Abstract
Recently, it was reported that mutations in the ubiqutin-like protein ubiquilin-2 (UBQLN2) are associated with X-linked amyotrophic lateral sclerosis (ALS), and that both wild-type and mutant UBQLN2 can co-localize with aggregates of C-terminal fragments of TAR DNA binding protein (TDP-43). Here, we describe a high affinity interaction between UBQLN2 and TDP-43 and demonstrate that overexpression of both UBQLN2 and TDP-43 reduces levels of both exogenous and endogenous TDP-43 in human H4 cells. UBQLN2 bound with high affinity to both full length TDP-43 and a C-terminal TDP-43 fragment (261-414 aa) with KD values of 6.2 nM and 8.7 nM, respectively. Both DNA oligonucleotides and 4-aminoquinolines, which bind to TDP-43, also inhibited UBQLN2 binding to TDP-43 with similar rank order affinities compared to inhibition of oligonucleotide binding to TDP-43. Inhibitor characterization experiments demonstrated that the DNA oligonucleotides noncompetitively inhibited UBQLN2 binding to TDP-43, which is consistent with UBQLN2 binding to the C-terminal region of TDP-43. Interestingly, the 4-aminoquinolines were competitive inhibitors of UBQLN2 binding to TDP-43, suggesting that these compounds also bind to the C-terminal region of TDP-43. In support of the biochemical data, co-immunoprecipation experiments demonstrated that both TDP-43 and UBQLN2 interact in human neuroglioma H4 cells. Finally, overexpression of UBQLN2 in the presence of overexpressed full length TDP-43 or C-terminal TDP-43 (170-414) dramatically lowered levels of both full length TDP-43 and C-terminal TDP-43 fragments (CTFs). Consequently, these data suggest that UBQLN2 enhances the clearance of TDP-43 and TDP-43 CTFs and therefore may play a role in the development of TDP-43 associated neurotoxicity.
Keywords: TDP-43, UBQLN2, 4-aminoquinolines, protein-protein interactions
1.0 Introduction
The ubiquilins (UBQLNs) are a family of 60-70 kDa ubiquitin-like proteins which include four distinct genes of UBQLNs (UBQLN1, 2, 3, 4) that have a high degree of homology between one another1, 2. While expression of UBQLN3 is confined to the testes, the other three isoforms are widely expressed throughout most other tissues3. UBQLNs are typically expressed in the cytosol, but can also be found in the nucleus as well as associated with membrane structures such as the endoplasmic reticulum and plasma membrane4-6. UBQLNs are composed of an N-terminal ubiquitin-like (UBL) domain and a C-terminal ubiquitin associated (UBA) domain7. The UBL domain interacts with the S5a component of the 19S regulatory cap complex of the 26S proteasome7, 8. The UBA domain interacts with ubiquitinated chains on proteins that have been covalently modified by ubiquitin ligases9-11. Therefore, one of the functions of UBQLNs is to provide a link between ubiquitinated proteins targeted for degradation and the protesome9, 12. UBQLNs are also involved in autophagy and augment maturation of autophagosomes leading to improved cell survival under conditions of nutrient deprivation13, 14. In addition, UBQLNs link integrin-associated protein (IAP, CD47) to the cytoskeleton (PLIC) and are involved in certain cell adhesion and migration processes4, 15 as well as in regulating some aspects of G protein signaling and G protein coupled receptor (GPCR) internalization16, 17.
Recently, mutations in UBQLN2 have been identified as a genetic marker for dominant X-linked juvenile and adult onset amyotrophic lateral sclerosis (ALS) and ALS with dementia18-21. Most of the mutations that have been identified are in a stretch of proline residues in the PXX domain of UBQLN218, 20, but more recently, some other mutations outside this domain have also been identified19, 21. The PXX domain is a proline-rich motif that is often important for protein-protein interactions22. Therefore, mutations in this domain might be expected to greatly affect UBQLN2 function. Similar to other proteins associated with neurodegeneration, mutations in UBQLN2 lead to aggregation and subsequent development of cytoplasmic inclusions in spinal cord and other neuronal tissues18, 19, 23. These aggregates of UBQLN2 can also be associated with other proteins such as trans-activating response (TAR) DNA binding protein (TDP-43) and fused in sarcoma protein (FUS)19, 23. Both TDP-43 and FUS are nucleic acid binding proteins that have also been implicated in ALS as well as frontotemporal lobar degeneration (FTLD)24-30. Typically, TDP-43 is predominantly localized to the nucleus, but in ALS and FTLD, TDP-43 is excessively translocated to the cytoplasm where it is metabolized by caspases leading to the accumulation of phosphorylated and ubiquitinylated TDP-43 C-terminal fragments (CTFs) 24, 31-33. Interestingly, mutations in UBQLN2 result in both UBQLN2 and TDP-43 positive cytoplasmic inclusions19, 23. However, UBQLN2 and TDP-43 are not always co-localized in the same aggregates, but the degree of co-localization appears to be dependent upon the location of the UBQLN2 mutation, being more common in patients where the mutation in UBQLN2 is just upstream of the PXX domain19. In addition, it has been demonstrated in cell culture models that both UBQLN2 and C-terminal TDP-43 (218-414 aa) are co-localized in cytoplasmic aggregates when both proteins are overexpressed. While the significance of these findings is not well understood, it does suggest that there may be a link between UBQLN2 and TDP-43 that warrants further investigation.
The structure of TDP-43 is typical of other heterogeneous nuclear ribonucleoproteins (hnRNPs). The N-terminal region is comprised of two RNA recognition motifs (RRMs), which are the sites of nucleic acid binding, while the C-terminal region is a glycine rich prion-like domain that is important for protein-protein interactions25, 34-36. In this report, we utilized homogenous time-resolved fluorescence (HTRF) technology to characterize a high-affinity interaction between UBQLN2 and TDP-43. We have determined that UBQLN2 binds to the C-terminal region of TDP-43, which is consistent with the findings of other investigators who demonstrated co-localization of cytoplasmic C-terminal TDP-43 fragments and UBQLN218, 19. Titrations of single stranded DNA oligonucleotides as well as 4-aminoquinlines (AAQs), a series of small molecules which we described previously as inhibitors of nucleic acid binding to TDP-4337, inhibited UBQLN2 binding to TDP-43 with the same rank order potency as inhibition of nucleic acid binding to TDP-43. In addition, we demonstrate that UBQLN2 is noncompetitive with oligonucleotide binding to TDP-43, while AAQ1 is competitive with UBQLN2 binding to TDP-43. These data support the hypotheses that the UBQLN2 binding site is distinct from the nucleic acid binding site and that the AAQs bind to the C-terminal region of TDP-43. To support the findings of the biochemical assay, we also demonstrate that TDP-43 is co-immunoprecipated in untransfected human H4 neuroglioma cells as well as H4 cells transfected with either TDP-43 or UBQLN2. Furthermore, we report that overexpression of UBQLN2 reduces levels of overexpressed full length or C-terminal TDP-43 in human neuroglioma H4 cells, suggesting that the interaction between UBQLN2 and TDP-43 is important for cellular clearance of excess full length and C-terminal fragments of TDP-43.
