Abstract
FosB is a divalent-metal-dependent thiol-S-transferase implicated in fosfomycin resistance among many pathogenic Gram-positive bacteria. In the present paper, we describe detailed kinetic studies of FosB from Staphylococcus aureus (SaFosB) that confirm that bacillithiol (BSH) is its preferred physiological thiol substrate. SaFosB is the first to be characterized among a new class of enzyme (bacillithiol-S-transferases), which, unlike glutathione transferases, are distributed among many low-G + C Gram-positive bacteria that use BSH instead of glutathione as their major low-molecular-mass thiol. The Km values for BSH and fosfomycin are 4.2 and 17.8 mM respectively. Substrate specificity assays revealed that the thiol and amino groups of BSH are essential for activity, whereas malate is important for SaFosB recognition and catalytic efficiency. Metal activity assays indicated that Mn2+ and Mg2+ are likely to be the relevant cofactors under physiological conditions. The serine analogue of BSH (BOH) is an effective competitive inhibitor of SaFosB with respect to BSH, but uncompetitive with respect to fosfomycin. Coupled with NMR characterization of the reaction product (BS– fosfomycin), this demonstrates that the SaFosB-catalysed reaction pathway involves a compulsory ordered binding mechanism with fosfomycin binding first followed by BSH which then attacks the more sterically hindered C-1 carbon of the fosfomycin epoxide. Disruption of BSH biosynthesis in S. aureus increases sensitivity to fosfomycin. Together, these results indicate that SaFosB is a divalent-metal-dependent bacillithiol-S-transferase that confers fosfomycin resistance on S. aureus.
Keywords: antibiotic detoxification, bacillithiol, metalloenzyme, substrate mimic
INTRODUCTION
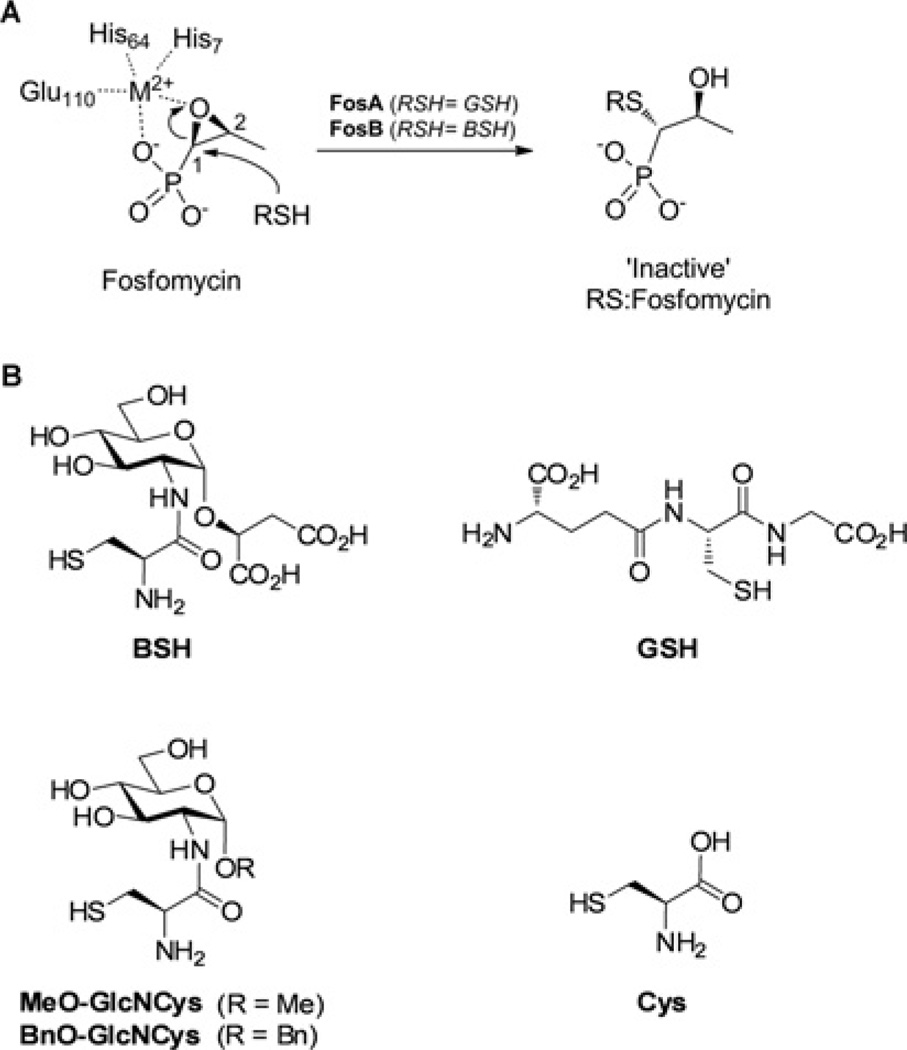
The antibiotic fosfomycin [(1R,2S)-epoxypropylphosphonic acid] possesses broad-spectrum activity against both Gram-positive and Gram-negative bacteria [1]. It is a covalent inhibitor of MurA, a key enzyme that mediates the first obligate step in peptidoglycan biosynthesis [2]. Covalent inhibition of MurA results from nucleophilic attack by an active-site cysteine thiol at the less hindered C-2 position of the fosfomycin epoxide [3]. To date, three different types of fosfomycin-resistance enzymes have been identified (FosA, FosB and FosX), which can inactivate fosfomycin by ring opening of the epoxide motif [4]. FosX is a metal-dependent hydrolase found in Mesorhizobium loti and Listeria monocytogenes, which catalyses the hydrolysis of the epoxide [5,6]. FosA has been characterized as a manganese-dependent glutathione transferase which catalyses glutathione (GSH)-dependent ring opening at the more sterically hindered C-1 position of the fosfomycin epoxide to form an inactive GS–fosfomycin conjugate (Figure 1A) [7–10]. Both plasmid- and chromosome-encoded fosA genes have been identified in various Gram-negative bacteria [9,11,12].
Figure 1. Thiol-S-transferase-catalysed inactivation of fosfomycin.
(A) Fosfomycin epoxide activation by M2+ (based on the Pseudomonas aeruginosa FosA structure numbering) and regioselective ring opening mechanism of FosA and FosB enzymes [42]. Conserved metal-chelating residues for SaFosB are His7, His66 and Glu115 (see Supplementary Figure S5 at http://www.biochemj.org/bj/451/bj4510069add.htm for the sequence alignment). (B) Structures of BSH, GSH, cysteine and the BSH derivatives MeO-GlcNCys and BnO-GlcNCys.
FosB is a thiol-S-transferase related to FosA. The fosfomycin-resistance gene fosB has been identified in the chromosomes and on plasmids in many low-G+C Gram-positive bacteria that do not produce GSH [4,13–16]. Until recently, the potential identity of the native thiol substrate for FosB had long been elusive. Previous kinetic studies of FosB from Bacillus subtilis (BsFosB) revealed negligible activity towards GSH and no apparent activity for CoA [16]. Cysteine also proved to be a poor thiol substrate with a very high Km of 35 mM, which is approximately 200-fold greater than its intracellular concentration [17].
In 2009, bacillithiol (BSH) (Figure 1B) was identified as a unique low-molecular-mass thiol among numerous low-G+ C Gram-positive bacteria (Firmicutes) that do not produce GSH. These include several pathogens, such as Staphylococcus aureus (bacterial sepsis), Staphylococcus saprophyticus (urinary tract infection), Bacillus cereus (food poisoning) and Bacillus anthracis (livestock pathogen and biowarfare agent), as well as B. subtilis (soil bacterium) and Deinococcus radiodurans (extremophile) [18]. All of these bacteria also harbour a fosB gene. BSH-deficient mutants of B. subtilis [19] and B. anthracis [20] exhibit increased sensitivity to fosfomycin. In B. subtilis, this increased fosfomycin sensitivity was comparable with that observed in the fosB mutant and in a double mutant for both fosB and BSH biosynthesis, which suggested that FosB utilizes BSH as its native thiol substrate [19]. A total chemical synthesis of BSH [21] recently provided sufficient material for preliminary activity assays, which, at fixed thiol substrate concentrations (1 mM), demonstrated S. aureusFosB (SaFosB) to be significantly more active with BSH than cysteine. This supported further the notion of FosB being a bacillithiol-S-transferase.
In the present paper, we report the detailed kinetic and mechanistic studies of SaFosB in terms of its metalion-dependence, substrate-binding order and comparison of its substrate efficiency with BSH, GSH, cysteine and some BSH analogues, which help to identify key features of BSH that are important for substrate recognition.
EXPERIMENTAL
General
BSH was chemically synthesized as described previously [21]. Cysteine, N-acetylcysteine, GSH, homocysteine, γ-Glu-Cys (γ-glutamylcysteine) and CoA were purchased from Sigma–Aldrich. The amine derivatization reagent AQC (6-aminoquinolyl-N-hydroxysuccinimidyl carbamate) was either purchased from Waters (AccQ-Fluor™) or synthesized as described previously [22]. The S. aureus USA300 JE2 (MRSA) wild-type strain and BSH-deficient transposon mutants were obtained from the ‘Network on Antimicrobial Resistance in Staphylococcus aureus’ (NARSA) Program. The B. subtilis CU1065 wild-type and bshA mutant were kindly provided by Professor John Helmann [19]. All kinetic data for substrate and inhibition assays were analysed (using the appropriate rate equations) by non-linear regression, and then transformed into double-reciprocal plots for graphical representation, using GraFit Version 5 (Erithacus Software).
