Abstract
Chronic obstructive pulmonary disease (COPD) is a clinically heterogeneous disease composed of variable degrees of airflow obstruction, emphysematous destruction, and small airway wall thickening. The natural history of this disease, although generally characterized by continued decline in lung function, is also highly variable. Novel transcriptomic approaches to study the airway and lung tissue in COPD hold the potential to improve our understanding of the molecular mechanisms underlying this heterogeneity and identify molecular subtypes of disease that have similar clinical manifestations. This new understanding can be leveraged to develop targeted COPD therapies and ultimately personalize treatment of COPD based on each patient’s specific molecular subphenotype.
Keywords: chronic obstructive pulmonary disease, airway gene expression, bioinformatics, class discovery, drug discovery
Chronic obstructive pulmonary disease (COPD) remains a leading cause of morbidity and mortality worldwide (1, 2). Although cigarette smoking is the major cause of COPD in developed countries, several other environmental exposures and chronic inflammatory states (3) also predispose to this disease, which is characterized by an incompletely reversible obstruction to the flow of air. Although this clinical definition of COPD can be assessed quickly and noninvasively using office spirometry (4, 5), COPD is in fact a very heterogeneous disease characterized by a wide range of symptoms, clinical findings, radiologic and pathologic abnormalities, and varied responses to treatment and outcomes.
There is a critical need to understand the molecular pathways that underlie the heterogeneity within COPD to better understand mechanisms of disease pathogenesis and personalize management of this chronic debilitating disease. Although the molecular heterogeneity underlying a number of cancers, including lung and breast cancer, has led to significant advances in targeted therapy for these conditions (6–10), we have yet to see these types of advances in COPD. In this review, we describe how transcriptomic profiling of airway and lung tissue within COPD can be used to provide insight into the molecular pathways underlying this heterogeneous disease and the potential to personalize its management.
The Clinical, Pathologic, and Radiographic Heterogeneity Underlying COPD
Although there is a definition of COPD used clinically, there is a significant amount of heterogeneity in disease presentation and clinical manifestations. A low ratio of FEV1 to FVC after an inhaled bronchodilator is used to define the presence of COPD (11). This represents an incompletely reversible obstruction to the flow of air through the airways. The grade, or severity, of COPD is based on level of impairment in FEV1 as a percentage of the predicted value (11), which reflects a decrease in the volume of air forcibly exhaled from the lungs during the beginning of exhalation. Beyond this simple clinical definition of COPD, however, there are other clinical, radiographic, and pathologic features associated with this disease that vary markedly across individuals with this disease.
Studies aimed at understanding molecular phenotypes of COPD have likely been limited by this underlying clinical heterogeneity. COPD can be associated with a varying degree of exacerbations, or acute episodes of worsening symptoms, such as increased cough, sputum production, and shortness of breath. The severity and frequency of these exacerbations vary across individuals (12).
Pathologic changes in COPD also vary across individuals, which further complicates studies aimed at uncovering molecular phenotypes using diseased tissue specimens. Small airway wall thickening is one of these pathologic phenotypes and is believed to be related to tissue repair and remodeling and a defect of mucociliary clearance (13, 14). Emphysema, or destruction of the alveolar septal walls without fibrosis (15) leading to overinflation and impairment in gas exchange, occurs in varying amounts among patients with airflow obstruction due to COPD. Furthermore, the degree of emphysema varies regionally within the lungs of smokers with COPD, representing the complex relationship between cigarette smoking and resultant lung disease related to emphysema (16). Additionally, there is cellular heterogeneity within lung tissue samples from smokers with COPD, with a larger extent of inflammation by neutrophils, macrophages, B lymphocytes, and CD4 cells in lung tissue from patients with more severely impaired FEV1.
Radiologic measurements can be used to estimate some of these pathologic parameters. For example, quantitative computerized tomography (CT) has been used to quantify the amount of emphysema and degree of small airway wall thickening (17). Because this approach is less invasive than sampling lung tissue, it might eventually be used on a larger number of patients with COPD. The clinical importance of these COPD subphenotypes remains unclear, as is the potential for therapies that might show preferential benefit in a particular subtype.
Beyond the symptomatic, radiographic, and pathologic manifestations of the disease, there is also considerable variability in disease progression and response to therapy. Although a hallmark of COPD is a more rapid decline in FEV1 compared with individuals without this disease, the rate of disease-associated decline is highly variable (18). Variable outcomes of clinical trials of inhaled therapies (19, 20) suggest that the response to specific therapy may be similarly heterogeneous. There is therefore a clear and present need to identify the molecular pathways that give rise to the complex clinical, radiographic, and pathologic diversity of COPD. Furthermore, it is also possible that there are distinct pathways that contribute to molecular heterogeneity independently of the observed clinical heterogeneity.