2.0 Materials and Methods
2.1 Materials
Single stranded DNA oligonucleotides: TG4 (TG x 4, 8 b), TG6, TG8, TG12, TAR32 (5′-CTG CTT TTT GCC TGT ACT GGG TCT CTG TGG TT-3′) and 5′-biotinylated TG12 (bt-TG12) were synthesized by Integrated DNA Technologies (Coralville, IA). 4-aminoquinolines: AAQ1 (3-{[2-(4-methoxyphenyl)quinolin-4-yl]amino}propyl)dimethylamine, compound 1 in ref. 37) and AAQ2 (N-[2-(dimethylamino)ethyl]-N-methyl-2-phenylquinolin-4-amine, compound 9 in ref. 37) were provided by Fox Chase Center for Chemical Diversity, Inc. (Doylestown, PA). Human N-terminal GST tagged full length TDP-43, N-terminal TDP-43 (1-260 aa) and C-terminal TDP-43 (261-414 aa) were purchased from ProteinTech Group (Chicago, IL). N-terminal 6XHIS tagged full length UBQLN2 was custom made by ProteinTech Group. The following anti-tag mouse monoclonal antibodies and streptavidin conjugated to homogenous time-resolved fluorescence (HTRF) donors and acceptors were purchased from Cis-Bio Inc (Bedford, MA): anti-GST terbium (Tb) HTRF donor, anti-HIS d2 HTRF acceptor, streptavidin Tb HTRF donor, anti-GST d2 HTRF acceptor. Human, wild-type, full length TDP-43 (NM_007375.3) was synthesized by Genewiz (South Plainfield, NJ) and inserted into pcDNA 3.2 His/V5 mammalian expression vector (Life Technologies, Grand Island, NY). The caspase-3 cleavage product of human wild-type TDP-43 (170-414aa) was generated PCR and inserted into pcDNA 3.1 mammalian expression vector (Life Technologies) and confirmed by sequencing analysis by Genewiz. Human, wild-type, full length UBQLN2 (NM_013444.3) inside a pCMV-XL5 mammalian expression vector was purchased from Origene (Rockville, MD). Human neuroglioma H4 cells were obtained from the ATCC (Manassas, VA). Control and UBQLN2 specific siRNA was purchased from Life Technologies. The co-immunoprecipation antibodies were rabbit polyclonal anti-TDP-43, N-terminus (ProteinTech Group, 10782-2-AP) and rabbit polyclonal anti-UBQLN2 (PAB1700, Abnova, Tapei City, Taiwan). The following primary antibodies were used for immunblotting: mouse monoclonal anti-TDP-43 for detection of immunoprecipates (60019-2-Ig, ProteinTech Group), rabbit polyclonal anti-TDP-43, C-terminus (12892-1-AP, ProteinTech Group), rabbit polyclonal anti-UBQLN2 (PAB1700, Abnova, Tapei City, Taiwan), and mouse monoclonal anti-GAPDH (Genscript, Piscataway, NJ).
2.2 UBQLN2 binding to TDP-43 using HTRF technology
A series of concentrations of N-terminal HIS-tagged full length UBQLN2 (0-100 ng/mL) was incubated for 2 hr at room temperature with a series of concentrations of N-terminal GST-tagged full length TDP-43 (0-100 ng/mL) in a total volume of 10 μL assay buffer (20 mM Tris, pH 7.0, 2 mM MgCl2, 0.05% CHAPS, 1% glycerol) in white, opaque, low volume 384-well plates. Nonspecific binding was determined in the absence of TDP-43. After incubation, anti-GST Tb HTRF donor (2.6 μg/mL) and anti-HIS d2 HTRF acceptor (2.6 μg/mL) in 5 μL of assay buffer was added to each well and the assays incubated an additional hr at room temperature. Time-resolved fluorescence measurements were obtained using a Synergy 2 plate reader (Biotek, Winooski, VT) using excitation wavelength of 330 nm and emission wavelengths of 620 nm and 665 nm, with a 250 μsec delay between excitation and emission. Data were expressed as a ratio of RFU665/RFU620 X 1000.
2.3 UBQLN2 binding to full length, N-terminal and C-terminal TDP-43 fragments
Assay mixtures for saturation binding experiments contained either full length, N-terminal (1-260 aa) or C-terminal (261-414 aa) GST-tagged TDP-43 (100 ng/mL, 1.6 nM, 2.2, nM, and 4.6 nM, respectively) and a series of concentrations of HIS-tagged UBQLN2 (0.1 nM – 60 nM) in 10 μL assay buffer. After a 2 hr incubation at room temperature, anti-GST and anti-HIS HTRF donor and acceptor antibodies were added and assays incubated an additional hr at room temperature as before. Nonspecific binding was determined in the absence of TDP-43 and was less than 20% of total binding. Time-resolved fluorescence was measured as before and equilibrium dissociation constants (KD) were determined by nonlinear regression fits of specific binding (total binding – nonspecific binding) data to a one-site binding model using Graph Pad Prism®.
For UBQLN2 or TG12 mediated inhibition of bt-TG12 binding to TDP-43, assay mixtures contained the same concentrations of either full length or N-terminal TDP-43 as before along with 1.0 nM of bt-TG12 and a series of concentrations of either unlabeled TG12 (1 pM to 1 μM) or UBQLN2 (0.3 pM to 0.3 μM) in 10 μL of assay buffer. After a 2 hr incubation at room temperature, streptavidin Tb HTRF donor and anti-GST d2 HTRF acceptor (2.6 μg/mL) antibodies were added and assays incubated for an additional hr at room temperature followed by measurement of time resolved fluorescence as before. Data were normalized to % bound, where 100% is the HTRF signal in the absence of TG12 and 0% is the HTRF signal in the presence of 1.0 μM TG12. IC50 values were obtained from nonlinear regression fits of the data to a three parameter dose response model in Graph Pad Prism®.
2.4 Inhibition of UBQLN2 binding and bt-TG12 binding to full length TDP-43
Series of concentrations of test compounds were incubated with full length GST-tagged TDP-43 (100 ng/mL, 1.6 nM) for 15-30 min at room temperature. Assays were initiated by the addition of either HIS-tagged UBQLN2 (100 ng/mL, 1.4 nM) or bt-TG12 (1.0 nM) and then incubated for 2 hr at room temperature. After incubation, anti-GST and anti-HIS HTRF donor and acceptor antibodies were added for assays containing UBQLN2 or streptavidin Tb HTRF donor and anti-GST d2 HTRF acceptor (2.6 μg/mL) for assays containing bt-TG12, followed by an additional 1 hr incubation and then measurement of time-resolved fluorescence as before. IC50 values were obtained from nonlinear regression fits of the data to a three parameter dose response model in Graph Pad Prism®.
