SUMMARY
The type II bacterial CRISPR/Cas system is a novel genome engineering technology with the ease of multiplexed gene targeting. Here we created reporter and conditional mutant mice by co-injection of zygotes with Cas9 mRNA, different guide RNAs (sgRNAs) as well as DNA vectors of different sizes. Using this one step procedure we generated mice carrying a tag or a fluorescent reporter construct in the Nanog, the Sox2 and the Oct4 gene as well as Mecp2 conditional mutant mice. In addition, using sgRNAs targeting two separate sites in the Mecp2 gene, we produced mice harboring the predicted deletions of about 700 bps. Finally, we analyzed potential off-targets of five sgRNAs in gene-modified mice and ESC lines and identified off-target mutations in only rare instances.
INTRODUCTION
Mice with specific gene modification are valuable tools for studying development and disease. Traditional gene targeting in embryonic stem (ES) cells, while suitable for generating sophisticated genetic modifications in endogenous genes, is complex and time-consuming (Capecchi, 2005). The production of genetically modified mice and rats has been greatly accelerated by novel approaches using direct injection of DNA or mRNA of site-specific nucleases into the one cell stage embryo, generating DNA double strand breaks (DSB) at specified sequences leading to targeted mutations (Carbery et al., 2010; Geurts et al., 2009; Shen et al., 2013; Sung et al., 2013; Tesson et al., 2011; Wang et al., 2013). Co-injection of a single stranded or double stranded DNA template containing homology to the sequences flanking the DSB can produce mutant alleles with precise point mutations or DNA inserts (Brown et al., 2013; Cui et al., 2011; Meyer et al., 2010; Wang et al., 2013; Wefers et al., 2013). Recently, pronuclear injection of two pairs of ZFNs and two double stranded donor vectors into rat fertilized eggs produced rat containing loxP-flanked (floxed) alleles (Brown et al., 2013). However, the complex and time-consuming design and generation of ZFNs and double stranded donor vectors limit the application of this method.
CRISPR (clustered regularly interspaced short palindromic repeat) and Cas (CRISPR-associated) proteins function as the RNA-based adaptive immune system in bacteria and archaea (Horvath and Barrangou, 2010; Wiedenheft et al., 2012). The type II bacterial CRISPR/Cas system has been demonstrated as an efficient gene targeting technology that facilitates multiplexed gene targeting (Cong et al., 2013; Wang et al., 2013). Because the binding of Cas9 is guided by the simple base-pair complementarities between the engineered single guide RNA (sgRNA) and a target genomic DNA sequence, it is possible to direct Cas9 to any genomic locus, by providing the engineered sgRNA (Cho et al., 2013; Cong et al., 2013; Gilbert et al.; Hwang et al., 2013; Jinek et al., 2012; Jinek et al., 2013; Mali et al., 2013b; Qi et al., 2013; Wang et al., 2013).
Previously, we used the type II bacterial CRISPR/Cas system as an efficient tool to generate mice carrying mutations in multiple genes in one step (Wang et al., 2013). However, this study left a number of issues unresolved. For example, the efficiency of using the CRISPR/Cas gene editing approach for the insertion of DNA constructs into endogenous genes was not clarified nor its utility to create conditional mutant mice. Here we report the one step generation of mice carrying reporter constructs in three different genes as well as the derivation of conditional mutant mice. In addition we performed an extensive off-target cleavage analysis and show that off-target mutations are rare in targeted mice and ES cells derived from CRISPR/Cas zygote injection.
RESULTS
Targeted insertion of short DNA fragments
In previous work, we introduced precise base pair mutations into the Tet1 and Tet2 genes through homology directed repair (HDR)-mediated genome editing following co-injection of single stranded mutant DNA oligos, sgRNAs and Cas9 mRNA (Wang et al., 2013). To test whether a larger DNA construct could be inserted at the same DSBs at Tet1 exon 4 and Tet2 exon 3, we designed oligos containing the 34bp loxP site and a 6bp EcoRI site flanked by 60bps sequences on each side adjoining the DSBs (Figure S1A). We co-injected Cas9 mRNA, sgRNAs, and single stranded DNA oligos targeting both Tet1 and Tet2 into zygotes. The restriction fragment length polymorphism (RFLP) assay shown in Figure S1B identified six out of 15 tested embryos carrying the loxP site at the Tet1 locus, eight carrying the loxP site at the Tet2 locus, and three had at least one allele of each gene correctly modified. The correct integration of loxP sites was confirmed by sequencing (Figure S1C). These results demonstrate that HDR-mediated repair can introduce targeted integration of 40bp DNA elements efficiently through CRISPR/Cas mediated genome editing (summarized in Table 1).