Airway Gene Expression Reflects Genomic Alterations within the Diseased Lung
Whole-genome gene expression profiling is a powerful tool for characterizing the molecular changes underlying COPD as well as the heterogeneity among patients with COPD (Table 1). Early studies of gene expression in COPD used profiling of lung tissue specimens obtained during surgical resections for suspicious lung nodules, lung cancer, or advanced COPD/emphysema in the setting of lung volume reduction surgery or lung transplant (21–25). Although these studies have identified a number of molecular processes contributing to COPD pathogenesis, their impact has been limited by a number of factors. First, they have relatively small sample sizes, due to the difficulty in obtaining large numbers of tissue specimens. Second, they are limited by confounding variables, such as differences in smoking status, cumulative smoke exposure, or comorbidities such as lung cancer. Third, they may be impacted by disease-associated differences in cellular composition between diseased and nondiseased lung tissue. Nevertheless, these studies have provided key insights into potential genes and pathways underlying COPD.
Table 1.
Overview of microarray studies of chronic obstructive pulmonary disease
| Tissue Studied | Reference | Study Design | Clinical Setting | Clinical Disease Phenotypes Studied |
|---|---|---|---|---|
| Lung tissue | GSE1122 (21) | Case vs. control | Lung tissue from patients undergoing lung transplantation and donors whose lungs could not be used for transplant | Severe emphysema and α1-antitrypsin deficiency |
| GSE8500 (24) | Case vs. control | Lung tissue resected for lung nodules | Airflow obstruction | |
| GSE8581 (25) | Case vs. control | Noncancerous resected lung tissue | Airflow obstruction | |
| GSE1650 (23) | Case vs. control | Lung tissue resected during lung volume reduction surgery or for suspicion of lung cancer | Airflow obstruction | |
| Ning et al. 2004 (22) | Case vs. control | Surgically resected lung tissue | Airflow obstruction | |
| GSE27597 (16) | Regional disease severity across multiple samples from each lung | Lung tissue from patients undergoing lung transplantation and donors whose lungs could not be used for transplant | Emphysema severity (mean linear intercept between alveolar walls) | |
| Airway epithelium | Pierrou et al. 2007 (36) | Case vs. control | Bronchial brushings | Smoking status, airflow obstruction |
| GSE7832 (35) | Case vs. control | Brushings of the 10th-12th generation bronchi | Smoking status, airflow obstruction | |
| GSE37147 (34) | Case vs. control | Brushings of 6th-8th generation bronchi | Airflow obstruction |
One approach to studying heterogeneity in COPD is to leverage the regional differences in emphysema severity within a single lung. Using multiple samples obtained from a lung, each taken from a region with different emphysema severity, a 127-gene expression signature of regional emphysema severity was developed (16). This signature reflects an emphysema-associated increase in B-cell abundance and a decrease in transforming growth factor (TGF)-β signaling (16). Similar approaches leveraging the clinical heterogeneity of COPD might lead to even more insights about the molecular mechanisms underlying these different disease-associated phenotypes and potential novel therapeutics targeted to these different subtypes of the disease.
Another approach to the study of COPD is to leverage the airway field of injury paradigm (26). Because bronchial brushings collect a relatively pure population of epithelial cells (27), this provides a method to collect a more uniform cell population from potentially larger cohorts than lung tissue collection. If the bronchial epithelium reflects disease-associated molecular alterations occurring more proximal to the site of disease, profiling these epithelial cells would provide an easy way to study disease-associated processes as well as provide an avenue for measuring the activity of these processes as part of the clinical management of COPD. The airway field of injury hypothesis, initially established in the setting of cigarette smoke exposure, posits that toxins and carcinogens from cigarette smoke bathe the entire respiratory tract. This exposure leads to measurable alterations in the small airway epithelium (28), large airway epithelium (27), and epithelium of the nose and mouth (29, 30). A disease-specific airway field of injury has been described in the setting of lung cancer, where gene expression profiling of the cytologically normal airway epithelium distal from the lung tumor can serve as a sensitive and specific biomarker of lung cancer (31) that performs independently of other clinical variables (32). These cancer-associated changes in the cytologically normal airway epithelium are similar to those in the distal lung cancers in independent datasets, suggesting that a field of transcriptomic alterations occurs throughout the respiratory tract that reflects the presence of lung cancer. This cancer-associated field of injury extends to the precancerous state, where cytologically normal airway epithelial cells reflect reversible oncogenic pathway activation in patients with airway dysplasia (33), a precancerous lesion.
