 |
0 |
0 |
Type II |
− |
+ |
− |
− |
Neg |
|
Pos |
Pos |
Neg |
Neg |
| 21 |
CCDC6 |
SOL |
0 |
0 |
1 |
0 |
9 |
Polygonal |
+ |
− |
+ |
− |
Pos |
C |
Pos |
Pos |
Neg |
Neg |
| 22 | CCDC6 | SOL | 0 | 1 | 4 | 0 | 5 | Columnar | + | − | + | + | Pos | C | Pos | Pos | Neg | Neg |
Abbreviations: ACI=acinar; C=cytoplasmic; CCDC6=coiled-coil domain containing 6; KIF5B=kinesin family member 5B; LEP=lepidic; M=membranous; M-Crib=mucinous cribriform, MPC=micropapillary; #N/A=not assessed; Neg=negative; PAP=papillary; PAX8=paired box gene 8; Pos=positive; RET=rearranged during transfection; SOL=solid; SRC=signet-ring cell; TTF-1=thyroid transcription factor-1.
Case numbers 2, 4, 7, 12, 14, and 15 were reported previously and have been highlighted in bold.
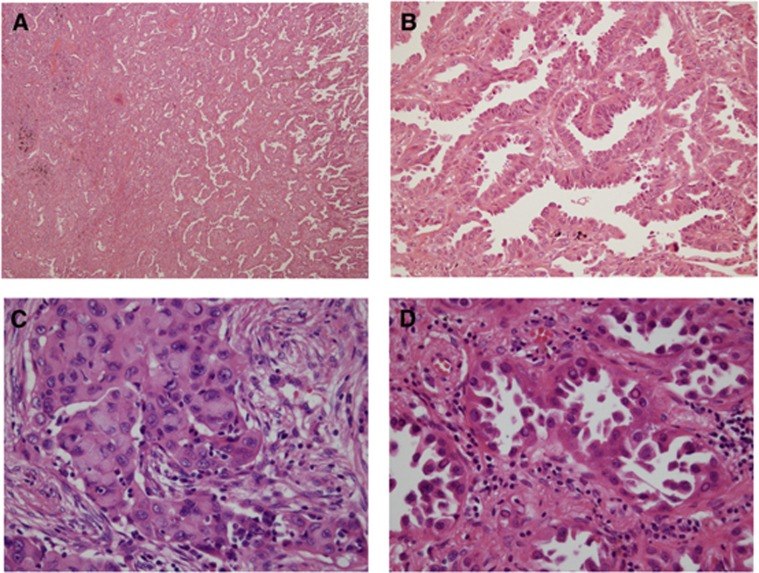
Figure 2.
Representative images of RET-rearranged adenocarcinoma of the lung. (A and B) Many RET-rearranged adenocarcinomas displayed a papillary growth pattern (A: low magnification, and B: high magnification). (C) Solid signet-ring cell pattern was observed in a minority of RET-rearranged adenocarcinoma (original magnification × 200). (D) Some tumour cells displayed homogeneously eosinophilic-to-pale inclusions in the nuclei (original magnification × 200).
The predominant cell type was Clara/type II in 13 cases, columnar in 4 cases, and polygonal in 5 cases. Cells with cytoplasmic mucin production were at least focally present in 12 of the 22 (54.5%) RET-rearranged ADC cases. The presence of signet-ring (Figure 2C) or mucinous cribriform patterns were only observed in 27.3% and 13.6% of cases, respectively. The nuclear inclusion was at least focally present in 9 of the 22 (40.9%) RET-rearranged ADC cases (Figure 2D).
RET protein expression by immunohistochemistry
The RET expression was observed in 261 of the 1795 (14.5%) NSCLCs evaluated (Figure 3). The RET-immunopositive cases were significantly associated with histological subtype (P<0.001); 255 of the 1543 ADCs (16.5%), 1 of the 201 SCCs (0.5%), none of the 8 LCCs, and 5 of the 43 SACs (11.6%). The RET-immunopositive tumours were more commonly associated with younger patients (P=0.006), patients with no history of smoking (P=0.007), lymph node metastasis (P<0.001), and higher pathological stages (P=0.001) compared with RET-immunonegative tumours.