Table 1.
Mice with reporters in the endogenous genes
| Donor | Blastocysts/Injected zygotes | Targeted blastocysts/Total | Targeted ESCs/Total | Transferred embryos (recipients) | Knock-in pre- and postnatal mice/Total | |
|---|---|---|---|---|---|---|
| Tet1-loxP + Tet2-loxP | 65/89 | Tex1-loxP | 6/15 | N/A | N/A | N/A |
| Tex2-loxP | 6/15 | |||||
| Both | 3/15 | |||||
|
| ||||||
| Sox2-V5 | 414/498 | ND | 7/16 | 200(10) | 12/35 | |
|
| ||||||
| Nanog-mCherry | 936/1262 | 86/936 | ND | 415 (21) | 7/86 | |
|
| ||||||
| Oct4-GFP | 254/345 | 47/254 | 3/9 | 100(4) | 3/10 | |
Cas9 mRNA, sgRNAs targeting Tet1, Tet2, Sox2, Nanog, or Oct4, and single stranded DNA oligos or double stranded donor vectors were injected into fertilized eggs. Targeted blastocysts were identified by RFLP or fluorescence of reporters. The blastocysts derived from injected embryos were derived ES cell lines or transplanted into foster mothers and E13.5 embryos and postnatal mice were obtained and genotyped.
ND, not determined.
Mice with reporters in the endogenous Nanog, Sox2 and Oct4 genes
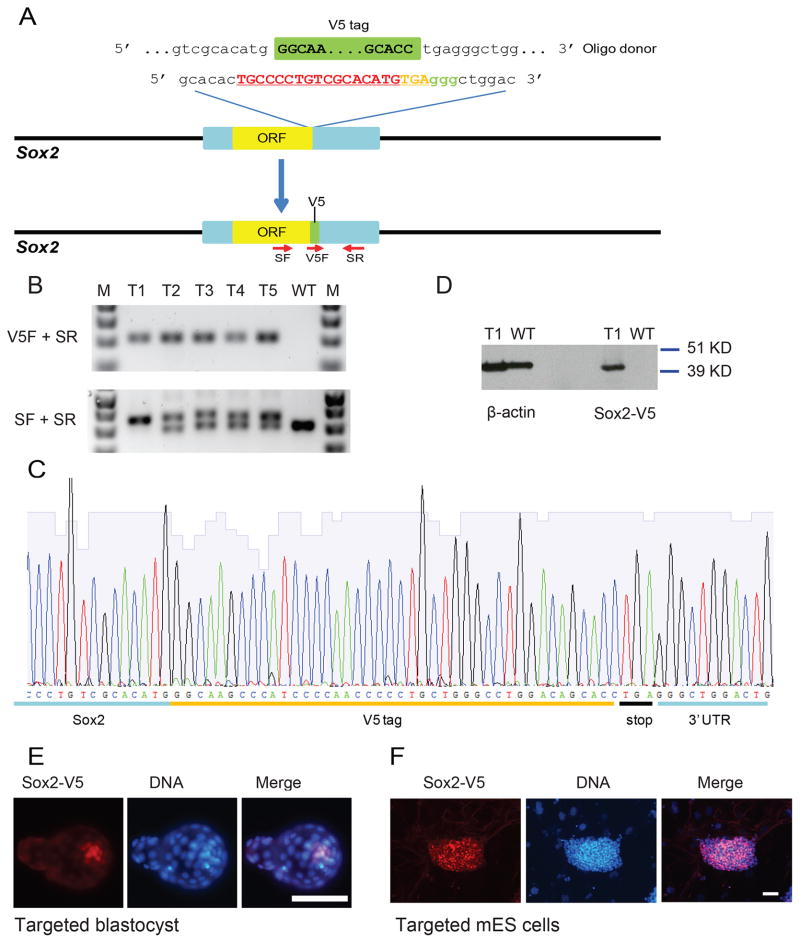
Since the study of many genes and their protein products are limited by the availability of high quality antibodies, we explored the potential of fusing a short epitope tag to an endogenous gene. We designed a sgRNA targeting the stop codon of Sox2 and a corresponding oligo to fuse the 42bp V5 tag into the last codon (Figure 1A). After injection of the sgRNA, Cas9 mRNA and the oligo into zygotes, in vitro differentiated blastocysts were explanted into culture to derive ES cells. PCR genotyping and sequencing identified seven out of 16 ES cell lines carrying a correctly targeted insert (Figures 1B and 1C). Western blot analysis revealed a protein band at the predicted size using V5 antibody in targeted ES cells but not in the control cells (Figure 1D). As expected from a correctly targeted and functional allele, Sox2 expression was seen in targeted blastocysts and ES cells using V5 antibody (Figures 1E and 1F). 12 of 35 E13.5 embryos and live born mice derived from injected zygotes carried the V5 tag correctly targeted into the Sox2 gene as indicated by PCR genotyping and sequencing (data not shown, Table 1).