A similar field of injury has recently been demonstrated for COPD (34), suggesting a potential method by which it may be possible to readily characterize the molecular heterogeneity of COPD in patients with impaired lung function. Early studies of airway gene expression in COPD focused specific pathways believed to be involved in COPD pathogenesis. Tilley and colleagues (35) described alterations in the NOTCH pathway associated with COPD, and Pierrou and colleagues (36) studied the expression of oxidant/antioxidant response genes using microarrays. More recently, analysis of gene expression profiling on a genomewide scale has revealed a 98-gene expression signature of COPD and lung function impairment (Figure 1) that mirrors distal disease-associated changes in independent small airway and lung tissue datasets (34). Genes increased in this bronchial airway signature of COPD appear to be regulated in part by activating transcription factor 4 (ATF4), a key mediator of the unfolded protein response (34).
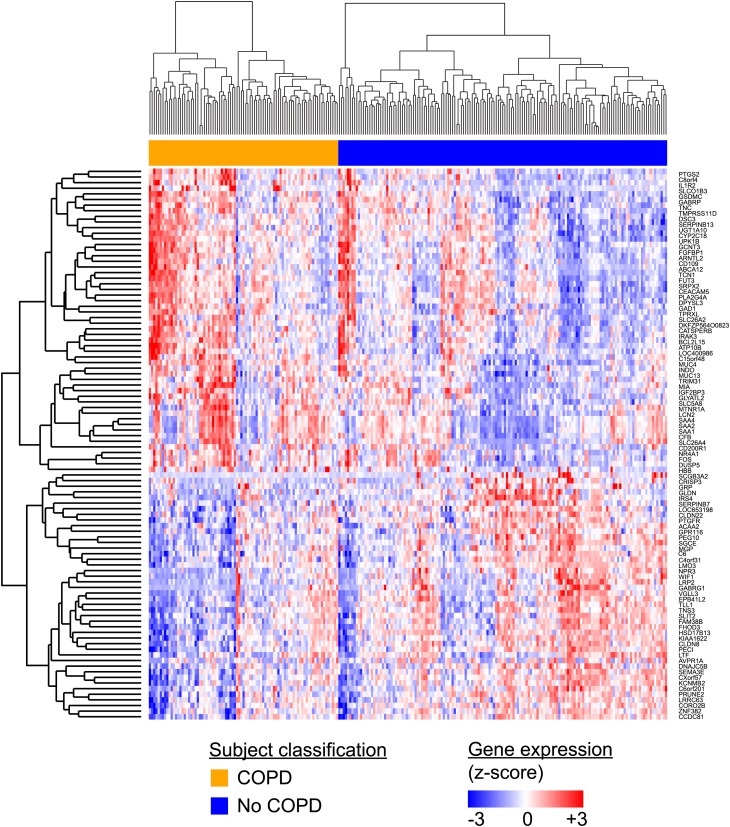
Figure 1.
Heterogeneity in an airway gene expression signature of chronic obstructive pulmonary disease (COPD). Using gene expression profiling of the airway epithelial samples obtained during bronchoscopy from 238 active and former smokers with and without COPD, Steiling and colleagues (34) described a 98-gene expression signature of COPD and lung function impairment that reflects disease-associated changes present in distal lung tissue from independent datasets. Despite the consistent association of this signature with disease, there remains substantial heterogeneity across individuals with and without COPD that may reflect both the clinical heterogeneity of this disease as well as distinct molecular subtypes of the disease. Reprinted by permission from Reference 34.
Airway Gene Expression as Therapeutic Biomarker in COPD
The observation that gene expression profiles in relatively accessible bronchial airways reflect disease-associated processes occurring deep in the lung raises the possibility of developing clinically relevant biomarkers to guide therapy. One of the more compelling findings of the airway gene expression study described above was the dynamic nature of the transcriptomic alterations post treatment with inhaled corticosteroids (34). Using Gene Set Enrichment Analysis (37), the 98-gene signature was found to be inversely correlated with genes that changed 30 months post treatment with fluticasone with or without salmeterol therapy within the GLUCOLD study, a cohort in which these therapies reduced the decline in lung function (19, 34). The dynamic nature of this “field of molecular injury” with treatment suggests that gene expression might serve as an intermediate marker of therapeutic efficacy. To that end, the GLUCOLD investigators have recently demonstrated that a more pronounced treatment-induced alteration in airway gene expression was significantly associated with a lower rate of decline in FEV1 as well as health status measured with the St. George’s Respiratory Questionnaire (38). Together, these studies emphasize the key components of deriving and validating disease and disease subtype signatures, including the critical step of validation in samples from independent cohorts. There are a number of computational approaches to the general problem of sample classification, and the field of cancer genomics has been at the forefront of adapting these computational approaches to gene expression data to develop clinically useful gene expression signatures (39). The possibility of more easily obtainable airway samples that reflect distal disease processes will facilitate the development of these types of clinical gene expression signatures for lung disease.
