Abstract
TGFβ plays a critical role in tendon formation and healing. While its downstream effector Smad3 has been implicated in the healing process, little is known about the role of Smad3 in normal tendon development or tenocyte gene expression. Using mice deficient in Smad3 (Smad3–/–), we show that Smad3 ablation disrupts normal tendon architecture and has a dramatic impact on normal gene and protein expression during development as well as in mature tendon. In developing and adult tendon, loss of Smad3 results in reduced protein expression of the matrix components Collagen 1 and Tenascin-C. Additionally, when compared to wild type, tendon from adult Smad3–/– mice shows a downregulation of key tendon marker genes. Finally, through in vitro work, we have established that Smad3 has the ability to physically interact with the critical transcriptional regulators Scleraxis and Mohawk. Together these results indicate a central role for Smad3 in normal tendon development and in the maintenance of mature tendon.
Keywords: Tendon, Smad3, Scleraxis, Mohawk, TGFβ
Introduction
Flexor tendon injuries cause significant morbidity in the working-age population with patients rarely regaining their pre-injury range of motion. Surgical repair of tendon injuries initially results in the formation of scar tissue rather than recreating true tendon tissue, leaving patients prone to both re-rupture and excessive scar formation that can impede motion. Elucidation of the pathways involved in normal tendon development may lead to the identification of factors that enhance surgical reconstruction of damaged tendons and allow us to recreate the normal biomechanical properties of tendon. Unfortunately, relatively little is known about the mechanisms directing the development of embryonic tendon progenitors into mature tendons. In addition, much remains to be learned about the factors that control tenocyte gene expression and maintain the unique material matrix properties of tendon.
The transforming growth factor-β (TGFβ) family of growth factors plays an essential role in tendon biology (1). TGFβ has been implicated in tendon development as well as in the pathogenesis of tendon injury and healing (2). Prior studies have revealed that simultaneous ablation of both TGFβ2 and TGFβ3 signaling in mouse embryos prevents development of almost all tendons and ligaments (3). Similar results were observed following inactivation of the TGFβ type II receptor (3). TGFβ also induces key markers of tenocyte differentiation including Scleraxis (Scx) and type I Collagen in vivo (3; 4). In addition, TGFβ regulates the expression of matrix metalloproteinases, which are essential for tendon function and maturation (4-7). Other studies have revealed a key role for myostatin (GDF-8), another member of the TGFβ family, in tendon development in mice (8). Finally, in a rabbit model of flexor tendon injury, TGFβ was found to be upregulated at the site of injury, implicating the growth factor in the processes of injury-repair and scar formation (2). Similar effects were found in a mouse model of tendon repair (9).
TGFβ acts by binding to a complex of type I and type II transmembrane serine/threonine kinases. The receptor/ligand complex then phosphorylates several intracellular effectors that regulate cell differentiation. Although both TGFβ and myostatin induce the phosphorylation of common downstream effectors, including Smad2, Smad3, Erk1/2 and p38 MAP kinases, which of these effectors is responsible for the tendon phenotypes of the TGFβ and myostatin-deficient mice remains unclear. In other tissues derived from the mesenchymal lineage, TGFβ acts via Smad3 to regulate the function of key lineage specific transcription factors. For example, in bone, Smad3 represses the transcriptional activity of the osteogenic transcription factor Runx2, which in turn regulates osteoblast differentiation and bone matrix material properties (10). In cartilage, Smad3 regulates the critical chondrogenic transcription factor Sox9, while in muscle it regulates the myogenic transcription factor MyoD (11; 12). In this way, TGFβ-activated Smad3 is a major regulator of cell differentiation in tissues of the musculoskeletal system including bone, cartilage, and muscle (10-13). Therefore, Smad3 may play a similar role in the TGFβ-dependent regulation of tenocyte differentiation.
TGFβ-activated Smad3 had already been shown to induce the mRNA expression of the tenocyte lineage-specific transcription factor Scx in avian mesenchymal cultures (14). However, the ability of Smad3 to regulate Scx activity in order to control tendon cell differentiation remains unknown. A second tenocyte lineage-specific transcription factor, Mohawk (Mkx), has recently been identified and found to be necessary for the development of tendons during embryogenesis as well as for tendon maturation (15). The ability of Smad3 to regulate Mkx expression also remains to be elucidated.
Smad3 signaling has been shown to play a role in tendon healing as well as in the maintenance of tendon mechanical properties (16). Although deletion of Smad3 decreases adhesion formation in repaired tendons, tendons in these same Smad3-deficient mice exhibit diminished tensile strength and abnormal collagen deposition following injury (16). The mechanisms by which Smad3 asserts these affects in normal and injured tendon remain unknown.