Fosfomycin resistance in S. aureus wild-type and BSH-deficient mutants
S. aureus Newman and MRSA wild-type strains, as well as the BSH-deficient MRSA strains, NE1728 (bshA mutant), NE1596 (bshB mutant) and NE230 (bshC mutant), were grown in TSB (trypticase soy broth) with 0, 1, 2.5, 5, 7.5, 10, 15, 20, 25, 30, 40, 50, 60, 70, 80, 100, 120, 140 or 160 µg·ml−1 fosfomycin. Growth was monitored at D600 and the experiment was performed in triplicate. LB (Luria–Bertani) agar plates supplemented with 0–20 mg·ml−1 fosfomycin were used to determine the MIC (minimum inhibitory concentration) values for Escherichia coli transformed with pET151:FosB, as well as the B. subtilis CU1065 wild-type and B. subtilis CU1065 bshA mutant.
Thiol quantification in S. aureus and the effect of fosfomycin on thiol content
S. aureus Newman was grown in 100 ml of TSB in two sets of triplicate cultures. When the D600 reached 0.7, one set of triplicate cultures was treated with a sub-lethal concentration of fosfomycin (5 µg · ml−1) and the remaining set was left untreated. The D600 was monitored and samples corresponding to approximately 5 mg of residual dry weight of cells were harvested from each culture at various times for analysis of thiol content. Cell pellets were frozen at − 20 °C until derivatization with mBBr (monobromobimane) and analysis by reverse-phase HPLC as described previously [18]. BSmB (bacillithiol-bimane) and CySmB (cysteinyl-bimane) were eluted at 8 and 10 min respectively, and were quantified by comparison with RSmB (methylbimane derivative of thiol) standards of known concentration.
SaFosB cloning, overexpression and purification
FosB was amplified from S. aureus strain ATCC25923 DNA with the primers SaFosBF2 (5′-CACCATGTTAAAATCTATTAAT-C-3′) and SaFosBR2 (5′-TTATTTGTAAAATGTCATATGTGG-TTT-3′), and cloned into pET151 (Invitrogen) with an additional stop codon and native promoter to prevent fusion with the His6 tag. The expression plasmid was transformed into BL21 Star™ (DE3) E. coli cells. The culture was grown to a D600 of 1.5 in Terrific broth at room temperature (22 °C) before induction with 25 µM IPTG (isopropyl β-d-thiogalactopyranoside) for 20 h. Cells were harvested and stored at − 80 °C.
The frozen cell pellet was thawed in lysis buffer (20 mM potassium phosphate, pH 6.8, 10% glycerol and 60 mM NaCl) and was disrupted by sonication. The supernatant was applied to a 5 ml SP Sepharose HiTrap ion-exchange column (GE Healthcare) equilibrated with lysis buffer, which was subsequently washed with lysis buffer supplemented with 40 mM NaCl. Proteins were eluted with 200 mM NaCl and were pooled and concentrated by centrifugation in an Amicon spin column (Millipore), before applying to a 5 ml hydroxyapatite column equilibrated with 60 mM potassium phosphate (pH 6.8) and 10% glycerol. The column was washed with 80 mM potassium phosphate, and proteins were eluted at 300 mM potassium phosphate. Purified SaFosB was treated with the Na+ -form of 100 mesh Chelex (3 g, Bio-Rad Laboratories) and 5 mM EDTA overnight. EDTA was then removed with a PD-10 desalting column (GE Healthcare) equilibrated with 10% glycerol. FosB was then concentrated to 280 µM in an Amicon spin column (Supplementary Figure S2 at http://www.biochemj.org/bj/451/bj4510069add.htm).
Synthesis of BSH analogues
The chemical syntheses of MeO-GlcNCys [O-methylglucos-amine cysteine (O-methylbacillithiol)], BnO-GlcNCys [O-benzylglucosamine cysteine (O-benzylbacillithiol)] and BOH (serine analogue of BSH) are described in the Supplementary Online Data and Supplementary Scheme S1 at http://www.biochemj.org/bj/451/bj4510069add.htm.
SaFosB thiol specificity assays by thiol consumption
Assays for BSH, cysteine, N-acetylcysteine, GSH, homocysteine, γ-Glu-Cys, CoA, MeO-GlcNCys and BnO-GlcNCys consisted of 50 mM Hepes (pH 7.0), 1 mM MgCl2, 5 mM fosfomycin and 10 µM SaFosB and mixtures were incubated at 22 °C for 5 min. Reactions were then initiated with the addition of 2 mM thiol, and aliquots were taken at various time points and mixed immediately with 10 mM DTNB [5,5′-dithiobis-(2-nitrobenzoic acid)] [23] in 50 mM sodium phosphate buffer (pH 7.5) with 20 mM KCl and 1 mM MgCl2. The absorbance of each reaction was measured at 412 nm to calculate the consumption of thiol using the molar absorption coefficient of 14150 M−1 [24]. Control reactions performed without enzyme determined the background (non-enzymatic) oxidation of each thiol.
HPLC-based SaFosB assays monitoring RS–fosfomycin formation
In general, SaFosB activity assays consisted of Chelex-treated Hepes (pH 7.0), fosfomycin, SaFosB and M2+ (divalent metal ions) at various concentrations (see the Figure legends for individual assay details). Assay mixtures were equilibrated at 22 °C for at least 5 min before initiating reactions by the addition of the thiol substrate. After 1 min, the reactions were stopped by heating at 85 °C for 10 min and then cooled to room temperature before derivatization for 1 min in sodium borate buffer (pH 8.8) with at least 4 molar excess of AQC with respect to the total amine content of the assay. After AQC derivatization, samples were diluted with 70 mM sodium acetate and 6.25 mM triethylamine, adjusted to pH 5.05 with phosphoric acid (solvent A) for analysis by HPLC. When product formation was below the limit of detection, AQC-labelled samples were first dried in vacuo and resuspended in a minimal volume of solvent A. Control experiments showed that all enzyme reactions were linear for at least 5 min.
HPLC analysis of AQC-labelled samples
RS–fosfomycin samples were analysed on a HiChrom ACE C18, 4.6 mm diameter×250 mm length, 5 µM particle size and 100 Å (1 Å = 0.1 nm) pore size column equilibrated to 37 °C with 90% solvent A and 10% solvent B (80%, v/v, acetonitrile). Samples were eluted with a flow rate of 1.5 ml · min−1 using the following gradient of solvent B: 0–4 min, 10%; 4–8 min, 10–12%; 8– 10 min, 12–15%; 10–13 min, 15–40%; 13–15 min, 40–100%. A Jasco fluorescence detector was used to analyse the samples, with excitation at 250 nm and emission at 395 nm. RS–fosfomycin was quantified using a standard curve of AQC-derivatized RS–fosfomycin standards of known concentration. HPLC retention times of the AQC-derivatized RS–fosfomycin conjugates were: BS–fosfomycin (bacillithiol–fosfomycin) (4.8 min), cysteine–fosfomycin (6.9 min), MeO-GlcNCys–fosfomycin (9.6 min) and BnO-GlcNCys–fosfomycin (12.7 min).
SaFosB substrate saturation assays
For the saturated substrate kinetics of SaFosB with BSH, a 7×7 matrix of initial rate reactions were constructed with various concentrations of BSH (0.1–2 mM) and fosfomycin (1–25 mM). Reaction mixtures consisted of 50 mM Hepes (pH 7.0), 5 mM MnCl2 and 50 nM SaFosB. Assays were performed as detailed above and initial reaction rates were determined using analytical HPLC. The results for various fosfomycin concentrations were fitted by non-linear regression to the velocity equation for a rapid equilibrium ternary complex system using GraFit version 5.
Inhibition assays
Duplicate assay mixtures consisted of 150 mM Hepes (pH 7.0), 5 mM MnCl2, 50 nM SaFosB and 0, 1, 3 or 8 mM BOH inhibitor. In the reactions with various BSH concentrations (0.125–6 mM), the fosfomycin concentration was fixed at 15 mM. For the reactions where fosfomycin concentration varied (2–75 mM), the BSH concentration was fixed at 2.5 mM. Reactions were carried out and analysed as described above.
Enzymatic synthesis of RS–fosfomycin standards
BS–fosfomycin was prepared by incubating BSH (6.8 mg) in 10 ml of 20 mM ammonium formate buffer (pH 7.0) containing MnCl2 (0.1 mM), fosfomycin (1.3 mM) and SaFosB (1 µM) at room temperature. Thiol consumption was monitored over time (by titrating aliquots with DTNB), until the reaction was complete after 2 h. The reaction mixture was freeze-dried (to remove excess ammonium formate) and then resuspended in 2 ml of water, adjusted to pH 3 with 0.1% formic acid. The solution was applied to a cation-exchange column (Isolute-SCX2 SPE, 0.5 g), eluted with water (10 ml) and dried. The resultant residue was resuspended in water (5 ml) and adjusted to pH 8.0 with 2% (v/v) aqueous ammonia, then purified further by anion-exchange chromatography using DEAE-cellulose (1 g, AX, fibrous fast flow, Sigma) packed in an SPE cartridge. The product was eluted with a 10–200 mM stepwise gradient of ammonium bicarbonate using 12 ml portions at 10–20 mM increments. Fractions containing the product (which eluted at 100–120 mM) were identified by spotting on a TLC plate and staining with ninhydrin or charring with 10% (v/v) ethanolic sulfuric acid.
Preparations of cysteine–fosfomycin, MeO-GlcNCys–fosfomycin and BnO-GlcNCys–fosfomycin were carried out in a similar manner, using 9.6 mg of cysteine, 7.1 mg of MeO-GlcNCys or 7.4 mg of BnO-GlcNCys with an equimolar amount of fosfomycin in 40 mM ammonium formate (pH 7.0) with 0.5 mM MnCl2 and 2 µM FosB. The reactions were allowed to proceed for 16 h at room temperature. Purification was performed as above. For BnO-GlcNCys–fosfomycin, an addition final purification step was required on an Isolute C18 (5 g) column by gradient elution using 0.1% formic acid in water and acetonitrile.