Beyond serving as an intermediate biomarker of therapeutic efficacy, the true clinical potential of airway gene expression may reside within the molecular heterogeneity found in the airway of smokers with COPD. Heterogeneity in transcriptomic profiles has been leveraged to identify novel molecular subclasses of breast cancer and lymphoma, which has enabled more personalized therapies for those diseases (6, 7, 40). The potential to assess COPD-associated heterogeneity in bronchial epithelium will enable the types of large studies needed to identify molecular subtypes of COPD. Recently, transcriptomic profiling of the airway epithelium has enabled this molecular subtype discovery paradigm to be applied to asthma (41), leading to the identification of a distinct Th-2 cell–driven subphenotype characterized by the increased expression of IL-13–responsive genes (42). Expression of one of these genes, POSTN, and expression of IL-13 itself were found to be higher in the bronchoalveolar lavage fluid and serum of a subset of patients with increased airway hyperresponsiveness and eosinophilia (43) and greater improvement in lung function after treatment with the anti–IL-13 medication lebrikizumab (43). The heterogeneity in the gene expression signature among smokers with COPD (Figure 1) may similarly reflect novel molecular subclasses of disease that can ultimately enable more targeted therapy.
Leveraging Lung Tissue Gene Expression to Repurpose Drug Therapy for COPD
There are a number of in silico approaches to predicting novel treatments for disease. Some strategies involve either network analysis of gene expression data (44) or eQTL analysis (45, 46) that combines gene expression data together with single-nucleotide polymorphism (SNP) genotyping data to identify potential regulatory nodes that are important for driving disease-associated patterns of gene expression. These regulatory nodes can then serve as candidate drug targets. A key advantage of the eQTL approach is that the unidirectional effect of SNPs on gene expression makes it easier to more confidently identify regulators of gene expression if there are SNPs affecting expression of that regulator (46), and this type of approach has led to the identification potential drug targets for traits related to obesity and diabetes (47).
Other strategies, which have been especially productive for rapidly identifying candidate therapies, involve comparing disease-associated patterns of gene expression to gene expression alterations that have been observed in other systems. One example of this type of strategy involves comparing disease-associated gene expression differences to the gene expression effects of bioactive compounds on cells in vitro. The Connectivity Map (48) is a compendium of several thousand microarray profiles generated from five human cancer cell lines treated with varying doses of more than 1,000 compounds (Figure 2). In addition to providing access to in vitro data, this resource also provides an online tool for interrogating these patterns of gene expression (48). By querying disease signatures against drug signatures, it is possible to identify existing compounds that cause a more “healthy-like” pattern of gene expression and are predicted therefore to potentially serve as new treatments for that disease. As an example, the Connectivity Map has been leveraged to predict valproic acid, an anti-seizure medication, as a potential therapy for triple-negative breast cancer (49). The ability of valproic acid to inhibit triple-negative breast cancer was subsequently validated in an in vivo tumor-specific xenograft model (49). The Connectivity Map has also been used to identify topiramate, another anti-seizure medication, as a potential therapy for inflammatory bowel disease, a prediction that was validated in a rat model of this disease (50).
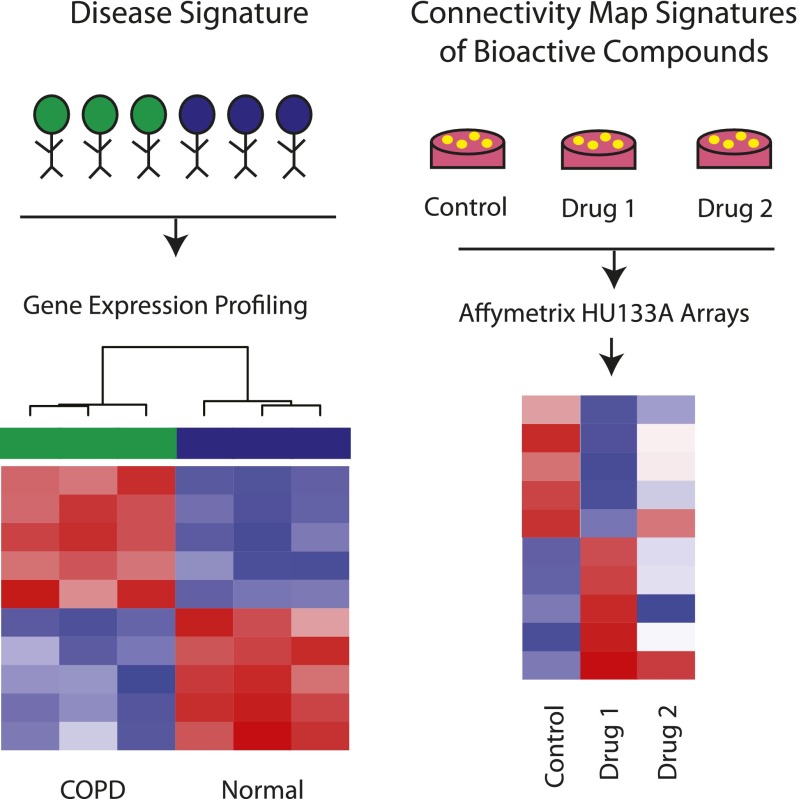
Figure 2.
