Abstract
A sensitive new plate reader assay has been developed showing that adult mammalian blood serum contains circulating soluble sulfhydryl oxidase activity that can introduce disulfide bonds into reduced proteins with the reduction of oxygen to hydrogen peroxide. The activity was purified 5000-fold to > 90% homogeneity from bovine serum and found by mass spectrometry to be consistent with the short isoform of Quiescin-sulfhydryl oxidase 1 (QSOX1). This FAD-dependent enzyme is present at comparable activity levels in fetal and adult commercial bovine sera. Thus cell culture media that are routinely supplemented with either fetal or adult bovine sera will contain this facile catalyst of protein thiol oxidation. QSOX1 is present at approximately 25 nM in pooled normal adult human serum. Examination of the unusual kinetics of QSOX1 towards cysteine and glutathione at low micromolar concentrations suggests that circulating QSOX1 is unlikely to significantly contribute to the oxidation of these monothiols in plasma. However the ability of QSOX1 to rapidly oxidize conformationally mobile protein thiols suggests a possible contribution to the redox status of exofacial and soluble proteins in blood plasma. Recent proteomic studies showing that plasma QSOX1 can be utilized in the diagnosis of pancreatic cancer and acute decompensated heart failure, together with the overexpression of this secreted enzyme in a number of solid tumors, suggest that the robust QSOX assay developed here may be useful in the quantitation of enzyme levels in a wide range of biological fluids.
Keywords: Amplex UltraRed, Assay, Blood, Cysteine, Disulfide, Dithiothreitol, Glutathione, Horseradish peroxidase, Hydrogen peroxide, Plasma, QSOX1, Quiescin, Ribonuclease, Serum, Sulfhydryl oxidase, Thiol
Introduction
In 1999 Hoober et al. recognized the existence of a family of flavin-dependent sulfhydryl oxidases [1] whose first member was partially purified from skin more than 50 years ago [2]. These QSOX enzymes (named after a human growth factor, Quiescin Q6 [3]) can catalyze the rapid and direct generation of disulfides in a wide range of thiol-containing species – from small monothiols and peptides to large unfolded reduced proteins [4–6]:
QSOX enzymes are found in one or more isoforms in all higher eukaryotes for which genome sequences are available [2, 7–9]. The enzymological properties of QSOX have been studied extensively in our laboratory (reviewed in [2, 7–9]). Only conformationally flexible proteins are facile substrates of QSOX; cysteine residues within well-structure proteins are inefficiently oxidized by the enzyme [4, 5]. Reduced protein disulfide isomerase is not a substrate of QSOX, but these two enzymes can cooperate in the efficient oxidative folding of proteins with complex disulfide connectivity [10].
While the substrate specificity of QSOX in vitro has been extensively investigated, the likely contribution of this oxidase to disulfide bond formation in biological contexts has only begun to emerge. Early studies revealed that QSOX is associated with cells with a heavy secretory load [7, 9, 11], and QSOX has been found in the endoplasmic reticulum [9, 12, 13], Golgi [9, 12–15], and secretory granules [12, 15]. QSOX is also found at the cell surface and is secreted [3, 13, 16–22]. Bulleid and coworkers have suggested that QSOX1 may function late in the secretory pathway or following secretion from the cell [13, 14]. In the latter role, Fass and coworkers have recently shown that QSOX1 is involved in the integration of laminin into the extracellular matrix generated by human lung fibroblasts [23]. QSOX1 has been found to be highly up-regulated in cancers of prostate [24, 25], pancreas [26] and breast [27, 28]. Depressing levels of QSOX1 by RNAi, or by using an inhibitory antibody, leads to a marked decline in invasiveness in cell migration assays [23, 27, 29, 30].
Two recent studies have also revealed the potential for QSOX to be a diagnostic marker for diseases including pancreatic cancer [26, 29] and heart failure [31]. In 2008 Lake and colleagues first identified a QSOX1 peptide in plasma that was strongly correlated with pancreatic cancer patients but not to patients without this disease [26, 29]. More recently, Mebazaa et al. reported that mass spectroscopic analysis of plasma QSOX1 peptide levels, in combination with quantitation of B-type natriuretic peptide, significantly improved the accuracy of diagnosis for acute decompensated heart failure [31]. While these studies reveal the potential of plasma QSOX, or peptides there from, as a diagnostic marker, it is unknown whether circulating QSOX is catalytically active, and what role(s) circulating QSOX may play in these diseases.
Here, we describe a simple and sensitive microplate assay for QSOX activity which is suitable for testing small samples of serum, plasma, or other biological fluids. While evaluating the performance of this assay, we discovered that normal adult bovine serum contained relatively high levels of sulfhydryl oxidase activity. Zanata et al. had already found QSOX1 in fetal bovine serum; they reported that the oxidase activity and protein levels declined rapidly after birth to become undetectable in sera from 60-day old animals [19]. Our finding of sulfhydryl oxidase activity in adult bovine serum prompted us to reinvestigate the ability of bovine sera to oxidize protein thiols, and our assay facilitated the purification of sulfhydryl oxidase activity to essential homogeneity from adult bovine serum. Peptide digests confirmed that this disulfide-generating activity was indeed due to QSOX1. We further observed comparable enzyme activity levels in adult human serum and investigated QSOX reactivity with cysteine and glutathione at concentrations relevant to those that might be encountered by the circulating enzyme. The presence of a facile catalyst of the oxidation of peptide and protein thiols in mammalian blood provides an additional dimension to the study of thiol/disulfide transformations in blood.
Materials and Methods
Materials
Bovine serum albumin, cysteine, dithiothreitol, glutathione, homovanillic acid, horseradish peroxidase type II, hydrogen peroxide and ribonuclease A were from Sigma-Aldrich. Amplex UltraRed was from Life Technologies. Tween 80 (Surfact-Amps, low peroxide) was from Thermo Fisher Scientific. Commercial sera were as follows. Defined fetal bovine serum, newborn calf serum (less than 10 days old) and donor adult bovine serum were from Hyclone (Thermo Fisher). Normal human serum and normal adult mouse serum were from Atlanta Biologicals. Balb C, C57BL6, CD-1 and non-Swiss albino mouse sera were from Innovative Research.