31P-NMR was used to confirm product purity and excess ammonium bicarbonate was removed by repeated freeze-drying. For use as HPLC standards, RS–fosfomycin concentrations were determined by quantitative 1H-NMR [25]. The final isolated yields of the products were: BS–fosfomycin, 3.5 mg, 44% yield; cysteine–fosfomycin, 6.6 mg, 32% yield; MeO-GlcNCys–fosfomycin, 1.2 mg, 11% yield; and BnO-GlcNCys–fosfomycin, 4.1 mg, 39% yield.
RESULTS AND DISCUSSION
Fosfomycin resistance in S. aureus wild-type and BSH-deficient mutants
The involvement of BSH in fosfomycin resistance in S. aureus was established by testing the fosfomycin-sensitivity of wild-type and mutant strains deficient in each of the BSH biosynthetic genes (bshA-C). The MIC for the wild-type S. aureus MRSA strain, grown in TSB, was 80 µg · ml−1, whereas the MICs for its BSH-deficient mutants were 10–20 µg · ml−1. To ensure that this resistance was not restricted to MRSA strains, the MIC of S. aureus Newman was also tested and was shown to be the same as the MRSA wild-type (80 µg · ml−1). These results are consistent with previous reports in B. subtilis [19] and B. anthracis [20], although the effect is less pronounced for S. aureus. We observed a 60-fold lower MIC in the B. subtilis BSH-deficient mutant compared with the wild-type, whereas there is only a 4– 8-fold change in S. aureus (Table 1). This difference could be due to variations in the intracellular concentrations and BSH-dependent activities of FosB and/or differences in fosfomycin-uptake rates and potency against MurA in the different bacteria. In B. anthracis, the bshB mutant is less sensitive to fosfomycin than the bshA mutant [20]. This is due to the presence of a separate bacillithiol-S-conjugate amidase (Bca) that is able to complement for the GlcNAc-malate N-deacetylase activity of BshB in BSH biosynthesis. S. aureus, however, does not encode a separate Bca enzyme and therefore the bshB mutant is equally as sensitive to fosfomycin as the bshA mutant (Table 1).
Table 1. MIC values for fosfomycin for strains used in the present study.
| Strain | MIC (µg · ml−1) |
|---|---|
| E. coli BL21(DE3) Star + pET151 (control) | 50 |
| E. coli BL21(DE3) Star + pET151:fosB | > 20 000 |
| S. aureus Newman | 80 |
| S. aureus MRSA | 80 |
| S. aureus MRSA bshA | 10–20 |
| S. aureus MRSA bshB | 10–20 |
| S. aureus MRSA bshC | 10–20 |
| B. subtilis strain CU1065 | >1500 |
| B. subtilis strain CU1065 bshA | 25 |
The increased sensitivity of the BSH-deficient mutants to fosfomycin implies that co-administration of a BSH biosynthesis inhibitor with fosfomycin could enhance the efficacy of fosfomycin against BSH-utilizing Gram-positive pathogens. The high incidence of MurA [26] and GlpT [12] mutations (also conferring resistance to fosfomycin) in clinical settings, however, may counter any efficacy of this therapeutic strategy. Interestingly, although E. coli does not produce BSH, an E. coli strain transformed with the SaFosB overexpression vector showed a 400-fold increase in fosfomycin resistance (Table 1). These data are consistent with similar observations of the BsFosB overexpression plasmid in E. coli [16]. Whereas cysteine is a much less efficient FosB substrate than BSH (see below), presumably the high levels of overexpressed SaForB in this E. coli strain are sufficient for it to effectively utilize cysteine in the absence of BSH to confer fosfomycin resistance.
Effect of fosfomycin on intracellular BSH and cysteine content in S. aureus
The changes in intracellular BSH and cysteine levels were compared when S. aureus was challenged with sub-lethal (5 µg· ml−1) quantities of fosfomycin (Figure 2). The D600 of the cultures was slightly affected by fosfomycin, reaching 2.0 in the treated cultures compared with 2.9 in the control. Whereas there was no immediate lag in growth after fosfomycin treatment, BSH levels decreased slightly; presumably due to conjugation of BSH to fosfomycin for detoxification. After 1 h, there was no difference between BSH levels in fosfomycin-treated and control cultures. However after 4 h, as the cells entered the stationary phase, intracellular BSH levels increased 2-fold in the treated cultures, whereas there was a 3-fold decrease in BSH content in the control. This difference may be due to the increased transcription of BSH biosynthetic genes, which could be up-regulated to compensate for the consumption of BSH in fosfomycin detoxification. The effect of fosfomycin on BSH, but not on cysteine, levels indicates a more prominent role for BSH in response to S. aureus being challenged by fosfomycin.
Figure 2. Effect of sub-lethal levels of fosfomycin on S. aureus growth and thiol content.
▲ Control D600; Δ, fosfomycin-treated D600; ●, control intracellular BSH content; ○, fosfomycin-treated intracellular BSH content; ■ control intracellular cysteine content; □, fosfomycin-treated intracellular cysteine content. Results are means ± S.E.M. of three replicates. rdw, residual dry weight.
Assay development
Previous assays for FosA and FosB utilized an assay buffer containing either Hepes [4,10,27] or sodium borate [9] at pH 8.0. As some thiols have the tendency to oxidize to their disulfides under aerobic conditions at alkaline pH, we tested the stability of cysteine and BSH in Hepes buffers of pH 7.0– 8.0. No oxidation of cysteine was detected over 1 h under any of the pH conditions tested. BSH was more prone to oxidation with 10% (at pH 7.5) and 20% (at pH 8.0) of the thiol being oxidized after 1 h, compared with only 3% oxidation at pH 7.0 (Supplementary Figure S1 at http://www.biochemj.org/bj/451/bj4510069add.htm). No significant difference in SaFosB activity was observed between pH 6.0 and pH 8.0, in contrast with FosA, which has an activity optimum at pH 8.0 [7]. Therefore, to minimize the potential for thiol oxidation, all of the SaFosB activity assays in the present study were carried out at pH 7.0. In addition, the thiol substrate was added last to initiate the 1 min reactions, before quenching and derivatization with AQC for HPLC analysis.
Divalent metal ion specificity
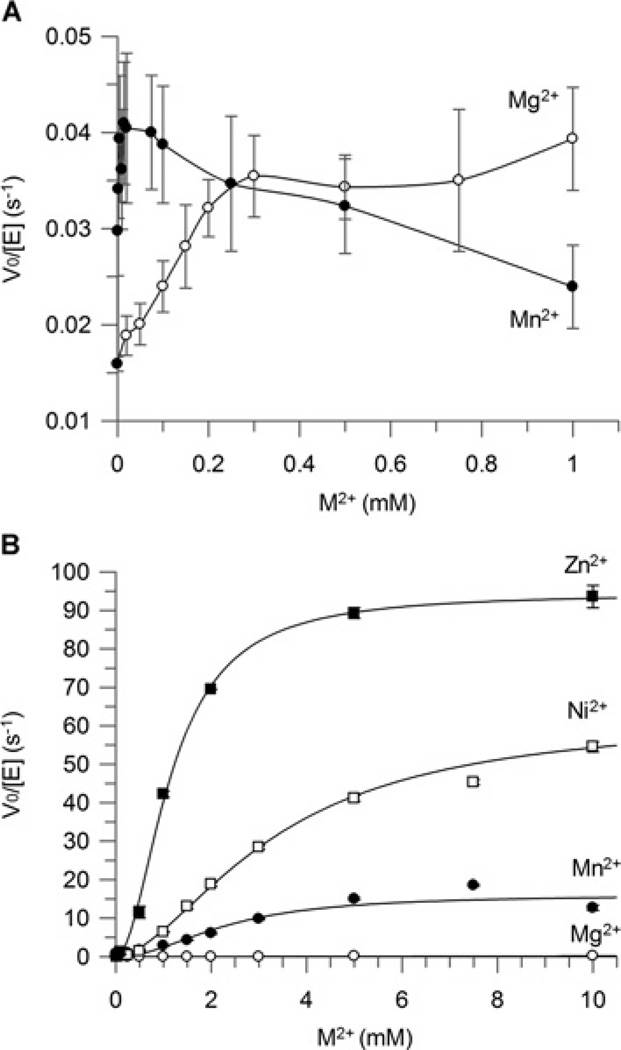
The divalent metal cation in FosA and FosB plays an essential role in bidentate co-ordination to the phosphonate motif of fosfomycin, as well as stabilizing the negative charge generated on the epoxide oxygen during the transition state [28,29]. FosA has previously been demonstrated to be a Mn2+-dependent enzyme [9]. However, previous studies on the activation of BsFosB with cysteine reported that the cysteine-S-transferase activity with 0.5 mM Mg2+ was almost 10-fold greater than with 0.5 mM Mn2+ [16], with an activation constant for Mg2+ of 200 µM that is well below cellular Mg2+ concentrations. This supported the likelihood of Mg2+ being the native metal ion cofactor in vivo.
In the present study, similar assays with SaFosB showed no difference in the cysteine-S-transferase activity with 0.5 mM concentrations of Mn2+ or Mg2+ (Figure 3A). In the presence of 1 mM cysteine, SaFosB activity displayed low S-transferase activity with fosfomycin, which reached a maximum at ~20 µM Mn2+ or 300 µM Mg2+ (Figure 3A). At higher Mn2+ concentrations, SaFosB activity decreased.
Figure 3. Metal specificity for FosB with (A) cysteine and (B) BSH as substrates.
■ Zn2+; □, Ni2+; ●, Mn2+; ○, Mg2+. Results are means ± S.E.M. for three replicates. Assay conditions for (A): 20 mM Hepes (pH 7.0), 2 mM fosfomycin, 2 µM FosB, 1 mM cysteine and 0–1 mM M2+. Assay conditions for (B): 50 mM Hepes (pH 7.0), 2 mM fosfomycin, 100 nM FosB, 0.5 mM BSH and 0–10 mM M2+. Sigmoidal metal activation curves for BSH with Zn2+, Ni2+ and Mn2+ were fitted using the Hill equation.