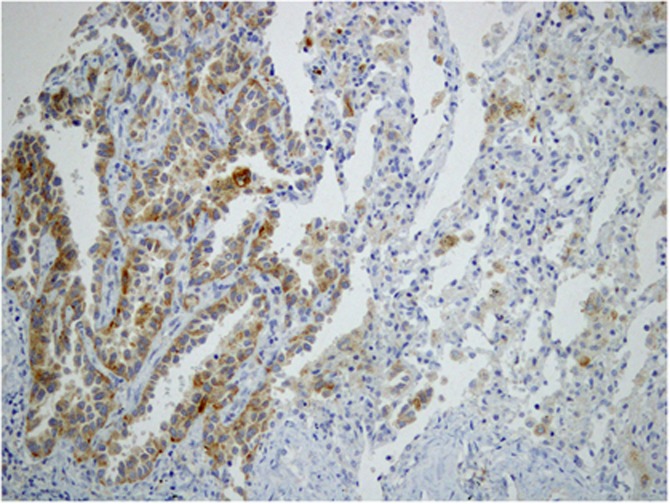
Figure 3.
Representative images of RET-immunostaining positivity in a RET-rearranged adenocarcinoma. Diffuse, fine granular cytoplasmic staining was observed in the adenocarcinoma component, as shown in the left part of the figure, whereas negative signals were observed in nontumourous areas, as shown in the right part of the figure (original magnification × 400).
Among RET-rearranged cases, RET immunopositivity was observed in 14 of the 21 cases (66.7%) analysed with immunohistochemistry. Although RET immunoreactivity was significantly associated with RET rearrangement (P<0.001), its test performance was poor with 66.7% sensitivity and 86.1% specificity (Table 1).
Upon classification of the staining pattern of 261 RET-immunopositive cases, we observed that 158 cases (60.5%) displayed cytoplasmic staining, irrespective of membrane staining, and 103 cases (39.5%) displayed only membrane staining. All RET-immunopositive cases with RET-rearrangement displayed cytoplasmic staining. Among the 151 cytoplasmic staining cases, the detection rate for RET-rearranged cases increased from 5.4 to 9.9%.
Survival analysis
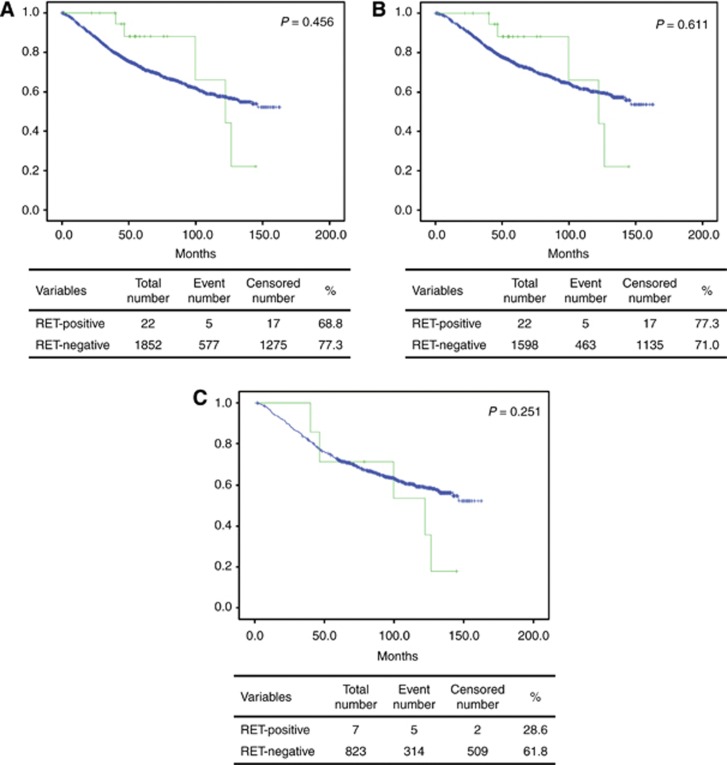
We investigated the existence of an association between patient OS and RET gene rearrangement. Follow-up data were available for all 1874 patients for a median of 62.3 months (range, 0.1−162.8 months). The RET gene rearrangement was not associated with OS in any of the cases analysed, among all cases (P=0.456; Figure 4A), among ADC-only cases, (P=0.611, Figure 4B), or among consecutively resected ADC cases (P=251, Figure 4C).
