Abstract
Objective:
To investigate the role of oxidative stress in the development of cisplatin resistance in epithelial ovarian cancer (EOC).
Methods:
Two parent EOC cell lines (MDAH-2774 and SKOV-3) and their chemoresistant counterparts (cisplatin, 50 µmol/L) were used. Total RNA was extracted and subjected to real-time reverse transcriptase polymerase chain reaction to evaluate the expression of glutathione reductase (GSR) and inducible nitric oxide synthase (iNOS), as well as nitrate/nitrite levels. Analysis of variance was used for main effects and Tukey for post hoc analysis at P < .05 for statistical significance.
Results:
Both cisplatin resistant cell lines displayed a significant decrease in GSR messenger RNA (mRNA) levels and activity (P < .01). As compared to sensitive controls, nitrate/nitrite levels were significantly higher in SKOV-3 cisplatin resistant cells while iNOS mRNA levels were significantly higher in MDAH-2774 cisplatin resistant cells (P < .05).
Conclusion:
Our data suggest that the development of cisplatin resistance tilts the balance toward a pro-oxidant state in EOC.
Keywords: epithelial ovarian cancer, cisplatin resistance, reactive oxygen species
Introduction
Ovarian cancer is the principal cause of death of all gynecologic cancers, with an estimated 22 240 new cases and 14 030 deaths expected in 2013 in the United States alone.1 Over the past several decades, the chemotherapeutic management of epithelial ovarian cancer (EOC) has significantly evolved from single alkylating agent to platinum-taxane combinations and bevacizumab, more recently. Unfortunately, between 60% and 80% of those who initially responded will relapse with a platinum-free interval at less than 6 months and therefore are considered platinum resistant.2
Reactive oxygen species (ROS) are known to be involved in many physiological processes such as redox signaling and immunity, as well as pathological processes such as cancer, through the promotion of genetic instability, abnormal cell proliferation, and angiogenesis.3–5 The nicotinamide adenine dinucleotide phosphate (NADPH) oxidase family, a membrane-bound enzyme complex, is a key player in the generation of ROS as well as inducible nitric oxide synthase (iNOS).5–8 Physiologic pathways involving crucial enzymes such as glutathione reductase (GSR), glutathione peroxidase, catalase, and superoxide dismutase consume free radicals and other oxidants, thus assisting in the maintenance of oxidative balance. When these latter processes are overwhelmed, a state of oxidative stress ensues. Ultimately, this pervasive environment can damage all components of the cell, including proteins, lipids, and DNA.9,10 Paradoxically, evidence suggests that cancer cells can take advantage of these processes to evade the immune system, avoid apoptosis, and become resistant to chemotherapy.11
The rationale of this research proposal is that cancer cells function under significant intrinsic oxidative stress using glycolytic metabolism even in the presence of high oxygen availability (Warburg effect).12,13 Previous studies have demonstrated that alterations in redox signaling pathways are involved in the pathophysiology of EOC.5,6,14,15 Reportedly, increasing iNOS activity can lead to S-nitrosylation of caspase-3 and decreased apoptosis in EOC cells.5 Moreover, many have evidence outlining the role of glutathione and GS-XlMRPP2 efflux pumps as one of the mechanisms of cisplatin resistance.16–18 Discovering the exact mechanisms involving the main players of ROS metabolism and their relationship to resistance to chemotherapy will provide new insights into tumor biology and allow the identification of new therapeutic targets that can be swiftly translated to clinical interventions. Our objective in this study was to investigate the role of oxidative stress in the development of cisplatin resistance in EOC cells.
Materials and Methods
Cell Culture
Two parent human EOC cell lines (SKOV-3 and MDAH-2774 from American Type Culture Collection, Manassas, Virginia) were utilized in developing cells resistant to cisplatin. Cells were exposed to a stepwise increase in cisplatin (Sigma Aldrich, St Louis, Missouri) over the course of a year, with final concentrations of 50 μmol/L. The Trypan Blue Dye Exclusion Method was used to confirm resistance as follows: once the desired level of resistance of 50 μmol/L was achieved, a 2-week incubation period without cisplatin and subsequent reintroduction of cisplatin was performed. Cells were cultured in 60-mm2 dishes with McCoy’s 5A medium (Invitrogen, Grand Island, New York) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, Georgia) and 1% penicillin and streptomycin (Invitrogen).