Given that Smad3 plays a key role in the differentiation of other musculoskeletal tissues as well as in tendon healing, we hypothesized that Smad3 is a key regulator of tendon development and of tenocyte gene expression. We used a combination of in vitro and in vivo approaches to determine the expression and function of Smad3 during embryonic development and the effect of Smad3 ablation on tendon development and tenocyte gene expression. Furthermore, we investigated the ability of Smad3 to interact directly with the tendon transcription factors Scx and Mkx.
Methods
Mice
All animal procedures were performed in accordance with University of California, San Francisco Institutional Care and Use Committee-approved protocols. Smad3-deficient (Smad3–/–) mice were obtained from Dr. X.-F. Wang (17). SBE-Luciferase mice express a luciferase reporter gene under control of 12 copies of a Smad2/3 binding sequence from the Smad7 promoter (18).
Histology, Immunohistochemistry, and In situ Hybridization
Achilles and flexor digitorum profundus (FDP) tendons were harvested from 5-8 week old male mice, while embryos were harvested at E16.5 and forelimbs were disarticulated for further processing. Harvested tissues were fixed in 4% paraformaldehyde at 4° C overnight and prepared for sectioning. Achilles tendons were dehydrated and embedded in paraffin while FDP tendons were embedded in OCT for frozen sectioning. E16.5 forelimbs were embedded in paraffin as well as OCT. FDP tendons were used for immunohistochemistry experiments because their morphology was better maintained during frozen sectioning, while Achilles tendons were used for paraffin sectioning.
To analyze tissue morphology, longitudinal sections of FDP tendon were stained with hematoxylin and eosin. Frozen sections of FDP tendons and E16.5 forelimbs were analyzed by immunohistochemistry using antibodies for Smad3 (Abcam, ab63577), Collagen 1 (Abcam, ab292), Tenascin-C (gift of H. Erickson, Duke University) and luciferase (Abcam, ab21176). Sections were rinsed, dehydrated, and mounted using standard procedures.
In situ hybridization was performed on paraffin sections of adult Achilles tendon and E16.5 forelimbs as described (19). 35S-labeled antisense riboprobes were generated to mouse Collαl (gift of E. Vuorio, University of Finland), Mkx (gift of R. Jiang, Cincinnati Children's Hospital Medical Center), and Scx and Tenomodulin (Tnmd) (gifts of R. Schweitzer, Shriner's Hospital for Children) (20-22). Sections were counterstained with Hoechst nuclear dye (Sigma). Hybridization signals were detected using darkfield illumination and the nuclear stain detected with epifluorescence. All figures show representative images of N ≥ 3 mice.
Quantitative Reverse Transcriptase-PCR (qPCR)
For analysis of gene expression, tail, flexor, and tibialis anterior tendons were isolated from 6-8 week old male mice. The tendon sheath and surrounding soft tissue were carefully removed with the aid of loupe magnification. The tendons were snap frozen and stored in liquid N2. Frozen tendons were placed into Trizol and homogenized with a dounce homogenizer. After homogenization, RNA was extracted using the Purelink RNA Mini Kit (Invitrogen). cDNA was reverse transcribed from the RNA using the iScript cDNA synthesis kit (BioRad). Transcripts were amplified using the following primers: Smad3 (Fwd: 5′-ACCAAGTGCATTACCATCC -3′; Rev: 5′-CAGTAGATAACGTGAGGGAGCCC -3′), Col1α1 (Fwd: 5′-GCATGGCCAAGAAGACATCC-3′; Rev: 5′-CCTCGGGTTTCCACGTCTC-3′), Tnc (Fwd: 5′-AGGCGATCCCAGCCAGTCAGT -3′; Rev: 5′-ATGGACGGGGCACCTCCTGTC-3′), Tnmd (Fwd: 5′-TGTACTGGATCAATCCCACTCT-3′; Rev: 5′-GCTCATTCTGGTCAATCCCCT-3′), Scx (Fwd: 5′-CCTTCTGCCTCAGCAACCAG-3′; Rev: 5′-GGTCCAAAGTGGGGCTCTCCGTGAC-3′) Mkx (Fwd: 5′-GACTCCGAGGCTCTGCCGCAA-3′; Rev: 5′-CAGGAGTCGCCATCGCTGCTCA-3′).
Results were detected based on amplicon binding of SYBR Green (BioRad) using the CFX96 Real-Time PCR Detection System (BioRad). Fold change was calculated using the delta-delta Ct method based on the average of technical duplicates (23). Significance was determined using a two-tailed student's t-test with a threshold of p=0.05.
