SUMMARY
Axonal myelination is essential for rapid saltatory impulse conduction in the nervous system, and malformation or destruction of myelin sheaths leads to motor and sensory disabilities. DNA methylation is an essential epigenetic modification during mammalian development, yet its role in myelination remains obscure. Here, using high-resolution methylome maps, we show that DNA methylation could play a key gene regulatory role in peripheral nerve myelination and that S-adenosylmethionine (SAMe), the principal methyl donor in cytosine methylation, regulates the methylome dynamics during this process. Our studies also point to a possible role of SAMe in establishing the aberrant DNA methylation patterns in a mouse model of diabetic neuropathy, implicating SAMe in the pathogenesis of this disease. These critical observations establish a link between SAMe and DNA methylation status in a defined biological system, and provides a novel mechanism that could direct methylation changes during cellular differentiation and in diverse pathological situations.
INTRODUCTION
Axonal myelination is essential for rapid saltatory impulse conduction, and malformation or destruction of myelin sheaths can lead to severe motor and sensory disabilities(Suter and Scherer, 2003). In the peripheral nervous system, genome-wide changes in gene expression characterize both the differentiation of myelinating Schwann cells(Nagarajan et al., 2002; Verheijen et al., 2003) and development of neuropathies, including diabetic neuropathy(Pande et al., 2011). The coordinated changes in gene expression patterns associated with Schwann cell myelination are controlled transcriptionally by an elaborate network of transcription factors (TFs), including Egr2, Sox10, Oct6, YY1 and NFκB(Jessen and Mirsky, 2005; Pereira et al., 2012), recruitment of chromatin-remodeling complexes to regulatory regions of key myelin genes(Hung et al., 2012; Weider et al., 2012), and possibly by histone deacetylation(Pereira et al., 2012). In addition, microRNAs and RNA-binding proteins play a crucial role in the post-transcriptional regulation of these global changes in gene expression(Iruarrizaga-Lejarreta et al., 2012; Pereira et al., 2012).
DNA methylation is a key epigenetic mechanism with essential functions, including repression of target promoters, genomic imprinting, silencing of transposable elements and maintenance of genomic integrity(Jaenisch and Bird, 2003). Methylation occurs on the fifth position of cytosine, mostly in the context of CpG dinucleotides, and is catalyzed by a family of three conserved DNA methyltransferase enzymes, Dnmt1, Dnmt3a and Dnmt3b(Smith and Meissner, 2013). During the methylation process, DNMTs transfer the methyl group from the principal methyl donor SAMe to cytosine residues, generating S-adenosylhomocysteine (SAH)(Lu and Mato, 2012). Mouse functional studies and, more recently, large-scale DNA methylation mapping have provided important insights into the regulatory role of DNA methylation in mammalian development(Smith and Meissner, 2013). In particular, DNA methylome characterization, often at single-base resolution, has enabled the genome-wide identification of changes in DNA methylation patterns during somatic lineage commitment with associated changes in gene expression at promoter regions and regulatory elements, including enhancers(Jones, 2012).
In this study, we used high-resolution genomic maps to examine the methylome dynamics during Schwann cell myelination in vivo. Like other somatic cellular differentiation paradigms(Calvanese et al., 2012; Lee et al., 2012; Shearstone et al., 2011), we found that Schwann cell myelination was characterized by global DNA demethylation, which was associated with activation of critical myelination-associated genes, particularly those involved in synthesis of lipid, a critical component of myelin. Importantly, our observations support the notion that levels of SAMe play a critical role in regulating the methylation status of myelinating Schwann cells. Peripheral nerves from mice with enforced elevated levels of SAMe were characterized by DNA hypermethylation globally, and at promoter and enhancer regions of several lipid synthesis genes. This correlated with an altered lipid profile, likely contributing to the peripheral myelin defects in these mice. Furthermore, we showed that reduced SAMe levels correlated with DNA demethylation in peripheral nerves of a mouse model of diabetic neuropathy, suggesting a possible role of SAMe in the pathogenesis of this disease.
Taken together, this study shows that Schwann cell myelination is characterized by widespread DNA methylation changes that likely play a key gene regulatory role. These methylation dynamics are dependent on a tight control of SAMe levels, and imbalances to its levels likely lead to aberrant DNA methylation patterns and consequently gene expression changes during development of neuropathies.
RESULTS
Genome-scale DNA methylation maps of Schwann cell myelination
To examine DNA methylation dynamics during Schwann cell myelination, we isolated sciatic nerves from mice at different ages that broadly correspond to the main stages of the myelination process(Jessen and Mirsky, 2005). Thus, nerves were obtained from new-born mice (NB), which are highly enriched for immature Schwann cells, post-natal day 10 (P10) nerves, which contain mostly actively myelinating Schwann cells, and P60 nerves, which contain terminally differentiated myelinating Schwann cells
DNA methylation mapping was performed by reduced representation bisulfite sequencing (RRBS), which enables quantitative methylation analysis at single-base resolution of relatively small cell numbers(Smith et al., 2012). Methylation levels of individual CpGs were combined over non-overlapping 1-kilobase (kb) tiling regions to increase statistical power for subsequent analyses(Bock, 2012). For each stage, 2–3 biological replicates were obtained from different mice with high reproducibility and expected genomic coverage (Figure S1A and S1B). RRBS, in our case, was particularly suitable since it allowed us to study DNA methylation at gene regulatory elements such as promoter regions, given its strong bias at detecting methylation changes in genomic regions of medium to high CpG density(Bock et al., 2010).
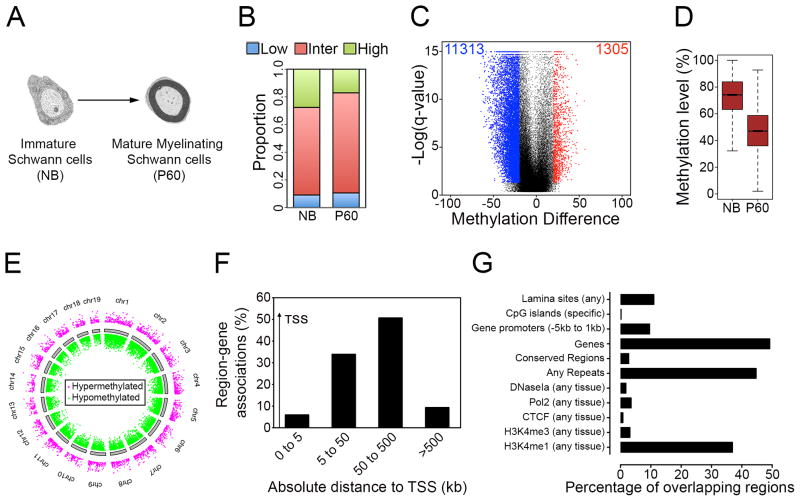
To examine the impact of DNA methylation in the overall process of myelination, we compared the DNA methylation patterns in the progenitor cells (immature Schwann cells) and the terminally differentiated cells (mature myelinating Schwann cells)(Figure 1A). From the pair-wise analysis, methylation levels of 73,317 1-kb tiles were obtained (Table S1). We found that there was a reduction in the proportion of regions with high methylation levels (≥80%) in P60 nerves compared to NB nerves (Figure 1B). Next, we compared changes in regional DNA methylation by classifying methylation levels of each 1-kb tile in P60 nerves relative to NB nerves as differentially methylated regions (DMRs) if the percentage methylation difference exceeded 20% and had a false discovery rate (FDR)-corrected t-test <0.05. We found that there was about a 9-fold higher number of hypomethylated than hypermethylated 1 kb-tiling regions (Figure 1C). The DMRs were distributed across all chromosomes (Figure 1E) and were present at promoter regions, although the large majority was found in gene bodies and intergenic regions (Figure 1F). Interestingly, putative enhancers (identified by presence of H3K4me1)(Shen et al., 2012) and repeat regions were significantly enriched (Figure 1G). Comparison of the methylation levels of the DMRs showed a substantial reduction in median methylation levels in P60 nerves in 1-kb tiling regions (Figure 1D), and in other genomic regions (Figure S1C and S1D).
Figure 1. Methylation dynamics during Schwann cell myelination.
(A) Stages of Schwann cell development used for RRBS analysis to show DNA methylation dynamics during Schwann cell myelination. Progenitor cells (immature Schwann cells) are enriched in NB peripheral nerves, and terminally differentiated cells (mature myelinating cells) are enriched in P60 peripheral nerves. (B) Proportion of 1-kb tiles with high (≥ 80%, green), intermediate (>20% and <80%, red) and low (≤20%, blue) percentage methylation levels. (C) Violin plot shows about a 9-fold greater number of hypomethylated DMRs (blue dots) than hypermethylated DMRs (red dots) in P60 nerves. Black dots represent regions with percentage methylation difference <20% and/or FDR>0.05. (D) Boxplot showing methylation levels of DMRs in 1-kb tiles. Boxplots correspond to center quartiles with black bar indicating median, and whiskers extend to the most extreme data point, which is no more than 1.5 times the interquartile range from the box. (E) Chromosome ideogram representing DMRs in P60 nerves compared to NB nerves (Magenta: hypermethylated DMRs; green: hypomethylated DMRs). (F) Histogram showing proportion of DMRs at relative distances from transcription start site (TSS), determined with GREAT software. (G) Histogram showing enrichment of DMRs in different genomic regions, determined with EpiExplorer software.