General Methods
UV-VIS spectra were recorded in self-masking microcells using Agilent 8452A or 8453 instruments. Reduced RNase was prepared, and conveniently stored, as a lyophilized powder [5]. Solutions of thiols were prepared daily either from concentrated stocks maintained at −20 °C, or freshly from solid reagents. Quantitation of reagents, including protein and small molecular weight thiols and hydrogen peroxide, was as described previously [5]. Protein concentrations were determined using the Bradford assay. Unless otherwise stated the buffer used in this work was 50 mM potassium phosphate, pH 7.5, containing 1 mM EDTA.
Sulfhydryl oxidase assays
Fluorescence assays using HVA were conducted in a fluorescence microcell in an SLM Aminco Bowman Series 2 luminescence spectrometer using 50 μM DTT in 50 mM phosphate buffer, pH 7.5, containing 1 mM EDTA [32]. The increase in fluorescence emission was followed at 420 nm with excitation at 320 nm. The Amplex UltraRed assay used a BMG POLARstar OMEGA plate reader with 96-well black flat-bottomed polystyrene plates from Corning. The following reagents were used in a total volume of 150 μL in phosphate buffer: 10 μM AUR, 50 nM HRP and 0.5% v/v low-peroxide Tween 80 (included for serum samples; see Results). Typically, 125 μL of a cocktail formed by mixing AUR, HRP and Tween 80 in buffer was delivered to each well, followed by 5 μL of the sample. The reaction was started by the addition of 20 μL of 0.75 mM thiols (e.g. 0.375 mM DTT, 94 μM reduced RNase, or 0.75 mM GSH) in buffer to give a final concentration of 100 μM thiols in each well. Care was taken throughout this procedure to minimize exposure of the AUR reagent to light [33] by wrapping stock solutions and the cocktail mixture with aluminum foil, as well as shading the 96-well plate with foil where practical. Because this assay provides very sensitive detection of hydrogen peroxide, the non-enzymatic oxidation of thiols by traces of copper and iron [34] provide a detectable background signal that can be suppressed, but not eliminated, by the inclusion of 1 mM EDTA. This background can be further minimized by the preparation of fresh thiol stock reagents, and by mixing the reagents for the plate-reader assay immediately before measurement. Assays were conducted in fluorescence intensity mode (using excitation filter 544 and emission filter 590-10) with measurement every 0.5 min for 10 min. Rates were typically determined over the first 3 min of data acquisition. The instrument sensitivity was set following the addition of excess hydrogen peroxide to 10 μM AUR in 150 μL of assay solution. The assay was calibrated by adding increasing concentrations of H2O2 (0–1.6 μM) per well in the presence of the assay cocktail and thiol substrate; the linear fluorescence increase with added peroxide enabled conversion of relative fluorescence to μM hydrogen peroxide.
It is important to note that while the plate reader assay represents a convenient way to measure sulfhydryl oxidase activity levels in a range of biological fluids, it is unsuited to determine Km values for thiol substrates. This is because thiols depress fluorophore generation (Supplementary Figure S1), likely by intercepting the radical intermediates in the generation of the resorufin-like fluorophore [32]. The concentrations of thiols used in these assays (100 μM) provides for sufficient linearity of hydrogen peroxide production while avoiding excessive non-enzymatic metal-catalyzed thiol oxidation (see above).
Purification of QSOX1 from bovine sera
Adult non heat-treated bovine donor serum from Atlanta Biologicals (500 mL) was diluted with 2 L of distilled water at 4 °C and centrifuged for 8 min in a GS3 rotor at 13,700 RCF (Sorvall RC-5B) to remove a small amount of flocculent precipitate. Two 5 mL HiTrap SP HP cation exchange columns were connected in series and equilibrated at 1.5 mL/min at 4 °C with a 5-fold diluted solution of phosphate buffered saline solution (0.2X PBS) containing 1.62 mM Na2HPO4, 0.38 mM K2HPO4 and 29.8 mM NaCl. The diluted serum was then applied to the columns at 2 mL/min using a peristaltic pump at 4 °C. The combined columns were then connected to an ÄKTA FPLC operated at room temperature and developed at 1 mL/min using a 200 min linear gradient from 0.2X PBS to 100% of buffer B (50 mM potassium phosphate, pH 7.5, containing 500 mM NaCl and 1 mM EDTA) followed by a further 20 min of buffer B. Sulfhydryl oxidase assays (using 5 μL from each 5 mL fraction) were performed using the plate reader as above, except that at this stage of the work, Tween 80 was not included in the wells. The peak oxidase fractions were analyzed by UV-VIS spectroscopy and by SDS-PAGE. Three fractions containing 77 % of activity recovered from the column were pooled and brought to 40% saturation with ammonium sulfate at room temperature. The solution was clarified by centrifugation (2 min at 2700 RCF) and the supernatant applied at 0.3 mL/min to a 1 mL HiTrap Butyl HP hydrophobic interaction column equilibrated with 40% saturated ammonium sulfate in 50 mM phosphate buffer, pH 7.5, containing 1 mM EDTA. The column was developed with a decreasing linear gradient to phosphate buffer alone at 1 mL/min over a total volume of 20 mL. Fractions (of 0.5 mL) were analyzed for sulfhydryl oxidase activity as before. The three peak fractions were pooled and dialyzed at 4 °C against 50 mM phosphate buffer, pH 7.5, containing 1 mM EDTA. The dialyzed pool was diluted with an equal volume of water and then applied at 0.5 mL/min to a 1 mL HiTrap SP XL cation exchange column equilibrated with 25 mM phosphate buffer, pH 7.5, containing 1 mM EDTA. The column was then developed at 0.5 mL/min with a 40 min linear gradient starting with 100% of 25 mM Tris buffer, pH 7.4, containing 1 mM EDTA, and ending with the same buffer adjusted to pH 9.4. Sulfhydryl oxidase activity was then eluted with a 4 mL wash using the pH 9.4 buffer supplemented with 200 mM NaCl and active fractions were adjusted to pH 7.5 by the addition of 50 μL of 1 M Tris buffer pH 7.2. Peak fractions were pooled, concentrated and washed into 50 mM phosphate buffer, pH 8.0, containing 1 mM EDTA using a Microcon YM-30 Ultrafiltration device (Millipore). The purified sulfhydryl oxidase was stored at 4 °C prior to characterization.