For measurement of GSR messenger RNA (mRNA) and activity and nitrate/nitrite levels, cells (2.5 × 106) were seeded in 150-mm dishes (Corning, Corning, New York) and allowed to rest for 24 hours followed by media replacement and cell collection 24 hours later.
For measurement of iNOS mRNA levels, cells (1 × 106) were seeded in 150 mm3 dishes (Corning) and cultured over 72 hours.
Real-Time Reverse Transcription Polymerase Chain Reaction
RNA isolation
Total RNA was extracted from EOC cells using the RNeasy Mini Kit (Qiagen, Valencia, California) according to the protocol provided by the manufacturer.
Reverse transcription
A 40-μL-complementary DNA (cDNA)-reaction volume utilizing 2 μg RNA was prepared using the QuantiTect Reverse Transcription Kit (Qiagen), as described by the manufacturer’s protocol.
Real-time RT-PCR primer design and controls
Optimal oligonucleotide primer pairs for real-time reverse transcription polymerase chain reaction (RT-PCR) amplification of reverse-transcribed cDNA were selected with the aid of the software program, Beacon Designer (Premier Biosoft Int, Palo Alto, California). Human oligonucleotide primers, which amplify variable portions of the protein coding regions, were used. Sequences of the oligonucleotides used for amplification of iNOS and GSR are described in Table 1.
Table 1.
Human Oligonucleotide Primers.
| Locus | Sense (5′-3′) | Antisense (3′-5′) | Size, bp |
|---|---|---|---|
| GSR | TCACCAAGTCCCATATAGAAATC | TGTGGCGATCAGGATGTG | 116 |
| iNOS | GGCACAGAACTTAAGGATGG | TTGTTAGGAGGTCAAGTAAAGG | 145 |
Abbreviations: bp, base pair; GSR, glutathione reductase; iNOS, inducible nitric oxide synthase.
Quantitative real-time RT-PCR was performed using a QuantiTect SYBR Green RT-PCR kit (Qiagen) and Cepheid 1.2f Detection System (Cepheid, Sunnyvale, California). The RT-PCR was performed in a 25 μL total reaction volume including 12.5 μL of 2× QuantiTect SYBR Green RT-PCR master mix, 5 μL of 10× diluted cDNA template for GSR and 2 μL of undiluted cDNA template for iNOS, and 0.2 μmol/L each of target specific primers designed to amplify each gene. Standards with known concentrations and lengths (base pairs [bp]) were designed specifically for iNOS [103 bp] and GSR [103 bp] using the Beacon Designer software (Premier Biosoft), allowing for construction of a standard curve using a 10-fold dilution series. A specific standard for each gene allows for absolute quantification of the gene in a number of copies, which can then be expressed per μg of RNA. The RT-PCR conditions were programmed as follows: an initial cycle was performed at 95°C for 1000 seconds for iNOS and 900 seconds for GSR. This was followed by 35 cycles of denaturation at 95°C for 15 seconds, annealing for 30 seconds at 58°C for iNOS, and 59°C for GSR. This was followed by a final cycle at 72°C for 30 seconds to allow completion of product synthesis. Following RT-PCR, a melting curve analysis was performed to demonstrate the specificity of the PCR product as a single peak. A control, containing all the reaction components except for the template, was included in all experiments. All experiments were performed in triplicate.
Protein extraction
Cell lysates were prepared utilizing cell lysis buffer (Cell Signaling Technology, Danvers, Massachusetts) supplemented with Protease Arrest (G-Biosciences, St Louis, Missouri). Total protein concentration of cell lysates was measured using the Pierce BCA Protein Assay Kit (Thermo Scientific, Rockford, Illinois) according to the manufacturer’s protocol.