2.5 Mode of inhibition of UBQLN2, TG12 and AAQ1 binding to TDP-43
Full length GST-tagged TDP-43 (100 ng/mL) was incubated for 2 hrs with a series of concentrations of HIS-tagged UBQLN2 (1.0 – 60 nM) in the presence of a range of concentrations (0 – 1 μM) of the DNA oligonucleotide TG12 or (0 – 32 μM) of AAQ1 followed by the addition of anti-GST and anti-HIS HTRF donor and acceptor antibodies. Alternatively, TDP-43 was incubated for 2 hrs with a series of concentrations of biotinylated TG12 (3 – 1300 pM) in the presence of a range of concentrations (0 – 32 nM) of UBQLN2, followed by the addition of streptavidin Tb HTRF donor and anti-GST d2 HTRF acceptor (2.6 μg/mL). After additional 1 hr incubation, time resolved fluorescence was measured as before and nonspecific binding was determined in the absence of TDP-43. Comparison of global nonlinear regression fits of the data to either a mixed inhibition model or noncompetitive inhibition model were evaluated using the F-test in Graph Pad Prism®.
2.6 Cell culture and transfection
H4 cells were maintained in DMEM supplemented with 4.5 g/L glucose, 1 mM sodium pyruvate, 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM L-gluatamine, and 10% FBS. Cells were plated in 12-well plates (105 cells/mL) the day before and then transfected with empty pCMV-XL5, His/V5-TDP-43, TDP-43 CTF or UBQLN2 alone or in combination using Transit 20/20 transfection reagent (Mirus Bio LLC, Madison, WI) followed by a 24 hr incubation at 37 °C, 5% CO2. For siRNA experiments, cells were plated at similar density and transfected with either control or UBQLN2 specific siRNA using siQuest transfection reagent (Mirus Bio LLC) followed by a 72 hr incubation under the same conditions. After transfection, cells were either processed for immunoprecipation as described below or prepared directly for immunoblotting through lysis in SDS sample buffer and 10 min incubation at 95 °C.
2.7 Immunoprecipation and immunoblotting
For co-immunoprecipation experiments, H4 cells were plated in 6-well plates and transfected with empty vector, His/V5-TDP-43 (1-414 aa), or full length UBQLN2. The following day after transfection, cells were washed with PBS and detached from the cell culture dish with 2.2 mM EDTA in PBS and centrifuged at 1000 x g for 5 min. After centrifugation, cells were lyzed with 700 μL of IP buffer (25 mM Tris, pH 7.5, 10 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.1% Igepal CA-630, protease inhibitor cocktail (Sigma-Aldrich) for 30 min at room temperature by end-over-end rotation. The cell lysate was incubated for 4 hr at room temperature by end-over-end rotation with either 0.5 μg control rabbit IgG, rabbit polyclonal anti-TDP-43, or rabbit polyclonal UBQLN2. After incubation, the tubes were centrifuged at 12,000 x g to pellet any insoluble material and the supernatant was transferred to a clean microcentrifuge tube containing 25 μL of protein A sepharose (Invitrogen) and incubated for an additional 2 hr at room temperature. After incubation, the tubes were centrifuged at 1000 x g for 2 min and the protein A sepharose was washed quickly 1 × 1 mL with PBS, centrifuged again, and then resuspended in 50 μL of 2X SDS sample buffer and heated to 95 °C for 10 min.
Denatured samples in SDS sample buffer were loaded onto 12% Tris-MOPS polyacrylamide gel (Genscript) and subject to electrophoresis followed by transfer onto Immobilon FL® PVDF membranes (Millipore, Billerica, MA) and blocked for 1 hr with Odyssey blocking buffer (Li-cor Biosciences, Lincoln, NE). Primary antibodies were then added to the blocking buffer at a final dilution of 1:1000 to 1:2000 depending on the antibody, along with 0.1% Tween 20, and the membranes incubated overnight at 4 °C. After incubation, the membranes were washed in PBS + 0.1% Tween 20 (PBST) and anti-mouse (1:20,000) and/or anti-rabbit (1:5000) secondary antibodies conjugated to IRDye® 680 nm or 800 nm (Li-cor) were added to the membranes in Odyssey blocking buffer + 0.2% Tween 20 and 0.01% SDS followed by a 1 hr incubation at room temperature. After incubation, the membranes were washed in PBST, then rinsed with PBS and scanned using an Odyseey CLx infared imaging system (Li-cor).
3.0 Results
3.1 UBQLN2 binding to TDP-43 is concentration dependent
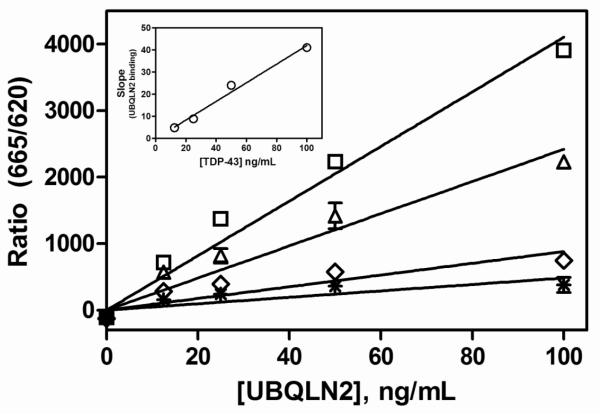
In order to determine if there was a direct interaction between TDP-43 and UBQLN2, we incubated increasing concentrations of full length, N-terminal GST tagged TDP-43 in the presence of increasing concentrations of full length, HIS tagged UBQLN2. Binding was detected by post incubation with specific anti-HIS and anti-GST monoclonal antibodies conjugated to HTRF donor and acceptor moieties, which generate a time-resolved fluorescent signal only when the donor and acceptor are in close proximity. As shown in Figure 1, UBQLN2 binding to TDP-43 was linearly proportional to the concentration of UBQLN2. In addition, plotting the slopes of UBQLN2 binding at each concentration of TDP-43 against the concentration of TDP-43 demonstrate that binding was also linear with the concentration of TDP-43 (Fig 1). Therefore, the binding of UBQLN2 to TDP-43 is dependent upon the concentration of both proteins.
Figure 1. Concentration dependence of UBQLN2 binding to TDP-43.
UBQLN2 (0-100 ng/mL) was incubated with several concentrations of TDP-43 [100 ng/mL (□), 50 ng/mL (△), 25 ng/mL (◇), 12.5 ng/mL ( )] for 1 hr at room temperature followed by the addition of HTRF antibodies with subsequent ratiometric measurement of time-resolved fluorescence as described in Section 2.2. Nonspecific binding was determined in the absence of TDP-43. Inset: Slopes of the lines from UBQLN2 binding plotted against TDP-43 concentration. Data are the means ± S.E.M. of triplicate determinations from a single experiment that was repeated three times with similar results.
)] for 1 hr at room temperature followed by the addition of HTRF antibodies with subsequent ratiometric measurement of time-resolved fluorescence as described in Section 2.2. Nonspecific binding was determined in the absence of TDP-43. Inset: Slopes of the lines from UBQLN2 binding plotted against TDP-43 concentration. Data are the means ± S.E.M. of triplicate determinations from a single experiment that was repeated three times with similar results.