Repositioning existing drugs for chronic obstructive pulmonary disease (COPD) using the Connectivity Map. The Connectivity Map is a freely available resource for interrogating disease-associated expression patterns against drug signatures. The Connectivity Map contains more than 7,000 microarray expression profiles generated from the treatment of five cancer cell lines with 1,309 bioactive compounds. After development of an in vivo disease signature (left), the Connectivity Map can be queried to find compounds that reverse the disease signature (right). In this example, the COPD disease signature is reversed by Drug 1, but not by Drug 2.
A related approach involving comparisons between gene expression patterns has been used to identify disease-specific pathway dysregulation that can be targeted with pathway-directed medications. Beyond treating cells in vitro with bioactive compounds, it is possible to specifically activate or repress different cellular pathways through targeted overexpression of key regulators or inhibitors. When this is followed by gene expression profiling relative to control cells, it is possible to develop gene expression signatures of pathway activation.
Following this strategy, a gene expression signature of PI3K pathway activation was developed in an epithelial cancer cell line and used to predict PI3K pathway activity in normal bronchial epithelium from patients with lung cancer or airway dysplasia (33). Using this approach, up-regulation of PI3K pathway activity was identified in the cytologically normal airway epithelial cells from individuals with lung cancer and those with precancerous airway dysplasia. Moreover, this activation was reversible among individuals with dysplasia treated with myoinositol, an inhibitor of the PI3K pathway, in whom airway dysplasia improved, suggesting that myoinositol might serve as a chemopreventive agent in lung cancer by reversing oncogenic pathway activation. This illustrates the potential usefulness of coupling compound-based and pathway-based approaches.
The combination of compound-based and pathway-based approaches to repositioning existing medications for new clinical indications have been recently leveraged in the search for new emphysema therapeutics. Using curated pathway lists from publicly available sources (37), gene expression profiles associated with regional emphysema severity were enriched in genes in the TGF-β pathway (16). In parallel analysis of the Connectivity Map, the gene expression signature of the tripeptide compound GHK was found to be the inverse of the emphysema gene expression signature, suggesting that it might serve as a potential therapy for this disease (16). GHK treatment of a disease-relevant in vitro model was found to induce TGF-β pathway gene expression and reverse emphysema-associated gene expression patterns. Furthermore, treatment with either GHK or TGF-β restored collagen I contraction and remodeling in cultured fibroblasts from patients with COPD (16). This example highlights the usefulness of coupling pathway-based and compound-based approaches to develop relevant therapeutics capable of impacting disease-related phenotypes.
Conclusions and Future Directions
Transcriptomic signatures of the airway and lung in COPD have begun to provide us with novel insights into the molecular pathways that underlie the clinical phenotypes associated with the disease and have begun to suggest novel therapeutic opportunities. Importantly, the molecular profiles from the airway of smokers with COPD may ultimately serve as clinically relevant biomarkers of therapeutic response and have the potential to reflect molecular subtypes of disease. Airway transcriptomic studies on larger numbers of patients enrolled in clinical trials with therapeutics targeting disease-specific pathways are needed to better assess this molecular heterogeneity and its potential to guide therapy. Airway epithelial cells represent a promising biospecimen for studying COPD, as molecular profiles in minimally invasively collected tissue specimens will be needed if these biomarkers are to ultimately translate into the clinic. Beyond the potential of bronchial epithelium, which we have highlighted in this review, recent work suggests that gene expression signatures in nasal and buccal epithelium may serve as surrogates for the bronchial airway response to cigarette smoke (29, 30), and future studies should evaluate whether COPD-associated gene expression changes can also be identified at these readily accessible sites. Technologic advances, such as single-cell sequencing (51), also hold the potential to improve characterization of disease heterogeneity by providing a detailed portrait of disease-associated processes occurring in different cell types within a tissue and the heterogeneity of those processes between different cells of the same cell type.