Protein digestion and analysis
Samples containing sulfhydryl oxidase activity (~15 μg of protein in 0.1 mL of phosphate buffer) were incubated in 1.5 mL polypropylene centrifuge tubes for 10 min in a boiling water bath. The samples were cooled on ice, incubated with 0.75 μg of Promega Sequencing Grade modified trypsin for 2 h at 37 °C, and stored frozen. The peptide digest was separated using a Waters Acquity UPLC system equipped with a BEH C18 100 × 1 mm, 1.7 micron particle size column utilizing a linear gradient composed of 0.1% formic acid in water (solvent A) and 0.1% formic acid in methanol (solvent B). Following a 3 min isocratic hold at 5% solvent B, the column was developed with an increasing gradient to 65% solvent B over an additional 67 min. Peptides were detected using a Thermo Scientific Orbitrap Velos mass spectrometer. The data were acquired using the data-dependent analysis mode which allowed for the collection of both full scan and MS-MS data of the peptides. The MS-MS data were further analyzed using the SEQUEST peptide search algorithm of Thermo Scientific Proteome Discoverer (Version 1.3.0.339).
Results and Discussion
Fluorescence assay for sulfhydryl oxidase activity
While oxygen electrode assays remain a reliable staple for determination of sulfhydryl oxidase levels [35], they are of limited sensitivity and relatively large sample volume requirement. For this reason we earlier developed an assay in which the generation of hydrogen peroxide is coupled to the horseradish peroxidase-mediated oxidation of homovanillic acid (HVA) to yield a fluorescent product [32]. This was the assay employed by Zanata et al. in their analysis of sulfhydryl oxidase activity in bovine serum samples [19]. Figure 1A shows our application of this assay to commercial fetal, newborn and adult bovine sera.
Fig. 1.
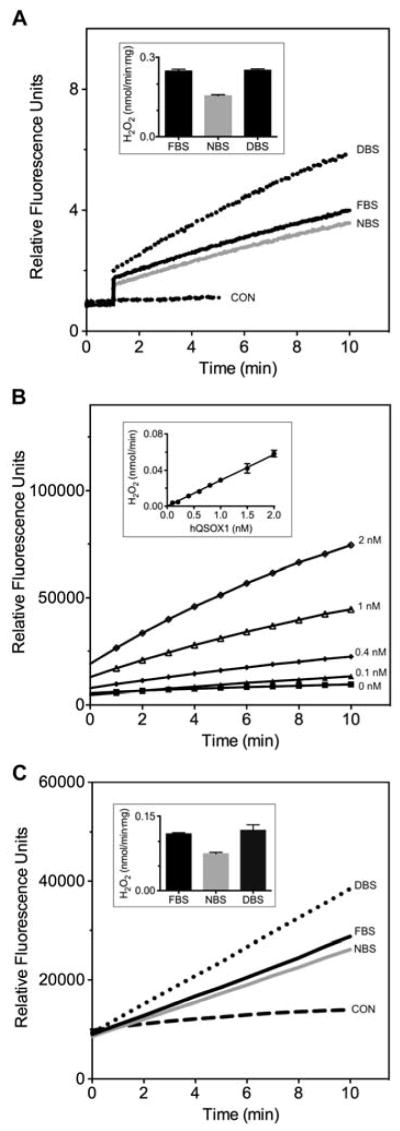
Sulfhydryl oxidase assays of bovine serum. Panel A shows the homovanillic acid (HVA) assay in phosphate buffer, pH 7.5, containing 1 mM EDTA using 5 μL of fetal (FBS), newborn (NBS) and adult bovine sera (DBS) in a total assay volume of 150 μL (see Methods). The inset provides rates corrected for the protein content of the samples (see Methods). Panel B represents the Amplex UltraRed assay with increasing concentrations of recombinant human QSOX1. The inset shows the linearity of initial rates, corrected for the non-enzymatic background oxidation of thiols, as a function enzyme concentration. Panel C shows the Amplex UltraRed assay using 5 μL of fetal, newborn and adult bovine sera in the presence of 0.5% Tween 80 (see Materials and Methods). Control assays without serum are represented by CON. Two additional controls, lacking either DTT, or both DTT and serum, showed no detectable increase in fluorescence over the measurement period (data not shown). The inset presents initial rates normalized to the protein concentration of the serum samples.
During these experiments, several limitations of our original HVA assay method became apparent. For example, because the HVA assay [32] generates a product that fluoresces in the blue region of the spectrum (excitation at 320 nm, emission at 420 nm), it is particularly prone to interference in media with high absorbance or scattering in the near UV. The HVA assay would also be problematic in high throughput screening applications because a significant fraction of compounds in many small-molecule libraries fluoresce in the blue region of the spectrum [36]. The new plate reader assay presented here couples hydrogen peroxide formation generated by sulfhydryl oxidases to the generation of the strong red fluorescence formed during HRP-catalyzed oxidation of Amplex UltraRed [37]. A demonstration of the readout from this assay with recombinant human QSOX1 is included in Figure 1B together with an indication of the linearity of the assay over the 0–2 nM range.
In preliminary work a variety of pH values were tested; the pH of 7.5 represents a suitable compromise between maintaining enzyme activity and minimizing the non-enzymatic oxidation of thiols by trace metals in the reagents (data not shown). At pH 7.5, the presence of the chelating agent EDTA proved critical in minimizing this background increase in fluorescence. With these as preliminaries, Figure 1C shows assays of bovine sera with the Amplex UltraRed method. Again, we found that adult serum has approximately the same level of activity as that found in fetal animals when the samples are normalized for protein content (inset Figure 1C).
When serum is analyzed the inclusion of Tween 80 (at 0.5% v/v; see Methods) largely prevented the suppression of the fluorescence signal in the presence of the high concentrations of bovine serum albumin (BSA) found in blood serum. Thus, Figure S2 shows that the detergent markedly improved the response of the plate reader assay with increasing volumes of sera. Figure S3 shows that the response of the AUR assay using recombinant human QSOX1 is markedly decreased by BSA at 2 mg/mL and that this apparent inhibition is largely reversed by the inclusion of Tween 80. Presumably this attenuation reflects the well known ability of albumin to bind a range of aromatic ligands [38] including, perhaps, the AUR substrate or its fluorescent product.