Detection of GSR activity
The Glutathione Reductase Assay Kit (Cayman Chemical, Ann Arbor, Michigan) determines GSR activity by measuring the rate of NADPH oxidation. The oxidation of NADPH to NADP+ is accompanied by a decrease in the absorbance at 340 nm. Since GSR is present at rate-limiting concentrations, the rate of decrease in the absorbance at 340 nm is directly proportional to the GSR activity in the sample. The same concentration of protein was utilized for each sample.
Measurement of Nitrate/Nitrite Levels
The nitrate/nitrite colorimetric assay (Cayman Chemical, Ann Arbor, Michigan) was used to measure the levels of stable nitric oxide (NO) by-products, nitrate (NO2 −), and nitrite (NO3 −) as an indication of NO production. Due to the fact that the proportion of NO2 − and NO3 − is variable and cannot be predicted with certainty, the sum of both NO by-products is a more accurate indicator of NO production. The assay was performed utilizing cell culture media according to the manufacturer’s protocol. Absorbance was detected at 540 nm, and a standard curve for nitrite was utilized to determine total NO2 − and NO3 −.
Measurement of Cellular Proliferation
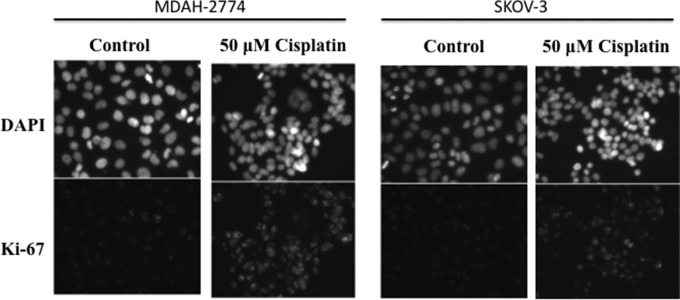
Sensitive and cisplatin-resistant MDAH-2774 and SKOV-3 (5 × 103) EOC cells were cultured in 96-well tissue culture plates in 200 μL of medium (Becton Dickinson, Lincoln Park, New Jersey) and assessed for immunofluorescence as reported previously.19 Nuclei expressing Ki-67 were labeled with a monoclonal antibody (Ki-S5; DAKO, Carpinteria, California). Briefly, after removing media from wells, the cells were fixed with 100 μL of formaldehyde for 30 minutes and then rinsed with phosphate-buffered saline (PBS). Subsequently, the cells were then treated with 100 μL of 0.01% Triton x-100 solution diluted in PBS. After administration of the primary antibody to wells and a 24-hour incubation period, the wells were washed 3 times with PBS, and then the FITC-conjugated secondary antibody was applied. Again, the wells were washed with PBS, and the nuclei were counterstained with 4′,6-diamidino-2-phenylindole. Finally, the wells were rinsed 3 times prior to visualization under the fluorescent microscope.
Statistical Analysis
Data were analyzed using SPSS 19.0. Unpaired t tests were used to compare controls and the resistant cell lines. P values are expressed at α < .05 for significance.
Results
Cisplatin-Resistant EOC Cells Have Lower GSR mRNA and Activity Levels
There was a significant decrease in the mRNA expression and activity of GSR in both cisplatin-resistant EOC cell lines when compared to their sensitive counterparts (Figure 1A and B). The GSR mRNA levels decreased in cisplatin-resistant MDAH-2774 (20.3 ± 0.7 pg/μg RNA) and SKOV-3 (11.6 ± 0.25 pg/μg RNA) when compared to control MDAH-2774 (27.7 ± 1.2 pg/μg RNA) and control SKOV-3 (30.1 ± 0.6 pg/μg RNA) EOC cells (P < .05, Figure 1A).
Figure 1.