The large amount of transcriptomic data available in the public domain has provided an unprecedented opportunity to identify drugs and other biological perturbations that are relevant to COPD. Although the Connectivity Map has enabled the identification of GHK as a drug candidate for emphysema, profiling of various lung cell lines with a large compendium of drugs/small molecules might help facilitate repositioning of drugs for chronic lung disease if they help capture relevant lung-specific effects. In vitro and animal models of COPD that recapitulate human disease pathogenesis, however, will be needed to test the large number of hypotheses generated by these types of experiments. Additionally, gene expression signatures of biological perturbations related to environmental stimuli (e.g., cytokines, environmental exposures) may provide insights into disease pathogenesis and help explain the mechanisms that give rise to disease heterogeneity.
Footnotes
Supported by National Institutes of Health/National Heart, Lung, and Blood Institute grants 1R01 HL095388 (A.S., M.E.L.) and KL2RR025770 (K.S.).
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.National Heart Lung and Blood Institute 2010NHLBI Fiscal Year 2010 Fact Book [Accessed August 26, 2013]Available from: http://www.nhlbi.nih.gov/about/factbook-10/Factbook_2010.pdf
- 2.Murphy SL, Xu J, Kochanek KD. Deaths: Preliminary Data for 2012. Natl Vital Stat Rep. 2012;60:1–8. [PubMed] [Google Scholar]
- 3.Barnes PJ. Chronic obstructive pulmonary disease. N Engl J Med. 2000;343:269–280. doi: 10.1056/NEJM200007273430407. [DOI] [PubMed] [Google Scholar]
- 4.American Thoracic Society. Standardization of spirometry, 1994 Update. Am J Respir Crit Care Med. 1995;152:1107–1136. doi: 10.1164/ajrccm.152.3.7663792. [DOI] [PubMed] [Google Scholar]
- 5.Miller MR, Hankinson J, Busasco V, Burgos F, Basaburi R, Coates A, Crapo R, Enright P, van der Grinen CPM, Gustafsson P, et al. Standardisation of spirometry. Eur Respir J. 2005;26:319–338. doi: 10.1183/09031936.05.00034805. [DOI] [PubMed] [Google Scholar]
- 6.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perou CM, Serlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumors. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 8.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 9.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 10.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13011. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Global Initiative for Chronic Obstructive Lung Disease 2011. Global Initiative for Chronic Obstructive Lung Disease: global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease [Accessed August 26, 2013]. Available from: http://www.goldcopd.org/uploads/users/files/GOLD_Report_2011_Feb21.pdf
- 12.Hurst JR, Vestbo J, Anzueto A, Locantore N, Mullerova H, Tal-Singer R, Miller B, Lomas DA, Agusti A, MacNee W, et al. Susceptibility to exacerbation in chronic obstructive pulmonary disease. N Engl J Med. 2010;363:1128–1138. doi: 10.1056/NEJMoa0909883. [DOI] [PubMed] [Google Scholar]
- 13.Hogg JC, Chu F, Utokaparch S, Woods R, Elliott M, Buzatu L, Cherniack RM, Rogers RM, Sciurba FC, Coxson HO, et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N Engl J Med. 2004;350:2645–2653. doi: 10.1056/NEJMoa032158. [DOI] [PubMed] [Google Scholar]
- 14.McDonough JE, Yuan R, Suzuki M, Seyednejad N, Elliott WM, Sanchez PG, Wright AC, Gefter WB, Litzky L, Coxson HO, et al. Small-airway obstruction and emphysema in chronic obstructive pulmonary disease. N Engl J Med. 2011;365:1567–1575. doi: 10.1056/NEJMoa1106955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hogg JC, Senior RM. Chronic obstructive pulmonary disease c2: pathology and biochemistry of emphysema. Thorax. 2002;57:830–834. doi: 10.1136/thorax.57.9.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell JD, McDonough JE, Zeskind JE, Hackett TL, Pechkovsky DV, Bradsma CA, Suzuki M, Gosselink JV, Liu G, Alekseyev YO, et al. A gene expression signature of emphysema-related lung destruction and its reversal by the tripeptide GHK. Genome Med. 2012;4:67. doi: 10.1186/gm367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coxson HO, Mayo J, Lam S, Santyr G, Parraga G, Sin DD. New and current clinical imaging techniques to study chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2009;180:588–597. doi: 10.1164/rccm.200901-0159PP. [DOI] [PubMed] [Google Scholar]