Purification of a sulfhydryl oxidase from bovine blood serum
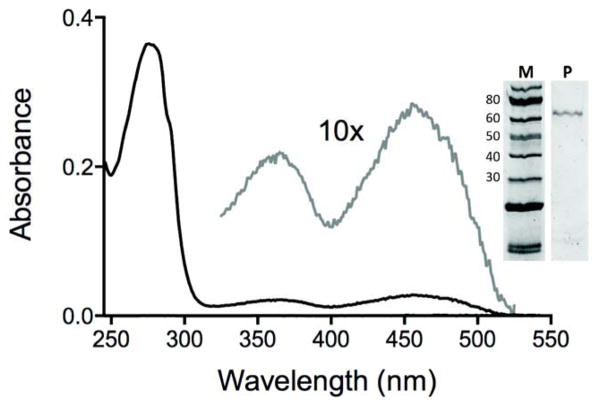
We then wished to purify the sulfhydryl oxidase activity from bovine serum to ascertain whether it reflected the presence of QSOX, or represented a hitherto unrecognized hydrogen peroxide generating catalyst. While the best characterized eukaryotic sulfhydryl oxidases are flavin-dependent (such as the Ero1, ERV/ALR and QSOX families of flavoproteins [2, 9, 16, 39–41]) a number of poorly-characterized metal-dependent sulfhydryl oxidases have been described in the earlier literature [2, 20, 39]. In developing a purification protocol from adult bovine sera, we needed an initial step that would conveniently separate the oxidase activity from the albumin and globulins that are present at an aggregate concentration of about 70 mg/mL in mammalian sera. The initial cation-exchange step at pH 7.5 (see Methods, supplementary Figure S4 and Table 1) resulted in a dramatic (~1200-fold) purification of activity leading to a single peak of enzyme activity as judged by the plate reader assay. The pooled pale yellow fractions were then applied to a hydrophobic interaction chromatography column with recovery of a single peak of enzyme activity. A second cation-exchange step, in which the protein was eluted from the column using Tris buffer/NaCl at pH 9.4 led to an overall 3-step purification of ~5300-fold (Table 1). The resulting preparation was ~ 90% pure protein and showed a visible spectrum with absorbance peaks at 365 nm and 456 nm consistent with that of mammalian flavin-linked QSOX enzymes (Figure 2; [20, 42]).
Table 1.
Purification of sulfhydryl oxidase from adult bovine serum
| Steps | Total protein (mg) | A280/A456 | Total activity (nmol H2O2/min) | Yield (%) | Specific activity (nmol/min·mg) | Fold purification |
|---|---|---|---|---|---|---|
| Serum (500 mL diluted 5-fold) | 31350 | 140 | 4148 | 100 | 0.132 | 1 |
| Cation exchange (salt gradient) | 10.3 | 31 | 1686 | 411 | 164 | 1240 |
| Hydrophobic interaction | 1.52 | 22 | 560 | 13.5 | 369 | 2793 |
| Cation exchange (pH gradient) | 0.57 | 12 | 402 | 10 | 706 | 5345 |
During loading the 2.5 L of diluted serum, the cation exchange column was intentionally overloaded as described in Methods. Without such overloading 82% of the sulfhydryl oxidase applied to the column could be recovered from the salt gradient in a single peak of enzyme activity.
Fig. 2.
UV-VIS spectrum of the sulfhydryl oxidase purified from adult bovine serum. The main panel shows the spectrum of the oxidase recorded in 50 mM phosphate buffer, pH 7.5, containing 1 mM EDTA. The dashed line highlights the flavin region of the spectrum. The inset shows an SDS-PAGE analysis of the purified protein.
The QSOX sulfhydryl oxidases are the only known enzymes that can directly oxidize unfolded reduced proteins with catalytic efficiencies similar to those observed with the small, highly reducing, model substrate DTT [2, 4, 18, 42]. While the assay data in Table 1 were collected using DTT, we also compared the rates of hydrogen peroxide production of the unfractionated serum enzyme with those from the purified enzyme using three substrates: DTT, reduced RNase, and GSH. These values, normalized to the activity with DTT, were 1.0: 0.68: 0.0012 in serum, and 1.0: 0.66: 0.0009 after purification, respectively, suggesting that the sulfhydryl oxidase activity in serum is substantially due to one QSOX-like enzyme. This was confirmed following trypsin digests of the purified activity (Figure 3 and Methods). The peptides were identified by MS/MS and were confined to the short-form of the enzyme lacking the ~150 residue C-terminal extension that terminates in a transmembrane helix [2, 16]. This conclusion is also supported by the apparent molecular weight deduced from SDS-PAGE analysis (~63 kDa estimated from Figure 2 compared to >82 kD expected for the full-length form of the oxidase [20].
Fig. 3.

Bovine QSOX1 sequence. The amino acid sequence of Quiescin sulfhydryl oxidase 1 precursor is shown. Peptides identified by MS/MS are underlined, yielding a total coverage of 59% over the 537 residues remaining after cleavage of the signal sequence (shown boxed). The cysteine residues from the two redox-active CxxC motifs are indicated in inverse font.
QSOX in mammalian blood
A survey of bovine fetal, newborn, and adult commercial sera revealed rather similar levels of QSOX activities irrespective of age (Table 2). While the sulfhydryl oxidase activities are generally rather comparable between bovine and human samples, mouse sera (Table 2 and Materials and Methods) reproducibly show an almost 10-fold higher level of activity. Commercial heat-treated bovine sera (56 °C for 30 min) retained 60–80% of the sulfhydryl oxidase activity found in untreated sera (data not shown).
Table 2.