These results show that the myelination program proceeds with marked DNA demethylation, and a substantial enrichment of DMRs at gene-regulatory regions (promoters and putative enhancers) and repeat elements.
DNA demethylation correlates with increased expression of myelination-specific genes
In contrast to the global DNA demethylation seen during the myelination process, gene expression changes showed about an equal number of upregulated and downregulated genes (Figure S2A). This is consistent with DNA methylation being only one of other concerted factors in regulating gene expression(Portela and Esteller, 2010). DNA demethylation was associated with both upregulated and downregulated genes (Figure S2B), suggesting that DNA demethylation is not always associated with gene expression This is similar to other somatic differentiation paradigms (Bock et al., 2012; Calvanese et al., 2012; Lee et al., 2012; Shearstone et al., 2011),
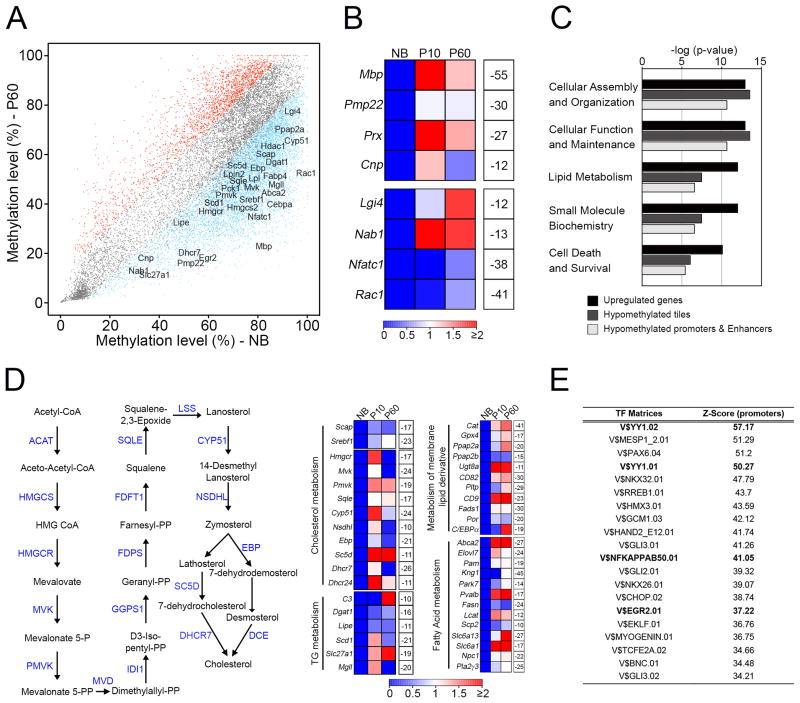
DNA demethylation, instead, has been shown to play an important role during somatic cell differentiation by regulating expression of a modest number of genes with cell-type specific functional roles (Bock et al., 2012; Calvanese et al., 2012; Lee et al., 2012). Consistent with these findings, we found that critical myelination-associated genes that were upregulated during development had a concomitant loss of methylation in gene-associated 1-kb tiles (Figure 2A) that were often annotated to gene-regulatory regions (promoters and putative enhancers)(Figure S2C and S2D). These included genes encoding structural components of myelin (Mbp, Pmp22, Prx and Cnp) and transcriptional regulators (Lgi4, Nab1 and Nfatc1) (Figure 2B). We found, on general, that there were a four-fold higher number of hypomethylated DMRs at different genomic regions associated with upregulated genes, including lipid metabolism genes (example of myelination-associated genes, see below). This led to an overall reduced methylation level (Figure S2E–G), confirming that upregulation of genes during development is more significantly associated with hypomethylation rather than hypermethylation.
Figure 2. Global demethylation during Schwann cell myelination.
(A) Scatterplot of percentage DNA methylation levels of 1-kb tiles, showing hypomethylated regions (blue) and hypermethylated regions (red) in P60 nerves, relative to NB nerves. Selected genes with important functions in Schwann cell myelination are highlighted. (B) Heatmap showing changes in expression of myelination-related genes from microarray data (Table S2) in P10 and P60 nerves, compared to NB nerves (red-blue color scale), and methylation difference in P60 nerves relative to NB nerves (white boxes). (C) GO analysis showing top Molecular and Cellular Function categories enriched in all up-regulated genes, up-regulated genes that become hypomethylated in tiling regions and up-regulated genes that become hypomethylated at promoter and putative enhancer regions. (D) Scheme of the cholesterol biosynthesis pathway in which the site of action of enzymes (blue) is shown (left). Heatmaps (right) showing changes in expression of genes from microarray experiments associated with cholesterol biosynthesis and distinct aspects of lipid metabolism in P10 and P60 nerves compared to NB nerves (red-blue color scale). Percentage methylation difference in P60 nerves relative to NB nerves is shown in white boxes. (E) Table showing enrichment of transcription factor recognition motifs in hypomethylated 1-kb tiling regions using Genomatix RegionMiner. Each row represents a cis-regulatory module with significant over-representation relative to a random set of mammalian promoters (Z-score).
Gene Ontology (GO) analysis has been an important tool to demonstrate quantitatively that genome-wide DNA methylation changes are significantly enriched for categories associated with specific cellular differentiation paradigms(Bock et al., 2012; Calvanese et al., 2012). Here, similarly, we found that upregulated genes with a concomitant decrease in methylation levels either in 1-kb tiling regions or gene-regulatory regions were enriched for functional categories associated with the transcriptomic signature of Schwann cell myelination (Figure 2C and Table S2). Among these enriched categories, lipid metabolism was particularly interesting since myelin consists of tightly compacted membranes, highly enriched in cholesterol and lipids, and PNS myelin is severely affected in several lipid metabolism disorders(Chrast et al., 2011). Here, we found that genes encoding many of the enzymes involved in lipid metabolism became demethylated during myelination (Figure 2D). These included key regulators (Scap and Srebf1) and individual members (e.g. Hmgcr, Mvk, Pmvk) of cholesterol biosynthetic pathways, as well as genes involved in triglyceride metabolism (Dgat1, Lipe, Scd1) and fatty acid metabolism (Abca2 and Elovl7).
Taken together, demethylation at gene-regulatory regions can be associated with activation of genes, which form part of the complex machinery of myelin formation, most notably the lipid biosynthetic process.
Demethylation is regulated by different mechanisms
Next, we examined the mechanisms that could regulate these methylation patterns. Establishment and maintenance of DNA methylation at promoter and enhancer regions are strongly influenced by transcription factor (TF) binding(Smith and Meissner, 2013). Here, we performed a TF motif enrichment analysis of all hypomethylated 1-kb tiles and found a significant enrichment of specific TF binding sites, including YY1, NFκB and Egr2, which are critical regulators of Schwann cell myelination(Pereira et al., 2012) (Figure 2E and Table S2). These results suggest that DNA demethylation at regulatory regions of myelination-associated genes could be mediated by binding of these TFs.
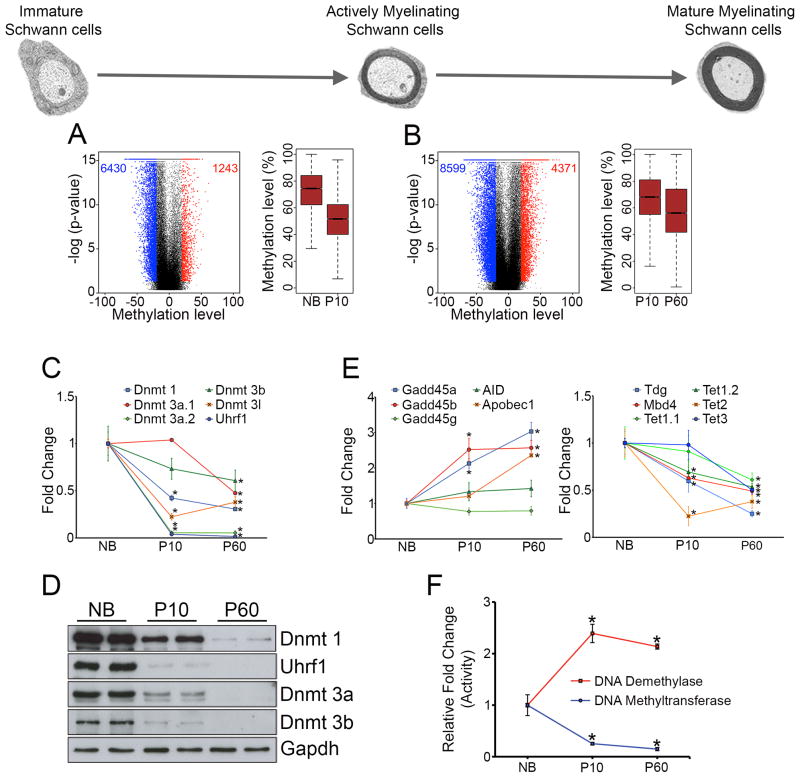
Generation of the mature myelinating Schwann cells can be conveniently divided into two overlapping phases. First, immature Schwann cells differentiate into pro-myelinating Schwann cells, which then undergo extensive membrane wrapping and compaction into myelin to generate mature myelinating Schwann cells. RRBS analysis (Figure S1A and S1B, and Table S3) showed that both phases were characterized by global DNA demethylation with a higher number of hypomethylated than hypermethylated regions, which resulted in a notable reduction in median methylation levels of DMRs, although the effect was more pronounced for the first developmental transition than for the second one (Figure 3A and 3B). Since DNA replication has been associated with DNA demethylation during somatic cellular differentiation(Shearstone et al., 2011) and the first phase is characterized by a high proliferation rate (Woodhoo and Sommer, 2008), we examined a possible contribution of cellular division to this demethylation pattern. We found that a 16h treatment of Schwann cells with the mitogen NRG1 led to a significant decrease in global DNA methylation levels, which, notably, was reduced by a block of cell cycle progression (Figure S3A and S3B). Similarly, comparison of DNA methylation levels in cAMP-treated cultures at 24h and 48h, an in vitro model of progressive myelin differentiation, showed a decrease in global DNA methylation levels in the more mature myelinated state (48h cAMP treatment)(Figure S3C). These results show that both cell division and myelin differentiation likely regulate the global DNA demethylation in the formation of mature myelinated Schwann cells.