3.2 UBQLN2 binds with high affinity to full length and C-terminal TDP-43
Saturation binding isotherms of UBQLN2 binding to full length TDP-43 demonstrated that the binding was saturable and of high affinity (Fig 2A). The equilibrium dissociation constant (KD) for UBQLN2 binding to full length TDP-43 was 6.2 nM (95% CI: 4.8-8.1, n=4). Previous investigators demonstrated co-localization of UBQLN2 with C-terminal fragments of TDP-43 in cell culture18. Therefore, in order to extend these observations, we conducted saturation binding experiments with C-terminal TDP-43 (261-414 aa) and N-terminal TDP-43 (1-260 aa). UBQLN2 bound with high affinity to C-terminal TDP-43, with a KD value of 8.7 nM (95% CI: 5.1-15, n=4), which is similar to its affinity for full length TDP-43 (Fig 2A). However, we were unable to detect appreciable UBQLN2 binding to N-terminal TDP-43 (data not shown). In order to confirm these observations and verify that our N-terminal TDP-43 protein preparation had functional activity, we measured the ability of both UBQLN2 and the DNA oligonucleotide TG12 (12 TG repeats) to inhibit the binding of biotinylated TG12 (bt-TG12) to both full length and N-terminal TDP-43. UBQLN2 inhibited bt-TG12 binding to TDP-43 with an IC50 value of 8.9 nM (95%CI: 3.2-25 nM, n=3), but was unable to inhibit bt-TG12 binding to N-terminal TDP-43 (Fig 2A). In contrast, unlabeled TG12 was able to inhibit bt-TG12 binding to both full length and N-terminal TDP-43 with similar affinities, with IC50 values of 1.6 nM (95% CI: 0.58-2.4 nM, n=3) and 0.88 nM (95% CI: 0.35-1.2 nM, n=3). Taken together, these data indicate that UBQLN2 binds to the glycine rich C-terminal tail of TDP-43 rather than the N-terminal RRM domains.
Figure 2. UBQLN2 binds to full length TDP-43 and C-terminal TDP-43 (261-414aa), but not N-terminal TDP-43 (1-260 aa).
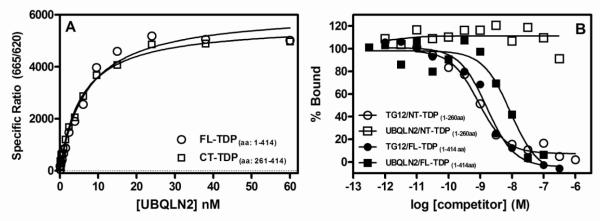
(A) A series of concentrations of UBQLN2 (0.1 to 60 nM) was incubated for 2 hr at room temperature with full length TDP-43 (100 ng/mL, 1.6 nM) or C-terminal TDP-43 fragment (100 ng/mL, 4.6 nM) followed by the addition of HTRF antibodies with subsequent ratiometric measurement of time-resolved fluorescence as described in Section 2.3. (B) A series of concentrations of unlabeled TG12 (1.0 pM to 1.0 μM) or UBQLN2 (0.30 pM to 0.3 μM) were incubated for 2 hr with either full length TDP-43 (100 ng/mL, 1.6 nM) or N-terminal TDP-43 (100 ng/mL, 2.2 nM) along with 1.0 nM bt-TG12 followed by the addition of HTRF antibodies with subsequent ratiometric measurement of time-resolved fluorescence as described in Section 2.3. Nonspecific binding was determined in the absence of TDP-43. Equilibrium dissociation rate constants (KD) (A) or IC50 values (B) were determined by nonlinear regression fits of the data to either a one-site binding equation or three parameter dose-response equation, respectively, using Graph Pad Prism®. Data are the means of duplicate determinations from a single experiment that was repeated three times with similar results.
3.3 Rank order affinities are similar for inhibition of UBQLN2 or inhibition of bt-TG12 binding to TDP-43
In order to determine if UBQLN2 binding to full length TDP-43 could be inhibited by nucleic acid binding to TDP-43, we generated dose-response curves for single stranded DNA oligonucleotides with increasing TG repeats as well as the 32 base pair TAR DNA sequence (TAR-32). In addition, we also tested these same nucleic acids and test compounds in bt-TG12 binding to TDP-43 using HTRF technology in order to obtain a direct comparison in the same system. For both inhibition of UBQLN2 binding and inhibition of nucleic acid binding, we observed generally similar rank order affinities where TG12 ≥ TAR32 > TG8 > TG6 > TG4 (Fig 3, Table 1). In addition, the 4-aminoquinoline AAQ1 inhibited both UBQLN2 and bt-TG12 binding to TDP-43, with IC50 values of 5.5 μM and 1.0 μM, respectively, while the structurally related negative control, AAQ2, had no activity (Fig 3, Table 1). Interestingly, the nucleic acids had substantially greater affinity for inhibiting nucleic acid binding, with 30-100 fold greater affinity for inhibition of bt-TG12 binding compared to inhibition of UBQLN2, while AAQ1 had similar affinities for inhibition of either UBQLN2 or bt-TG12 (Fig 3, Table 1). Furthermore, the IC50 value for TAR32 was ~10 fold greater than that of TG12 for inhibition of nucleic acid binding, but it exhibited similar affinity as TG12 for inhibition of UBQLN2 binding. Despite these differences, the similar rank order affinities for inhibition of UBQLN2 binding and nucleic acid binding is consistent with a specific interaction between UBQLN2 and TDP-43.
Figure 3. Comparison of inhibition of UBQLN2 binding and inhibition of bt-TG12 binding to full length TDP-43 using HTRF technology.
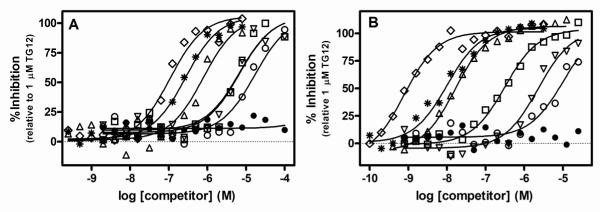
Dilutions of TG4 (○), TG6 (□), TG8 (△), TG12 (◇), TAR32 ( ), AAQ1 (▽), AAQ2 (•) were incubated with TDP-43 (1.6 nM) and either 1.4 nM UBQLN2 (A) or 1 nM bt-TG12 (B) for 2 hr at room temperature followed by the addition of HTRF antibodies and subsequent ratiometric measurement of time resolved fluorescence as described in Section 2.4. Nonlinear regression fits of the data to a three parameter dose response equation were performed using Graph Pad Prism®. Data points and curves are an example of a single experiment that was repeated four times with similar results. IC50 values are the geometric means and 95% confidence intervals of four determinations.