QSOX activity levels towards DTT in bovine, human and mouse commercial sera. Assays were conducted as described in Materials and Methods. Errors represent standard deviations of 3 determinations.
| Serum type | H2O2 production (nmol/min·mg) |
|---|---|
| Human normal | 0.082 ± 0.011 |
| Mouse1 | 0.687 ± 0.056 |
| Fetal bovine | 0.099 ± 0.002 |
| Newborn bovine | 0.070 ± 0.003 |
| Adult donor bovine | 0.119 ± 0.011 |
Normal mouse serum; sera from 4 additional mouse strains (see Methods) yield an average of 0.893 ± 0.197 nmol/min per mg.
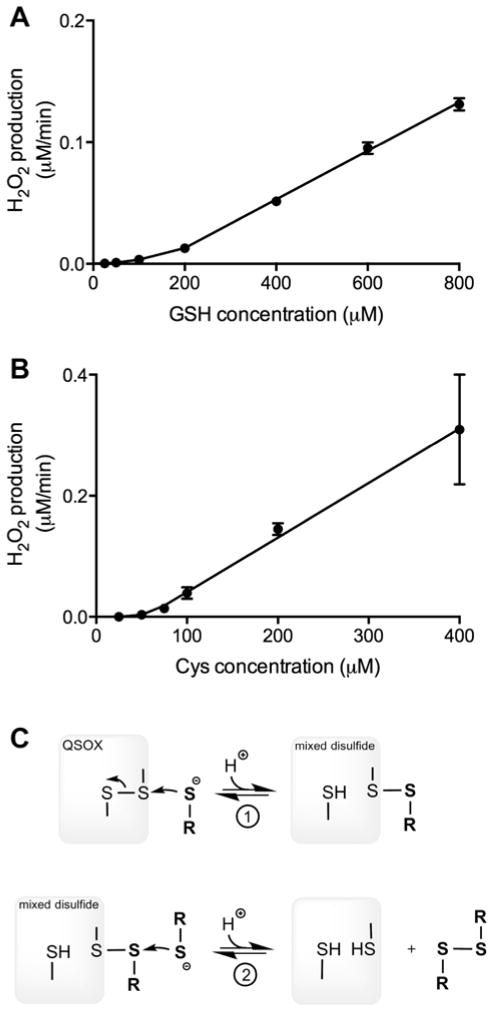
The confirmation of QSOX1 activity in mammalian sera raises several issues of general relevance. One point is that the sulfhydryl oxidase activity of conditioned media derived from mammalian cell culture will likely include a contribution from the serum used in the preparation of the growth medium. Thus, any evaluation of secreted QSOX activity in mammalian cell culture will require accounting for such background activity. A second issue is that experiments in which small molecular weight thiols, such as β-mercaptoethanol, cysteine or glutathione, are added to cell culture media containing animal serum may lead to an exogenous QSOX-dependent depletion of thiols and to the corresponding appearance of hydrogen peroxide. A third aspect concerns the potential of QSOX to oxidize the low concentrations of the monothiols cysteine and glutathione that are normally present in blood plasma [43, 44]. With Km and kcat values for vertebrate QSOX1 enzymes towards these thiols of about 10 mM and about 2000 thiols/min respectively [18, 20, 42] application of the Michaelis-Menten equation would predict an apparent turnover number for QSOX1 at 50 μM thiol of approximately 10/min. Hence at a concentration of ~25 nM QSOX (the concentration of QSOX1 deduced by comparing the activity of human blood serum with that of the recombinant human enzyme under standard assay conditions using DTT) this would imply that there could be significant generation of low molecular weight disulfides and H2O2 by this circulating oxidase. Since the steady state parameters for cysteine and glutathione were previously determined using the relatively insensitive oxygen electrode, the rates at very low thiol concentrations could not be investigated. Figure 4 documents the behavior of QSOX at these low thiol concentrations revealed with the new assay. Here, the activity of QSOX with GSH and cysteine is barely detectable but becomes significant as the thiol concentration is raised into the mM range. Hence GSH and cysteine at 50 μM show turnover numbers of ~ 0.19 thiols/min and ~ 0.73 thiols/min respectively. This apparently discrepant behavior is not unique to QSOX; precedent for this type of upward curvature has recently been observed during the oxidation of β-mercaptoethanol by another flavin-linked sulfhydryl oxidase, augmenter of liver regeneration [45], and in the oxidation of glutathione by protein disulfide isomerase [46]. Since QSOX functions in a hit-and-run mode without a detectable substrate binding site [5], a monothiol substrate must first reversibly generate a mixed-disulfide intermediate followed by capture of this species with a second monothiol (Figure 4C; equilibrium 1). The requirement for the sequential intervention of two thiols leads to the observed upward curvature and to the unexpectedly modest reactivity of QSOX at low micromolar levels of monothiols. Thus circulating soluble QSOX1 is unlikely to significantly influence the cysteine or glutathione redox pools in blood directly, although it could contribute to the accumulation of hydrogen peroxide in stored plasma [47].
Fig. 4.
Comparison of the rates of hydrogen peroxide generation by recombinant human QSOX1 in the presence of sub-millimolar concentrations of glutathione and cysteine. The data were corrected for background non-enzymatic peroxide generation and for the attenuation of fluorescence signal that is observed in the presence of increasing thiol concentration (supplementary Figure S1; see the Text). Panels A and B refer to GSH and cysteine respectively. Panel C shows that reduction of QSOX by monothiols involves capture of the mixed disulfide intermediate formed in equilibrium 1 with a second molecule of monothiol depicted in reaction 2.
Conclusions
This work shows that murine, bovine and human sera contain significant levels of sulfhydryl oxidase activity. We found similar levels of enzymatic activity between fetal and adult bovine sera. A three-step purification protocol using adult bovine serum showed that this activity reflects circulating, soluble QSOX1. While the levels of QSOX1 in blood are unlikely to significantly contribute to the oxidation of circulating cysteine and glutathione, there is a wealth of data showing that the redox poise of thiols and disulfides located in exofacial protein domains on a range of blood cells, including platelets and lymphocytes, are key modulators of biological function [48–57]. For example, protein secretion, adhesion and integrin-mediated association in platelets are regulated by the thiol/disulfide exchange and redox transformations that are modulated by membrane-bound thiol/disulfide oxidoreductases/isomerases [48–50, 54]. Since QSOX is a direct and facile oxidant of conformationally mobile protein thiols, some of these proteins may be effective substrates of QSOX1 and the activities of this oxidase may oppose those of circulating or membrane-bound reductants.