Cell Culture and Immunoprecipitation
C3TH10T1/2 cells (ATCC) were cultured and maintained in Basal Medium Eagle supplemented with 10% FBS and 2 mM L-Glutamine at 37°C and 5% CO2. Cells were transfected using lipofectamine with expression vectors for Scx, Mkx, and/or Smad3. Smad3 tagged with either Flag or HA epitopes at the N-terminus were cloned into a pRK5 expression vector. Scx tagged with both HA and Flag tags at the N-terminus was cloned into a pRK5 expression vector. Mkx-Myc expression vector was a gift of A. Rawls, Arizona State University (24). Cells grew for 18 hours following transfection before treatment with TGFβ (5ng/ml) for 3 hours. Cells were rinsed with PBS and lysed in 50 mM Tris pH 7.5, 150 mM NaCl, and 0.5% Triton X-100 for 15 minutes at 4° C. Lysates were collected and briefly sonicated before centrifugation. Complexes were immunoprecipitated by Flag-M2 affinity gel (Sigma, A2220) before analysis by western blot. Images are representative of three independent experiments.
Results
Smad3 is required for normal tendon formation
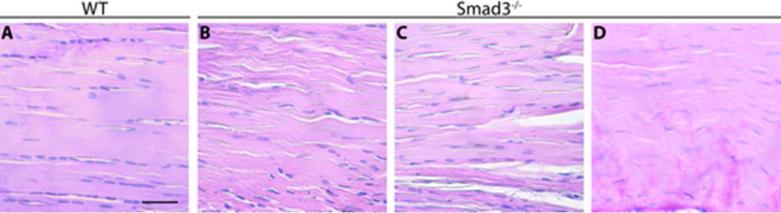
Given that Smad3 is highly involved in tendon formation and healing, we assessed the requirement for Smad3 in normal tendon formation using a Smad3–/– mouse model (3; 16). Histologic analysis revealed that while tendons from wild type mice (Fig. 1A) displayed normal matrix organization, tendons from Smad3–/– mice displayed crimped fibers (Fig. 1B), large gaps within the tendon (Fig. 1C), and a high degree of cellular disorganization (Fig. 1D). These results suggest that Smad3 is essential for the formation of normal tendon architecture.
Figure 1. Loss of Smad3 disrupts normal tendon development.
H&E staining of flexor digitorum profundus (FDP) tendon from 8-week-old mice reveals that relative to wild type (WT) (A), Smad3–/– mice display crimped fibers (B), gaps (C), and cellular disorganization (D). Images are representative of N≥3 WT and Smad3–/– mice. Scale bar=20 μm.
Smad3 is required for normal gene and protein expression in adult tendon
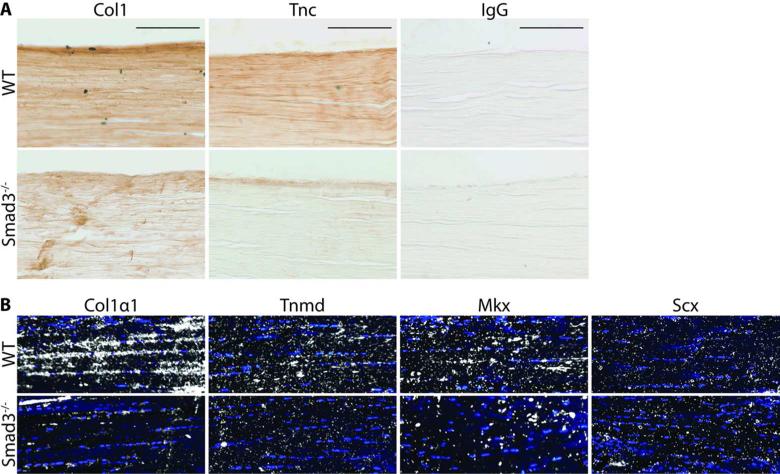
Given the impact of Smad3 ablation on normal tendon matrix organization, we next examined whether the fibrillar disorganization observed in Smad3–/– tendons was associated with abnormal tendon extracellular matrix protein expression, such as Collagen 1 (Col1) and Tenascin-C (Tnc). Immunohistochemistry of longitudinal sections of FDP tendons revealed significant reductions in the expression of Col1 and Tnc in Smad3–/– tendons relative to wild type tendons (Fig. 2A). This finding suggests that Smad3 regulates tendon matrix protein expression.
Figure 2. Loss of Smad3 disrupts normal tendon matrix protein and gene expression in mature tendon.
(A) Protein expression of Col1 and Tnc in WT and Smad3–/– FDP tendons. The specificity of staining is indicated by the IgG control. Scale bar=100 μm. (B) In situ hybridization experiments show mRNA expression of Collαl, Tnmd, Mkx and Scx in Achilles tendon from adult WT and Smad3–/– mice. Images are representative of N≥3 mice.