), AAQ1 (▽), AAQ2 (•) were incubated with TDP-43 (1.6 nM) and either 1.4 nM UBQLN2 (A) or 1 nM bt-TG12 (B) for 2 hr at room temperature followed by the addition of HTRF antibodies and subsequent ratiometric measurement of time resolved fluorescence as described in Section 2.4. Nonlinear regression fits of the data to a three parameter dose response equation were performed using Graph Pad Prism®. Data points and curves are an example of a single experiment that was repeated four times with similar results. IC50 values are the geometric means and 95% confidence intervals of four determinations.
Table 1.
Comparison of the affinities of ssDNA oligonucleotides and 4-aminoquinolines for inhibition of UBQLN2 binding or inhibition of nucleic acid binding to full length TDP-43.
| IC50 (95% CI: nM) | ||
|---|---|---|
| Compound | UBQLN2 | bt-TG12 |
| TG12 | 110 (44-270) | 1.2 (0.46-3.0) |
| TAR32 | 160 (63-430) | 16 (6.0-43) |
| TG8 | 1200 (410-3300) | 30 (16-56) |
| TG6 | 6500 (3400-12000) | 370 (250-540) |
| TG4 | 24000 (14000-41000) | 4600 (2500-8300) |
| AAQ1 | 5.5 (3.0-10) | 1.0 (0.45-2.2) |
|
| ||
| AAQ2 | > 100 | > 100 |
Values represent the geometric means and 95% confidence intervals of four determinations.
3.4 UBQLN2 and TG12 interact noncompetitively with TDP-43
We postulated that if UBQLN2 binds to the C-terminal region of TDP-43, then UBQLN2 binding should be noncompetitive with nucleic acid binding. In order to test this hypothesis, we characterized TG12 mediated inhibition of UBQLN2 binding to TDP-43 as well as UBQLN2 mediated inhibition of bt-TG12 binding to TDP-43. Increasing concentrations of TG12 reduced the Bmax values of UBQLN2 binding to TDP-43, but had little effect on the KD values for UBQLN2 binding (Fig 4A). In addition, increasing concentrations of UBQLN2 also had a similar effect on bt-TG12 binding to TDP-43 (Fig 4B). These data are consistent with noncompetitive inhibition model, where UBQLN2 and TG12 bind to different sites on TDP-43. Indeed, comparison of global nonlinear regression fits of the data to either a mixed model inhibition equation or a noncompetitive inhibition equation indicated that the simpler, noncompetitive inhibition equation was the preferred model for describing the data. The dissociation constant for inhibition (Ki) values derived from the global noncompetitive inhibition model were 330 nM (95%CI: 250-440 nM, n=4) for TG12 inhibition of UBQLN2 binding to TDP-43 and 5.8 nM (95%CI: 2.6-12 nM, n=4) for UBQLN2 inhibition of bt-TG12 binding to TDP-43. Both of these Ki values are consistent with affinities determined by displacement and saturation of UBQLN2 binding to TDP-43 discussed previously.
Figure 4. TG12 and UBQLN2 interact noncompetitively with TDP-43.
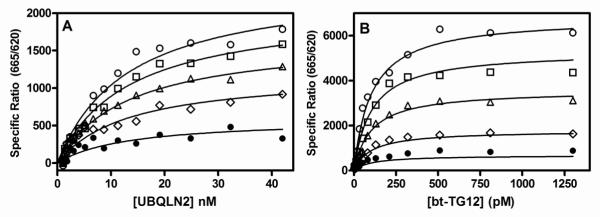
Various concentrations of TG12 and UBQLN2 were incubated with TDP-43 (1.6 nM) for 2 hrs at room temperature followed by the addition of HTRF antibodies and subsequent ratiometric measurement of time resolved fluorescence as described in Section 2.5. (A) Saturation binding isotherms for UBQLN2 binding to TDP-43 in the absence (○) or presence of 32 nM (□), 100 nM ( △), 320 nM (◇), 1000 nM (•) TG12. (B) Saturation binding isotherms of biotinylated TG12 binding to TDP-43 in the absence (○) or presence of 1.0 nM (□), 3.2 nM (△), 10 nM (◇), 32 nM (•) UBQLN2. Global nonlinear regression fits of the data to a noncompetitive inhibition model were obtained using Graph Pad Prism®. Data are the means of duplicate determinations from a single experiment that was repeated at least three times with similar results.
3.5 AAQ1 competitively inhibits UBQLN2 binding to both full length and C-terminal TDP-43
Since our previous work demonstrated that the AAQs bound to TDP-43 at a site distinct from the nucleic acid binding site on TDP-43, we hypothesized that the characterization of AAQ1 mediated inhibition of UBQLN2 binding to TDP-43 might yield some useful information about the binding site of the AAQs. Increasing concentrations of AAQ1 reduced the affinity of UBQLN2 for full length TDP-43 (Fig 5A), which is consistent with competitive inhibition, where both ligands compete for the same binding site on the protein. Comparison of global nonlinear regression fits of the data to either a mixed model inhibition equation or competitive inhibition equation indicated that the competitive inhibition equation was preferred. The Ki value for AAQ1 mediated inhibition of UBQLN2 binding to full length TDP-43 was 3.4 μM (95% CI: 1.8 – 6.5 μM, n=4). These data suggest that AAQ1 also binds to the C-terminal region of TDP-43. This hypothesis was further confirmed by characterizing AAQ1 mediated inhibition of UBQLN2 binding to the C-terminal fragment of TDP-43, where AAQ1 also exhibited competitive inhibition, with a Ki value of 4.2 μM (95% CI: 2.3 – 7.8 μM, Fig 5B).
Figure 5. AAQ1 competitively inhibits UBQLN2 binding to both full length and C-terminal TDP-43.
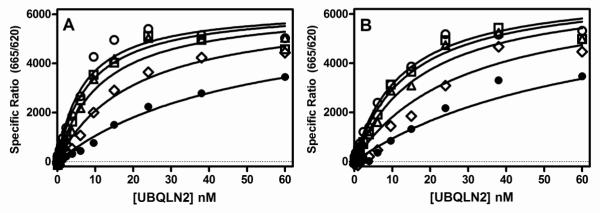
Various concentrations of AAQ1 and UBQLN2 were incubated with either full length TDP-43 (1.6 nM) (A) or C-terminal TDP-43 (261-414 aa, 4.6 nM) (B) for 2 hrs at room temperature followed by the addition of HTRF antibodies and subsequent ratiometric measurement of time resolved fluorescence as described in Section 2.5. Saturation binding isotherms for UBQLN2 binding to full length TDP-43 (A) or C-terminal TDP-43 (261-414 aa) (B) in the absence (○) or presence of 1 μM (□), 3.2 μM (△), 10 μM (◇), 32 μM (•) AAQ1. Global nonlinear regression fits of the data to a competitive inhibition model were obtained using Graph Pad Prism®. Data are the means of duplicate determinations from a single experiment that was repeated at least three times with similar results.