The finding from mass spectroscopic analyses that the levels of QSOX1 peptides and/or protein in serum have diagnostic applications in pancreatic cancer [26] and in acute decompensated heart failure [31] suggests that these approaches may be complemented by the simple and cost-effective QSOX assay described here. More generally, the observation that QSOX1 enzyme activity is found in blood plasma of all developmental stages suggests that renewed consideration should be directed towards the origins, substrate specificity and physiological roles of this catalytically most proficient stand-alone oxidant of protein and peptide thiols.
Supplementary Material
A sensitive new plate-reader assay for Quiescin-sulfhydryl oxidase was developed
Assays of serum demonstrated suitability for evaluating biological samples
Purification from adult bovine serum shows that oxidase activity is due to QSOX1
QSOX1 activity towards glutathione and protein thiols in human serum is discussed
Acknowledgments
This work was supported in part by NIH GM26642 (CT), USPHS Training Grant 1-T32-GM08550 (BAI). We thank Dr. Douglas F. Lake for helpful comments.
Abbreviations
- AUR
Amplex UltraRed
- BSA
bovine serum albumin
- HVA
homovanillic acid
- HRP
horseradish peroxidase
- PBS
phosphate buffered saline
- RNase
ribonuclease A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hoober KL, Glynn NM, Burnside J, Coppock DL, Thorpe C. Homology between egg white sulfhydryl oxidase and quiescin Q6 defines a new class of flavin-linked sulfhydryl oxidases. J Biol Chem. 1999;274:31759–31762. doi: 10.1074/jbc.274.45.31759. [DOI] [PubMed] [Google Scholar]
- 2.Kodali VK, Thorpe C. Oxidative protein folding and the Quiescin-sulfhydryl oxidase family of flavoproteins. Antioxid Redox Signal. 2010;13:1217–1230. doi: 10.1089/ars.2010.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coppock DL, Cina-Poppe D, Gilleran S. The Quiescin Q6 gene (QSCN6) is a fusion of two ancient gene families: thioredoxin and ERV1. Genomics. 1998;54:460–468. doi: 10.1006/geno.1998.5605. [DOI] [PubMed] [Google Scholar]
- 4.Hoober KL, Sheasley SS, Gilbert HF, Thorpe C. Sulfhydryl oxidase from egg white: a facile catalyst for disulfide bond formation in proteins and peptides. J Biol Chem. 1999;274:22147–22150. doi: 10.1074/jbc.274.32.22147. [DOI] [PubMed] [Google Scholar]
- 5.Codding JA, Israel BA, Thorpe C. Protein Substrate Discrimination in the Quiescin Sulfhydryl Oxidase (QSOX) Family. Biochemistry. 2012;51:4226–4235. doi: 10.1021/bi300394w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cline DJ, Thorpe C, Schneider JP. Structure Based Design of a Fluorimetric Redox Active Peptide Probe. Anal Biochem. 2003;325:144–150. doi: 10.1016/j.ab.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 7.Coppock DL, Thorpe C. Multidomain flavin-dependent sulfhydryl oxidases. Antioxid Redox Signal. 2006;8:300–311. doi: 10.1089/ars.2006.8.300. [DOI] [PubMed] [Google Scholar]
- 8.Thorpe C, Coppock DL. Generating disulfides in multicellular organisms: Emerging roles for a new flavoprotein family. J Biol Chem. 2007;282:13929–13933. doi: 10.1074/jbc.R600037200. [DOI] [PubMed] [Google Scholar]
- 9.Thorpe C, Hoober K, Raje S, Glynn N, Burnside J, Turi G, Coppock D. Sulfhydryl oxidases: emerging catalysts of protein disulfide bond formation in eukaryotes. Arch Biochem Biophys. 2002;405:1–12. doi: 10.1016/s0003-9861(02)00337-5. [DOI] [PubMed] [Google Scholar]
- 10.Rancy PC, Thorpe C. Oxidative Protein Folding in vitro: a Study of the Cooperation between Quiescin-sulfhydryl Oxidase and Protein Disulfide Isomerase. Biochemistry. 2008;47:12047–12056. doi: 10.1021/bi801604x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tury A, Mairet-Coello G, Esnard-Feve A, Benayoun B, Risold PY, Griffond B, Fellmann D. Cell-specific localization of the sulphydryl oxidase QSOX in rat peripheral tissues. Cell Tissue Res. 2006;323:91–103. doi: 10.1007/s00441-005-0043-x. [DOI] [PubMed] [Google Scholar]
- 12.Tury A, Mairet-Coello G, Poncet F, Jacquemard C, Risold PY, Fellmann D, Griffond B. QSOX sulfhydryl oxidase in rat adenohypophysis: localization and regulation by estrogens. J Endocrinol. 2004;183:353–363. doi: 10.1677/joe.1.05842. [DOI] [PubMed] [Google Scholar]
- 13.Rudolf J, Pringle MA, Bulleid NJ. Proteolytic Processing of QSOX1A Ensures Efficient Secretion of a Potent Disulfide Catalyst. Biochem J. 2013 doi: 10.1042/BJ20130360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakravarthi S, Jessop CE, Willer M, Stirling CJ, Bulleid NJ. Intracellular catalysis of disulphide bond formation by the human sulphydryl oxidase, QSOX1. Biochem J. 2007;404:403–411. doi: 10.1042/BJ20061510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mairet-Coello G, Tury A, Esnard-Feve A, Fellmann D, Risold PY, Griffond B. FAD-linked sulfhydryl oxidase QSOX: topographic, cellular, and subcellular immunolocalization in adult rat central nervous system. J Comp Neurol. 2004;473:334–363. doi: 10.1002/cne.20126. [DOI] [PubMed] [Google Scholar]