Figure 3. Methylation dynamics and methyltransferase expression during different stages of Schwann cell myelination.
(A, B) DNA methylation changes (refer to Figure 1C, D) between (A) actively myelinating Schwann cells (P10) relative to immature Schwann cells (NB) and, (B) mature myelinating Schwann cells (P60) relative to actively myelinating Schwann cells (P10). (C, D) DNA methyltransferases are downregulated during Schwann cell myelination, as shown by (C) qPCR analysis and (D) Western blotting (WB). (E) qPCR analysis of regulators associated with active DNA demethylation. (F) Assay of total DNA methyltransferase and demethylase activity during Schwann cell myelination. Data is mean ± sem, *p < 0.01, n=3, Student’s t-test.
Next, we examined the expression of DNA methylation regulators during Schwann cell myelination. Both by qPCR and Western blotting, we found that levels of all DNMTs were downregulated during development, an effect more pronounced between P10 and NB nerves than between P60 and P10 nerves (Figure 3C and 3D). Of the regulators implicated in active demethylation(Bhutani et al., 2011), Gadd45a, Gadd45b and Apobec1 were upregulated (Figure 3E). Measurement of total DNMT and DNA demethylase activity broadly correlated with these observations (Figure 3F). Interestingly, we found that there was a significant difference in the activity of these enzymes between the P10 and NB stages, but not between the P60 and P10 stages. The downregulation of DNMTs expression between NB and P10 nerves is likely mediated by the transition from the highly proliferative immature Schwann cells in NB nerves to the quiescent cells in P10 nerves, since we found that high DNMT expression in cultured Schwann cells maintained under proliferative conditions is reduced after cell cycle block (Figure S3D). Similarly, downregulation of DNMTs expression between P10 and P60 nerves is likely mediated by the myelin differentiation process since we found significant downregulation of DNMT expression in vitro in the more mature myelinated state (48h cAMP treatment) (Figure S3E).
These results show that both formation and maturation of myelinated Schwann cells are characterized by DNA demethylation, likely established by cell division and myelin differentiation.
Demethylation correlates with decreased SAMe levels
SAMe donates its methyl group to a large variety of acceptor molecules, including histones, DNA and proteins(Lu and Mato, 2012). Recent studies have shown that SAMe levels can regulate histone methylation patterns in several systems(Shyh-Chang et al., 2013; Towbin et al., 2012; Ulanovskaya et al., 2013).
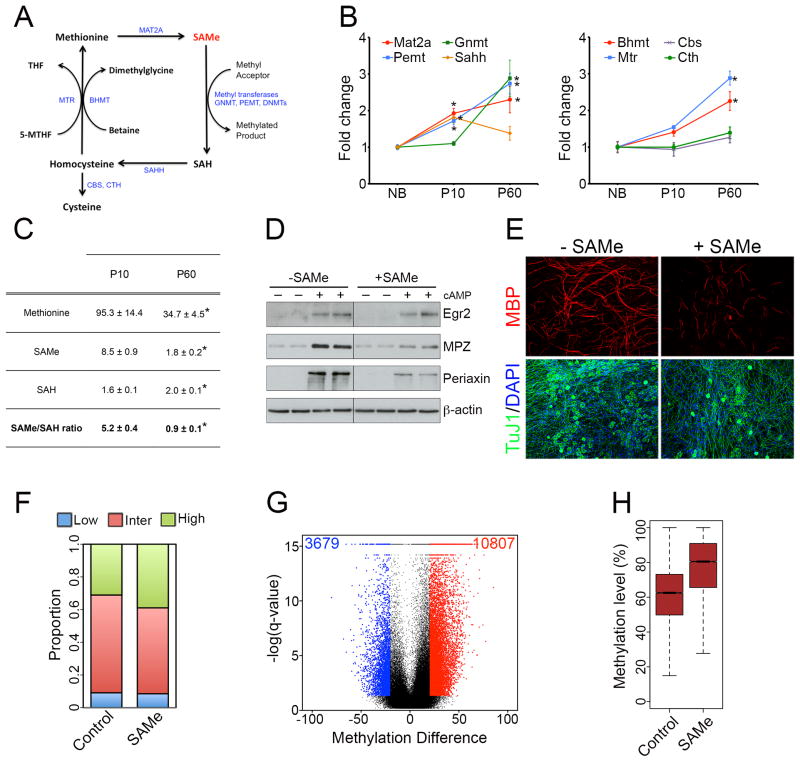
To investigate whether availability of SAMe could also determine the DNA methylation pattern during Schwann cell myelination, we first examined expression of the genes encoding the enzymes involved in the biosynthesis and catabolism of SAMe, which form part of the methionine cycle(Lu and Mato, 2012) (Figure 4A). We found that all the main enzymes were expressed and were differentially regulated during the myelination process (Figure 4B). Importantly, we found that the levels of SAMe were significantly reduced in P60 nerves, compared to P10 nerves (Figure 4C). In transmethylation reactions, SAH is produced and acts as a competitive inhibitor of methyltransferases. The SAMe/SAH ratio provides an important metabolic indicator of cellular methylation status(Ulanovskaya et al., 2013) and global hypomethylation has been associated with a decrease in this ratio(Lu and Mato, 2012). Here, we found about a 5-fold decrease in the SAMe/SAH ratio in P60 nerves compared with P10 nerves. These metabolites could not be measured in NB nerves since an unrealistically high number of nerves (> 250) was required for each out of five replicates.
Figure 4. SAMe levels are reduced during Schwann cell myelination, and SAMe supplementation blocks Schwann cell myelination in vitro.
(A) Schematic representation of the methionine cycle, in which the site of action of different enzymes involved (blue) is shown. (B) qPCR analysis showing differential expression of the main genes involved in the methionine cycle during Schwann cell myelination. Data is mean ± sem, *p < 0.01, n=3, Student’s t-test. (C) Table showing levels of the metabolites methionine, SAMe and SAH (pmol mg−1 of tissue), and the SAMe/SAH ratio in P10 and P60 nerves. Data is mean ± sem, *p < 0.05, n=5, Student’s t-test. (D) WB showing that exogenous SAMe supplementation prevents up-regulation of the myelin proteins Mpz and Periaxin under myelinogenic conditions (cAMP treatment). β-actin is shown as a loading control. (E) Immunocytochemistry showing fewer MBP+ myelinated segments (red) in SAMe-treated DRG-Schwann cell co-cultures. Note that axonal network (Tuj1+ cells, green) appear normal in both control and SAMe-treated cultures. (F) Proportion of 1-kb tiles with high (≥80%, green), intermediate (>20% and <80%, red) and low (≤20%, blue) percentage methylation levels in control and SAMe-treated cultures. (G) Violin plot showing about a 3-fold greater number of hypermethylated tiling regions (red) in SAMe-treated cultures than hypomethylated tiling regions. Black dots represent regions with percentage methylation difference <20% and/or FDR>0.05. (H) Boxplot showing an increased median methylation level in differentially-methylated tiling regions.
These results raised the possibility that SAMe could play a role in establishing the DNA methylation patterns during Schwann cell myelination, at least for the second phase of the process. To test this, we examined the effects of exogenous SAMe treatment on Schwann cell myelination in vitro. Treatment of purified Schwann cell cultures using cAMP induces upregulation of several myelin proteins, including Egr2, Mpz and Periaxin(Iruarrizaga-Lejarreta et al., 2012). We found that SAMe treatment prevented upregulation of Mpz and Periaxin (Figure 4D). A more physiologic model of in vitro Schwann cell myelination is co-culture of Schwann cells with dorsal root ganglion (DRG) axons, in which Schwann cells associate with and form myelin segments around axons. Similar to above, we found that SAMe treatment significantly reduced the number of MBP+ myelin sheaths (Figure 4E)(63 ± 8 MBP+ myelin sheaths in control cultures compared to 24 ± 3 MBP+ myelin sheaths in SAMe-treated cells, n=6, p<0.01, Student’s t-test), and led to reduced internodal length (Figure S4A).