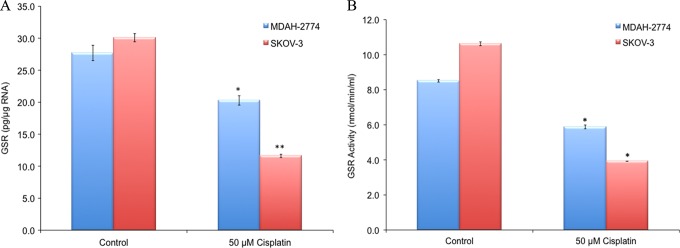
A, Real-time RT-PCR for GSR and (B) GSR activity in EOC cells. Sensitive (control) and resistant (50 µmol/L) cisplatin is correlated with GSR (A) mRNA levels and (B) activity. (*P < .05). EOC indicates epithelial ovarian cancer; GSR, glutathione reductase; mRNA, messenger RNA; RT-PCR, real-time reverse transcription polymerase chain reaction.
Correspondingly, GSR protein activity also decreased in cisplatin-resistant MDH-2774 (from 8.5 ± 0.07 to 5.9 ± 0.1 nmol/min/mL) and SKOV-3 (from 10.6 ± 0.1 to 3.9 ± 0.0 nmol/min/mL) when compared to respective sensitive controls (P < .05, Figure 1B).
Cisplatin-Resistant EOC Cells Have Higher iNOS and Nitrate/Nitrite Levels
Nitrate/nitrite levels were significantly increased in SKOV-3 cisplatin-resistant EOC cells (13.7 + 0.2 μmol/L) when compared to its sensitive counterpart (8.2 + 0.0 μmol/L; P < .05, Figure 2A). There was no change observed in nitrate/nitrite levels in MDAH-2774-sensitive EOC cells when compared to its cisplatin-resistant counterpart.
Figure 2.
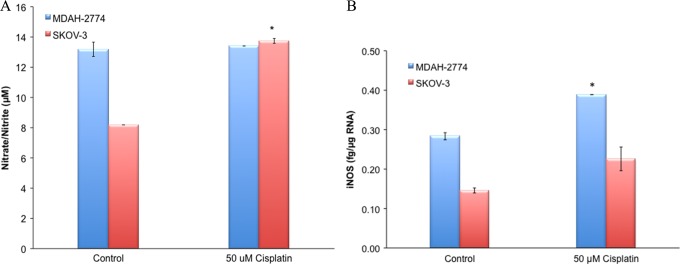
A, Real-time RT-PCR for iNOS and (B) nitrate/nitrate levels in EOC cells. Sensitive (control) and resistant (50 µmol/L) cisplatin is correlated with iNOS (A) mRNA levels and (B) nitrate/nitrite levels (*P < .05). EOC indicates epithelial ovarian cancer; iNOS, inducible nitric oxide synthase; mRNA, messenger RNA; RT-PCR, real-time reverse transcription polymerase chain reaction.
The iNOS mRNA levels were significantly increased in MDAH-2774 cisplatin-resistant EOC cells (0.28 ± 0.01 fg/µg RNA) when compared to its sensitive counterpart (0.39 ± 0.00 fg/µg RNA; P < .05, Figure 2B). There was an increased trend, however not significant, observed in iNOS mRNA levels in SKOV-3 when compared to its cisplatin-resistant counterpart.
Cisplatin-Resistant EOC Cells Have Increased Proliferation
The Ki-67 immunofluorescence staining revealed increased proliferation in cisplatin-resistant EOC cell lines when compared to their sensitive controls for both MDAH-2774 and SKOV-3 (Figure 3).
Figure 3.
Proliferation of epithelial ovarian cancer cells lines (A) MDAH-2774 and (B) SKOV-3 cultured in the presence of cisplatin (50 µmol/L). The columns represent control and high dose (50 µmol/L) of cisplatin treatments and the rows illustrate anti-Ki-67 antibody and 4′,6-diamidino-2-phenylindole counterstains.