- 16.Heckler EJ, Rancy PC, Kodali VK, Thorpe C. Generating disulfides with the Quiescin-sulfhydryl oxidases. Biochim Biophys Acta. 2008;1783:567–577. doi: 10.1016/j.bbamcr.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benayoun B, Esnard-Feve A, Castella S, Courty Y, Esnard F. Rat seminal vesicle FAD-dependent sulfhydryl oxidase. Biochemical characterization and molecular cloning of a member of the new sulfhydryl oxidase/quiescin Q6 gene family. J Biol Chem. 2001;276:13830–13837. doi: 10.1074/jbc.M010933200. [DOI] [PubMed] [Google Scholar]
- 18.Hoober KL, Joneja B, White HB, III, Thorpe C. A Sulfhydryl Oxidase from Chicken Egg White. J Biol Chem. 1996;271:30510–30516. doi: 10.1074/jbc.271.48.30510. [DOI] [PubMed] [Google Scholar]
- 19.Zanata SM, Luvizon AC, Batista DF, Ikegami CM, Pedrosa FO, Souza EM, Chaves DF, Caron LF, Pelizzari JV, Laurindo FR, Nakao LS. High levels of active quiescin Q6 sulfhydryl oxidase (QSOX) are selectively present in fetal serum. Redox Rep. 2005;10:319–323. doi: 10.1179/135100005X83699. [DOI] [PubMed] [Google Scholar]
- 20.Jaje J, Wolcott HN, Fadugba O, Cripps D, Yang AJ, Mather IH, Thorpe C. A flavin-dependent sulfhydryl oxidase in bovine milk. Biochemistry. 2007;46:13031–13040. doi: 10.1021/bi7016975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin DB, Gifford DR, Wright ME, Keller A, Yi E, Goodlett DR, Aebersold R, Nelson PS. Quantitative proteomic analysis of proteins released by neoplastic prostate epithelium. Cancer Res. 2004;64:347–355. doi: 10.1158/0008-5472.can-03-2062. [DOI] [PubMed] [Google Scholar]
- 22.Kulasingam V, Diamandis EP. Proteomic analysis of conditioned media from three breast cancer cell lines: A mine for biomarkers and therapeutic targets. Mol Cell Proteomics. 2007;6:1997–2011. doi: 10.1074/mcp.M600465-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Grossman I, Alon A, Ilani T, Fass D. An Inhibitory Antibody Blocks the First Step in the Dithiol/Disulfide Relay Mechanism of the Enzyme QSOX1. J Mol Biol. 2013;425:4366–4378. doi: 10.1016/j.jmb.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 24.Ouyang X, DeWeese TL, Nelson WG, Abate-Shen C. Loss-of-function of Nkx3.1 promotes increased oxidative damage in prostate carcinogenesis. Cancer Res. 2005;65:6773–6779. doi: 10.1158/0008-5472.CAN-05-1948. [DOI] [PubMed] [Google Scholar]
- 25.Song H, Zhang B, Watson MA, Humphrey PA, Lim H, Milbrandt J. Loss of Nkx3.1 leads to the activation of discrete downstream target genes during prostate tumorigenesis. Oncogene. 2009;28:3307–3319. doi: 10.1038/onc.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antwi K, Hostetter G, Demeure MJ, Katchman BA, Decker GA, Ruiz Y, Sielaff TD, Koep LJ, Lake DF. Analysis of the plasma peptidome from pancreas cancer patients connects a peptide in plasma to overexpression of the parent protein in tumors. J Proteome Res. 2009;8:4722–4731. doi: 10.1021/pr900414f. [DOI] [PubMed] [Google Scholar]
- 27.Katchman BA, Ocal IT, Cunliffe HE, Y-HC, Hostetter G, Watanabe A, LoBello J, Lake DF. Expression of Quiescin Sulfhydryl Oxidase 1 is associated with a highly invasive phenotype and correlates with a poor prognosis in Luminal B breast cancer. Breast Cancer Res. 2013;15:R28. doi: 10.1186/bcr3407. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soloviev M, Esteves MP, Amiri F, Crompton MR, Rider CC. Elevated Transcription of the Gene QSOX1 Encoding Quiescin Q6 Sulfhydryl Oxidase 1 in Breast Cancer. PloS one. 2013;8:e57327. doi: 10.1371/journal.pone.0057327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katchman BA, Antwi K, Hostetter GH, Demeure MJ, Watanabe A, Decker GA, Miller LJ, Von Hoff DD, Lake DF. Quiescin Sulfhydryl Oxidase 1 Promotes Invasion of Pancreatic Tumor cells Mediated by Matrix Metalloproteinases. Mol Cancer Res. 2011 doi: 10.1158/1541-7786.MCR-11-0018. [DOI] [PubMed] [Google Scholar]
- 30.Ilani T, Alon A, Grossman I, Horowitz B, Kartvelishvily E, Cohen SR, Fass D. A Secreted Disulfide Catalyst Controls Extracellular Matrix Composition and Function. Science. 2013 doi: 10.1126/science.1238279. [DOI] [PubMed] [Google Scholar]
- 31.Mebazaa A, Vanpoucke G, Thomas G, Verleysen K, Cohen-Solal A, Vanderheyden M, Bartunek J, Mueller C, Launay JM, Van Landuyt N, D’Hondt F, Verschuere E, Vanhaute C, Tuytten R, Vanneste L, De Cremer K, Wuyts J, Davies H, Moerman P, Logeart D, Collet C, Lortat-Jacob B, Tavares M, Laroy W, Januzzi JL, Samuel JL, Kas K. Unbiased plasma proteomics for novel diagnostic biomarkers in cardiovascular disease: identification of quiescin Q6 as a candidate biomarker of acutely decompensated heart failure. Eur Heart J. 2012;33:2317–2324. doi: 10.1093/eurheartj/ehs162. [DOI] [PubMed] [Google Scholar]
- 32.Raje S, Glynn N, Thorpe C. A continuous fluorescence assay for sulfhydryl oxidase. Anal Biochem. 2002;307:266–272. doi: 10.1016/s0003-2697(02)00050-7. [DOI] [PubMed] [Google Scholar]
- 33.Zhao B, Summers FA, Mason RP. Photooxidation of Amplex red to resorufin: implications of exposing the Amplex red assay to light. Free Radic Biol Med. 2012;53:1080–1087. doi: 10.1016/j.freeradbiomed.2012.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munday R, Munday CM, Winterbourn CC. Inhibition of copper-catalyzed cysteine oxidation by nanomolar concentrations of iron salts. Free Radic Biol Med. 2004;36:757–764. doi: 10.1016/j.freeradbiomed.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 35.Hoober KL, Thorpe C. Flavin-dependent sulfhydryl oxidases in protein disulfide bond formation. Methods Enzymol. 2002;348:30–34. doi: 10.1016/s0076-6879(02)48622-3. [DOI] [PubMed] [Google Scholar]