Figure 1.
One step generation of the Sox2-V5 allele. (A) Schematic of the Cas9/sgRNA/oligo targeting site at the Sox2 stop codon. The sgRNA coding sequence is underlined, capitalized, and labeled in red. The protospacer-adjacent motif (PAM) sequence is labeled in green. The stop codon of Sox2 is labeled in orange. The oligo contained 60bp homologies on both sides flanking the DSB. In the oligo donor sequence, the V5 tag sequence is labeled as a green box. PCR primers (SF, V5F, and SR) used for PCR genotyping are shown as red arrowheads. (B) Upper panel, PCR genotyping using primers V5F and SR produced bands with correct size in targeted ES samples T1 to T5, but not in WT sample. Lower panel, PCR genotyping using primers SF and SR produced slightly larger products, indicating the 42bp V5 tag sequence was integrated. T1 only contain larger product, suggesting either both alleles were targeted, or one allele failed to amplify. (C) PCR products using primers SF and SR were cloned into plasmid and sequenced. Sequence across the targeting region confirmed correct fusion of V5 tag to the last codon of Sox2. (D) Western blot analysis identified Sox2-V5 protein using V5 antibody in ES cells containing Sox2-V5 allele. Beta-actin was shown as the loading control. Because beta-actin and Sox2-V5 run at the same size, the samples for the V5 signal and beta-actin were run in parallel on the same gel. (E) Immunostaining of targeted blastocyst using V5 antibody showed signal in ICM. Scale bar, 50μm. (F) Immunostaining of targeted ES cells using V5 antibody showed uniform Sox2 expression. Scale bar, 100μm.
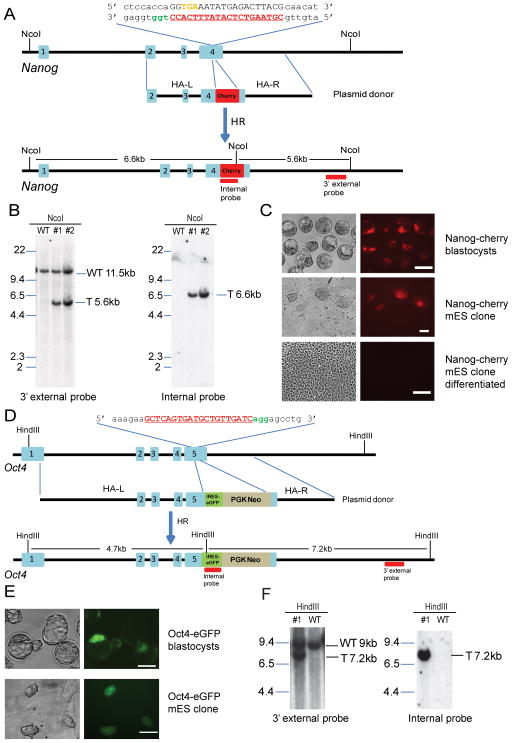
To assess whether a marker transgene could be inserted into an endogenous locus, we co-injected Cas9 mRNA, sgRNA and a double stranded donor vector which was designed to fuse a p2A-mCherry reporter with the last codon of the Nanog gene (Figure 2A). A circular donor vector was used to minimize random integrations. To assess toxicity and to optimize the concentration of donor DNA, we microinjected different amounts of Nanog-2A-mCherry vector. Injection with a high concentration of donor DNA (500 ng/ul) yielded mCherry-positive embryos with high efficiency with most blastocysts being retarded, whereas injection with a lower donor DNA concentration (10 ng/ul) yielded mostly healthy blastocysts most of which were mCherry-negative. When 200ng/ul donor DNA was used, 75% (936/1262) of the injected zygotes developed to blastocysts, 9% (86/936) of which were mCherry-positive (Figure 2C, Table S1). mCherry was mainly expressed in the inner cell mass (ICM) consistent with targeted integration of the mCherry transgene into the Nanog gene. We derived six ES cell lines from mCherry positive blastocysts, four of which uniformly expressed mCherry with the signal disappearing upon cellular differentiation (Figure 2C). The other two lines showed variegated mCherry expression with some colonies being mCherry positive and others negative (Figure S2A, Table S2) consistent with mosaic donor embryos, which would be expected if transgene insertion occurred later than the zygote stage, as has been previously observed with ZNF and TALEN-mediated targeting (Brown et al., 2013; Cui et al., 2011; Wefers et al., 2013). Correct transgene integration in ES cell lines was confirmed by Southern blot analysis (Figure 2B). We also generated