With cysteine as the thiol substrate, the effect of different divalent metal ions on the activation of BsFosB was reported previously as Ni2+ ~Mg2+ >Mn2+ >Fe2+ >Cu2+>Ca2+ ~Co2+ >Zn2+ [16]. In the presence of 0.5 mM BSH, the order of 2 mM metal ion activation of SaFosB is significantly different showing that Zn2+ >Ni2+ >Mn2+ >Mg2+ ~Fe2+ ~Co2+ ~Cu2+ ~Ca2+ . The addition of K+ to the reaction did not affect the activity of SaFosB in vitro (Supplementary Figure S3 at http://www.biochemj.org/bj/451/bj4510069add.htm), which is consistent with previous reports on FosB activity for cysteine [4,16]. Metal activation studies identified increased bacillithiol-S-transferase activity for SaFosB in the presence of increasing concentrations of Zn2+, Ni2+ and Mn2+ (Figure 3B). These metal activation curves followed sigmoidal kinetics with Kact values of 1.2, 3.1 and 3.7 mM for Zn2+ , Ni2+ and Mn2+ respectively. The rates of reaction with 10 mM Mn2+ or Ni2+ were 25- and 60-fold greater than that for equivalent concentrations of Mg2+ . The high Kact values for SaFosB suggest that the metal cofactor is not tightly bound to the enzyme.
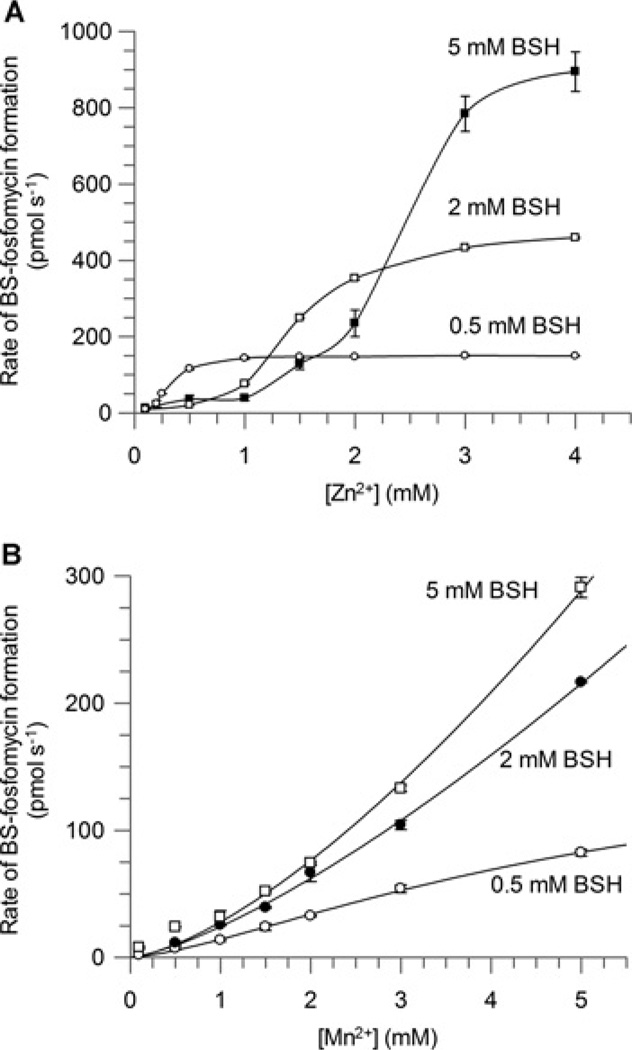
The cellular concentrations of free Ni2+ are well below the activation constant observed for this metal cation [30]. This suggests that Ni2+ is unlikely to be a physiologically relevant metal that contributes to SaFosB activity in vivo and it was therefore not pursued further. Increasing concentrations of BSH appeared to inhibit the activation of SaFosB by Zn2+ (Figure 4A), as activity only increased when concentrations of Zn2+ were higher than those of BSH. This indicates that BSH is competing with SaFosB for complexation with Zn2+ , consistent with observations that BSH is a strong Zn2+ chelator (Z. Ma and J.D. Helmann, personal communication). The fact that excess BSH is able to inactivate such pre-equilibrated mixtures of SaFosB and Zn2+ implies that BSH is a strong enough Zn2+ chelator to rapidly demetallate SaFosB. Total intracellular concentrations of Zn2+ are estimated to be approximately 100 µM [30], although the amount of free cytosolic Zn2+ is suggested to be maintained in the femtomolar range by the zinc-uptake regulator Zur [31]. As BSH levels in S. aureus are thought to be ~200 µM [18], it would appear that, under these conditions, Zn2+ is also unlikely to be the physiological metal for in vivo activation of SaFosB.
Figure 4. Effect of BSH concentration on (A) Zn2+ and (B) Mn2+ activation of FosB.
■ 5 mM BSH; □, 2 mM BSH; ○, 0.5 mM BSH. Results are means ± S.E.M. of three replicates. Assay conditions: 50 mM Hepes (pH 7.0), 25 mM fosfomycin, 100 nM FosB, 0.5 mM BSH and 0.1–10 mM M2+.
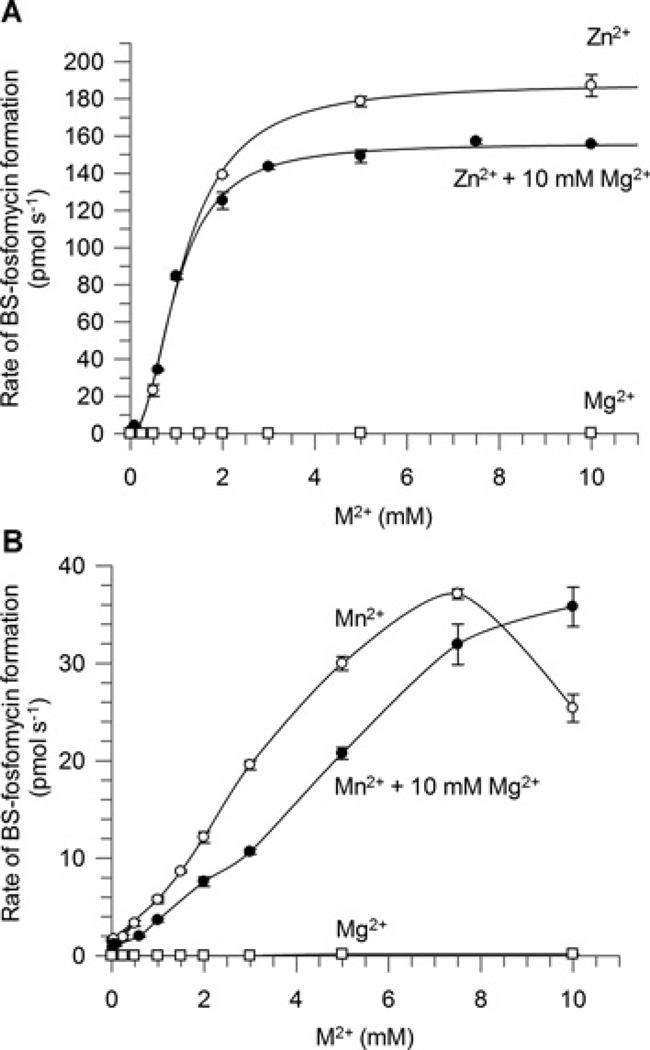
In contrast, the BSH concentration did not notably inhibit the activation of SaFosB by Mn2+ , and the sigmoidal nature of the activation curves did not become prominent until at least 1.5 mM Mn2+ , which is much greater than cellular Mn2+ concentrations (Figure 4B). Interestingly, the activity for SaFosB at physiological Mn2+ concentrations (~10 µM) [30] is only approximately 5-fold greater than in the presence of physiological Mg2+ concentrations (~10 mM) [30] (Figure 3B). Mg2+ -addition experiments were used to determine the effect of this cation on metal activation by Mn2+ and Zn2+ . In both cases, the addition of 10 mM Mg2+ decreased the activity observed with Mn2+ or Zn2+ alone (Figure 5), suggesting that physiological concentrations of Mg2+ are able to compete with Mn2+ and Zn2+ for activation of SaFosB, albeit with lower reaction rates. Together, these results suggest that physiological concentrations of both Mn2+ and Mg2+ could plausibly activate SaFosB with a similar rate of activity in vivo. When Mn2+ -uptake systems are overexpressed, concentrations of up to 1–3 mM Mn2+ have been observed in E. coli and Salmonella enterica serovar Typhimurium [32]. If similar cellular concentrations of Mn2+ could occur in BSH-utilizing bacteria, then higher levels of SaFosB activation by Mn2+ could be physiologically relevant. It is probable that in S. aureus, however, intracellular Mn2+ would be maintained at low-micromolar levels [33] by the Mn2+ -dependent transcriptional regulator MntR [34,35].
Figure 5. Effect of 10 mM Mg2+ on(A)Zn2+ (B) Mn2+ activation of FosB.
○, Zn2+ or Mn2+ only; ●, Zn2+ or Mn2+ with 10 mM Mg2+; □, Mg2+ only. Results are means ± S.E.M. of three replicates. Assay conditions: 50 mM Hepes (pH 7.0), 2 mM fosfomycin, 100 nM FosB, 0.5 mM BSH, 0 or 10 mM MgCl2 and 0.025–10 mM MnCl2 or ZnCl2.