- 36.Simeonov A, Jadhav A, Thomas CJ, Wang YH, Huang RL, Southall NT, Shinn P, Smith J, Austin CP, Auld DS, Inglese J. Fluorescence spectroscopic profiling of compound libraries. J Med Chem. 2008;51:2363–2371. doi: 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- 37.Zhou M, Diwu Z, Panchuk-Voloshina N, Haugland RP. A stable nonfluorescent derivative of resorufin for the fluorometric determination of trace hydrogen peroxide: applications in detecting the activity of phagocyte NADPH oxidase and other oxidases. Anal Biochem. 1997;253:162–168. doi: 10.1006/abio.1997.2391. [DOI] [PubMed] [Google Scholar]
- 38.Fasano M, Curry S, Terreno E, Galliano M, Fanali G, Narciso P, Notari S, Ascenzi P. The extraordinary ligand binding properties of human serum albumin. IUBMB Life. 2005;57:787–796. doi: 10.1080/15216540500404093. [DOI] [PubMed] [Google Scholar]
- 39.Ramming T, Appenzeller-Herzog C. The physiological functions of mammalian endoplasmic oxidoreductin 1: on disulfides and more. Antioxid Redox Sign. 2012;16:1109–1118. doi: 10.1089/ars.2011.4475. [DOI] [PubMed] [Google Scholar]
- 40.Sevier CS, Kaiser CA. Conservation and diversity of the cellular disulfide bond formation pathways. Antioxid Redox Signal. 2006;8:797–811. doi: 10.1089/ars.2006.8.797. [DOI] [PubMed] [Google Scholar]
- 41.Fass D. The Erv family of sulfhydryl oxidases. Biochim Biophys Acta. 2008;1783:557–566. doi: 10.1016/j.bbamcr.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 42.Heckler EJ, Alon A, Fass D, Thorpe C. Human quiescin-sulfhydryl oxidase, QSOX1: probing internal redox steps by mutagenesis. Biochemistry. 2008;47:4955–4963. doi: 10.1021/bi702522q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turell L, Radi R, Alvarez B. The thiol pool in human plasma: The central contribution of albumin to redox processes. Free Radic Biol Med. 2013;65C:244–253. doi: 10.1016/j.freeradbiomed.2013.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones DP, Liang Y. Measuring the poise of thiol/disulfide couples in vivo. Free Radic Biol Med. 2009;47:1329–1338. doi: 10.1016/j.freeradbiomed.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaefer-Ramadan SA, Gannon SA, Thorpe C. Human Augmenter of Liver Regeneration; probing the catalytic mechanism of a flavin-dependent sulfhydryl oxidase. Biochemistry. 2013 doi: 10.1021/bi401305w. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lappi AK, Ruddock LW. Reexamination of the role of interplay between glutathione and protein disulfide isomerase. J Mol Biol. 2011;409:238–249. doi: 10.1016/j.jmb.2011.03.024. [DOI] [PubMed] [Google Scholar]
- 47.Halliwell B, Clement MV, Ramalingam J, Long LH. Hydrogen peroxide. Ubiquitous in cell culture and in vivo? IUBMB Life. 2000;50:251–257. doi: 10.1080/713803727. [DOI] [PubMed] [Google Scholar]
- 48.Essex DW. Redox control of platelet function. Antioxid Redox Sign. 2009;11:1191–1225. doi: 10.1089/ars.2008.2322. [DOI] [PubMed] [Google Scholar]
- 49.Jordan PA, Gibbins JM. Extracellular disulfide exchange and the regulation of cellular function. Antioxid Redox Sign. 2006;8:312–324. doi: 10.1089/ars.2006.8.312. [DOI] [PubMed] [Google Scholar]
- 50.Matthias LJ, Hogg PJ. Redox Control on the Cell Surface: Implications for HIV-1 Entry. Antioxid Redox Signal. 2003;5:133–138. doi: 10.1089/152308603321223621. [DOI] [PubMed] [Google Scholar]
- 51.Sahaf B, Heydari K, Herzenberg LA. Lymphocyte surface thiol levels. Proc Natl Acad Sci USA. 2003;100:4001–4005. doi: 10.1073/pnas.2628032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan Z, Garg SK, Kipnis J, Banerjee R. Extracellular redox modulation by regulatory T cells. Nature Chem Biol. 2009;5:721–723. doi: 10.1038/nchembio.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Go YM, Jones DP. Cysteine/cystine redox signaling in cardiovascular disease. Free Radic Biol Med. 2011;50:495–509. doi: 10.1016/j.freeradbiomed.2010.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swiatkowska M, Padula G, Michalec L, Stasiak M, Skurzynski S, Cierniewski CS. Ero1alpha is expressed on blood platelets in association with protein-disulfide isomerase and contributes to redox-controlled remodeling of alphaIIbbeta3. J Biol Chem. 2010;285:29874–29883. doi: 10.1074/jbc.M109.092486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Acosta-Serrano A, Vassella E, Liniger M, Kunz Renggli C, Brun R, Roditi I, Englund PT. The surface coat of procyclic Trypanosoma brucei: programmed expression and proteolytic cleavage of procyclin in the tsetse fly. Proc Natl Acad Sci USA. 2001;98:1513–1518. doi: 10.1073/pnas.041611698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Metcalfe C, Cresswell P, Ciaccia L, Thomas B, Barclay AN. Labile disulfide bonds are common at the leucocyte cell surface. Open Biol. 2011;1:110010. doi: 10.1098/rsob.110010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stegmann M, Metcalfe C, Barclay AN. Immunoregulation through membrane proteins modified by reducing conditions induced by immune reactions. Eur J Immunol. 2013;43:15–21. doi: 10.1002/eji.201242849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.