mice from injected zygotes. Southern blot analysis (Figures S2B and S2C) revealed that seven out of 86 E13.5 embryos and live born mice carried the mCherry transgene in the Nanog locus. One targeted mouse was mosaic (TableS2), since the intensity of targeted allele was lower than the wild type allele (Figure S2B, # 6). Two of the mice carried an additional randomly integrated transgene (Figure S2C, # 3). As summarized in Tables 1 and S1, the efficiency of targeted insertion of the transgene was about 10% in blastocysts and mice derived from injected zygotes.
Figure 2.
One step generation of an endogenous reporter allele. (A) Schematic overview of strategy to generate a Nanog-mCherry knock-in allele. The sgRNA coding sequence is underlined, capitalized, and labeled in red. The protospacer-adjacent motif (PAM) sequence is labeled in green. The stop codon of Nanog is labeled in orange. The homologous arms of the donor vector are indicated as HA-L (2kb) and HA-R (3kb). The restriction enzyme used for Southern blot analysis is shown, and the Southern blot probes are shown as red boxes. (B) Southern analysis of Nanog-mCherry targeted allele. NcoI-digested genomic DNA was hybridized with 3′external probe. Expected fragment size: WT (wild type) = 11.5 kb, T (targeted) = 5.6 kb. The blot was then stripped and hybridized with mCherry internal probe. Expected fragment size: WT = N/A, T = 6.6 kb. (C) Nanog-mCherry targeted blastocysts showed expression in ICM. Mouse ES cell lines derived from targeted blastocysts remain mCherry positive, and the mCherry expression disappear upon differentiation. Scale bar, 100μm. (D) Schematic overview of strategy to generate an Oct4-eGFP knock-in allele. The sgRNA coding sequence is underlined, capitalized, and labeled in red. The PAM sequence is labeled in green. The homologous arms of the donor vector are indicated as HA-L (4.5kb) and HA-R (2kb). The IRES-eGFP transgene is indicated as a green box, and the PGK-Neo cassette is indicated as a grey box. The restriction enzyme used for Southern blot analysis is shown, and the Southern blot probes are shown as red boxes. (E) Oct4-eGFP targeted blastocysts showed expression in ICM. Scale bar, 50μm. Mouse ES cell lines derived from targeted blastocysts remain GFP positive. Scale bar, 100μm. (F) Southern analysis of Oct4-eGFP targeted allele. Southern analysis of Oct4-eGFP targeted allele. HindIII-digested genomic DNA was hybridized with 3′external probe. Expected fragment size: WT = 9 kb, Targeted = 7.2 kb. The blot was then stripped and hybridized with eGFP internal probe. Expected fragment size: WT = N/A, Targeted = 7.2 kb. See also Figure S2.
Table 2.
Conditional Mecp2 mutant mice
| Donor | Blastocyst/Injected zygotes | Transferredembryos (recipients) | Sex | Pre- and postnatal Mice with loxP/Total | ||||
|---|---|---|---|---|---|---|---|---|
| Totala | L2b | R1c | Two loxP in two alleles | Two loxP in one allele | ||||
| Mecp2-L2 + Mecp2-R1 | 367/451 | 360(18) | Male | 28/60 | 26/60 | 12/60 | 2d/60 | 8/60 |
| Female | 21/38 | 19/38 | 13/38 | 3/38 | 8/38 | |||
| Total | 49/98 | 45/98 | 25/98 | 5/98 | 16/98 | |||
Cas9 mRNA, sgRNAs targeting Mecp2-L2 and Mecp2-R1, and single stranded DNA oligos were injected into fertilized eggs. The blastocysts derived from the injected embryos were transplanted into foster mothers and pre- and postnatal mice were genotyped.