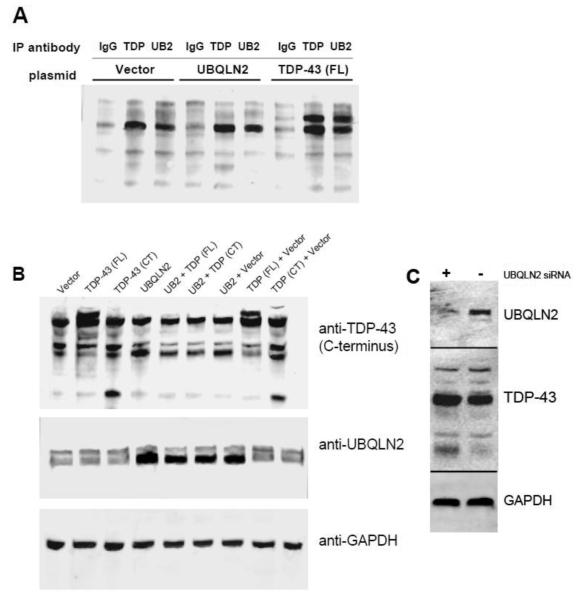
3.6 UBQLN2 co-immunoprecipates with TDP-43 in H4 cells
We then conducted co-immunoprecipation (IP) experiments in both transfected and untransfected human neuroglioma H4 cells to determine if the interaction between UBQLN2 and TDP-43 is present in the cellular environment. We used control rabbit IgG as a negative control and rabbit anti-TDP-43 as a positive control to validate the IP method. In cell lysate from untransfected H4 cells, IP with either rabbit anti-UBQLN2 or rabbit anti-TDP-43 yielded substantially greater amounts of TDP-43 compared to IP with control rabbit IgG (Fig 6A). Similar results were also obtained in lysates from H4 cells transfected with UBQLN2 (Fig 6A). In addition, IP experiments from H4 cells transfected with His/V5 full length TDP-43 yielded even greater amounts of TDP-43 compared to the results from either untransfected or UBQLN2 transfected cells when the IP antibody was either anti-TDP-43 or anti-UBQLN2 (Fig 6A). Therefore, these data suggest that UBQLN2 and TDP-43 do indeed interact in H4 cells.
Figure 6. UBQLN2 co-immunoprecipates with TDP-43 and regulates levels of both full length and C-terminal TDP-43 in human neuroglioma H4 cells.
(A) H4 cells were transfected with empty pCMV-XL5 vector, his/V5-TDP-43 or UBQLN2 as described in Section 2.6. After 24 hrs, the cells were lyzed and immunoprecipated with control rabbit IgG, rabbit anti-TDP-43 or rabbit anti-UBQLN2 followed by immunblotting using mouse anti-TDP-43 as described in Section 2.7. (B) H4 cells were transfected with either empty pCMV-XL5 vector, his/V5-TDP-43, UBQLN2 alone or in combination as described in Section 2.6 and incubated for 24 hrs followed by immunoblotting for TDP-43, UBQLN2, or GAPDH as described in Section 2.7. (C) H4 cells were transfected with either control siRNA or UBQLN2 siRNA using siQuest transfection reagent as described in Section 2.6 and incubated for 72 hrs. Cells were then lyzed in SDS sample buffer and subject to immunoblotting for TDP-43, UBQLN2, or GAPDH. Blots are an example of a single experiment that was repeated at least three times with similar results.
3.7 UBQLN2 modulates levels of TDP-43 in H4 cells
In order to explore the functional consequences of the interaction between UBQLN2 and TDP-43, we overexpressed UBQLN2 in the presence and absence of overexpressed full length TDP-43 or C-terminal TDP-43 (170-414aa) in human neuroglioma H4 cells. Overexpression of UBQLN2 alone had little effect on the levels of TDP-43 compared to transfection with the control vector (Fig 6B). However, when UBQLN2 and TDP-43 were co-transfected into H4 cells, it resulted in a substantial reduction in TDP-43 levels compared to co-transfection of TDP-43 and control vector (Fig 6B). In addition, co-transfection of UBQLN2 and C-terminal TDP-43 also substantially reduced levels of C-terminal TDP-43 compared to co-transfection of C-terminal TDP-43 with control vector. These observations led us to investigate whether or not reducing levels of UBQLN2 using siRNA would therefore increase levels of TDP-43. However, transfection of H4 cells with UBQLN2 specific siRNA resulted in a very small decrease in endogenous TDP-43 that was not statistically significant (Fig 6C). Nonetheless, the co-transfection experiments suggest that UBQLN2 may play a role in maintaining the appropriate levels of both full length TDP-43 and TDP-43 CTFs in H4 cells.
4.0 Discussion
We have demonstrated that there is a high affinity interaction between TDP-43 and UBQLN2 which may be important for the regulation of TDP-43 levels in cells. Based on this biochemical assay using HTRF technology, UBQLN2 binds to both full length and a C-terminal fragment of TDP-43 (261-414 aa) with similar affinities, but not N-terminal TDP-43 (1-260 aa), suggesting that UBQLN2 interacts with TDP-43 at the C-terminus. Single stranded DNA oligonucleotides and 4-aminoquinolines which bind to TDP-4337, also inhibit UBQLN2 binding to TDP-43 with similar rank order affinities. Inhibitor characterization experiments demonstrated that the TDP-43 oligonucleotide TG12 and UBQLN2 interact noncompetitively with TDP-43, thereby supporting the hypothesis that UBQLN2 interacts with TDP-43 at a site distinct from oligonucleotide binding. We also demonstrated that AAQ1 competitively inhibits UBQLN2 binding to both full length and C-terminal TDP-43, suggesting that these small molecules also bind to the C-terminal region of TDP-43. IP experiments in lysates from human neuroglioma H4 cells, either untransfected or transfected with TDP-43 or UBQLN2, demonstrated that TDP-43 and UBQLN2 interact in cells. One potential role of this interaction may involve the clearance of TDP-43, since overexpression of UBQLN2 in the presence of either full length or C-terminal TDP-43 results in a substantial reduction in both full length and TDP-43 CTF. Taken together, these data suggest that UBQLN2 interacts with TDP-43 in the C-terminal region and plays a role in the regulation of TDP-43 and TDP-43 CTFs in cells.
The C-terminal binding site for UBQLN2 on TDP-43 is distinct from the prototypical nucleic acid binding sites, RRM1 and RRM2, located in the N-terminal region of the protein. Therefore nucleic acid binding is allosteric to UBQLN2 binding. The data reported here suggest that binding of UBQLN2 to TDP-43 inhibits nucleic acid binding, and vice versa, albeit with different affinities. While the rank order affinities were similar for inhibition of UBQLN2 binding and inhibition of bt-TG12 binding to TDP-43 for the DNA oligonucleotides, there was a substantial difference in absolute affinity, where these compounds had 30-100 lower affinities for inhibition of UBQLN2 binding to TDP-43 (Table 1). These data suggest that the binding of UBQLN2 to TDP-43 reduces the affinity of TDP-43 for nucleic acids. Consequently, UBQLN2 may act as a negative regulator of TDP-43 function. Indeed, the C-terminal region of TDP-43 is the site where other proteins such as the heterogeneous ribonucleoproteins (hnRNP) A/B have been shown to interact and modulate TDP-43 function38, 39. Interestingly, interactions between TDP-43 and hnRNP A1 and hnRNP A2 via the C-terminal tail of TDP-43 actually promote the function of TDP-43 in silencing the inclusion of exon-9 of the CFTR gene.39 Therefore, the C-terminal tail of TDP-43 is an important regulatory site for protein-protein interactions which can promote or potentially inhibit the function of TDP-43 in cells.