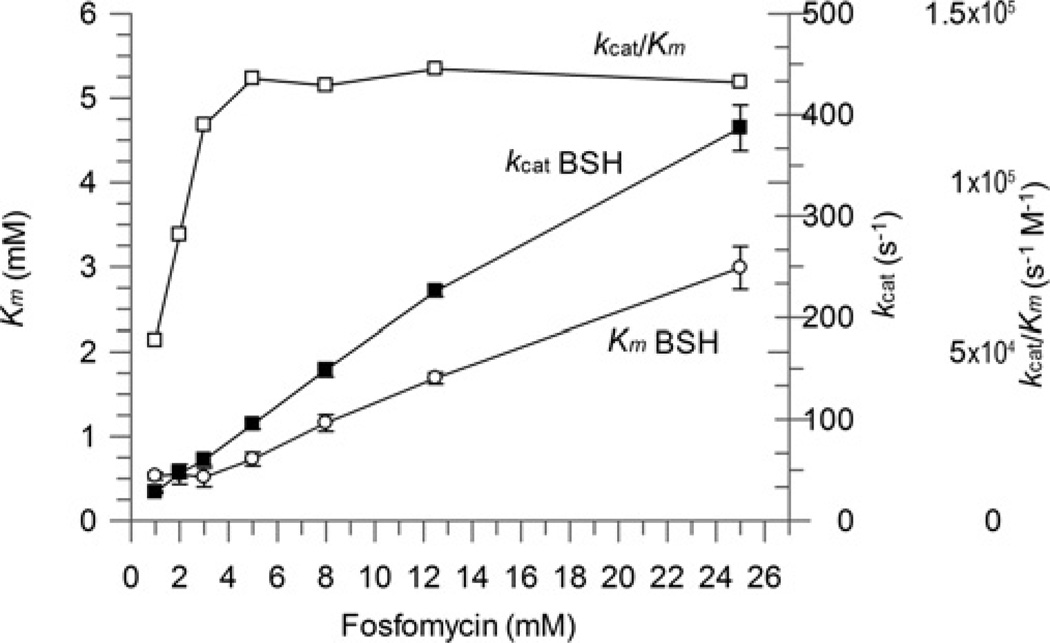
Detailed substrate saturation kinetics for BSH and fosfomycin
Detailed substrate kinetic assays were performed for BSH in the presence of 5 mM Mn2+ to determine the kinetic constants under saturating substrate conditions. The reciprocal plots obtained from these data at various fosfomycin concentrations gave a family of converging lines consistent with a rapid equilibrium ternary substrate complex mechanism (Figure 6A). It was initially surprising to observe that the saturation substrate kinetics extrapolated a high Km value for BSH of 4.2 mM (Table 2) relative to the cytosolic concentrations of BSH in S. aureus (~200 µM) [18]. A high Km value of 17.8 mM was also obtained for fosfomycin, which is similar to that reported for the E. coli FosA (9.4 mM) [7]. Closer analysis revealed how Km (app) values for both BSH (Table 2) and fosfomycin (Table 3) decrease with decreasing co-substrate concentration, although there is no significant change in the overall catalytic efficiency (kcat/Km) (Figure 7). These results suggest elements of negative co-operativity between the binding of these two substrates to the SaFosB homodimer. Others have previously alluded that the Km of fosfomycin with FosA can also increase in the presence of increasing thiol substrate concentrations [10], so such cooperative substrate interactions appear to be present in both FosA and FosB enzymes alike.
Figure 6. FosB reaction mechanism analysis.
(A) The FosB reaction with BSH and fosfomycin follows an ordered mechanism as predicted by the double-eciprocal plots of the substrate saturation assay. Fosfomycin concentrations: ○, 1 mM; ●, 2 mM; □, 3 mM; ■ 5 mM; Δ, 8 mM; ▲ 12.5 mM; ∇ 25 mM. Inhibition studies with BOH (structure pictured) show that the SaFosB reaction is compulsory ordered with fosfomycin binding first, and that (B) BOH is competitive with respect to BSH and (C) uncompetitive with respect to fosfomycin. BOH concentrations: ○, 0 mM; ●, 1 mM; □, 3 mM; ■ 8 mM. Results are means ± S.E.M. of three replicates. Assay conditions for (A): 50 mM Hepes (pH 7.0), BSH (0.1, 0.2, 0.3, 0.5, 0.7, 1 and 2 mM), fosfomycin (1, 2, 3, 5, 8, 12.5 and 25 mM), 5 mM MnCl2 and 50 nM SaFosB. Assay conditions for (B): 150 mM Hepes (pH 7.0), 5 mM MnCl2, 15 mM fosfomycin, 50 nM FosB, 0.125–6 mM BSH and 0, 1, 3 or 8 mM BOH. Assay conditions for (C): 150 mM Hepes (pH 7.0), 5 mM MnCl2, 2.5 mM BSH, 50 nM FosB, 2–75 mM fosfomycin and 0, 1, 3 or 8 mM BOH.
Table 2. Thiol substrate kinetics data.
Data were determined by HPLC analysis (except where specified). Experimental details for the substrate saturation assays are described in the Supplementary Online Data at http://www.biochemj.org/bj/451/bj4510069add.htm.
| Organism | Thiol | Fosfomycin concentration (mM) | Metal | Km (mM) | kcat (s−1) | kcat/Km (s−1 ·M−1) | Reference |
|---|---|---|---|---|---|---|---|
| S. aureus | BSH | Saturating* | 5 mM Mn2+† | 4.2 ± 0.7 | 604 ± 86 | (1.4 ± 0.5)×105 | The present study |
| BSH | 25 | 5 mM Mn2+† | 3.0 ± 0.3 | 390 ± 23 | (1.3 ± 0.2)×105 | The present study | |
| BSH | 1 | 5 mM Mn2+† | 0.53 ± 0.04 | 28.2 ± 0.9 | (5.3 ± 0.6)×104 | The present study | |
| Cysteine | 2 | 50 µM Mn2+ † | 13 ± 2 | 2.3 ± 0.2 | 175 ± 44 | The present study | |
| BSH | 50 | 10 µM Mn2+ | 12 ± 3 | 51±7 | (4 ± 2)×103 | The present study | |
| BSH | 50 | 5 mM Mn2+ | 5.1 ± 0.2 | 380 ± 9 | (7.4 ± 0.5)×104 | The present study | |
| BSH | 50 | 10 mM Mg2+ | 9 ± 2 | 62±6 | (7 ± 2)×103 | The present study | |
| BSH | 50 | 10 µM Mn2+ /10 mM Mg2+ | 4.2 ± 0.5 | 38 ± 2 | (9 ± 1)×103 | The present study | |
| MeO-GlcNCys | 50 | 10 µM Mn2+ /10 mM Mg2+ | 32 ± 7 | 5.3 ± 0.4 | 163 ± 46 | The present study | |
| BnO-GlcNCys | 50 | 10 µM Mn2+ /10 mM Mg2+ | 73 ± 12 | 19 ± 1 | 266 ± 59 | The present study | |
| Cysteine | 50 | 10 µM Mn2+ /10 mM Mg2+ | 157 ± 9 | 49 ± 1 | 311±25 | The present study | |
| BOH | 2–75 | 5 mM Mn2+† | Ki = 3.9 ± 0.3 | – | – | The present study | |
| Homocysteine | 5 | 10 µM Mn2+ /10 mM Mg2+ | –‡ | –‡ | –‡ | The present study | |
| N-acetylcysteine | 5 | 10 µM Mn2+ /10 mM Mg2+ | –‡ | –‡ | –‡ | The present study | |
| GSH | 5 | 10 µM Mn2+ /10 mM Mg2+ | –‡ | –‡ | –‡ | The present study | |
| γ-Glu-Cys | 5 | 10 µM Mn2+ /10 mM Mg2+ | –‡ | –‡ | –‡ | The present study | |
| CoA | 5 | 10 µM Mn2+ /10 mM Mg2+ | –‡ | –‡ | –‡ | The present study | |
| Water | 50 | Various§ | –§ | –§ | –§ | The present study | |
| B. subtilis∥ | Cysteine | Mg2+ | 35 | 6.3 | 180 | [16] | |
| GSH | Mg2+ | 15 | 0.027 | 1.8 | [16] | ||
| CoA | Mg2+ | >50 | – | 0.4 | [16] | ||
| Cysteine | Mn2+ | >200 | – | 6.9 | [16] | ||
| GSH | Mn2+ | >50 | – | 0.093 | [16] | ||
| CoA | Mn2+ | >100 | – | 0.0009 | [16] | ||
| Pseudomonas aeruginosa (FosA) | GSH | 0.5 mM¶ | 200 µM Mn2+ | 6.2 | 1.07×103 | 1.73×105 | [10] |
Saturating fosfomycin concentrations extrapolated from detailed substrate saturation assay.
Optimized in vitro metal concentrations (BSH, 5 mM Mn2+ ; cysteine, 50 µM Mn2+).
No activity observed with 2 mM thiol with 10 µM FosB, as determined by DTNB assay.
No hydrolase activity observed with 2 µM FosB, as determined by 31P-NMR. Various metal cofactors were tested, including 100 µM Zn2+, 10 mM Mg2+ and 10 µM Mn2+ with 10 mM Mg2+.
Fixed fosfomycin concentration (concentration not specified, but possibly up to 4 mM).
The Km for fosfomycin is 0.5 mM when GSH is 50 mM [10]
Table 3. Fosfomycin substrate kinetics data.
| Organism | Thiol | Thiol concentration (mM) | Metal | Km (mM) | kcat (s−1) | kcat/Km (s−1·M−1) | Reference |
|---|---|---|---|---|---|---|---|
| S. aureus | BSH | Saturating* | 5 mM Mn2+ | 17.8 ± 4.2 | 604 ± 86 | (1.9 ± 0.2)×104 | The present study |
| 2 | 5 mM Mn2 + | 12.4 ± 0.7 | 237 ± 6 | (1.9 ± 0.2)×104 | The present study | ||
| 0.3 | 5 mM Mn2+ | 3.4 ± 0.3 | 42 ± 1 | (1.2 ± 0.1) × 104 | The present study | ||
| B. subtilis† | Cysteine | 1 mM Mg2+ | –‡ | 4.8 | 4×103 | [4] | |
| S. aureus† | Cysteine | 1 mM Mg2+ | –‡ | 0.99 | 9.2×103 | [4] |
Saturating BSH concentrations extrapolated from detailed substrate saturation assay.