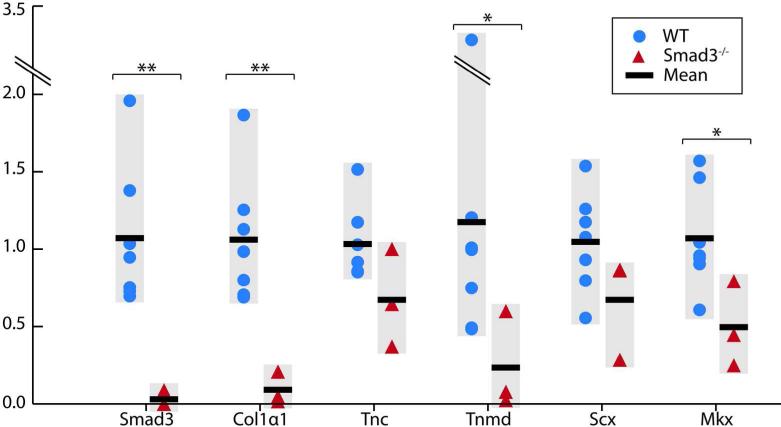
In order to determine whether this decrease in protein expression was accompanied by a corresponding reduction in gene expression, in situ hybridization analyses were performed on adult Achilles tendon, revealing reduced expression of genes encoding Collagen 1 (Collαl) and the collagen packaging regulator Tenomodulin (Tnmd) (Fig. 2B). In addition, while we could not detect a difference in Scx expression, we did observe a decrease in expression of the tendon transcription factor Mkx in Smad3–/– tendon relative to wild type (Fig. 2B). In order to quantify this reduction, we examined gene expression by qPCR. The expression of Collαl (p≤0.001), Tnmd (p≤0.05), and Mkx (p≤0.05) in Smad3–/– tendon was significantly reduced relative to wild type (Fig. 3). While the expression of Tnc and Scx appeared to be slightly reduced in Smad3–/– tendon, these changes were not statistically significant. The dramatically reduced expression of key tendon marker genes in the absence of Smad3 strongly suggests that Smad3 is required for the maintenance of adult tendon.
Figure 3. Expression of key tendon genes is dramatically reduced with the loss of Smad3.
Gene expression analysis by qPCR of Smad3, Collαl, Tnc, Tnmd, Scx, and Mkx in WT and Smad3–/– tendon. Gene expression data for individual mice (WT: N=7, Smad3–/–: N=3) is shown alongside the mean for each genotype. * and ** indicate significant differences of p≤0.05 and p≤0.001 respectively.
Smad3 is requiredfor normal gene and protein expression during tendon development
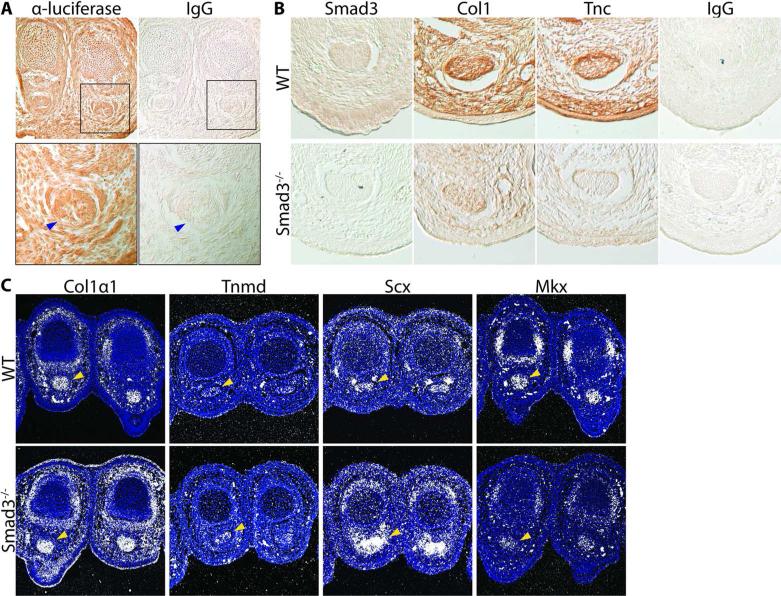
Given the effects of Smad3 deletion in adult tendon, we investigated the role of Smad3 during tendon development. First we assessed overall Smad3 activity during embryonic development using a reporter mouse model. Embryos carrying a luciferase reporter gene driven by a Smad2/3 responsive element were harvested at E16.5. Immunohistochemistry against luciferase revealed broad Smad2/3 activity throughout the limb bud including in the FDP tendon (Fig. 4A).