Total mice containing loxP site integration in the genome.
Mice containing loxP site integrated at L2 site.
Mice containing loxP site integrated at R1 site.
These male mice were mosaic.
Finally, we designed sgRNA targeting the Oct4 3′ UTR, which was co-injected with a published donor vector designed to integrate the 3kb transgene cassette (IRES-eGFP-loxP-Neo-loxP; Figure 2D) at the 3′ end of the Oct4 gene (Lengner et al., 2007). Blastocysts were derived from injected zygotes, inspected for GFP expression and explanted to derive ES cells. About 20% (47/254) of the blastocysts displayed uniform GFP expression in the ICM region. Three of nine derived ES cell lines expressed GFP (Figure 2E), including one showed mosaic expression (Table S2). Three out of ten live born mice contained the targeted allele (Table 1). Correct targeting in mice and ES cell lines was confirmed by Southern blot analysis (Figure 2F).
Conventionally, transgenic mice are generated by pronuclear instead of cytoplasmic injection of DNA. To optimize the generation of CRISPR/Cas9 targeted embryos we compared different concentrations of RNA and the Nanog-mCherry or the Oct4-GFP DNA vectors as well as three different delivery modes: (i) simultaneous injection of all constructs into the cytoplasm, (ii) simultaneous injection of the RNA and the DNA into the pronucleus and (iii) injection of Cas9/sgRNA into the cytoplasm followed 2 hours later by pronuclear injection of the DNA vector. Table S1 shows that simultaneous injection of all constructs into the cytoplasm at a concentration of 100 ng/μl Cas9 RNA, 50 ng/μl of sgRNA and 200 ng/μl of vector DNA was optimal, resulting in 9% (86/936) to 19% (47/254) of targeted blastocysts. Similarly, the simultaneous injection of 5 ng/μl Cas9 RNA, 2.5 ng/μl of sgRNA and 10 ng/μl of DNA vector into the pronucleus yielded between 9% (7/75) and 18% (13/72) targeted blastocysts. In contrast, the two step procedure with Cas9 and sgRNA simultaneous injected into the cytoplasm followed 2 hours later by pronuclear injection of different concentrations of DNA vector yielded no or at most 3% (1/34) positive blastocysts. Thus, our results suggest that simultaneous injection of RNA and DNA into the cytoplasm or nucleus is the most efficient procedure to achieve targeted insertion.
Conditional Mecp2 mutant mice
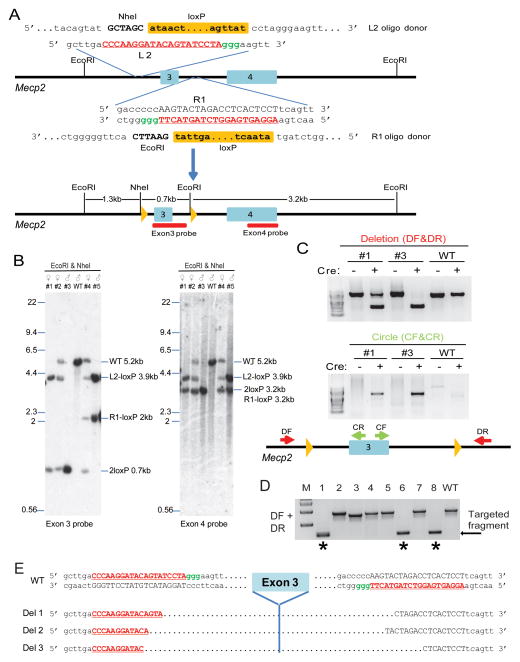
We investigated whether conditional mutant mice can be generated in one step by insertion of two loxP sites into the same allele of the Mecp2 gene. To derive conditional mutant mice similar to those previously described using traditional homologous recombination methods in ES cells (Chen et al., 2001), we designed two sgRNAs targeting Mecp2 intron 2 (L1, L2), and two sgRNAs targeting intron 3 (R1, R2) as well as the corresponding loxP site oligos with 60bp homology to sequences on each side surrounding each sgRNA mediated DSB (Figure S3A). To facilitate detection of correct insertions, the oligos targeting intron 2 were engineered to contain a NheI restriction site and the oligos targeting intron 3 to contain an EcoRI site in addition to the LoxP sequences (Figures 3A and S3A). To determine the efficiency of single loxP site integration at the Mecp2 locus, we injected Cas9 mRNA and each single sgRNA and corresponding oligo into zygotes, which were cultured to the blastocyst stage and genotyped by the RFLP assay. As shown in Figure S3B, the L2 and R1 sgRNAs were more efficient in integrating the oligos with 4 out of 8 embryos carrying the L2 oligo and 2 out of 6 embryos carrying the R1 oligo. Therefore, L2 and R1 sgRNAs and the corresponding oligos were chosen for the generation of a floxed allele (Figure 3A).