Fixed cysteine concentration (concentration not specified, but possibly up to 15 mM).
Km values not reported, presumably due to subsaturating thiol concentrations in these assays.
Figure 7. Effect of increasing substrate concentrations on FosB Km and kcat values.
○, Km BSH; ■, kcat BSH; □, kcat/Km. Results are means ± S.E.M. of three replicates. Assay conditions: 50 mM Hepes (pH 7.0), 5 mM MnCl2, 50 nM FosB, 1–25 mM fosfomycin and 0.1–5 mM BSH.
Although the Mn2+ and/or Mg2+ concentrations do not significantly affect the Km value for BSH (Table 2), the kcat value was much lower for 10 µM Mn2+, 10 mM Mg2+ or a 10 µM Mn2+ + 10 mM Mg2+ mixture than in the presence of 5 mM Mn2+ (Table 2). We propose that turnover rates derived in the presence of both 10 µM Mn2+ and 10 mM Mg2+ are more representative of SaFosB’s intracellular activity. It is unlikely that intracellular fosfomycin concentrations would ever reach saturating levels, so the Km (app) for BSH should be much closer to its intracellular concentration under physiological conditions presented when cells are challenged with fosfomycin. Although previous substrate kinetics for FosA reported relatively high Km values for GSH (6–11 mM) [7,10], these concentrations are comparable with intracellular GSH levels. The relatively high saturating Km value for BSH, compared with its cellular concentrations, and its lower kcat value (compared with that of FosA) indicate that SaFosB is a less efficient S-transferase for fosfomycin detoxification than the GSH-dependent FosA.
No S-transferase activity was observed between BSH and the epoxide-containing antibiotic cerulenin in the presence of a large excess of SaFosB (2 µM). This suggests that FosB does not have broad-range activity against epoxide-containing electrophiles in general.
Substrate-binding order
The serine analogue of BSH (BOH) was synthesized (Supplementary Scheme S1) to establish whether the SaFosB reaction pathway proceeds via a random or compulsory ordered substrate-binding mechanism (Figure 6). When BOH (1 mM) and fosfomycin (25 mM) were incubated with high concentrations of SaFosB (1 µM), no BO–fosfomycin product formation was detected. This indicates that BOH is not a substrate for SaFosB and that the thiol motif of BSH is essential for FosB-catalysed ring opening of the fosfomycin epoxide.
Inhibition assays showed BOH to be a competitive inhibitor of BSH with a Ki of 3.9 mM compared with a Km (app) of 1.6 mM for BSH (Figure 6B), demonstrating that BOH is an effective substrate mimic of BSH. The effective inhibition of FosB by BOH also reveals that the thiol group has little impact on substrate recognition compared with the malate moiety (see below).
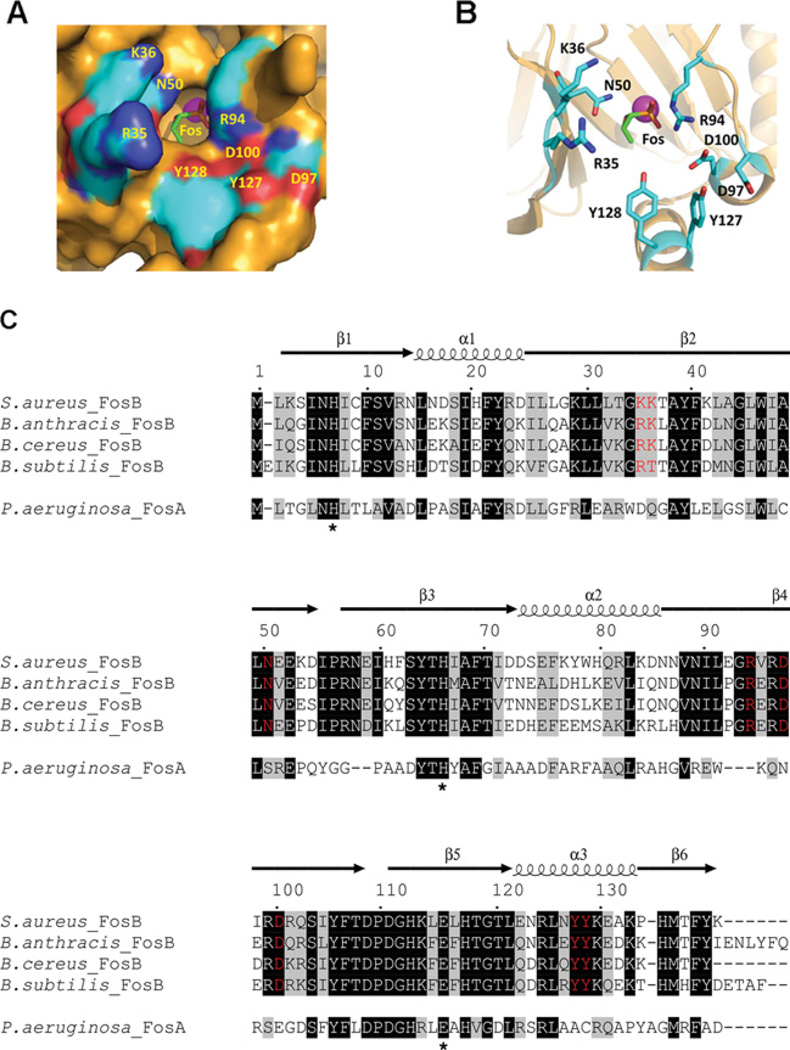
BOH also proved to be an uncompetitive inhibitor with respect to fosfomycin (Figure 6C). Taken together, these data indicate that the FosB-catalysed reaction proceeds via a rapid equilibrium compulsory ordered binding mechanism with fosfomycin binding first followed by BSH, and where the binding of either substrate increases the Km (app) for the second substrate (Figure 7). High BSH concentrations also appear to inhibit SaFosB in a metal-independent manner, but only in the presence of low fosfomycin concentrations (Supplementary Figure S4 at http://www.biochemj.org/bj/451/bj4510069add.htm). Under these conditions, if BSH binds first, the active site will be blocked thereby preventing fosfomycin binding and subsequent catalysis. This inhibition is overcome when fosfomycin concentrations are high enough to outcompete BSH, enabling fosfomycin to bind to FosB first. A fosfomycin-bound structure of FosB from B. anthracis strain Ames has recently been deposited in the PDB (PDB code 4IR0). This structure shows how the enzyme-bound fosfomycin is buried at the bottom of a narrow binding pocket which would clearly be inaccessible if BSH were to bind first (Figure 8A, and Supplementary Figure S6 at http://www.biochemj.org/bj/451/bj4510069add.htm).
Figure 8. The BaFosB-binding pocket with M2+ (pink sphere) and fosfomycin (Fos) bound (PDB code 4IR0).
(A) Surface diagram. (B) Ribbon diagram highlighting residues lining the binding pocket that are conserved or semi-conserved in FosB, but not FosA. (C) Sequence alignment of FosA from Pseudomonas aeruginosa with the characterized FosB sequences from S. aureus, B. subtilis, B. cereus and B. anthracis. The α-helices and β-sheets are indicated by spirals and arrows respectively. Amino acids highlighted in black indicate fully conserved residues. Grey amino acids correspond to residues with ≥70% similarity in their physicochemical properties. Red residues indicate the conserved or semi-conserved amino acids in the FosB active site that may be involved in BSH recognition. Conserved residues involved in M2+ binding are His7, His64 and Glu110 (Pseudomonas aeruginosa numbering [29]), as indicated by asterisks.
Regiochemistry of the BS–fosfomycin product
To confirm the regiochemistry of the FosB-catalysed reaction with BSH, SaFosB was used for the enzymatic synthesis of milligram quantities of BS–fosfomycin. The regiochemistry of the BS–fosfomycin product resulting from nucleophilic attack of BSH at the C-1 carbon of fosfomycin (Figure 1A) was confirmed by two-dimensional NMR spectroscopy, with a three-bond HMBC (heteronuclear multiple bond correlation) between the 13C resonance of C-1″′ on the fosfomycin moiety and the methylene protons (H-3″) on the cysteine side chain (Figure 9). A similar correlation was also observed between cysteine C-3″ and the fosfomycin H-1″′ nuclei. The same HMBCs were also observed with the enzymatically synthesized cysteine–, MeO-GlcNCys– and BnO-GlcNCys–fosfomycin adducts (Supplementary Figure S8 at http://www.biochemj.org/bj/451/bj4510069add.htm). This regioselectivity has been reported previously for FosB-mediated fosfomycin conjugation with cysteine [16], and for the analogous FosA-catalysed reaction with GSH [10], confirming the conserved mechanism among this family of thiol-S-transferases.
Figure 9. BS-fosfomycin HMBC spectrum (1H-13C long-range correlation NMR).
The highlighted peaks indicate the mechanism of conjugation of BSH to fosfomycin. Continuous arrows show the three-bond HMBC between the C-1″′ adjacent to the fosfomycin phosphate group and the H-3″ adjacent to the cysteinyl-amine of BSH. Broken arrows show the reciprocal correlation, between the H-1″′ adjacent to the fosfomycin phosphate group and the C-3″ adjacent to the cysteinyl-amine of BSH.