Figure 4. Loss of Smad3 results in reduced tendon matrix protein and gene expression in the developing tendon.
(A) Mice with a Smad2/3 responsive element driving a luciferase reporter gene display broad luciferase expression across the limb bud and in the FDP tendon (blue arrow) as analyzed by immunohistochemistry at E16.5. (B) Immunohistochemistry of axial sections of the mouse embryonic forelimb at E16.5 show protein expression of Smad3, Coll, and Tnc, in WT and Smad3–/– FDP tendon. The IgG control indicates the specificity of staining. (C) In situ hybridization of axial limb sections of WT and Smad3–/– E16.5 embryos show expression of Collαl, Tnmd, Scx, and Mkx mRNA in the FDP tendon (yellow arrow). Images are representative of N=3 mice.
To further assess the functional role of Smad3 activity in tendon development, the expression of key tendon matrix proteins Coll and Tnc was analyzed in Smad3–/– embryos. As expected, immunohistochemistry of E16.5 forelimbs revealed an absence of Smad3 expression in Smad3–/– relative to wild type (Fig. 4B). In wild type mice, we observed broad expression of Coll and Tnc throughout the limb bud with enrichment in the tendon. In contrast, in Smad3–/– limbs, we observed a reduction in Col1 and Tnc expression across the limb as well as within the tendon (Fig. 4B).
Next, the expression of genes encoding key tendon transcription factors and extracellular matrix proteins in the absence of Smad3 was analyzed by in situ hybridization. These experiments revealed an upregulation of RNA encoding Collαl and Scx in Smad3–/– mice relative to wild type, in contrast to the reduced levels of Coll protein expression observed in Smad3–/– tendon (Fig. 4C). In addition, Tnmd expression appeared slightly elevated in the Smad3–/– tendon relative to wild type. However, mRNA expression for the gene Mkx was found to be decreased in Smad3–/– embryos (Fig. 4C).
Smad3 physically interacts with the tendon transcription factors Scleraxis and Mohawk
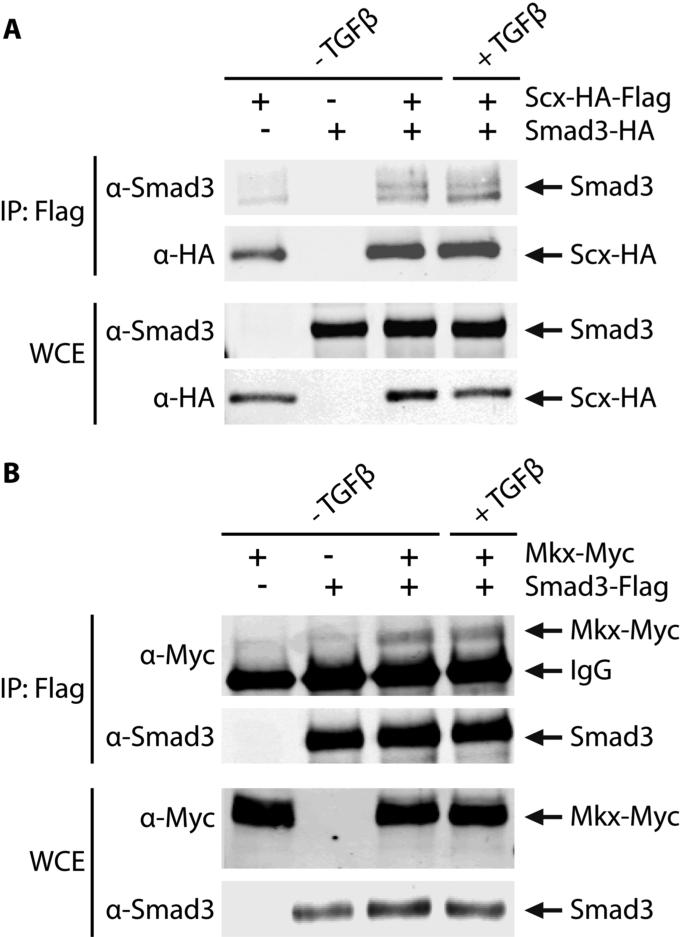
Given the profound impact that Smad3 ablation has on gene expression in tendon, we sought to understand the mechanism by which Smad3 exerts its effects. In other tissues of the musculoskeletal system, including bone and cartilage, Smad3 has been shown to regulate differentiation through direct interaction with tissue-specific transcription factors (13; 25). We therefore hypothesized that Smad3 may impact the expression of key tendon marker genes in a similar manner by interacting directly with the tendon-specific transcriptional regulators Scx and Mkx. Co-immunoprecipitation experiments were performed to analyze the capability of Smad3 to physically interact with both Scx and Mkx.