Figure 4.
Overall survival analysis in RET-rearranged lung carcinoma. (A) The overall survival curves for patients with RET-rearranged (green line) and RET-wild-type (blue line) non-small-cell lung carcinomas (P=0.9613). (B) The overall survival curves for patients with RET-rearranged (green line) and RET-wild-type (blue line) adenocarcinoma (P=0.9665). (C) The overall survival curves for patients with RET-rearranged (green line) and RET-wild-type (blue line) consecutively resected adenocarcinoma (P=0.547). A full colour version of this figure is available at the British Journal of Cancer journal online.
Discussion
We observed that ∼1.2% of NSCLC cases harboured RET rearrangements, all of which were ADCs, whereas neither SCCs, SACs, nor LCCs harboured this rearrangement. The prevalence of RET rearrangements (1.2%) in our cohort was in line with the range of results reported previously (0.6–10%) (Ju et al, 2012; Kohno et al, 2012; Lipson et al, 2012; Suehara et al, 2012; Takeuchi et al, 2012; Wang et al, 2012). The prevalence rate is considered to be affected by the specific case selection, such as that collected by known gene alterations in the negative cohort. In the subgroup of consecutively resected ADC cases, the frequency of RET rearrangements was only 0.9%.
Patients with RET rearrangements displayed nearly equivalent gender distributions, were relatively younger in age, and had no history of smoking compared with patients without RET rearrangements. The young age of onset and non-smoking history in RET-rearranged NSCLCs is reminiscent of the patient characteristics of ALK-rearranged NSCLCs (Shaw et al, 2009). However, other investigators have observed no statistical differences in age, gender, smoking history, or tumour stage between RET-rearranged ADC and wild-type RET ADCs (Wang et al, 2012). As is well known in PTC, RET gene fusions are associated with radiation exposure (Nikiforov and Nikiforova, 2011). However, no exposure to radioactivity was detected in either the current or previously reported RET-rearranged NSCLCs (Suehara et al, 2012).
Consistent with previous studies, RET rearrangements were more common in ADCs. Wang et al (2012) also reported RET rearrangements in two cases of adenosquamous carcinomas. There have been no reported RET rearrangements in SQC, LCC, or SAC tumours. Based on IASLC/ATS/ERS classification, we and other investigators have reported on the association between papillary growth pattern and RET rearrangement (Suehara et al, 2012; Yokota et al, 2012). Recently, Wang et al (2012) reported that a solid pattern was most prevalent in RET-rearranged ADCs, with signet-ring cells also frequently observed (36.4%). In this study, although cytoplasmic mucin was present, at least focally, in the majority (59%) of cases, signet-ring cells were observed in only 27% of cases. Of note, the mucinous cribriform pattern – another characteristic morphology associated with ALK- and ROS1-rearranged lung cancers (Yoshida et al, 2011, 2013) – was also infrequently observed (13.6%) in the present cohort.
Determining the gold standard for FISH specificity is challenging, because there may be an unknown fusion partner in FISH-positive cases that could be detectable with RT–PCR or FISH for fusion probes. Therefore, in order to yield a more precise cutoff value for break-apart FISH probes, we used previously reported RNA sequence data as the gold standard. In the present study, FISH analysis with a break-apart probe of the RET gene is highly sensitive (100% sensitivity), but unlikely to be sufficient to define RET-positive cases because of the potential for false positivity (58%) based on RT–PCR or break-apart FISH probes for known partners. Compared with the false-positive rate in our study, a slightly lower rate (41%) for RET break-apart FISH results has been reported with an alternate probe design (Takeuchi et al, 2012). The advantages of using a break-part FISH probe for gene translocation are that it can detect the translocations irrespective of fusion partners. Therefore, the current false-positive rate may be overestimated because it may include an unknown fusion partner for the RET gene. However, the possible effect of unknown fusion partners was not likely because the recently reported frequency of novel fusion partners of RET-rearranged lung carcinoma (i.e., NCOA4 and TRIM33) was extremely low (Wang et al, 2012; Drilon et al, 2013). Similar to EML4–ALK translocation, there exists intrachromosomal proximity (10.6 Mbp) of KIF5B and RET genes that complicates the generation of a proper break-apart probe that is easily resolvable by contemporary in situ technology.