Recently, we described a series of small molecules, the 4-aminoquinolines (AAQs), which inhibit nucleic acid binding to TDP-43.37 While we observed a substantial decrease in affinity for nucleic acids for TDP-43 upon UBQLN2 binding, the affinity of AAQ1 for TDP-43 was much less effected, being only 5-fold less potent for inhibition of UBQLN2 binding to TDP-43 compared to inhibition of nucleic acid binding to TDP-43. Further characterization of AAQ1 mediated inhibition of UBQLN2 binding indicated that AAQ1 was competitive with UBQLN2 binding, therefore suggesting that AAQ1 also binds to the C-terminal region of TDP-43. Given the emerging evidence concerning the importance of the C-terminal region of TDP-43, both for normal TDP-43 function38, 39, as well as mislocalization and aggregation of TDP-43 CTFs in ALS and FTLD24, 31-33, compounds which interact with this region may be useful tool compounds to further understand both normal and pathological functions of TDP-43.
Other investigators have demonstrated that UBQLN2 plays an important role in protein clearance and degradation. Not only is UBQLN2 involved in transporting ubiquitinated proteins to the proteasome for degradation9, 12, but it is also involved in autophagy, specifically in the maturation of autophagosomes13, 14. Here, we observed that overexpression of UBQLN2 reduces levels of TDP-43 when either full length TDP-43 or C-terminal TDP-43 is overexpressed. Therefore, UBQLN2 may be an important mediator of the clearance of TDP-43 and TDP-43 CTFs. In normal cells, TDP-43 clearance is mediated by both the proteasome and autophagy pathways40, 41. We tried to overexpress UBQLN2 and TDP-43 in the presence of either the proteasome inhibitor MG-132, or the autophagy inhibitor thapsigargin in order to determine if one or both pathways was involved in UBQLN2 mediated clearance of TDP-43, but the results were inconclusive (unpublished observations). Further experiments need to be done to elucidate the mechanisms involved in the clearance of TDP-43 mediated by UBQLN2. In addition, it should also be noted that the effect of UBQLN2 on the cellular levels of overexpressed TDP-43 may not necessarily be the result of a direct interaction between TDP-43 and UBQLN2, but it could involve an intermediary pathway(s) involving interactions with other proteins. Therefore, further studies exploring the functional consequences of UBQLN2 binding to TDP-43 are required to validate the importance of this interaction in cells.
The C-terminal region of TDP-43 is proving to be an important site of regulation, where protein-protein interactions can promote or inhibit TDP-43 function. However, the role of these protein-protein interactions with respect to the TDP-43 C-terminal aggregates present in ALS and FTLD is not well understood. Additional studies that further our understanding of these interactions with both normal and pathological TDP-43 may lead to the discovery of novel points of therapeutic intervention, leading to improved treatments for neurodegenerative diseases associated with TDP-43 proteinopathy.
Highlights.
UBLQN2 binds with high affinity to C-terminal region of TDP-43
Nucleic acids and 4-aminoquinolines (AAQs) inhibit UBQLN2 binding to TDP-43
UBQLN2 noncompetitively inhibits DNA oligonucleotide binding to TDP-43
AAQs bind to the C-terminus of TDP-43 and competitively inhibit UBQLN2 binding
UBQLN2 modulates levels of TDP-43 in human H4 cells.
Acknowledgements
We wish to acknowledge the financial support of the National Institutes of Health which supported work described here (1R21NS072749-01). We also wish to thank Drs. Vyacheslav Andrianov, Venkat Velvadapu and Mark McDonnell for helpful discussions and technical support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee DY, Brown EJ. Ubiquilins in the crosstalk among proteolytic pathways. Biol Chem. 2012;393(6):441–7. doi: 10.1515/hsz-2012-0120. [DOI] [PubMed] [Google Scholar]
- 2.Rothenberg C, Monteiro MJ. Ubiquilin at a crossroads in protein degradation pathways. Autophagy. 2010;6(7):979–80. doi: 10.4161/auto.6.7.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conklin D, Holderman S, Whitmore TE, Maurer M, Feldhaus AL. Molecular cloning, chromosome mapping and characterization of UBQLN3 a testis-specific gene that contains an ubiquitin-like domain. Gene. 2000;249(1-2):91–8. doi: 10.1016/s0378-1119(00)00122-0. [DOI] [PubMed] [Google Scholar]
- 4.Wu AL, Wang J, Zheleznyak A, Brown EJ. Ubiquitin-related proteins regulate interaction of vimentin intermediate filaments with the plasma membrane. Mol Cell. 1999;4(4):619–25. doi: 10.1016/s1097-2765(00)80212-9. [DOI] [PubMed] [Google Scholar]
- 5.Ko HS, Uehara T, Nomura Y. Role of ubiquilin associated with protein-disulfide isomerase in the endoplasmic reticulum in stress-induced apoptotic cell death. J Biol Chem. 2002;277(38):35386–92. doi: 10.1074/jbc.M203412200. [DOI] [PubMed] [Google Scholar]
- 6.Persson P, Stockhausen MT, Pahlman S, Axelson H. Ubiquilin-1 is a novel HASH-1-complexing protein that regulates levels of neuronal bHLH transcription factors in human neuroblastoma cells. Int J Oncol. 2004;25(5):1213–21. [PubMed] [Google Scholar]
- 7.Walters KJ, Kleijnen MF, Goh AM, Wagner G, Howley PM. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry. 2002;41(6):1767–77. doi: 10.1021/bi011892y. [DOI] [PubMed] [Google Scholar]
- 8.Walters KJ, Goh AM, Wang Q, Wagner G, Howley PM. Ubiquitin family proteins and their relationship to the proteasome: a structural perspective. Biochim Biophys Acta. 2004;1695(1-3):73–87. doi: 10.1016/j.bbamcr.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Kleijnen MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, et al. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol Cell. 2000;6(2):409–19. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- 10.Feng P, Scott CW, Cho NH, Nakamura H, Chung YH, Monteiro MJ, et al. Kaposi’s sarcoma-associated herpesvirus K7 protein targets a ubiquitin-like/ubiquitin-associated domain-containing protein to promote protein degradation. Mol Cell Biol. 2004;24(9):3938–48. doi: 10.1128/MCB.24.9.3938-3948.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang D, Raasi S, Fushman D. Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J Mol Biol. 2008;377(1):162–80. doi: 10.1016/j.jmb.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ko HS, Uehara T, Tsuruma K, Nomura Y. Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett. 2004;566(1-3):110–4. doi: 10.1016/j.febslet.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 13.N’Diaye EN, Debnath J, Brown EJ. Ubiquilins accelerate autophagosome maturation and promote cell survival during nutrient starvation. Autophagy. 2009;5(4):573–5. doi: 10.4161/auto.5.4.8312. [DOI] [PubMed] [Google Scholar]