Next, we examined whether SAMe could regulate DNA methylation patterns in Schwann cells in vitro. To show this, we performed RRBS analysis of Schwann cells after exogenous SAMe application (Figure S4B and S4C, and Table S4). We found that SAMe-treated cells were characterized by a higher proportion of regions with high methylation levels (Figure 4F) and a greater number of hypermethylated 1-kb tiles (Figure 4G). DMRs showed an increased methylation level for 1-kb tiles (Figure 4H) and all genomic regions analyzed (Figure S4D). These results indicate that exogenous SAMe application in Schwann cell cultures lead to global and locus-specific DNA hypermethylation.
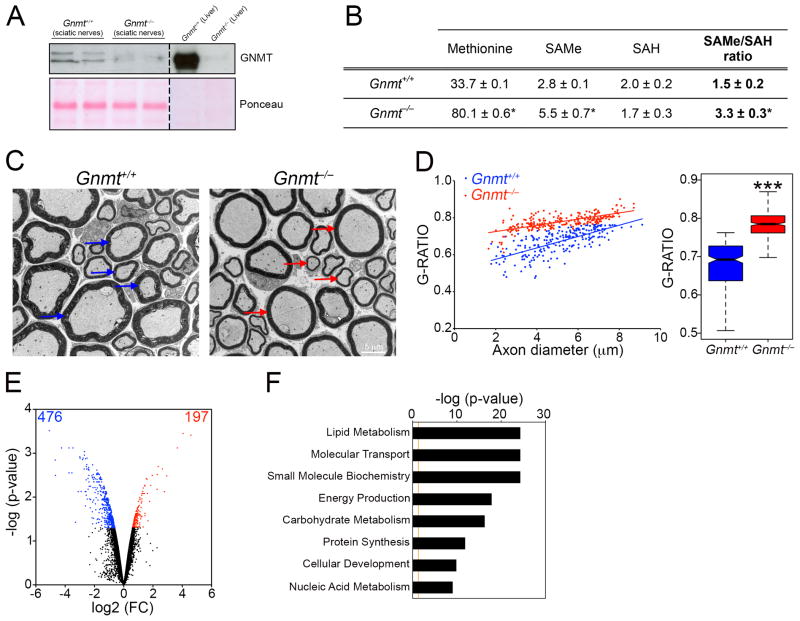
We next examined whether elevated SAMe levels could also affect Schwann cell myelination and DNA methylation patterns in vivo. For this, we used mice lacking the Gnmt gene (Gnmt−/− mice)(Martinez-Chantar et al., 2008). GNMT is an enzyme that is expressed in several tissues, including the liver, where it metabolizes SAMe to maintain its levels within a tight range and prevent aberrant methylation reactions(Lu and Mato, 2012; Martinez-Chantar et al., 2008). As shown above, Gnmt was also expressed in Schwann cells, and notably it was significantly upregulated in P60 nerves compared to P10 nerves (Figure 4B), correlating with a decrease in SAMe levels (Figure 4C). P90 sciatic nerves from Gnmt−/− mice did not express GNMT (Figure 5A), and this resulted in a two-fold increase in SAMe concentration and SAMe/SAH ratio (Figure 5B).
Figure 5. Elevated SAMe levels in Gnmt−/− mice lead to hypomyelination and gene expression changes.
(A) WB showing that GNMT is absent in sciatic nerves from Gnmt−/− mice. Liver, where GNMT is highly expressed (representing 1% of total protein)(Lu and Mato, 2012), is shown as a control. Ponceau is used as a loading control, since we found that GAPDH and β-actin were respectively downregulated and upregulated in Gnmt−/− nerves. (B) Table showing levels of the metabolites methionine, SAMe and SAH (pmol mg−1 of tissue), and the SAMe/SAH ratio in Gnmt+/+ and Gnmt−/− mice. Data is mean ± sem, *p < 0.01, n=5, Student’s t-test. (C) Electron micrographs showing thinner myelin sheaths (red arrows) in nerves from Gnmt−/− mice, compared to normal-sized myelin sheaths in Gnmt+/+ mice (blue arrows). (D) Morphometric analysis of myelinated axons in nerves from Gnmt+/+ and Gnmt−/− mice, showing G-ratio measurements, expressed as individual measurements in scatterplot (left), or boxplot (right)(***p<0.01, n=5, Student’s t-test). (E) Violin plot showing gene expression changes in Gnmt−/− mice compared to Gnmt+/+ mice, with a higher proportion of downregulated (blue) than upregulated genes (red). (F) GO analysis showing top Molecular and Cellular Function categories enriched in downregulated genes in Gnmt−/− mice.
See also Table S5.
Peripheral nerves of Gnmt−/− mice were hypomyelinated (thinner myelin sheaths), a characteristic feature of several peripheral neuropathies(Suter and Scherer, 2003) (Figure 5C), which was confirmed quantitatively using G-ratio measurements (Figure 5D). The hypomyelination phenotype was unlikely to be due to defects in radial sorting (Woodhoo and Sommer, 2008), since we did not find any differences in the ratio of myelinating to pro-myelinating Schwann cells in P5 nerves (Woodhoo et al., 2009) or defects in axons, which did not show differences in diameter in P90 nerves (data not shown).
Microarray analyses showed substantial differential gene expression profiles, between nerves from Gnmt+/+ and Gnmt−/− mice (Figure 5E and Table S5), with a greater number of downregulated than upregulated genes. GO analyses showed that the most significant category associated with the downregulated genes was lipid metabolism, which could likely explain the hypomyelination profile in the Gnmt−/− nerves (Figure 5F and Table S5).
Elevated SAMe levels in vivo leads to DNA hypermethylation
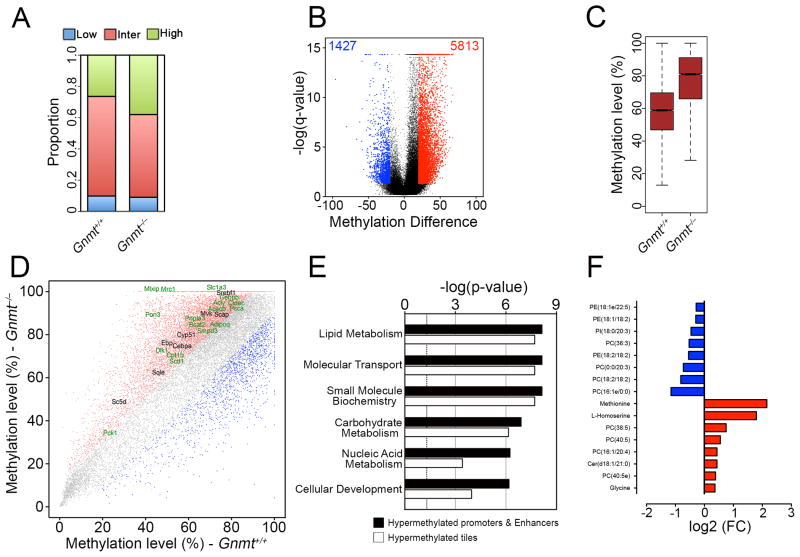
The elevated SAMe concentration in the nerves of the Gnmt−/− mice raised the interesting possibility that the myelination defects could be due to DNA hypermethylation, as shown for SAMe-treated cultures above. To show this, we performed RRBS analysis (Figure S5A–C), from which methylation levels of 61,916 1-kb tiles were obtained (Table S6). We found that Gnmt−/− nerves were characterized by a higher proportion of highly methylated regions (Figure 6A) and a greater number of hypermethylated 1-kb tiles (Figure 6B). DMRs showed an increased methylation level for 1-kb tiles (Figure 6C) and all genomic regions analyzed (Figure S5D)
Figure 6. Elevated SAMe levels in Gnmt−/− mice lead to global and locus-specific DNA hypermethylation.
(A) Proportion of 1-kb tiles with high (≥80%, green), intermediate (>20% and <80%, red) and low (≤20%, blue) percentage methylation levels. (B) Violin plot showing about a 3-fold greater number of hypermethylated tiling regions (red) in Gnmt−/− mice than hypomethylated tiling regions. Black dots represent regions with percentage methylation difference <20% and/or FDR>0.05. (C) Boxplot showing an increased median methylation level in differentially-methylated tiling regions in Gnmt−/− mice. (D) Scatterplot of DNA methylation of 1-kb tiles showing hypomethylated regions (blue) and hypermethylated regions (red) in Gnmt−/− mice. Selected hypermethylated genes are highlighted (genes which are downregulated are shown in green). (E) GO analysis showing top Molecular and Cellular Function categories enriched in all downregulated genes hypermethylated at tiling regions or gene-regulatory regions. (F) Histogram showing most-significantly changed lipid species and amino acids in Gnmt−/− mice compared to Gnmt+/+ mice.