In order to assess physical interactions between Smad3 and Scx, tagged versions of Smad3 and Scx were overexpressed in 10T1/2 cells. Immunoprecipitation of lysates with anti-Flag antibodies resulted in the co-precipitation of Smad3 with Flag-tagged Scx (Fig. 5A). The presence of Smad3 following the immunoprecipitation of Scx indicates that these two proteins can physically interact. To determine if Smad3 interacted similarly with the tendon transcription factor, Mkx, Myc-tagged Mkx was overexpressed in 10T1/2 cells along with Flag-tagged Smad3. Immunoprecipitation of lysates with anti-Flag antibodies and detection by western blot analysis revealed a similar interaction between Smad3 and Mkx in both the presence and absence of exogenous TGFβ (Fig. 5B). Neither of these complexes appeared to be affected by the addition of exogenous TGFβ under these conditions; however, this does not preclude the possibility that TGFβ may regulate complex formation in vivo.
Figure 5. Smad3 binds Scx and Mkx.
10T1/2 cells were transfected with the indicated plasmids (A: Smad3 and/or Scx; B: Smad3 and/or Mkx) and treated with TGFβ or vehicle control. Lysates were immunoprecipitated with anti-Flag antibodies. Smad3 binds Scx (A) and Mkx (B) as indicated by the coRprecipitating band in the immunoprecipitated (IP) fraction. Whole cell extract (WCE) shows total protein expression.
Discussion
Our work has elucidated a critical role for Smad3 in normal tendon development and in the regulation of tenocyte marker genes. Loss of Smad3 affects not only the expression of key tenocyte proteins, but also the organization of collagen fibrils within the extracellular matrix. The collagen fibers in mature tendons of Smad3–/– mice showed increased crimping and gap formation. Furthermore, immunohistochemistry revealed that expression of the tendon matrix proteins Col1 and Tnc are also diminished. This change in the normal collagen architecture and tendon matrix protein expression may at least in part explain the findings of previous studies that loss of Smad3 affects the biomechanical properties of tendon (16).
In mature tendon, Smad3 ablation resulted in diminished expression of the mRNA of major tenocyte marker genes including the tendon transcription factor Mkx. The effect of Smad3 deficiency on transcription factor expression suggests that Smad3 may impact the expression of tenocyte specific genes not only in a direct fashion but also indirectly via its regulation of the expression of Scx and Mkx. Furthermore, we found that Smad3 has the ability to directly interact with both Scx and Mkx. Thus, Smad3 may exert its key regulatory role in tendon via its physical interaction with these tendon-specific transcription factors.
Smad3 plays a critical role in the differentiation of other tissues of the musculoskeletal system. In bone, Smad3 binds the osteogenic transcription factor Runx2 to confer TGFβ-mediated inhibition of terminal osteoblast differentiation (25). Smad3 is also crucial to TGFβ-mediated regulation of bone mineral matrix properties (10). Similarly, during chondrocyte differentiation, Smad3 binds Sox9 to promote chondrogenic gene expression (13). Likewise, Smad3 participates in the regulation of myocyte and adipocyte differentiation by interactions with the lineage-specific transcription factors MyoD and C/EBPb respectively (12; 26). Although the functional role of this interaction remains to be defined, our findings strongly suggest that Smad3 functions in a similar manner in tendon by binding the tenocyte-specific transcription factors Scx and Mkx. In other mesenchymally-derived cells, Smad3 exerts its effects on the expression of lineage specific genes by recruitment of transcriptional co-activators such as CREB binding protein during chondrogenesis or co-repressors such as class IIa histone deacytylases in osteogenesis (13; 27). However, the effect of Smad3 interaction with Scx or Mkx on transcriptional co-regulator recruitment remains to be elucidated.
Embryonic immunohistochemistry studies reveal that Smad2/3 activity is widespread throughout the developing limb bud. Interestingly, in situ hybridization analysis of the developing limb revealed an upregulation of Scx and Collαl mRNA in the absence of Smad3 in contrast to our adult in situ data. The mechanism and significance of this apparent discrepancy are unclear. It is possible that the mechanisms controlling expression of the tenocyte genes are differentially sensitive to Smad3 in embryonic and adult tissue at different times. Alternatively, upregulation of an alternative pathway during development may be able compensate for the loss of Smad3. Although our data show that expression of Collαl mRNA in the developing limb is upregulated in the absence of Smad3, immunohistochemistry reveals that Collagen1 protein levels are reduced with loss of Smad3 during development. It is therefore possible that the post-transcriptional regulation of tenocyte genes during embryonic development is Smad3-dependent. For example, Smad3 is known to regulate the expression and activity of several miRNAs as well as of enzymes responsible for matrix protein modification and secretion in other tissues (10; 28). Smad3 may similarly play a post-transcriptional regulatory role in the developing limb.