- 14.N’Diaye EN, Kajihara KK, Hsieh I, Morisaki H, Debnath J, Brown EJ. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep. 2009;10(2):173–9. doi: 10.1038/embor.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orazizadeh M, Lee HS, Groenendijk B, Sadler SJ, Wright MO, Lindberg FP, et al. CD47 associates with alpha 5 integrin and regulates responses of human articular chondrocytes to mechanical stimulation in an in vitro model. Arthritis Res Ther. 2008;10(1):R4. doi: 10.1186/ar2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.N’Diaye EN, Brown EJ. The ubiquitin-related protein PLIC-1 regulates heterotrimeric G protein function through association with Gbetagamma. J Cell Biol. 2003;163(5):1157–65. doi: 10.1083/jcb.200307155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.N’Diaye EN, Hanyaloglu AC, Kajihara KK, Puthenveedu MA, Wu P, von Zastrow M, et al. The ubiquitin-like protein PLIC-2 is a negative regulator of G protein-coupled receptor endocytosis. Mol Biol Cell. 2008;19(3):1252–60. doi: 10.1091/mbc.E07-08-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477(7363):211–5. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams KL, Warraich ST, Yang S, Solski JA, Fernando R, Rouleau GA, et al. UBQLN2/ubiquilin 2 mutation and pathology in familial amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33(10):2527, e3–10. doi: 10.1016/j.neurobiolaging.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Daoud H, Suhail H, Szuto A, Camu W, Salachas F, Meininger V, et al. UBQLN2 mutations are rare in French and French-Canadian amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33(9):2230, e1–e5. doi: 10.1016/j.neurobiolaging.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 21.Synofzik M, Maetzler W, Grehl T, Prudlo J, Vom Hagen JM, Haack T, et al. Screening in ALS and FTD patients reveals 3 novel UBQLN2 mutations outside the PXX domain and a pure FTD phenotype. Neurobiol Aging. 2012;33(12):2949, e13–7. doi: 10.1016/j.neurobiolaging.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Ball LJ, Kuhne R, Schneider-Mergener J, Oschkinat H. Recognition of proline-rich motifs by protein-protein-interaction domains. Angew Chem Int Ed Engl. 2005;44(19):2852–69. doi: 10.1002/anie.200400618. [DOI] [PubMed] [Google Scholar]
- 23.Morris HR, Waite AJ, Williams NM, Neal JW, Blake DJ. Recent advances in the genetics of the ALS-FTLD complex. Curr Neurol Neurosci Rep. 2012;12(3):243–50. doi: 10.1007/s11910-012-0268-5. [DOI] [PubMed] [Google Scholar]
- 24.Kwong LK, Neumann M, Sampathu DM, Lee VM, Trojanowski JQ. TDP-43 proteinopathy: the neuropathology underlying major forms of sporadic and familial frontotemporal lobar degeneration and motor neuron disease. Acta Neuropathol. 2007;114(1):63–70. doi: 10.1007/s00401-007-0226-5. [DOI] [PubMed] [Google Scholar]
- 25.Buratti E, Baralle FE. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci. 2008;13:867–78. doi: 10.2741/2727. [DOI] [PubMed] [Google Scholar]
- 26.Buratti E, Baralle FE. The molecular links between TDP-43 dysfunction and neurodegeneration. Adv Genet. 2009;66:1–34. doi: 10.1016/S0065-2660(09)66001-6. [DOI] [PubMed] [Google Scholar]
- 27.Geser F, Martinez-Lage M, Kwong LK, Lee VM, Trojanowski JQ. Amyotrophic lateral sclerosis, frontotemporal dementia and beyond: the TDP-43 diseases. J Neurol. 2009;256(8):1205–14. doi: 10.1007/s00415-009-5069-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strong MJ. The syndromes of frontotemporal dysfunction in amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2008;9(6):323–38. doi: 10.1080/17482960802372371. [DOI] [PubMed] [Google Scholar]
- 29.Pawlyk AC, Cassel JA, Reitz AB. Current nervous system related drug targets for the treatment of amyotrophic lateral sclerosis. Curr Pharm Des. 2010;16(18):2053–73. doi: 10.2174/138161210791293024. [DOI] [PubMed] [Google Scholar]
- 30.Lanson NA, Jr., Pandey UB. FUS-related proteinopathies: lessons from animal models. Brain Res. 2012;1462:44–60. doi: 10.1016/j.brainres.2012.01.039. [DOI] [PubMed] [Google Scholar]
- 31.Banks GT, Kuta A, Isaacs AM, Fisher EM. TDP-43 is a culprit in human neurodegeneration, and not just an innocent bystander. Mamm Genome. 2008;19(5):299–305. doi: 10.1007/s00335-008-9117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lagier-Tourenne C, Cleveland DW. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136(6):1001–4. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liscic RM, Grinberg LT, Zidar J, Gitcho MA, Cairns NJ. ALS and FTLD: two faces of TDP-43 proteinopathy. Eur J Neurol. 2008;15(8):772–80. doi: 10.1111/j.1468-1331.2008.02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuo PH, Doudeva LG, Wang YT, Shen CK, Yuan HS. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 2009;37(6):1799–808. doi: 10.1093/nar/gkp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ou SH, Wu F, Harrich D, Garcia-Martinez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69(6):3584–96. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem. 2001;276(39):36337–43. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- 37.Cassel JA, McDonnell ME, Velvadapu V, Andrianov V, Reitz AB. Characterization of a series of 4-aminoquinolines that stimulate caspase-7 mediated cleavage of TDP-43 and inhibit its function. Biochimie. 2012;94(9):1974–81. doi: 10.1016/j.biochi.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buratti E, Brindisi A, Giombi M, Tisminetzky S, Ayala YM, Baralle FE. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J Biol Chem. 2005;280(45):37572–84. doi: 10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- 39.D’Ambrogio A, Buratti E, Stuani C, Guarnaccia C, Romano M, Ayala YM, et al. Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res. 2009;37(12):4116–26. doi: 10.1093/nar/gkp342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bose JK, Huang CC, Shen CK. Regulation of autophagy by the neuropathological protein TDP-43. J Biol Chem. 2011;286(52):44441–8. doi: 10.1074/jbc.M111.237115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Fan H, Ying Z, Li B, Wang H, Wang G. Degradation of TDP-43 and its pathogenic form by autophagy and the ubiquitin-proteasome system. Neurosci Lett. 2010;469(1):112–6. doi: 10.1016/j.neulet.2009.11.055. [DOI] [PubMed] [Google Scholar]