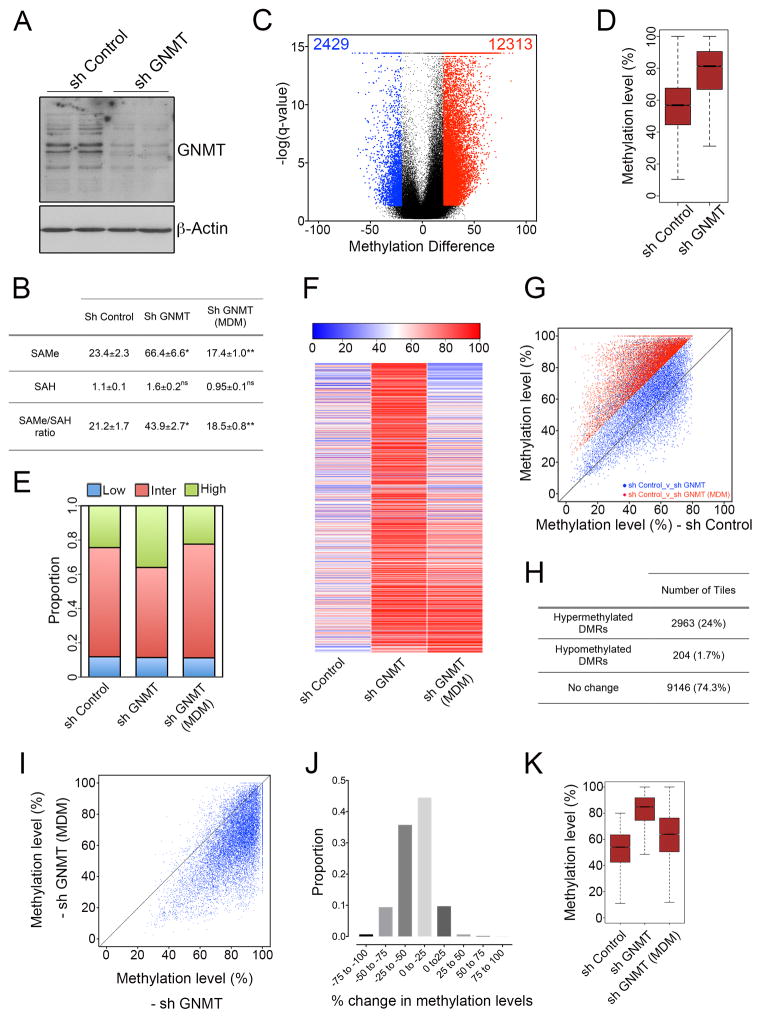
Similar to Gnmt−/− nerves in vivo, we found that GNMT silencing in vitro led to an increase in SAMe levels and SAMe/SAH ratio (Figure 7A and 7B). RRBS analysis (Figure S6A and S6B, and Table S7) showed that GNMT-silenced cells were also characterized by a predominantly DNA hypermethylation phenotype compared to control cultures (Figure 7C and 7D), suggesting that the DNA methylation changes in vivo are due to the absence of GNMT itself.
Figure 7. DNA hypermethylation induced by GNMT silencing in vitro is reverted by culture in low methionine medium.
(A) Western blot showing reduced GNMT levels in sh GNMT-infected Schwann cells compared to sh Control-infected cells. β-Actin is used as a loading control. (B) Table showing the levels of the metabolites SAMe and SAH (pmol per 2×106 cells), and the SAMe/SAH ratio in sh Control, sh GNMT and sh GNMT (MDM) cells. Data is mean ± sem, *p<0.05, n=5, Student’s t-test. Increase in SAMe levels, and the SAMe/SAH ratio induced by GNMT silencing is prevented by culture in low methionine medium. (C) Violin plot showing about a 5-fold greater number of hypermethylated tiling regions (red) than hypomethylated tiling regions (blue) in GNMT-silenced cells compared to control cells. Black dots represent regions with percentage methylation difference <20% and/or FDR>0.05. (D) Boxplot showing an increased median methylation level in differentially-methylated tiling regions. (E–K) Increased methylation induced in GNMT-silenced cells is prevented by culture in low methionine medium. (E) Proportion of 1-kb tiles with high (≥80%, green), intermediate (>20% and <80%, red) and low (≤20%, blue) percentage methylation levels. (F) Heatmap showing the absolute methylation levels of all hypermethylated 1kb tiles in the comparison sh GNMT_v_sh Control. When the GNMT-silenced cells were cultured in low methionine medium, there was a significant decrease in the methylation levels of many of the tiles. (G) Scatterplot showing that the large majority of tiles classified as hypermethylated DMRs (q<0.05, methylation increase of 20%) in the comparison sh GNMT_v_sh Control (red) did not show significant differences in methylation levels (methylation difference of 20%) in the comparison sh GNMT (MDM)_v_sh Control (blue). (H) Counts of the tiles (hypermethylated DMRs in the comparison sh GNMT_v_sh Control) classified as hypermethylated DMRs, hypomethylated DMRs, or with no change (methylation difference < 20%) in the comparison sh GNMT (MDM)_v_sh Control. Out of the 12,313 tiles hypermethylated in the comparison sh GNMT_v_sh Control, about 75% of them (9146) did not show significant methylation differences (methylation difference of 20% or q-value > 0.05) in the comparison sh GNMT (MDM)_v_sh Control. (I) Scatterplot of percentage DNA methylation levels of 1-kb tiles (12,313 tiles hypermethylated in the comparison sh GNMT_v_sh Control) in sh GNMT and sh GNMT (MDM) cultures. sh GNMT (MDM) cultures have an overall lower methylation level than sh GNMT cultures. (J) Graph showing the proportion of tiles with different changes in methylation levels in the comparison sh GNMT (MDM)_v_sh GNMT. A large number of tiles have a decreased methylation level of at least 25%, with a minimum number of tiles showing increased methylation. (K) Boxplot showing increased methylation levels of all differentially-methylated 1-kb tiling regions (q<0.05, methylation difference of 20%) in GNMT-silenced cells cultures (sh GNMT) compared to control cultures (sh Control). This effect is abolished when sh GNMT cells were cultured in low methionine medium (sh GNMT_MDM).
To directly show that GNMT-induced DNA hypermethylation was due to the elevated SAMe levels, we cultured the GNMT-silenced cells in medium containing low methione levels (MDM). As shown previously for cancer cells with reduced GNMT expression(Wang et al., 2011), culture of Schwann cells in this medium prevented the increase in SAMe levels in cells with lower GNMT expression (Figure 7B). RRBS analysis showed that the hypermethylation induced by GNMT knockdown was significantly blocked by culture of these cells in low methionine medium (Figure 7E J). As a result, the increased median methylation level induced by GNMT knockdown was strikingly lowered for 1-kb tiles (Figure 7K) and all genomic regions analyzed (Figure S6C). These data strongly suggest that elevated SAMe levels, induced by GNMT knockdown, are responsible for the DNA hypermethylation in Schwann cells.
We found that several downregulated genes in Gnmt−/− mice had increased methylation levels in 1-kb tiles (Figure 6D), several of which were annotated to promoter regions and putative enhancer elements (Figure S5E). GO analysis of these genes showed that the most significant category was lipid metabolism (Figure 6E and Table S5). A multi-platform metabolomic profiling was performed to examine differences in amino acid and lipid composition between Gnmt+/+ and Gnmt−/− sciatic nerves. Principal component analysis (PCA) of all metabolites analyzed showed clear sample segregation according to mice genotype (Figure S5F), and we identified several lipid species that significantly contributed to these differences (Figure 6F and Figure S5G and Table S6).
These results indicate that elevated SAMe concentration in Gnmt−/− mice leads to global and locus-specific DNA hypermethylation, which correlates with a downregulation of several lipid metabolism genes. This likely leads to the altered lipid profile and peripheral nerve defects in these mice.
Reduced SAMe levels could be responsible for pathogenesis of diabetic mice
Our results above argue for a key role of DNA methylation during Schwann cell myelination, and suggest that aberrant DNA methylation patterns can lead to defects in peripheral nerve myelination. There are several pathological conditions, which can cause malformation or destruction of myelin sheaths. Diabetic neuropathy is the most common and debilitating complication of diabetes, with injury to axons and Schwann cells both contributing to the pathogenesis of the disease(Vincent et al., 2011). It is a distinct possibility that peripheral nerves in this pathological situation could be characterized by aberrant DNA methylation patterns.