Discussion
In this study, we have shown that cisplatin-resistant EOC cells manifested a significant decrease in the mRNA expression and activity levels of GSR when compared to their sensitive counterparts (Figure 1A and B). Glutathione (GSH), a tripeptide, is a powerful antioxidant that plays an important role in preventing ROS-induced damages to vital cellular functions.20–22 However, it was shown that once cancer is established, high levels of GSH could be deleterious by preventing the cytotoxic effect of various chemotherapeutic agents through detoxification and increased activity of efflux pump.23–25 Conversely, it has also been reported that low levels of GSH are linked to impaired immune response and tumor progression.26–28 The latter is more aligned with our findings, because low expression of GSR is indicative of low GSH and increased oxidative stress, since GSH will not be adequately regenerated from its oxidized form (GSSG). Together, the antioxidant mechanisms involving GSR among others, when impaired, may adversely impact the response to cisplatin by failing to restore the oxidative balance.
On the other hand, cisplatin-resistant MDAH-2774 EOC cells manifested an increase in iNOS expression when compared to their sensitive counterpart. Moreover, nitrate/nitrite levels were significantly increased in cisplatin-resistant SKOV-3 EOC cells when compared to its sensitive counterparts. Our results suggest that increased expression of iNOS and nitrate/nitrite may be associated with resistance to cisplatin. The NO synthase family includes the calcium/calmodulin-mediated isoenzymes endothelial NOS (eNOS) and neuronal NOS (nNOS), in addition to the noncalcium-dependent iNOS.29 Earlier studies have shown that NO donors can induce apoptosis at high concentrations, whereas preventing it can be achieved at low physiologic levels.30,31 Moreover, when compared to normal cells, cancer cells have significantly increased level of iNOS.32 Previously, we have reported that silencing iNOS can lead to a decrease in S-nitrosylation of caspase-3 with subsequent increased apoptosis in EOC cells.14 Also, we have demonstrated that high levels of iNOS in EOC cells are associated with high levels of vascular endothelial growth factor (VEGF) production and angiogenesis induction.6 Additionally, other groups have reported an inverse relationship between the expression of eNOS/nNOS and iNOS that suggests elevated expression of iNOS coupled with decreased eNOS/nNOS expression may be associated with p53-mediated cisplatin resistance in EOC.33 Conversely, low expression of iNOS in head and neck squamous cell carcinoma was shown to be associated with resistance to cisplatin/taxol-induced apoptosis mediated through survivin.34 The physiology of the NO synthase family is fairly complex and not fully elucidated; however, they appeared to be involved in cisplatin resistance in many cancers.
Finally, utilizing Ki-67, we observed a significant increase in proliferation of cisplatin-resistant EOC cells when compared to their sensitive counterparts.
Several possible mechanisms of cisplatin resistance have been previously described, including decreased intracellular aquation, decreased uptake, and accelerated efflux due to increased activity of adenosine triphosphate (ATP)-binding cassette and soluble carrier transporter families.35–37 Additionally, poor tissue perfusion, increased proliferation, and decreased apoptosis altogether contribute to chemoresistance. The present study suggests a potential role for oxidative stress in the development of cisplatin resistance in ovarian cancer. Understanding the precise mechanism by which oxidative stress contributes to the development of cisplatin resistance will significantly impact survival of patients with ovarian cancer.
Footnotes
Authors’ Note: Jimmy Belotte and Nicole M. Fletcher contributed equally to this work.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This project was funded by NIH/NICHD grant K12HD001254.