Figure 3.
One step generation of a Mecp2 floxed allele. (A) Schematic of the Cas9/sgRNA/oligo targeting sites in Mecp2 intron 2 and intron 3. The sgRNA coding sequence is underlined, capitalized, and labeled in red. The PAM sequence is labeled in green. In the oligo donor sequence, the loxP site is indicated as an orange box, and the restriction site sequences are in bold and capitalized. The oligo contained 60bp homologies on both sides flanking the DSB. Restriction enzymes used for RFLP and Southern blot analysis are shown, and the Southern blot probes are shown as red boxes. (B) Southern analysis of targeted alleles. Data of five mice are shown. EcoRI/NheI-digested genomic DNA was hybridized with the exon3 probe. Expected fragment size: WT = 5.2 kb, 2loxP = 0.7 kb, L2-loxP = 3.9kb, R1-loxP = 2kb. The blot was then stripped and hybridized with the exon4 probe. Expected fragment size: WT = 5.2 kb, 2loxP = 3.2 kb. L2-loxP = 3.9kb, R1-loxP = 3.2kb. The sequence of the floxed allele is shown in Fig. S4b. (C) In vitro Cre-mediated recombination of the floxed Mecp2 allele. The genomic DNA of targeted mice #1 and #3 was incubated with Cre recombinase, and used as PCR template. Primers DF and DR flanking the floxed allele produce shorter products upon Cre-dependent excision. Primers CF and CR detect the circular molecule, which only form upon Cre-loxP recombination. The position of each primer is shown at the bottom cartoon. The deletion and circular PCR products were sequenced and the sequences are shown in Fig. S4c. (D) Injection of Cas9 mRNA and both L2 and R1 sgRNA generated Mecp2 mutant allele with deletion of exon 3. PCR genotyping using primers DF and DR identified defined deletion events in mice #1, #6, and #8 (indicated by stars). (E) Sequences of three mutant alleles with exon 3 deletions in three mice. R2 and L1 sgRNA coding sequences were underlined, capitalized, and labeled in red. The PAM sequence is labeled in green. See also Figures S3 and S4.
A total of 98 E13.5 embryos and mice were generated from zygotes injected with Cas9 mRNA, sgRNAs, and DNA oligos targeting the L2 and R1 sites. Genomic DNA was digested with both NheI and EcoRI, and analyzed by Southern blot using exon 3 and exon 4 probes (Figures 3A and 3B). The L2 and R1 oligos contained, in addition to the loxP site, different restriction sites (NheI or EcoRI). Thus, single loxP site integration at L2 or R1 will produce either a 3.9kb or a 2kb band, respectively, when hybridized with the exon3 probe (Figures 3A and B). We found that about 50% (45/98) of the embryos and mice carried a loxP site at the L2 site and about 25% (25/98) at the R1 site. Importantly, integration of both loxP sites on the same DNA molecule, generating a floxed allele, produces a 700bp band as detected by exon 3 probe hybridization (Figures 3A and 3B). RFLP analysis, sequencing (Figures S4A and S4B) and Southern blot analysis (Figure 3B) showed that 16 out of the 98 mice tested contained two loxP sites flanking exon 3 on the same allele. Table 2 summarizes the frequency of all alleles and shows that the overall insertion frequency of an L2 or R1 insertion was slightly higher in females (21/38) than in males (28/60) consistent with the higher copy number of the X-linked Mecp2 gene in females. To confirm that the floxed allele was functional, we used genomic DNA for in vitro Cre-mediated recombination. Upon Cre treatment, both the deletion and circular products were detected by PCR in targeted mice, but not in DNA from wild type mice (Figure 3C). The PCR products were sequenced and confirmed the precise Cre-loxP mediated recombination (Figure S4C).