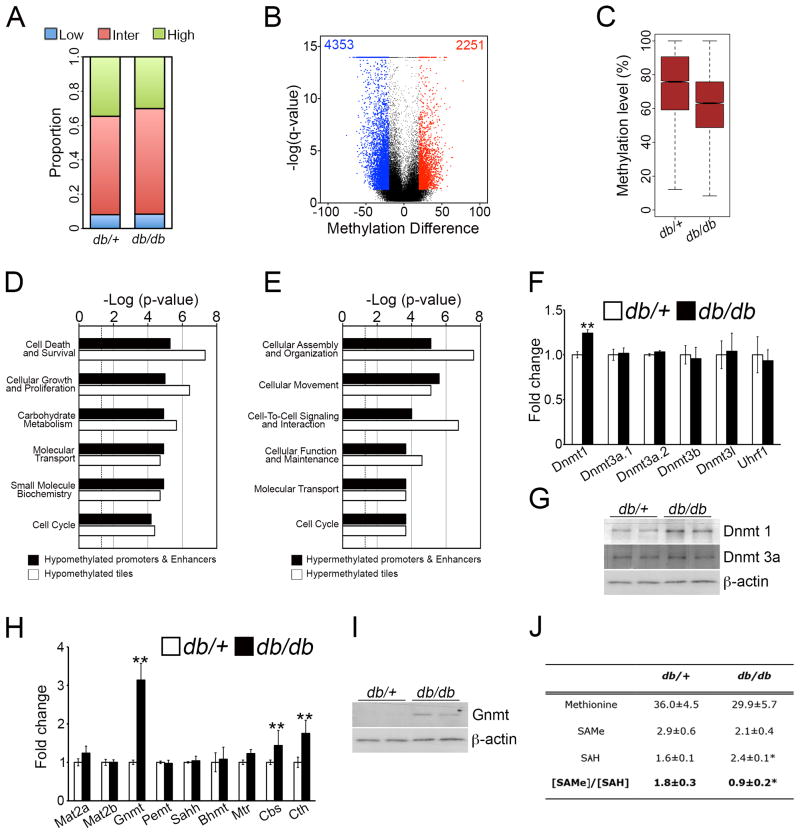
To show this, we performed a genome-wide analysis of the methylation pattern in 6-months old db/db mice, a mouse model of diabetic neuropathy, which closely resembles the human condition(Pande et al., 2011). RRBS analysis was performed (Figure S7A–C), from which methylation levels of 65,534 1-kb tiles were obtained for comparison (Table S8). There was a small reduction in proportion of highly methylated regions (Figure 8A) and analysis of DMRs showed twice as many hypomethylated 1-kb tiles as hypermethylated regions (Figure 8B). DMRs showed a decreased methylation level for 1-kb tiles (Figure 8C) and all genomic regions analyzed (Figure S7D).
Figure 8. DNA is globally hypomethylated in db/db mice at P180, correlating with reduced SAMe levels.
(A) Proportion of 1-kb tiles with high (≥80%, green), intermediate (>20% and <80%, red) and low (≤20%, blue) percentage methylation levels. (B) Violin plot showing about a 2-fold greater number of hypomethylated tiling regions (blue) in db/db mice compared to db/+ mice. (C) Boxplot showing a decreased methylation level in differentially-methylated tiling regions in db/db mice. (D) GO analysis showing top Molecular and Cellular Function categories enriched in upregulated genes that are hypomethylated at tiling regions or gene-regulatory regions. (E) GO analysis showing top Molecular and Cellular Function categories enriched in all downregulated genes that are hypermethylated at tiling regions or gene-regulatory regions. (F, G) Dnmt1 is upregulated in nerves from db/db mice, as shown by (F) qPCR analysis and (G) Western blotting. (H) qPCR analysis showing relative transcript levels of enzymes involved in methionine cycle. Data is mean ± sem, *p < 0.01, n=3, Student’s t-test. (I) Western blot analysis confirming higher GNMT expression in db/db mice. (J) Table showing levels of the metabolites methionine, SAMe and SAH (pmol mg−1 of tissue), and the SAMe/SAH ratio in db/+ and db/db mice. Data is mean ± sem, *p<0.01, n=5, Student’s t-test.
Using gene expression datasets published previously(Pande et al., 2011), we found that genes with concomitant changes in DNA methylation and transcript levels were enriched for functional categories (Figure 8D and 8E, and Table S8) that were associated with the gene expression signature of these mice (Figure S7E–G). For example, for upregulated genes, there was a significant enrichment in the cellular proliferation category, which was also one of the most significant categories associated with hypomethylated genes. Lipid metabolism was another significantly enriched category (Table S8), similar to what we found during developmental myelination and in the Gnmt−/− mice.
Next, we examined the mechanisms that could contribute to these aberrant methylation patterns. We found that expression of the Dnmt1 mRNA and protein were both upregulated in db/db mice, whereas the expression of other DNA methyltransferases (Figure 8F and 8G) and demethylases (data not shown) were unaffected. This was consistent with DNA hypermethylation of several regions, but not with hypomethylation (Figure 8B and 8C). We thus examined if the predominantly hypomethylated phenotype could instead be associated with SAMe levels. We found that db/db mice expressed more GNMT (Figure 8H and 8I), and importantly, we found that the nerves from these mice had a lower SAMe content and SAMe/SAH ratio than control mice (Figure 8J). Interestingly, we found that this reduced SAMe content and SAMe/SAH ratio seen in the aged mice, was already present in 8 weeks old db/db mice, when neuropathy had developed, but not in 4 weeks old mice, which are characterized by diabetes but have not yet developed neuropathy (Figure S7H).
Based on our previous results, we propose that the global DNA demethylation seen in this model of diabetic neuropathy could result from the reduced SAMe levels and lower SAMe/SAH ratio in these mice, due to the increased GNMT expression. This could play a role in the pathogenesis of the disease, possibly even at the onset of the disease. It would be an interesting possibility, in future studies, to examine whether administering SAMe to db/db mice could help prevent the DNA hypomethylation, and consequently, the nerve pathology in these mice.
DISCUSSION
Schwann cell myelination is a highly orchestrated and complex developmental process, which requires coordinated and widespread changes in gene expression. The establishment and maintenance of this gene expression pattern is principally regulated by an elaborate network of TFs(Jessen and Mirsky, 2005; Pereira et al., 2012). Recently, several studies have shown that another level of transcriptional control is provided by recruitment of the NuRD and BAF chromatin-remodeling complexes to regulatory regions of key myelin genes(Hung et al., 2012; Weider et al., 2012), and possibly by histone deacetylation by HDAC1 and HDAC2(Pereira et al., 2012). In this study, using high-resolution genome-scale maps of methylation patterns, we found that DNA methylation also is likely to be an essential component of the global regulatory mechanisms regulating Schwann cell myelination.
The role of DNA methylation during Schwann cell myelination
We found that Schwann cell myelination was characterized by global demethylation, whereas gene expression changes showed about an equal number of upregulated and downregulated genes. This is consistent with DNA methylation being only one of other concerted factors in regulating gene expression(Portela and Esteller, 2010). Instead, similarly to other somatic cell differentiation systems(Bock et al., 2012; Calvanese et al., 2012; Lee et al., 2012), DNA demethylation was associated with activation of myelination-specific genes, notably at promoter regions and putative enhancer elements. This suggests that DNA methylation could prevent their aberrant expression in progenitor cells, and loss of methylation could promote their transcription. These demethylated genes were significantly enriched for lipid metabolism, which has a critical function in the generation of the lipid-rich myelin(Chrast et al., 2011).
In addition to DNA methylation, several different mechanisms, including TF binding and occupancy, and histone modifications, act coordinately to regulate gene transcription during Schwann cell myelination (Jessen and Mirsky, 2005; Pereira et al., 2012). Moreover, microRNAs and regulation of mRNA stability by RNA-binding proteins also can regulate mRNA levels (Iruarrizaga-Lejarreta et al., 2012; Pereira et al., 2012). It is likely that the selective activation of myelination-specific genes by DNA demethylation is regulated by a unique concerted action with several of these factors. This argues for a key function of DNA methylation as part of the complex network of transcriptional, post-transcriptional and epigenetic factors that drive the myelination program.
To perform these studies, we used the sciatic nerve as a model to profile DNA methylation changes during the Schwann cell myelination, a model that has previously been used to generate gene expression or microRNA profiles(Gokey et al., 2012; Verheijen et al., 2003). In addition to Schwann cells, the sciatic nerve consists of other cell types, including fibroblasts, and our data would likely include DNA methylation profiles of these cells. However, this effect could be minimal since Schwann cells represent the majority of the endoneurial cells of the nerve(Verheijen et al., 2003).
Genomic enrichment classification of DMRs showed that repeat elements were one of the most significant groups, similarly to erythropoesis, another model of somatic cell differentiation(Shearstone et al., 2011), and in the early embryo and during germline development(Smith et al., 2012). Endogenous transposable elements consist of three major classes: LINEs, SINEs and LTRs and constitute about 40% of the mammalian genome. DNA methylation is one of the primary mechanisms used for silencing repeat elements(Beck et al., 2011). There is strong evidence that transposable elements can influence host gene expression(Rebollo et al., 2012). Interestingly, in the brain, LINE-1 retrotransposition, which is controlled by DNA methylation, can occur and has been suggested to influence neuronal transcriptomes and function(Muotri et al., 2010). Although much remains to be learned in this area, it is an interesting possibility that demethylation of repeat elements during Schwann cell myelination could lead to a similar increase in retrotransposition that could play an important role in regulation of gene expression.