References
- 1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. [DOI] [PubMed] [Google Scholar]
- 2. Markman M, Rothman R, Hakes T, et al. Second-line platinum therapy in patients with ovarian cancer previously treated with cisplatin. J Clin Oncol. 1991;9(3):389–393. [DOI] [PubMed] [Google Scholar]
- 3. Diplock AT, Rice-Evans CA, Burdon RH. Is there a significant role for lipid peroxidation in the causation of malignancy and for antioxidants in cancer prevention? Cancer Res. 1994;54(7 suppl):1952s–1956s. [PubMed] [Google Scholar]
- 4. Kamata H, Hirata H. Redox regulation of cellular signalling. Cell Signal. 1999;11(1):1–14. [DOI] [PubMed] [Google Scholar]
- 5. Jiang Z, Fletcher NM, Ali-Fehmi R, et al. Modulation of redox signaling promotes apoptosis in epithelial ovarian cancer cells. Gynecol Oncol. 2011;122(2):418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Malone JM, Saed GM, Diamond MP, Sokol RJ, Munkarah AR. The effects of the inhibition of inducible nitric oxide synthase on angiogenesis of epithelial ovarian cancer. Am J Obstet Gynecol. 2006;194(4):1110–1116; discussion 1116-1118. [DOI] [PubMed] [Google Scholar]
- 7. Dvoriantchikova G, Grant J, Santos AR, Hernandez E, Ivanov D. Neuronal NAD(P)H oxidases contribute to ROS production and mediate RGC death after ischemia. Invest Ophthalmol Vis Sci. 2012;53(6):2823–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Montezano AC, Touyz RM. Molecular mechanisms of hypertension-reactive oxygen species and antioxidants: a basic science update for the clinician. Can J Cardiol. 2012;28(3):288–295. [DOI] [PubMed] [Google Scholar]
- 9. Valko M, Morris H, Cronin MT. Metals, toxicity and oxidative stress. Curr Med Chem. 2005;12(10):1161–1208. [DOI] [PubMed] [Google Scholar]
- 10. Evans MD, Cooke MS. Factors contributing to the outcome of oxidative damage to nucleic acids. Bioessays. 2004;26(5):533–542. [DOI] [PubMed] [Google Scholar]
- 11. Halliwell B. Oxidative stress and cancer: have we moved forward? Biochem J. 2007;401(1):1–11. [DOI] [PubMed] [Google Scholar]
- 12. Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. [DOI] [PubMed] [Google Scholar]
- 13. Lopez-Lazaro M. The warburg effect: why and how do cancer cells activate glycolysis in the presence of oxygen? Anticancer Agents Med Chem. 2008;8(3):305–312. [DOI] [PubMed] [Google Scholar]
- 14. Saed GM, Ali-Fehmi R, Jiang ZL, et al. Myeloperoxidase serves as a redox switch that regulates apoptosis in epithelial ovarian cancer. Gynecol Oncol. 2010;116(2):276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fletcher NM, Jiang Z, Ali-Fehmi R, et al. Myeloperoxidase and free iron levels: potential biomarkers for early detection and prognosis of ovarian cancer. Cancer Biomark. 2011;10(6):267–275. [DOI] [PubMed] [Google Scholar]
- 16. Akimaru K, Kuo MT, Furuta K, Suzuki M, Noyori R, Ishikawa T. Induction of MRP/GS-X pump and cellular resistance to anticancer prostaglandins. Cytotechnology. 1996;19(3):221–227. [DOI] [PubMed] [Google Scholar]
- 17. Fujii R, Mutoh M, Sumizawa T, Chen ZS, Yoshimura A, Akiyama S. Adenosine triphosphate-dependent transport of leukotriene C4 by membrane vesicles prepared from cisplatin-resistant human epidermoid carcinoma tumor cells. J Natl Cancer Inst. 1994;86(23):1781–1784. [DOI] [PubMed] [Google Scholar]
- 18. Ishikawa T, Ali-Osman F. Glutathione-associated cis-diamminedichloroplatinum(II) metabolism and ATP-dependent efflux from leukemia cells. Molecular characterization of glutathione–platinum complex and its biological significance. J Biol Chem. 1993;268(27):20116–20125. [PubMed] [Google Scholar]
- 19. Armant DR, Kilburn BA, Petkova A, et al. Human trophoblast survival at low oxygen concentrations requires metalloproteinase-mediated shedding of heparin-binding EGF-like growth factor. Development. 2006;133(4):751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Han YH, Moon HJ, You BR, Kim SZ, Kim SH, Park WH. The effects of N-acetyl cysteine on the MG132 proteasome inhibitor-treated lung cancer cells in relation to cell growth, reactive oxygen species and glutathione. Int J Mol Med. 2010;25(4):657–662. [DOI] [PubMed] [Google Scholar]