Although previous studies have suggested that Smad3 activity increases the formation of pathologic adhesions after tendon injury, our findings suggest that Smad3 is also critical for normal tendon development (16). TGFβ similarly plays a role in both pathologic adhesion formation and in normal tendon development. Therefore, TGFβ may exert its crucial regulation of both tendon differentiation and adhesion formation via a Smad3-dependent pathway. The formation of flexor tendon adhesions after tendon injury and repair may be induced by the expression of factors from the tissues of the surrounding tendon sheath rather than from within the tendon itself. Thus, it is possible for Smad3 activity to induce scar and adhesion formation when expressed by the tissues of the surrounding tendon sheath while promoting tendon healing when expressed within the tendon itself. Our study adds to the growing body of evidence indicating that the same pathways may regulate tendon healing and tendon adhesion formation. Further studies differentiating the effects of Smad3 expression in cells of the surrounding tendon sheath from those of the tenocytes themselves may provide some clue in how to modulate this pathway to prevent adhesion formation while promoting tendon healing.
Acknowledgements
We thank Melissa Ehlers and Brian Black for plasmids and Ashton Mortazavi for assistance with histology. This work was supported in part by research grants from the NIH (NIDCR R01 DE019284 TA, NIDCR R01 DE016402 RS, NIDCR COHORT T32 CC), the American Society for Surgery of the Hand (MA), and the Resource Allocation Program at the University of California, San Francisco (MA).
References
- 1.Derynck R, Schneider RA, Piek E, Alliston T. TGF-β family signaling in mesenchymal differentiation. In: Derynck R, Miyazono K, editors. The TGF-β Family. Cold Spring Harbor Press; Woodbury, NY: 2008. pp. 613–655. [Google Scholar]
- 2.Chang J, Most D, Stelnicki E, et al. Gene Expression of Transforming Growth Factor Beta-1 in Rabbit Zone II Flexor Tendon Wound Healing: Evidence for Dual Mechanism of Repair. Plastic and Reconstructive Surgery. 1997;100:937–944. doi: 10.1097/00006534-199709001-00016. [DOI] [PubMed] [Google Scholar]
- 3.Pryce BA, Watson SS, Murchison ND, et al. Recruitment and maintenance of tendon progenitors by TGF(beta) signaling are essential for tendon formation. Development. 2009;136:1351–1361. doi: 10.1242/dev.027342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan KM, Fu SC, Wong YP, et al. Expression of transforming growth factor beta isoforms and their roles in tendon healing. Wound Repair Regen. 2008;16:399–407. doi: 10.1111/j.1524-475X.2008.00379.x. [DOI] [PubMed] [Google Scholar]
- 5.Harris SE, Bonewald LF, Harris MA, et al. Effects of transforming growth factor beta on bone nodule formation and expression of bone morphogenetic protein 2, osteocalcin, osteopontin, alkaline phosphatase, and type I collagen mRNA in long-term cultures of fetal rat calvarial osteoblasts. J Bone Miner Res. 1994;9:855–863. doi: 10.1002/jbmr.5650090611. [DOI] [PubMed] [Google Scholar]
- 6.Rydziel S, Varghese S, Canalis E. Transforming growth factor beta1 inhibits collagenase 3 expression by transcriptional and post-transcriptional mechanisms in osteoblast cultures. J Cell Physiol. 1997;170:145–152. doi: 10.1002/(SICI)1097-4652(199702)170:2<145::AID-JCP6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 7.Jung J, Wang P, Zhang G, et al. Collagen fibril growth during chicken tendon development: matrix metalloproteinase-2 and its activation. Cell Tissue Research. 2009;336:79–89. doi: 10.1007/s00441-009-0755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendias CL, Bakhurin KI, Faulkner JA. Tendons of myostatin-deficient mice are small, brittle, and hypocellular. Proc Natl Acad Sci U S A. 2008;105:388–393. doi: 10.1073/pnas.0707069105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsubone T, Moran S, Subramaniam M, et al. Effect of TFG-B Inducible Early Gene Deficiency on Flexor Tendon Healing. Journal of Orthopaedic Research. 2006;24:569575. doi: 10.1002/jor.20101. [DOI] [PubMed] [Google Scholar]