SAMe levels as a critical determinant of the methylation landscape
We found that DNA demethylation during Schwann cell myelination could be regulated by several mechanisms, including binding of critical TFs associated with Schwann cell myelination(Jessen and Mirsky, 2005; Pereira et al., 2012) to gene-regulatory regions, as established in classical studies(Smith and Meissner, 2013), or cell division, as shown for erythropoiesis(Shearstone et al., 2011), and by expression of DNMTs and DNA demethylases. In addition, we provide evidence that levels of SAMe, the principal methyl donor in methylation reactions, could also be a critical determinant in establishing DNA methylation patterns. Several studies have associated aberrant DNA methylation patterns with the availability of SAMe(Feil and Fraga, 2011). However, these studies have focused on locus-specific methylation changes. Our study, instead, provides a comprehensive analysis of genome-wide DNA methylation changes induced by alterations in SAMe levels in a biological process. We found that DNA demethylation during Schwann cell myelination correlates with a decrease in SAMe concentration, and importantly, elevated SAMe levels in vitro and in vivo lead to DNA hypermethylation globally and in several genomic regions, including repeat elements, gene promoters and bodies, and enhancers. Hypermethylation at gene-regulatory regions in vivo correlates with suppression of several genes, which were notably enriched for the lipid metabolism category. This was likely the cause of the abnormal lipidomic profile and myelination defects in these mice. The functional importance of lipid metabolism in peripheral myelination has been shown with mouse mutants and in human genetic conditions(Chrast et al., 2011). In one such study, genetic ablation of Scap, the transcriptional activator of SREBPs, which regulates lipogenesis, leads to congenital hypomyelination and an abnormal lipid profile, similar to the Gnmt−/− mice.
Post-translational modification of histones is another major component of the regulatory network that controls gene expression. Histone methylation can occur at different residues, and can lead to either gene activation or repression to control various biological processes(Greer and Shi, 2012). Several recent studies point to an important role of SAMe in regulating histone methylation(Shyh-Chang et al., 2013; Towbin et al., 2012; Ulanovskaya et al., 2013). Histone methylation is likely to play an important role in regulating gene expression in Schwann cells. However, further studies are required to determine the contribution of SAMe to establishing histone methylation patterns during Schwann cell myelination.
SAMe as part of the pathogenetic mechanisms of common diseases
Epigenetic mechanisms play a fundamental role in the pathogenesis of a growing list of human diseases, including cancer and neurological diseases(Portela and Esteller, 2010). Recently, it was shown that the leprosy bacterium can induce reprogramming of Schwann cells to cells with stem cell like properties, by altering DNA methylation status of critical genes(Masaki et al., 2013). Here, we show that diabetic neuropathy is characterized by an aberrantly demethylated DNA profile, possibly determined by the reduced SAMe concentration and SAMe/SAH ratio as a result of increased GNMT expression. SAMe levels are influenced by a variety of environmental factors, in addition to diabetes, including diet(Choi and Friso, 2010) and viral infections(Bottiglieri, 2002), which are also known to cause peripheral neuropathies. It is a distinct possibility that altered SAMe levels could contribute to the pathogenesis of these acquired neuropathies, through aberrant methylation of DNA and histones. Such a mechanism could also contribute to other neurological pathologies. For example, Alzheimer’s disease is also characterized by changes in DNA methylation patterns(Cyr and Domann, 2011), and recently decreased levels of SAMe were found in cerebrospinal fluid of patients with Alzheimer’s disease(Linnebank et al., 2010).
In conclusion, we show that myelination is characterized by DNA demethylation, which is associated with activation of differentiation-specific genes, and that local availability of SAMe is likely to play a critical role in regulating the methylation pattern during this process. Our results also suggest that SAMe could play a role in the establishment of the aberrant DNA methylation patterns in a mouse model of diabetic neuropathy. This study, together with other studies on the influence of SAMe on histone methylation, supports the view that the methylation landscape of cells could be critically dependent on concentrations of SAMe.
EXPERIMENTAL PROCEDURES
Animals
Mice and rats were housed at the Animal unit at CIC bioGUNE (AAALAC-accredited facility). All procedures were approved by the institutional committee on animal use. Sciatic nerves were isolated from C57 BL6/J mice at newborn (NB), post-natal day 10 (P10) and P60 ages. Gnmt−/− mice were described previously(Martinez-Chantar et al., 2008) and nerves isolated at P90. Db/db mice, as described previously(Pande et al., 2011), were purchased from Jackson Laboratories and nerves extracted at P180. Nerves from mice of either sex were used. Immediately after isolation, nerves were desheathed and flash-frozen in liquid nitrogen.
Reduced Representation Bisulfite sequencing
Preparation of RRBS libraries was adapted from published protocols(Gu et al., 2011), and libraries were sequenced by the CIC bioGUNE Genome Analysis Platform on a HiScanSQ platform (Illumina Inc.). Details of RRBS analysis and bioinformatic analysis are provided in Supplemental Experimental Procedures.
Gene Expression microarray analysis
Total RNA was isolated from different sets of nerves using standard techniques and cRNA libraries hybridized to the MOUSEWG-6 V2 BeadChips (Illumina Inc.). Data were processed as described(Iruarrizaga-Lejarreta et al., 2012). Adjustment of p-values was done by the determination of false discovery rates (FDR) using Benjamini-Hochberg procedure.
Primary Schwann cell culture & cAMP myelination assay
Schwann cells were isolated from P5 sciatic nerves of Wistar rats or P5-P8 sciatic nerves from C57 BL6/J mice, and purified and cultured as described before(Iruarrizaga-Lejarreta et al., 2012), except that horse serum was used instead of FBS for mouse Schwann cells. Only the first 5 passages were used. For myelination assays, a cAMP analogue, dibutryl cAMP (10−3 M), was added to cultures for 2 days. For mouse Schwann cells, the medium was also supplemented with NRG1 (10 ng ml−1).
Viral infection
Cells were treated with control lentiviral particles (pLKO.1) or short-hairpin lentiviral particles against GNMT (Sigma Mission clone: TRCN0000097601) in the presence of hexadimethrine bromide (8 μg ml−1). After 24h transduction, the cells were selected using puromycin (1.25 μg ml−1).
S-Adenosylmethionine treatment
SAMe, in the stable form of sulfate-p-toluensulfonate (Samyr) was obtained from Abbott or Sigma, and used freshly prepared at a concentration of 2mM.
In vitro RRBS analysis
(1) Purified mouse Schwann cells cultured under myelinogenic conditions were either treated or untreated for 2 days with SAMe, and then genomic DNA extracted for RRBS analysis. (2) Puromycin-selected shControl and shGNMT-silenced cells were switched to myelinogenic conditions for 2 days, and then genomic DNA extracted. For MDM treatment, shGNMT cells were switched to myelinogenic conditions for 2 days (medium containing 10 μm methionine) and then genomic DNA extracted.
Electron Microscopy and morphometric analyses
Processing of nerve samples was carried out as described(Woodhoo et al., 2009) and sections were viewed in a Jeol 1010 transmission electron microscope (TEM). See Supplemental Experimental Procedures for further details.
Metabolite measurements and profiling
For quantitative metabolite measurements (SAMe, SAH or methionine), several sets of nerves or Schwann cell cultures were pooled together and analysis performed(Martinez-Chantar et al., 2008) at CIC bioGUNE’s Metabolomics Platform. Metabolite profiling and analysis were performed at OWL metabolomics. In brief, three separate ultra-performance liquid chromatography–mass spectrometry based platforms were used for optimal profiling of: (1) Fatty acyls, bile acids and lysoglycerophospholipids, (2) Amino acids, and (3) Glycerolipids, cholesteryl esters, sphingolipids, and glycerophospholipids as described(Barr et al., 2012).
Supplementary Material
Acknowledgments
This work was supported by grants PI09/00094 and PI12/00005, cofinanced by the ISCIII-Subdirección General de Evaluación and Fondo Europeo de Desarrollo Regional (FEDER), Fundación Científica de la Asociación Española Contra el Cáncer (Cancer Infantil 2011), Departamento de Educación Política Lingüística y Cultura del Gobierno Vasco (PI2013-46) and the Ministerio de Economía y Competitividad – Plan Nacional de I+D+I 2008-2011 Subprograma Ramón y Cajal RYC2010-06901 to AW. International Joint Project grant from the Royal Society of Great Britain (to KRJ and AW), NIH AT-1576 (to SCL, MLMC and JMM), SAF 2011-29851 (to JMM), Sanidad Gobierno Vasco 2012 (to MVR), Wellcome Trust Program Grants (to KRJ. and RM), ETORTEK-2011 (to MLMC and AMA), Educación Gobierno Vasco 2011 (to MLMC), PI11/01588 (to MLMC), Innovation Technology Department of the Bizkaia County grant 2010 (to MLMC, JMM and AMA). DMA is supported by the Government of Navarra through the grant “Ayuda predoctoral para realizar una tesis doctoral y obtener el título de doctor (Plan de Formación y de I+D 2010/2011)”. CIBERehd is funded by the Instituto de Salud Carlos III. We thank C. Bock for expert advice on preparation of RRBS libraries, A. Carracedo for providing shGNMT lentiviral vectors, CIC bioGUNE’s Metabolomics Platform and OWL Metabolomics for metabolomic profiling, and N. Rodríguez-Ezpeleta for her contributions to the early parts of the project. José M Mato is a founder of OWL Metabolomics (www.owlmetabolomics.com) and a member of its scientific advisory board. None of the remaining authors have a financial interest related to this work.