Although RET immunoreactivity was significantly associated with RET rearrangement, its test performance was poor with only 66.7% sensitivity and 86.1% specificity. Other investigators have also reported a slightly lower positivity rate (54%) of RET antibody for RET-rearranged NSCLCs (Wang et al, 2012). Therefore, we conclude that RET immunohistochemistry possesses limited value in detecting RET-rearranged NSCLCs, unlike, for example, immunohistochemistry for human epidermal growth factor 2 status for breast carcinoma (Jacobs et al, 1999). Interestingly, cytoplasmic staining was more specific to gene rearrangement than membranous staining, likely because the KIF5B–RET chimeric proteins (except fusion partner of K24;R8) lack a transmembrane domain.
The present survival analysis indicated that RET rearrangement was not associated with OS. Even when the analysis was limited to ADCs or cases of consecutively resected ADC, no association was observed, consistent with previous reports (Wang et al, 2012; Yokota et al, 2012). However, the number of cases that have been investigated has been too small (spanning stages I–III) to draw any definitive conclusions regarding survival of RET-rearranged NSCLCs.
In summary, RET rearrangements were observed in 1.2% of NSCLC cases. All RET rearrangements in NSCLC were observed in ADCs. The RET rearrangements were observed in younger patients, patients with no smoking history, and papillary-predominant tumours. Although, cytoplasmic mucin production was at least focally present in 54.5% of RET-rearranged ADCs, distinct histological features were not detected. Furthermore, immunohistochemistry for RET protein has limited value to detect RET-rearranged NSCLC. Finally, these fusion genes did not coexist with EGFR and ALK alterations.
Acknowledgments
We thank Yuko Adegawa, Shouichi Harada, and Susumu Wakai for their skillful technical assistance. The National Cancer Center Biobank is supported by the National Cancer Center Development Fund, Japan. This work was supported in part by the National Cancer Center Research and Development Fund (23-A-2), (23-A-11), (23-A-35), and (24-A-1), and Grant-in-Aid for Scientific Research (C) Grant Number 25460446.
Author Contributions
Conception and design: Koji Tsuta and Takashi Kohno; financial support: Koji Tsuta and Takashi Kohno; administrative support: Koji Tsuta, Hisao Asamura, Takashi Kohno, and Ryoji Kushima; provision of study materials or patients: Koh Furuta and Hisao Asamura; data analysis and interpretation: Koji Tsuta, Yoko Shimada, Takashi Kohn, Koh Furuta, and Ryoji Kushima; manuscript writing: all authors; final approval of manuscript: all authors.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Drilon A, Wang L, Hasanovic A, Suehara Y, Lipson D, Stephens PJ, Ross J, Miller VA, Ginsberg MS, Zakowski MF, Kris MG, Ladanyi M, Rizvi NA. Response to cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov. 2013;3:630–635. doi: 10.1158/2159-8290.CD-13-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui T, Ohe Y, Tsuta K, Furuta K, Sakamoto H, Takano T, Nokihara H, Yamamoto N, Sekine I, Kunitoh H, Asamura H, Tsuchida T, Kaneko M, Kusumoto M, Yamamoto S, Yoshida T, Tamura T. Prospective study of the accuracy of EGFR mutational analysis by high-resolution melting analysis in small samples obtained from patients with non-small cell lung cancer. Clin Cancer Res. 2008;14 (15:4751–4757. doi: 10.1158/1078-0432.CCR-07-5207. [DOI] [PubMed] [Google Scholar]
- Gautschi O, Zander T, Keller FA, Strobel K, Hirschmann A, Aebi S, Diebold J. A patient with lung adenocarcinoma and RET fusion treated with vandetanib. J Thorac Oncol. 2013;8 (5:e43–e44. doi: 10.1097/JTO.0b013e31828a4d07. [DOI] [PubMed] [Google Scholar]
- Goldstraw P.2009International Association for the Study of Lung Cancer Staging Manual in Thoracic OncologyEditorial Rx Press: Florida.