- 10.Balooch G, Balooch M, Nalla RK, et al. TGF-beta regulates the mechanical properties and composition of bone matrix. Proc Natl Acad Sci U S A. 2005;102:18813–18818. doi: 10.1073/pnas.0507417102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furumatsu T, Tsuda M, Yoshida K, et al. Sox9 and p300 cooperatively regulate chromatin-mediated transcription. J Biol Chem. 2005;280:35203–35208. doi: 10.1074/jbc.M502409200. [DOI] [PubMed] [Google Scholar]
- 12.Liu D, Black BL, Derynck R. TGF-beta inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. 2001. submitted. [DOI] [PMC free article] [PubMed]
- 13.Furumatsu T, Tsuda M, Taniguchi N, et al. Smad3 induces chondrogenesis through the activation of SOX9 via CREB-binding protein/p300 recruitment. J Biol Chem. 2005;280:8343–8350. doi: 10.1074/jbc.M413913200. [DOI] [PubMed] [Google Scholar]
- 14.Lorda-Diez C, Montero J, Martinez-Cue C, et al. Transforming growth factors beta coordinate cartilage and tendon differentiation in the developing limb mesenchyme. Journal of Biological Chemistry. 2009 doi: 10.1074/jbc.M109.014811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu W, Watson S, Lan Y, et al. The Atypical Homeodomain Transcription Factor Mohawk Controls Tendon Morphogenesis. Molecular and Cellular Biology. 2010;30:4797–4807. doi: 10.1128/MCB.00207-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katzel EB, Wolenski M, Loiselle AE, et al. Impact of Smad3 Loss of Function on Scarring and Adhesion Formation During Tendon Healing. Journal of Orthopaedic Research. 2010 doi: 10.1002/jor.21235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borton AJ, Frederick JP, Datto MB, et al. The loss of Smad3 results in a lower rate of bone formation and osteopenia through dysregulation of osteoblast differentiation and apoptosis. J Bone Miner Res. 2001;16:1754–1764. doi: 10.1359/jbmr.2001.16.10.1754. [DOI] [PubMed] [Google Scholar]
- 18.Lin AH, Luo J, Mondshein LH, et al. Global analysis of Smad2/3-dependent TGF-beta signaling in living mice reveals prominent tissue-specific responses to injury. J Immunol. 2005;175:547–554. doi: 10.4049/jimmunol.175.1.547. [DOI] [PubMed] [Google Scholar]
- 19.Albrecht UEG. Visualization of gene expression patterns by in situ hybridization. In: Gaston GP, editor. Molecular and Cellular Methods in Developmental Toxicology. CRC Press; Boca Raton: 1997. pp. 23–48. [Google Scholar]
- 20.Metsaranta M, Toman D, De Crombrugghe B, Vuorio E. Specific hybridization probes for mouse type I, II, III and IX collagen mRNAs. Biochim Biophys Acta. 1991;1089:241–243. doi: 10.1016/0167-4781(91)90014-d. [DOI] [PubMed] [Google Scholar]
- 21.Liu W, Watson SS, Lan Y, et al. The atypical homeodomain transcription factor Mohawk controls tendon morphogenesis. Mol Cell Biol. 2010;30:4797–4807. doi: 10.1128/MCB.00207-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murchison ND, Price BA, Conner DA, et al. Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development. 2007;134:2697–2708. doi: 10.1242/dev.001933. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using realtime quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Anderson DM, Beres BJ, Wilson-Rawls J, Rawls A. The homeobox gene Mohawk represses transcription by recruiting the sin3A/HDAC co-repressor complex. Dev Dyn. 2009;238:572–580. doi: 10.1002/dvdy.21873. [DOI] [PubMed] [Google Scholar]
- 25.Alliston T, Choy L, Ducy P, et al. TGF-beta-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. The EMBO journal. 2001;20:2254–2272. doi: 10.1093/emboj/20.9.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choy L, Derynck R. Transforming Growth Factor-B Inhibits Adipocyte Differentiation by Smad3 Interacting with CCAAT/Enhancer-binding Protein (C/EBP) and Repressing C/EBP Transactivation Function. Journal of Biological Chemistry. 2003;278:9609–9619. doi: 10.1074/jbc.M212259200. [DOI] [PubMed] [Google Scholar]
- 27.Kang JS, Alliston T, Delston R, Derynck R. Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. Embo J. 2005;24:2543–2555. doi: 10.1038/sj.emboj.7600729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin W, Chung A, Huang X, et al. TGF-b/Smad3 signaling promotes renal fibrosis by inhibiting miR-39. Journal of the American Society of Nephrology. 2011;22:1462–1474. doi: 10.1681/ASN.2010121308. [DOI] [PMC free article] [PubMed] [Google Scholar]