Thiol substrate specificity
Substrate kinetics for BSH, and a number of other potential thiol substrates, were measured in the presence of fosfomycin (50 mM) and physiological metal concentrations (10 µM Mn2+ and 10 mM Mg2+) [30] (Table 2). None of GSH, CoA, N-acetylcysteine or homocysteine was turned over by SaFosB; neither was γ-Glu-Cys (a biosynthetic precursor of GSH, which serves as a GSH surrogate in some lactic acid bacteria) [36]. In addition, no fosfomycin hydrolase activity (with water) was observed (Table 2 and Supplementary Figure S7 at http://www.biochemj.org/bj/451/bj4510069add.htm). Cysteine was the only other biologically relevant thiol that was recognized, with a Km (app) value of 157 mM (Table 2), which is approximately three orders of magnitude greater than its intracellular concentration. Interestingly, the kcat values for BSH and cysteine are comparable. Therefore the 30-fold higher substrate specificity (kcat/Km) for BSH is attributed to substrate-binding affinity. In the presence of cellular thiol concentrations, BSH will clearly be much more efficient than cysteine as a SaFosB substrate in vivo.
Substituting the malate motif on BSH for an O-methyl group in MeO-GlcNCys led to a 10-fold increase in Km (40 mM) (Table 2). This increased further to 70 mM for BnO-GlcNCys, which was furnished with a much bulkier O-benzyl aglycone. These observations exemplify how the malate motif of BSH plays a prominent role in its preferential recognition and binding as a SaFosB substrate.
Analysis of the FosA/FosB sequence alignments indicates several amino acid residues that are not found in FosA, but are conserved among the four FosB enzymes that have been shown to recognize BSH as a substrate [21,37] (Figure 8C). The fosfomycin-bound BaFosB (B. anthracisFosB) structure reveals how several of these amino acids (Asn50, Tyr127 and Tyr128, as well as the Arg94, Asp97 and Asp100 residues of a conserved GRXRDXRD motif) line the active site in proximity to where BSH must bind in order to conjugate with fosfomycin (Figures 8A and 8B). In the same vicinity are two partially conserved residues in SaFosB (Lys35/Lys36 ), BaFosB/BcFosB (B. cereusFosB) (Arg35/Lys36) and BsFosB (Arg35/Thr36), which could plausibly be involved in recognition and binding of the negatively charged malate aglycone of BSH. Attempts to dock BSH into this FosB-fosfomycin structure using both Autodock Vina and Swiss Dock were unsuccessful, despite optimization of various parameters such as ligand energy minimization, ligand protonation state and flexibility in ligand and enzyme residues. The inability to successfully model BSH into the FosB structure suggests that a conformational change in FosB may occur during BSH binding, as has been observed previously in structural studies of some glutathione transferases [38]. In the BSH-binding pocket, Lys35, Lys36 and Asn50 are on the opposite chain of the homodimer to the other conserved residues (Supplementary Figure S6). Hence any BSH-induced conformational changes that might be propagated across the protein could account for the elements of negative substrate co-operativity between substrates that was observed in the present study. In future, structural studies of BSH-bound FosB would provide more detailed insights into such substrate-binding interactions.
Conclusions
In the present study, mechanistic details of the bacillithiol-S-transferase activity of SaFosB have been established in terms of thiol substrate and metal cofactor preference. SaFosB clearly prefers BSH, rather than cysteine, as its thiol substrate, and probably utilizes both Mn2+ and Mg2+ as its physiologically relevant metal cofactors. As the present paper was being submitted, Armstrong and co-workers published a brief communication comparing the effects of Mg2+ (1 mM) and Ni2+ (10 µM) on the S-transferase activities of FosB enzymes from B. subtilis, B. anthracis, B. cereus and S. aureus in the presence of a single BSH concentration (2 mM) [37]. Under these conditions, they showed that, whereas Ni2+ is a better activator of SaFosB than Mg2+ , the differences are less distinct with the FosB enzymes from the other Gram-positive bacteria. They also commented that Zn2+ appears to be a potent inhibitor of FosB, but details of these experiments were not reported. In the present study, detailed metal activation studies have shown that Zn2+ is the most effective for SaFosB activation in vitro. However, when BSH concentrations are greater than Zn2+ (as they are likely to be in vivo), SaFosB appears to be inactive due to the Zn2+ being sequestered by BSH.
Considering the virtual absence of free Ni2+ within the cytosol, it is likely that Mn2+ and Mg2+ are the more physiologically relevant metals in vivo. Whether the same metal preferences are reflected among other FosBs [37] remains to be seen and warrants further investigation. The present study demonstrates that the SaFosB-catalysed reaction pathway proceeds through a compulsory ordered mechanism, where fosfomycin binds before BSH. Also, whereas the malate group of BSH is particularly important for substrate recognition, the thiol motif, although essential for catalysis, is not important for ligand binding. These structure–activity relationships may help to facilitate the design of simplified BSH substrate mimics as FosB inhibitors. It will be interesting to see whether similar substrate-binding criteria are mirrored with other BSH-utilizing enzymes, such as the recently discovered DinB class of bacillithiol-S-transferases [39]. In addition, the catalytically inert BSH substrate mimic BOH could potentially prove useful for structural and mechanistic studies of FosB and other BSH-dependent enzymes.
In GSH-utilizing bacteria such as E. coli, GSH plays a sacrificial role in electrophile detoxification, as the glutathione S-conjugates are excreted directly from the cell and into the medium [40].
Early evidence of a more efficient detoxification pathway has recently been reported for BSH-utilizing bacteria, where the GlcN–Cys amide bond of bacillithiol S-conjugates is hydrolysed by a conjugate amidase allowing the GlcN–Mal motif to be recycled back into BSH biosynthesis [41]. Access to pure BS– fosfomycin will now provide an opportunity to establish whether such a mechanism is involved in the BSH-dependent fosfomycin-detoxification pathway.
Supplementary Material
ACKNOWLEDGEMENTS
Professor John Helmann (Cornell University, Ithaca, NY, U.S.A.) kindly provided the Bacillus subtilis CU1065 wild-type and BSH-deficient mutant strains. We thank the Engineering and Physical Sciences Research Council (EPSRC) Mass Spectrometry Service Centre, Swansea, for invaluable mass spectrometry support.
FUNDING
This work was supported by the Biotechnology and Biological Sciences Research Council [grant number BB/H013504/1 (to C.J.H.)] and the National Institutes of Health [grant number 1SC3GM100855 (to M.R.)].
Abbreviations used
- AQC
6-aminoquinolyl-N-hydroxysuccinimidyl carbamate
- BaFosB
Bacillus anthracisFosB
- Bca
bacillithiol-S-conjugate amidase
- BnO-GlcNCys
O-benzylglucosamine cysteine (O-benzylbacillithiol)
- BsFosB
Bacillus subtilisFosB
- BS–fosfomycin
bacillithiol–fosfomycin
- BSH
bacillithiol
- BOH
serine analogue of BSH
- DTNB
5,5′-dithiobis-(2-nitrobenzoic acid)
- γ-Glu-Cys
γ-glutamylcysteine
- HMBC
heteronuclear multiple bond correlation
- MeO-GlcNCys
O-methylglucosamine cysteine (O-methylbacillithiol)
- MIC
minimum inhibitory concentration
- SaFosB
Staphylococcus aureusFosB
- TSB
trypticase soy broth
Footnotes
AUTHOR CONTRIBUTION
Alexandra Roberts cloned and purified FosB, and developed and performed all of the kinetic studies and thiol analyses. Sunil Sharma synthesized the AQC, thiol substrates, inhibitor and enzyme products. Andrew Strankman performed the preliminary thiol analysis. Shayla Duran determined the S. aureus fosfomycin MICs. Chris Hamilton, Alexandra Roberts, Sunil Sharma and Mamta Rawat developed the project and interpreted the results and contributed to the writing.