We noticed that some pups carried large deletions but no loxP insertions, raising the possibility that two cleavage events may generate defined deletions. To confirm this notion, we co-injected Cas9 mRNA, Mecp2-L2 and R1 sgRNAs but without oligos. PCR genotyping and sequencing (Figures 3D and 3E) revealed that eight out of 23 mice carried deletions of about 700bp spanning the L2 and R1 sites removing exon 3. This was confirmed by Southern analysis (data not shown). Because DNA breaks are repaired through the non-homologous end joining (NHEJ) pathway, the ends of the breaks are different in different deletion alleles (Figure 3E).
Mosaicism
As mentioned above, we noted that some animals were mosaic for the targeted insertion. We decided to characterize the frequency of mosaicism in Mecp2 targeted mice by Southern blot analysis. Since Mecp2 is an X-linked gene, in males more than one allele and in females more than two different alleles suggest mosaicism, which would be expected if integration occurred at a later than the zygote stage. For example, as shown in Figure 3B, female mouse #2 contained three different alleles (one WT allele, one floxed allele, and one L2-loxP allele), and female mouse #4 contained four different alleles (one WT allele, one floxed allele, one L2-loxP allele, and one R1-loxP allele). Male mouse #5 contained two different alleles, with each allele carrying a single loxP site (Figure 3B). We identified eight mosaics out of 16 mice containing a Mecp2 floxed allele. The frequency of mosaicism among 49 embryos and mice containing loxP site was about 40% (20/49) (Table S2). Since Southern blot analysis cannot detect small in-del mutations caused by NHEJ repair, it is possible that this underestimates the overall mosaicism frequency.
Off-target analysis
Two recent studies identified a high level of off-target cleavage in human cell lines using the CRISPR/Cas system, with Cas9 targeting specificity being shown to tolerate small numbers of mismatches between sgRNA and target DNA in a sequence and position dependent manner (Fu et al., 2013; Hsu et al., 2013). Similarly, using a transcription-based method, Mali et al reported that Cas9/sgRNA binding could tolerate up to three mismatches (Mali et al., 2013a)
We characterized potential off-target (OT) mutations in mice and ES cell lines derived from zygotes injected with Cas9 and sgRNAs targeting the Sox2, the Nanog, the Oct4 and the Mecp2 gene. We identified all genomic loci containing up to three or four base pair mismatches compared to the 20bp sgRNA coding sequence (Table S3). We amplified all 13 potential OT sites of Sox2 sgRNA in six mice and four ES cell lines carrying the Sox2-V5 allele and tested for potential off target mutations using the Surveyor assay. No mutation was detected in any locus. When nine Nanog sgRNA potential OT sites (including seven genomic loci containing four base pair mismatches in the PAM distal region) were tested in five correctly targeted mice and four targeted ES cell lines, mutations were found in seven samples at OT1 (Table S3). Since Nanog OT1 has only one base pair difference at the very 5′ end of the sgRNA (position 20, numbered 1–20 in the 3′ to 5′ direction of sgRNA target site), it may not be surprising to find such a high frequency of mutations at this locus. In contrast, no off-target mutation was seen in any other Nanog OTs, which contain three or four base pair difference. For Oct4, we tested all eleven OT sites containing up to three base pair mismatches in three targeted mice and three targeted ES cell lines. Mutations were found in four out of six samples at OT1, which has only one base pair mismatch at position 19, while no off-target mutation was identified in any other Oct4 OTs, which contain two or three base pair mismatches (Table S3). Finally, four potential off-targets sites for Mecp2 L2, and ten sites for Mecp2 R1 were analyzed in ten mice carrying a Mecp2 floxed allele. Only one off-target mutation was identified in one mouse at the Mecp2 R1 OT2 (Table S3). In summary, we tested all potential off-target sites differing up to three or four base pairs in 35 mice or ES cell lines and only identified mutations in one off-target site for Nanog (OT1 7/9 samples, one mismatch at position 20), Oct4 (OT1 4/6 samples, 1 mismatch at position 19), and Mecp2 (R1-OT2 1/10 samples, two mismatches at positions 7 and 20). No off target mutation was identified in any genomic locus containing three base pair mismatches. Thus, although the off-target mutation rate was lower than what had been observed in the previous studies using cultured human cancer cell lines, our results are consistent with the conclusion that two or more interspaced mismatches dramatically reduce Cas9 cleavage (Fu et al., 2013; Hsu et al., 2013).