- 18.Vestbo J, Edwards L, Scanlon PD, Yates JC, Agusti A, Bakke P, Calverley PMA, Celle B, Coxson HO, Crim C, et al. Changes in forced expiratory volume in 1 second over time in COPD. N Engl J Med. 2011;365:1184–1192. doi: 10.1056/NEJMoa1105482. [DOI] [PubMed] [Google Scholar]
- 19.Lapperre TS, Snoeck-Stroband JB, Gosman MM, Jansen DF, van Schadewijk A, Thiadens HA, Vonk JM, Boezen HM, Ten Hacken NH, Sont JK, et al. Groningen Leiden Universities Corticosteroids in Obstructive Lung Disease Study Group. Effect of fluticasone with and without salmeterol on pulmonary outcomes in chronic obstructive pulmonary disease: a randomized trial. Ann Intern Med. 2009;151:517–527. doi: 10.7326/0003-4819-151-8-200910200-00004. [DOI] [PubMed] [Google Scholar]
- 20.Calverley PM, Anderson JA, Celli B, Ferguson GT, Jenkins C, Jones PW, Yates JC, Vestbo J. Salmeterol and fluticasone propionate and survival in chronic obstructive pulmonary disease. N Engl J Med. 2007;356:775–789. doi: 10.1056/NEJMoa063070. [DOI] [PubMed] [Google Scholar]
- 21.Golpon HA, Coldren CD, Zamora MR, Cosgrove GP, Moore MD, Tuder RM, Geraci MW, Voelkel NF. Emphysema lung tissue gene expression profiling. Am J Respir Cell Mol Biol. 2004;31:595–600. doi: 10.1165/rcmb.2004-0008OC. [DOI] [PubMed] [Google Scholar]
- 22.Ning W, Li CJ, Kaminski N, Feghali-Bostwick CA, Alber SM, Di YP, Otterbein SL, Song R, Hayashi S, Zhou Z, et al. Comprehensive gene expression profiles reveal pathways related to the pathogenesis of chronic obstructive pulmonary disease. Proc Natl Acad Sci USA. 2004;101:14895–14900. doi: 10.1073/pnas.0401168101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spira A, Beane J, Pinto-Plata V, Kadar A, Liu G, Shah V, Celli B, Brody JS. Gene expression profiling of human lung tissue from smokers with severe emphysema. Am J Respir Cell Mol Biol. 2004;31:601–610. doi: 10.1165/rcmb.2004-0273OC. [DOI] [PubMed] [Google Scholar]
- 24.Wang IM, Stepaniants S, Boie Y, Mortimer JR, Kennedy B, Elliott M, Hayashi S, Loy L, Coulter S, Cervino S, et al. Gene expression profiling in patients with chronic obstructive pulmonary disease and lung cancer. Am J Respir Crit Care Med. 2008;177:411. doi: 10.1164/rccm.200703-390OC. [DOI] [PubMed] [Google Scholar]
- 25.Bhattacharya S, Srisuma S, DeMeo DL, Shapiro SD, Bueno R, Silverman EK, Reilly JJ, Mariani TJ. Molecular biomarkers for quantitative and discrete COPD phenotyes. Am J Respir Cell Mol Biol. 2009;40:359–367. doi: 10.1165/rcmb.2008-0114OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steiling K, Ryan J, Brody JS, Spira A. The field of tissue injury in the lung and airway. Cancer Prev Res (Phila) 2008;1:396–403. doi: 10.1158/1940-6207.CAPR-08-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spira A, Beane J, Shah V, Liu G, Schembri F, Yang X, Palma J, Brody JS. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci USA. 2004;101:10143–10148. doi: 10.1073/pnas.0401422101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harvey BG, Heguy A, Leopold PL, Carolan BJ, Ferris B, Crystal RG. Modification of gene expression of the small airway epithelium in response to cigarette smoking. J Mol Med. 2007;85:39–53. doi: 10.1007/s00109-006-0103-z. [DOI] [PubMed] [Google Scholar]
- 29.Sridhar S, Schembri F, Zeskind J, Shah V, Gustafson AM, Steiling K, Liu G, Dumas YM, Zhang X, Brody JS, et al. Smoking-induced gene expression changes in the bronchial airway are reflected in nasal and buccal epithelium. BMC Genomics. 2008;9:259. doi: 10.1186/1471-2164-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Sebastiani P, Liu G, Schembri F, Zhang X, Dumas YM, Langer EM, Alekseyev Y, O'Connor GT, Brooks DR, et al. Similarities and differences between smoking-related gene expression in the nasal and bronchial epithelium. Physiol Genomics. 2010;41:1–8. doi: 10.1152/physiolgenomics.00167.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spira A, Beane JE, Shah V, Steiling K, Liu G, Schembri F, Gilman S, Dumas YM, Calner P, Sebastiani P, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;13:361–366. doi: 10.1038/nm1556. [DOI] [PubMed] [Google Scholar]