Footnotes
Accession numbers. RRBS data is deposited at the NCBI Gene Expression Omnibus under accession number GSE45343. Microarray data has been deposited at GEO: developmental Schwann cell myelination (GSE45700) and Gnmt−/− mice (GSE45701). Link: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=hfszlwemwgqmkbi&acc=GSE45702
AUTHOR CONTRIBUTIONS
M.V.R., M.I.L. and A.W. performed the experiments and analysed the data. A.M.A. prepared RRBS libraries and J.J.L performed RRBS bioinformatics analysis. A.F.F., J.L.L., D.M., and A.W. performed bioinformatic analyses. M.T. helped with electron microscopy. M.B., S.C.L., M.E., R.M., K.R.J., M.F.F., Z.L., C.W., M.L.M.C. and J.M.M. gave technical support and conceptual advice. A.W. designed and supervised the study, wrote the manuscript and prepared figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barr J, Caballeria J, Martinez-Arranz I, Dominguez-Diez A, Alonso C, Muntane J, Perez-Cormenzana M, Garcia-Monzon C, Mayo R, Martin-Duce A, et al. Obesity-dependent metabolic signatures associated with nonalcoholic fatty liver disease progression. J Proteome Res. 2012;11:2521–2532. doi: 10.1021/pr201223p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C. Analysing and interpreting DNA methylation data. Nat Rev Genet. 2012;13:705–719. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- Bock C, Beerman I, Lien WH, Smith ZD, Gu H, Boyle P, Gnirke A, Fuchs E, Rossi DJ, Meissner A. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell. 2012;47:633–647. doi: 10.1016/j.molcel.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Tomazou EM, Brinkman AB, Muller F, Simmer F, Gu H, Jager N, Gnirke A, Stunnenberg HG, Meissner A. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010;28:1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottiglieri T. S-Adenosyl-L-methionine (SAMe): from the bench to the bedside--molecular basis of a pleiotrophic molecule. Am J Clin Nutr. 2002;76:1151S–1157S. doi: 10.1093/ajcn/76/5.1151S. [DOI] [PubMed] [Google Scholar]
- Calvanese V, Fernandez AF, Urdinguio RG, Suarez-Alvarez B, Mangas C, Perez-Garcia V, Bueno C, Montes R, Ramos-Mejia V, Martinez-Camblor P, et al. A promoter DNA demethylation landscape of human hematopoietic differentiation. Nucleic Acids Res. 2012;40:116–131. doi: 10.1093/nar/gkr685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SW, Friso S. Epigenetics: A New Bridge between Nutrition and Health. Adv Nutr. 2010;1:8–16. doi: 10.3945/an.110.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrast R, Saher G, Nave KA, Verheijen MH. Lipid metabolism in myelinating glial cells: lessons from human inherited disorders and mouse models. J Lipid Res. 2011;52:419–434. doi: 10.1194/jlr.R009761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr AR, Domann FE. The redox basis of epigenetic modifications: from mechanisms to functional consequences. Antioxid Redox Signal. 2011;15:551–589. doi: 10.1089/ars.2010.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2011;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- Gokey NG, Srinivasan R, Lopez-Anido C, Krueger C, Svaren J. Developmental regulation of microRNA expression in Schwann cells. Mol Cell Biol. 2012;32:558–568. doi: 10.1128/MCB.06270-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 2011;6:468–481. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- Hung H, Kohnken R, Svaren J. The nucleosome remodeling and deacetylase chromatin remodeling (NuRD) complex is required for peripheral nerve myelination. J Neurosci. 2012;32:1517–1527. doi: 10.1523/JNEUROSCI.2895-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iruarrizaga-Lejarreta M, Varela-Rey M, Lozano JJ, Fernandez-Ramos D, Rodriguez-Ezpeleta N, Embade N, Lu SC, van der Kraan PM, Blaney Davidson EN, Gorospe M, et al. The RNA-binding protein human antigen R controls global changes in gene expression during Schwann cell development. J Neurosci. 2012;32:4944–4958. doi: 10.1523/JNEUROSCI.5868-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R. The origin and development of glial cells in peripheral nerves. Nat Rev Neurosci. 2005;6:671–682. doi: 10.1038/nrn1746. [DOI] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Lee ST, Xiao Y, Muench MO, Xiao J, Fomin ME, Wiencke JK, Zheng S, Dou X, de Smith A, Chokkalingam A, et al. A global DNA methylation and gene expression analysis of early human B-cell development reveals a demethylation signature and transcription factor network. Nucleic Acids Res. 2012;40:11339–11351. doi: 10.1093/nar/gks957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnebank M, Popp J, Smulders Y, Smith D, Semmler A, Farkas M, Kulic L, Cvetanovska G, Blom H, Stoffel-Wagner B, et al. S-adenosylmethionine is decreased in the cerebrospinal fluid of patients with Alzheimer’s disease. Neurodegener Dis. 2010;7:373–378. doi: 10.1159/000309657. [DOI] [PubMed] [Google Scholar]
- Lu SC, Mato JM. S-adenosylmethionine in liver health, injury, and cancer. Physiol Rev. 2012;92:1515–1542. doi: 10.1152/physrev.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Chantar ML, Vazquez-Chantada M, Ariz U, Martinez N, Varela M, Luka Z, Capdevila A, Rodriguez J, Aransay AM, Matthiesen R, et al. Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology. 2008;47:1191–1199. doi: 10.1002/hep.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki T, Qu J, Cholewa-Waclaw J, Burr K, Raaum R, Rambukkana A. Reprogramming adult Schwann cells to stem cell-like cells by leprosy bacilli promotes dissemination of infection. Cell. 2013;152:51–67. doi: 10.1016/j.cell.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan R, Le N, Mahoney H, Araki T, Milbrandt J. Deciphering peripheral nerve myelination by using Schwann cell expression profiling. Proc Natl Acad Sci U S A. 2002;99:8998–9003. doi: 10.1073/pnas.132080999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande M, Hur J, Hong Y, Backus C, Hayes JM, Oh SS, Kretzler M, Feldman EL. Transcriptional profiling of diabetic neuropathy in the BKS db/db mouse: a model of type 2 diabetes. Diabetes. 2011;60:1981–1989. doi: 10.2337/db10-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JA, Lebrun-Julien F, Suter U. Molecular mechanisms regulating myelination in the peripheral nervous system. Trends Neurosci. 2012;35:123–134. doi: 10.1016/j.tins.2011.11.006. [DOI] [PubMed] [Google Scholar]
- Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, Mager DL. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet. 2012;46:21–42. doi: 10.1146/annurev-genet-110711-155621. [DOI] [PubMed] [Google Scholar]
- Shearstone JR, Pop R, Bock C, Boyle P, Meissner A, Socolovsky M. Global DNA demethylation during mouse erythropoiesis in vivo. Science. 2011;334:799–802. doi: 10.1126/science.1207306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Suter U, Scherer SS. Disease mechanisms in inherited neuropathies. Nat Rev Neurosci. 2003;4:714–726. doi: 10.1038/nrn1196. [DOI] [PubMed] [Google Scholar]
- Towbin BD, Gonzalez-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- Ulanovskaya OA, Zuhl AM, Cravatt BF. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat Chem Biol. 2013 doi: 10.1038/nchembio.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheijen MH, Chrast R, Burrola P, Lemke G. Local regulation of fat metabolism in peripheral nerves. Genes Dev. 2003;17:2450–2464. doi: 10.1101/gad.1116203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent AM, Callaghan BC, Smith AL, Feldman EL. Diabetic neuropathy: cellular mechanisms as therapeutic targets. Nat Rev Neurol. 2011;7:573–583. doi: 10.1038/nrneurol.2011.137. [DOI] [PubMed] [Google Scholar]
- Wang YC, Tang FY, Chen SY, Chen YM, Chiang EP. Glycine-N methyltransferase expression in HepG2 cells is involved in methyl group homeostasis by regulating transmethylation kinetics and DNA methylation. J Nutr. 2011;141:777–782. doi: 10.3945/jn.110.135954. [DOI] [PubMed] [Google Scholar]
- Weider M, Kuspert M, Bischof M, Vogl MR, Hornig J, Loy K, Kosian T, Muller J, Hillgartner S, Tamm ER, et al. Chromatin-remodeling factor Brg1 is required for Schwann cell differentiation and myelination. Dev Cell. 2012;23:193–201. doi: 10.1016/j.devcel.2012.05.017. [DOI] [PubMed] [Google Scholar]
- Woodhoo A, Alonso MB, Droggiti A, Turmaine M, D’Antonio M, Parkinson DB, Wilton DK, Al-Shawi R, Simons P, Shen J, et al. Notch controls embryonic Schwann cell differentiation, postnatal myelination and adult plasticity. Nat Neurosci. 2009;12:839–847. doi: 10.1038/nn.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhoo A, Sommer L. Development of the Schwann cell lineage: from the neural crest to the myelinated nerve. Glia. 2008;56:1481–1490. doi: 10.1002/glia.20723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.