- 21. Chow HH, Hakim IA, Vining DR, et al. Modulation of human glutathione s-transferases by polyphenon e intervention. Cancer Epidemiol Biomarkers Prev. 2007;16(8):1662–1666. [DOI] [PubMed] [Google Scholar]
- 22. Balendiran GK, Dabur R, Fraser D. The role of glutathione in cancer. Cell Biochem Funct. 2004;22(6):343–352. [DOI] [PubMed] [Google Scholar]
- 23. Batist G, Behrens BC, Makuch R, et al. Serial determinations of glutathione levels and glutathione-related enzyme activities in human tumor cells in vitro. Biochem Pharmacol. 1986;35(13):2257–2259. [DOI] [PubMed] [Google Scholar]
- 24. Suzuki T, Nishio K, Tanabe S. The MRP family and anticancer drug metabolism. Curr Drug Metab. 2001;2(4):367–377. [DOI] [PubMed] [Google Scholar]
- 25. de Bittencourt Junior PI, Curi R, Williams JF. Glutathione metabolism and glutathione S-conjugate export ATPase (MRP1/GS-X pump) activity in cancer. I. Differential expression in human cancer cell lines. Biochem Mol Biol Int. 1998;45(6):1227–1241. [DOI] [PubMed] [Google Scholar]
- 26. Suthanthiran M, Anderson ME, Sharma VK, Meister A. Glutathione regulates activation-dependent DNA synthesis in highly purified normal human T lymphocytes stimulated via the CD2 and CD3 antigens. Proc Natl Acad Sci USA. 1990;87(9):3343–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Multhoff G, Meier T, Botzler C, et al. Differential effects of ifosfamide on the capacity of cytotoxic T lymphocytes and natural killer cells to lyse their target cells correlate with intracellular glutathione levels. Blood. 1995;85(8):2124–2131. [PubMed] [Google Scholar]
- 28. Droge W, Schulze-Osthoff K, Mihm S, et al. Functions of glutathione and glutathione disulfide in immunology and immunopathology. FASEB J. 1994;8(14):1131–1138. [PubMed] [Google Scholar]
- 29. Forstermann U, Gath I, Schwarz P, Closs EI, Kleinert H. Isoforms of nitric oxide synthase. Properties, cellular distribution and expressional control. Biochem Pharmacol. 1995;50(9):1321–1332. [DOI] [PubMed] [Google Scholar]
- 30. Fiscus RR. Involvement of cyclic GMP and protein kinase G in the regulation of apoptosis and survival in neural cells. Neurosignals. 2002;11(4):175–190. [DOI] [PubMed] [Google Scholar]
- 31. Fiscus RR, Yuen JP, Chan SL, Kwong JH, Chew SB. Nitric oxide and cyclic GMP as pro- and anti-apoptotic agents. J Card Surg. 2002;17(4):336–339. [DOI] [PubMed] [Google Scholar]
- 32. Wink DA, Vodovotz Y, Cook JA, et al. The role of nitric oxide chemistry in cancer treatment. Biochemistry (Mosc). 1998;63(7):802–809. [PubMed] [Google Scholar]
- 33. Leung EL, Fraser M, Fiscus RR, Tsang BK. Cisplatin alters nitric oxide synthase levels in human ovarian cancer cells: involvement in p53 regulation and cisplatin resistance. Br J Cancer. 2008;98(11):1803–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fetz V, Bier C, Habtemichael N, et al. Inducible NO synthase confers chemoresistance in head and neck cancer by modulating survivin. Int J Cancer. 2009;124(9):2033–2041. [DOI] [PubMed] [Google Scholar]
- 35. Liu J, Cristea MC, Frankel P, et al. Clinical characteristics and outcomes of BRCA-associated ovarian cancer: genotype and survival. Cancer Genet. 2012;205(1-2):34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ali AY, Farrand L, Kim JY, et al. Molecular determinants of ovarian cancer chemoresistance: new insights into an old conundrum. Ann N Y Acad Sci. 2012;1271:58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kalinina EV, Berezov TT, Shtil AA, et al. Expression of peroxiredoxin 1, 2, 3, and 6 genes in cancer cells during drug resistance formation. Bull Exp Biol Med. 2012;153(6):878–881. [DOI] [PubMed] [Google Scholar]