- 32.Beane J, Sebastiani P, Whitfield TH, Steiling K, Dumas YM, Lenburg ME, Spira A. A prediction model for lung cancer diagnosis that integrates genomic and clinical features. Cancer Prev Res (Phila) 2008;1:56–64. doi: 10.1158/1940-6207.CAPR-08-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gustafson AM, Soldi R, Anderlind C, Scholand MB, Qian J, Zhang X, Cooper K, Walker D, McWilliams A, Liu G, et al. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci Transl Med. 2010;2:26ra25. doi: 10.1126/scitranslmed.3000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steiling K, van den Berge M, Hijazi K, Florido R, Campbell J, Liu G, Xiao J, Zhang X, Duclos G, Drizik E, et al. A dynamic bronchial airway gene expression signature of COPD and lung function impairment. Am J Respir Crit Care Med. 2013;187:933–942. doi: 10.1164/rccm.201208-1449OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tilley AE, Harvey BG, Heguy A, Hackett NR, Wang R, O’Connor TP, Crystal RG. Down-regulation of the Notch pathway in human airway epithelium in association with smoking and chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2009;179:457–466. doi: 10.1164/rccm.200705-795OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pierrou S, Broberg P, O’Donnell RA, Pawlowski K, Virtala R, Lindqvist E, Richter A, Wilson SJ, Angco G, Moller S, et al. Expression of genes involved in oxidative stress responses in airway epithelial cells of smokers with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2007;175:577–586. doi: 10.1164/rccm.200607-931OC. [DOI] [PubMed] [Google Scholar]
- 37.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van den Berge M, Steiling K, Timens W, Hiemstra PS, Sterk PJ, Heijink IH, Liu G, Alekseyev Y, Lenburg ME, Spira A, et al. Airway gene expression in COPD is dynamic with inhaled corticosteroid treatment and reflects biological pathways associated with disease activity. Thorax. 2013;187:933–942. doi: 10.1136/thoraxjnl-2012-202878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon R. Diagnostic and prognostic prediction using gene expression profiles in high-dimensional microarray data. Br J Cancer. 2003;89:1599–1604. doi: 10.1038/sj.bjc.6601326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monti S, Savage KJ, Kutok JL, Feuerhake F, Kurtin P, Mihm M, Wu B, Pasqualucci L, Neuberg D, Aguiar RCT, et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust sybtypes including one characterized by host inflammatory response. Blood. 2005;105:1851–1861. doi: 10.1182/blood-2004-07-2947. [DOI] [PubMed] [Google Scholar]
- 41.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, Ellwanger A, Sidu SS, Dao-Pick TP, Pantoja C, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci USA. 2007;104:15858–15863. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, Arron JR, Koth LL, Fahy JV. T-helper type2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180:388–395. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corren J, Lemanske RF, Hanania NA, Korenblat PE, Parsey MV, Aaron JR, Harris JM, Schreerens H, Wu LC, Su Z, et al. Lebrikizumab treatment in adults with asthma. N Engl J Med. 2011;365:1088–1098. doi: 10.1056/NEJMoa1106469. [DOI] [PubMed] [Google Scholar]
- 44.Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, Kasif S, Collins JJ, Gardner TS. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang K, Narayanan M, Zhong H, Tompa M, Schadt EE, Zhu J. Meta-analysis of inter-species liver co-expression networks elucidates traits associated with common human diseases. PLOS Comput Biol. 2009;5:e1000616. doi: 10.1371/journal.pcbi.1000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Millstein J, Zhang B, Zhu J, Schadt EE. Disentangling molecular relationships with a causal inference test. BMC Genet. 2009;10:23. doi: 10.1186/1471-2156-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 49.Cohen AL, Soldi R, Zhang H, Gustafson AM, Wilcox R, Welm BE, Chang JT, Johnson E, Spira A, Jeffrey SS, et al. A pharmacogenomic method for individualize prediction of drug sensitivity. Mol Syst Biol. 2011;7:513. doi: 10.1038/msb.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dudley JT, Sirota M, Shenoy M, Pai RK, Roedder S, Chiang AP, Morgan AA, Sarwal MM, Pasricha PJ, Butte AJ. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med. 2011;3:96ra76. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdury R, Schwartz S, Yosef N, Malboeuf C, Lu D, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]