- Grieco M, Santoro M, Berlingieri MT, Melillo RM, Donghi R, Bongarzone I, Pierotti MA, Della Porta G, Fusco A, Vecchio G. PTC is a novel rearranged form of the ret proto-oncogene and is frequently detected in vivo in human thyroid papillary carcinomas. Cell. 1990;60 (4:557–563. doi: 10.1016/0092-8674(90)90659-3. [DOI] [PubMed] [Google Scholar]
- Jacobs TW, Gown AM, Yaziji H, Barnes MJ, Schnitt SJ. Specificity of HercepTest in determining HER-2/neu status of breast cancers using the United States Food and Drug Administration-approved scoring system. J Clin Oncol. 1999;17 (7:1983–1987. doi: 10.1200/JCO.1999.17.7.1983. [DOI] [PubMed] [Google Scholar]
- Ju YS, Lee WC, Shin JY, Lee S, Bleazard T, Won JK, Kim YT, Kim JI, Kang JH, Seo JS. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res. 2012;22 (3:436–445. doi: 10.1101/gr.133645.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles PP, Murray-Rust J, Kjaer S, Scott RP, Hanrahan S, Santoro M, Ibanez CF, McDonald NQ. Structure and chemical inhibition of the RET tyrosine kinase domain. J Biol Chem. 2006;281 (44:33577–33587. doi: 10.1074/jbc.M605604200. [DOI] [PubMed] [Google Scholar]
- Kohno T, Ichikawa H, Totoki Y, Yasuda K, Hiramoto M, Nammo T, Sakamoto H, Tsuta K, Furuta K, Shimada Y, Iwakawa R, Ogiwara H, Oike T, Enari M, Schetter AJ, Okayama H, Haugen A, Skaug V, Chiku S, Yamanaka I, Arai Y, Watanabe S, Sekine I, Ogawa S, Harris CC, Tsuda H, Yoshida T, Yokota J, Shibata T. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18 (3:375–377. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen JP, Mermel C, Zejnullahu K, Murphy C, Lifshits E, Holmes AJ, Choi HG, Kim J, Chiang D, Thomas R, Lee J, Richards WG, Sugarbaker DJ, Ducko C, Lindeman N, Marcoux JP, Engelman JA, Gray NS, Lee C, Meyerson M, Janne PA. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14 (13:4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Janne PA, Costa DB, Varella-Garcia M, Kim WH, Lynch TJ, Fidias P, Stubbs H, Engelman JA, Sequist LV, Tan W, Gandhi L, Mino-Kenudson M, Wei GC, Shreeve SM, Ratain MJ, Settleman J, Christensen JG, Haber DA, Wilner K, Salgia R, Shapiro GI, Clark JW, Iafrate AJ. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363 (18:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson D, Capelletti M, Yelensky R, Otto G, Parker A, Jarosz M, Curran JA, Balasubramanian S, Bloom T, Brennan KW, Donahue A, Downing SR, Frampton GM, Garcia L, Juhn F, Mitchell KC, White E, White J, Zwirko Z, Peretz T, Nechushtan H, Soussan-Gutman L, Kim J, Sasaki H, Kim HR, Park SI, Ercan D, Sheehan CE, Ross JS, Cronin MT, Janne PA, Stephens PJ. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18 (3:382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350 (21:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Nikiforov YE, Nikiforova MN. Molecular genetics and diagnosis of thyroid cancer. Nat Rev Endocrinol. 2011;7 (10:569–580. doi: 10.1038/nrendo.2011.142. [DOI] [PubMed] [Google Scholar]
- Pachnis V, Mankoo B, Costantini F. Expression of the c-ret proto-oncogene during mouse embryogenesis. Development. 1993;119 (4:1005–1017. doi: 10.1242/dev.119.4.1005. [DOI] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304 (5676:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. EGF receptor gene mutations are common in lung cancers from ‘never smokers' and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101 (36:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro M, Dathan NA, Berlingieri MT, Bongarzone I, Paulin C, Grieco M, Pierotti MA, Vecchio G, Fusco A. Molecular characterization of RET/PTC3; a novel rearranged version of the RETproto-oncogene in a human thyroid papillary carcinoma. Oncogene. 1994;9 (2:509–516. [PubMed] [Google Scholar]
- Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, Heist RS, Solomon B, Stubbs H, Admane S, McDermott U, Settleman J, Kobayashi S, Mark EJ, Rodig SJ, Chirieac LR, Kwak EL, Lynch TJ, Iafrate AJ. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27 (26:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448 (7153:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- Suehara Y, Arcila M, Wang L, Hasanovic A, Ang D, Ito T, Kimura Y, Drilon A, Guha U, Rusch V, Kris MG, Zakowski MF, Rizvi N, Khanin R, Ladanyi M. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin Cancer Res. 2012;18:6599–6608. doi: 10.1158/1078-0432.CCR-12-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S, Asaka R, Hamanaka W, Ninomiya H, Uehara H, Lim Choi Y, Satoh Y, Okumura S, Nakagawa K, Mano H, Ishikawa Y. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18 (3:378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- Travis WD, Brambilla E, Mu¨ller-Hermelink HK, Harris CC.2004Tumors of the lungIn: Kleihues P, Sobin LH, (eds).WHO Classification of Tumors. Pathology and Genetics of Tumors of the Lung, Pleura, Thymus and Heart IARC Press: Lyon, France, pp9–124. [Google Scholar]
- Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ, Van Schil PE, Garg K, Austin JH, Asamura H, Rusch VW, Hirsch FR, Scagliotti G, Mitsudomi T, Huber RM, Ishikawa Y, Jett J, Sanchez-Cespedes M, Sculier JP, Takahashi T, Tsuboi M, Vansteenkiste J, Wistuba I, Yang PC, Aberle D, Brambilla C, Flieder D, Franklin W, Gazdar A, Gould M, Hasleton P, Henderson D, Johnson B, Johnson D, Kerr K, Kuriyama K, Lee JS, Miller VA, Petersen I, Roggli V, Rosell R, Saijo N, Thunnissen E, Tsao M, Yankelewitz D. International association for the study of lung cancer/American Thoracic Society/European Respiratory Society International multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6 (2:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Hu H, Pan Y, Li Y, Ye T, Li C, Luo X, Wang L, Li H, Zhang Y, Li F, Lu Y, Lu Q, Xu J, Garfield D, Shen L, Ji H, Pao W, Sun Y, Chen H. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol. 2012;30:4352–4359. doi: 10.1200/JCO.2012.44.1477. [DOI] [PubMed] [Google Scholar]
- Yokota K, Sasaki H, Okuda K, Shimizu S, Shitara M, Hikosaka Y, Moriyama S, Yano M, Fujii Y. KIF5B/RET fusion gene in surgically-treated adenocarcinoma of the lung. Oncol Rep. 2012;28 (4:1187–1192. doi: 10.3892/or.2012.1908. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Kohno T, Tsuta K, Wakai S, Arai Y, Shimada Y, Asamura H, Furuta K, Shibata T, Tsuda H. ROS1-rearranged lung cancer: a clinicopathologic and molecular study of 15 surgical cases. Am J Surg Pathol. 2013;37 (4:554–562. doi: 10.1097/PAS.0b013e3182758fe6. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Tsuta K, Nakamura H, Kohno T, Takahashi F, Asamura H, Sekine I, Fukayama M, Shibata T, Furuta K, Tsuda H. Comprehensive histologic analysis of ALK-rearranged lung carcinomas. Am J Surg Pathol. 2011;35 (8:1226–1234. doi: 10.1097/PAS.0b013e3182233e06. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.