REFERENCES
- 1.Hendlin D, Stapley EO, Jackson M, Wallick H, Miller AK, Wolf FJ, Miller TW, Chaiet L, Kahan FM, Foltz EL, et al. Phosphonomycin, a new antibiotic produced by strains of Streptomyces . Science. 1969;166:122–123. doi: 10.1126/science.166.3901.122. [DOI] [PubMed] [Google Scholar]
- 2.Kahan PM, Kahan JS, Cassidy J, Kropp H. The mechanism of action of fosfomycin (phosphonomycin) Ann. N.Y. Acad. Sci. 1974;235:364–386. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- 3.Marguardt JL, Brown ED, Lane WS, Haley TM, Ichikawa Y, Wong CH, Walsh CT. Kinetics, stoichiometry, and identification of the reactive thiolate in the inactivation of UDP-GlcNAc enolpyruvoyl transferase by the antibiotic fosfomycin. Biochemistry. 1994;33:10646–10651. doi: 10.1021/bi00201a011. [DOI] [PubMed] [Google Scholar]
- 4.Rigsby RE, Fillgrove KL, Beihoffer LA, Armstrong RN. Fosfomycin resistance proteins: a nexus of glutathione transferases and epoxide hydrolases in a metalloenzyme superfamily. Methods Enzymol. 2005;401:367–379. doi: 10.1016/S0076-6879(05)01023-2. [DOI] [PubMed] [Google Scholar]
- 5.Fillgrove KL, Pakhomova S, Newcomer ME, Armstrong RN. Mechanistic diversity of fosfomycin resistance in pathogenic microorganisms. J. Am. Chem. Soc. 2003;125:15730–15731. doi: 10.1021/ja039307z. [DOI] [PubMed] [Google Scholar]
- 6.Fillgrove KL, Pakhomova S, Schaab MR, Newcomer ME, Armstrong RN. Structure and mechanism of the genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes . Biochemistry. 2007;46:8110–8120. doi: 10.1021/bi700625p. [DOI] [PubMed] [Google Scholar]
- 7.Arca P, Hardisson C, Suárez JE. Purification of a glutathione S-transferase that mediates fosfomycin resistance in bacteria. Antimicrob. Agents Chemother. 1990;34:844–848. doi: 10.1128/aac.34.5.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arca P, Rico M, Bran˜a AF, Villar CJ, Hardisson C, Suárez JE. Formation of an adduct between fosfomycin and glutathione: A new mechanism of antibiotic resistance in bacteria. Antimicrob. Agents Chemother. 1988;32:1552–1556. doi: 10.1128/aac.32.10.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernat BA, Laughlin LT, Armstrong RN. Fosfomycin resistance protein (FosA) is a manganese metalloglutathione transferase related to glyoxalase I and the extradiol dioxygenases. Biochemistry. 1997;36:3050–3055. doi: 10.1021/bi963172a. [DOI] [PubMed] [Google Scholar]
- 10.Bernat BA, Laughlin LT, Armstrong RN. Regiochemical and stereochemical course of the reaction catalyzed by the fosfomycin resistance protein, FosA. J. Org. Chem. 1998;63:3778–3780. [Google Scholar]
- 11.Mendoza C, Garcia JM, Llaneza J, Mendez FJ, Hardisson C, Ortiz JM. Plamid-determined resistance to fosfomycin in Serratia marcescens . Antimicrob. Agents Chemother. 1980;18:215–219. doi: 10.1128/aac.18.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arca P, Reguera G, Hardisson C. Plasmid-encoded fosfomycin resistance in bacteria isolated from the urinary tract in a multicentre survey. J. Antimicrob. Chemother. 1997;40:393–399. doi: 10.1093/jac/40.3.393. [DOI] [PubMed] [Google Scholar]
- 13.Etienne J, Gerbaud G, Courvalin P, Fleurette J. Plasmid-mediated resistance to fosfomycin in Staphylococcus epidermidis . FEMS Microbiol. Lett. 1989;61:133–137. doi: 10.1016/0378-1097(89)90184-5. [DOI] [PubMed] [Google Scholar]
- 14.Etienne J, Gerbaud G, Fleurette J, Courvalin P. Characterization of staphylococcal plasmids hybridizing with the fosfomycin resistance gene fosB . FEMS Microbiol. Lett. 1991;84:119–122. doi: 10.1016/0378-1097(91)90406-z. [DOI] [PubMed] [Google Scholar]
- 15.Zilhao R, Courvalin P. Nucleotide sequence of the fosB gene conferring fosfomycin resistance in Staphylococcus epidermidis . FEMS Microbiol. Lett. 1990;68:267–272. doi: 10.1016/s0378-1097(05)80052-7. [DOI] [PubMed] [Google Scholar]
- 16.Cao M, Bernat BA, Wang Z, Armstrong RN, Helmann JD. FosB, a cysteine-dependent fosfomycin resistance protein under the control of σW, an extracytoplasmic-function σ factor in Bacillus subtilis . J. Bacteriol. 2001;183:2380–2383. doi: 10.1128/JB.183.7.2380-2383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park S, Imlay JA. High levels of intracellular cysteine promote oxidative DNA damage by driving the Fenton reaction. J. Bacteriol. 2003;185:1942–1950. doi: 10.1128/JB.185.6.1942-1950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newton GL, Rawat M, La Clair JJ, Jothivasan VK, Budiarto T, Hamilton CJ, Claiborne A, Helmann JD, Fahey RC. Bacillithiol is an antioxidant thiol produced in Bacilli. Nat. Chem. Biol. 2009;5:625–627. doi: 10.1038/nchembio.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaballa A, Newton GL, Antelmann H, Parsonage D, Upton H, Rawat M, Claiborne A, Fahey RC, Helmann JD. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc. Natl. Acad. Sci. U.S.A. 2010;107:6482–6486. doi: 10.1073/pnas.1000928107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parsonage D, Newton GL, Holder RC, Wallace BD, Paige C, Hamilton CJ, Dos Santos PC, Redinbo MR, Reid SD, Claiborne A. Characterization of the N-acetyl-α-d-glucosaminyl L-malate synthase and deacetylase functions for bacillithiol biosynthesis in Bacillus anthracis . Biochemistry. 2010;49:8398–8414. doi: 10.1021/bi100698n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma SV, Jothivasan VK, Newton GL, Upton H, Wakabayashi JI, Kane MG, Roberts AA, Rawat M, La Clair JJ, Hamilton CJ. Chemical and chemoenzymatic syntheses of bacillithiol: a unique low-molecular-weight thiol amongst low G+C Gram-positive bacteria. Angew. Chem. Int. Ed. 2011;50:7101–7104. doi: 10.1002/anie.201100196. [DOI] [PubMed] [Google Scholar]
- 22.Cohen SA, Michaud DP. Synthesis of a fluorescent derivatizing reagent, 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate, and its application for the analysis of hydrolysate amino acids via high-performance liquid chromatography. Anal. Biochem. 1993;211:279–287. doi: 10.1006/abio.1993.1270. [DOI] [PubMed] [Google Scholar]
- 23.Ellman GL. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 24.Riddles PW, Blakeley RL, Zerner B. Reassessment of Ellman’s reagent. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- 25.Pauli GF, Go¨decke T, Jaki BU, Lankin DC. Quantitative 1H NMR. Development and potential of an analytical method: an update. J. Nat. Prod. 2012;75:834–851. doi: 10.1021/np200993k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horii T, Kimura T, Sato K, Shibayama K, Ohta M. Emergence of fosfomycin-resistant isolates of shiga-like toxin-producing Escherichia coli O26. Antimicrob. Agents Chemother. 1999;43:789–793. doi: 10.1128/aac.43.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernat BA, Laughlin LT, Armstrong RN. Elucidation of a monovalent cation dependence and characterization of the divalent cation binding site of the fosfomycin resistance protein (FosA) Biochemistry. 1999;38:7462–7469. doi: 10.1021/bi990391y. [DOI] [PubMed] [Google Scholar]
- 28.Bernat BA, Armstrong RN. Elementary steps in the acquisition of Mn2+ by the fosfomycin resistance protein (FosA) Biochemistry. 2001;40:12712–12718. doi: 10.1021/bi0114832. [DOI] [PubMed] [Google Scholar]
- 29.Rife CL, Pharris RE, Newcomer ME, Armstrong RN. Crystal structure of a genomically encoded fosfomycin resistance protein (FosA) at 1.19 angstrom resolution by MAD phasing off the L-III edge of TI. J. Am. Chem. Soc. 2002;124:11001–11003. doi: 10.1021/ja026879v. [DOI] [PubMed] [Google Scholar]
- 30.Finney LA, O'Halloran TV. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science. 2003;300:931–936. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- 31.Ma Z, Gabriel SE, Helmann JD. Sequential binding and sensing of Zn(II) by Bacillus subtilis . Zur. Nucleic Acids Res. 2011;39:9130–9138. doi: 10.1093/nar/gkr625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kehres DG, Maguire ME. Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria. FEMS Microbiol. Rev. 2003;27:263–290. doi: 10.1016/S0168-6445(03)00052-4. [DOI] [PubMed] [Google Scholar]
- 33.Anjem A, Varghese S. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli . Mol. Microbiol. 2009;72:844–858. doi: 10.1111/j.1365-2958.2009.06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horsburgh MJ, Wharton SJ, Cox AG, Ingham E, Peacock S, Foster SJ. MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol. Microbiol. 2002;44:1269–1286. doi: 10.1046/j.1365-2958.2002.02944.x. [DOI] [PubMed] [Google Scholar]
- 35.Que Q, Helmann JD. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol. Microbiol. 2000;35:1454–1468. doi: 10.1046/j.1365-2958.2000.01811.x. [DOI] [PubMed] [Google Scholar]
- 36.Kim E-K, Cha C-J, Cho Y-J, Cho Y-B, Roe J-H. Synthesis of γ-glutamylcysteine as a major low-molecular-weight thiol in lactic acid bacteria Leuconostoc spp. Biochem. Biophys. Res. Commun. 2008;369:1047–1051. doi: 10.1016/j.bbrc.2008.02.139. [DOI] [PubMed] [Google Scholar]
- 37.Lamers AP, Keithly ME, Kim K, Cook PD, Stec DF, Hines KM, Sulikowski GA, Armstrong RN. Synthesis of bacillithiol and the catalytic selectivity of FosB-type fosfomycin resistance proteins. Org. Lett. 2012;14:5207–5209. doi: 10.1021/ol302327t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wongsantichon J, Robinson RC, Ketterman AJ. Structural evidence for conformational changes of Delta class glutathione transferases after ligand binding. Arch. Biochem. Biophys. 2012;521:77–83. doi: 10.1016/j.abb.2012.03.023. [DOI] [PubMed] [Google Scholar]
- 39.Newton GL, Leung SS, Wakabayashi JI, Rawat M, Fahey RC. The DinB superfamily includes novel mycothiol, bacillithiol, and glutathione S-transferases. Biochemistry. 2011;50:10751–10760. doi: 10.1021/bi201460j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaluzna A, Bartosz G. Transport of glutathione S-conjugates in Escherichia coli . Biochem. Mol. Biol. Int. 1997;43:161–171. doi: 10.1080/15216549700203931. [DOI] [PubMed] [Google Scholar]
- 41.Newton GL, Fahey RC, Rawat M. Detoxification of toxins by bacillithiol in Staphylococcus aureus . Microbiology. 2012;158:1117–1126. doi: 10.1099/mic.0.055715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rigsby RE, Rife CL, Fillgrove KL, Newcomer ME, Armstrong RN. Phosphonoformate: a minimal transition state analogue inhibitor of the fosfomycin resistance protein, FosA. Biochemistry. 2004;43:13666–13673. doi: 10.1021/bi048767h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.