DISCUSSION
In this study we demonstrate that CRISPR/Cas technology can be used for efficient one-step insertions of a short epitope or longer fluorescent tags into precise genomic locations, which will facilitate the generation of mice carrying reporters in endogenous genes. Mice and embryos carrying reporter constructs in the Sox2, the Nanog and the Oct4 gene were derived from zygotes injected with Cas9 mRNA, sgRNAs and DNA oligo or vectors encoding a tag or a fluorescent marker. Moreover, microinjection of two Mecp2 specific sgRNAs, Cas9 mRNA and two different oligos encoding loxP sites into fertilized eggs allowed for the one-step generation of conditional mutant mice. In addition, we show that the introduction of two spaced sgRNAs targeting the Mecp2 gene can produce mice carrying defined deletions of about 700 bp. Though all RNA and DNA constructs were injected into the cytoplasm or nucleus of zygotes, the gene modification events could happen at the one cell stage or later. Indeed, Southern analyses revealed mosaicism in 17% (1/6) to 40% (20/49) of the targeted mice and ES cell lines indicating that the insertion of the transgenes had occurred after the zygote stage (Table S2).
Our previous experiments (Wang et al., 2013) demonstrated an efficiency of CRISPR/Cas sgRNA mediated cleavage that was high enough to allow for the one-step production of engineered mice up to 90% of which carried homozygous mutations in two genes (4 mutant alleles). The results reported here show that the sgRNA mediated DSBs occur at a significantly higher frequency than insertion of exogenous DNA sequences. Therefore, the allele not carrying the insert will likely be mutated as a consequence of NHEJ-based gene disruption. Thus, the reporter allele would need to be segregated away from the mutant allele in order to produce mice carrying a reporter as well as a wild type allele.
Two recent studies reported a high off-target mutation rate in CRISPR/Cas9 transfected human cell lines (Fu et al., 2013; Hsu et al., 2013). We analyzed the off-target rate for five different sgRNAs and identified cleavage of Nanog OT1, Oct4 OT1, and Mecp2 R1 OT2. Nanog OT1 has only one base pair difference from the targeting sequence at the extreme 5′ end (position 20), while Oct4 OT1 contains one base pair mismatch at position 19, and Mecp2 R1 OT2 has one base pair mismatch at position 20, and one mismatch at position 7. Thus, the only off-target mutations in 2bp mismatch targets were seen when one of the mismatches was at the distal 5′ end. This result is consistent with previous findings that Cas9 can catalyze DNA cleavage in the presence of single-base mismatches in the PAM-distal region (Cong et al., 2013; Hsu et al., 2013; Jiang et al., 2013; Jinek et al., 2012). No mutations were detected in 42 potential OTs of Sox2, Nanog, Oct4 or Mecp2 containing 3 or 4 bp mismatches in a total of 35 mice and ES cell lines tested, consistent with the observation that three or more interspaced mismatches dramatically reduce Cas9 cleavage (Hsu et al., 2013). Thus, for designing the most suitable target sequences for generating Cas9 cleavage-mediated genetically modified mice, it is important to avoid targeting sites that have only one or two mismatches in other genomic loci. This is particularly important for mismatches that are at the PAM distal region.
We consider several possibilities to explain the lower off-target cleavage rate seen in animals derived from manipulated zygotes and the results reported for CRISPR/Cas treated human cell lines (Fu et al., 2013; Hsu et al., 2013). (i) In our study the off-target mutagenesis was based on the analysis of a “clonal genome” in animals derived from a single manipulated zygote in contrast to the previous reports that analyzed heterogeneous cell populations. The surveyor assay, based upon extensive PCR amplification, may identify any mutation, even very rare alleles that may be present in the heterogeneous population. (ii) The transformed human cell lines may have different DNA damage responses resulting in a different mutagenesis rate than the normal one cell embryo. (iii) In our experiments CRISPR/Cas was injecting as short-lived RNA in contrast to Fu et al. and Hsu et al. who used DNA plasmid transfection, which may express the Cas9/sgRNA for longer time periods leading to more extensive cleavage. Thus, our data suggest high specificity of the CRISPR/Cas9 system for gene editing in early embryos aimed at generating gene-modified mice. Nevertheless, future characterization of off-target mutagenesis of CRISPR/Cas system using whole genome sequencing would be highly informative and may allow designing sgRNAs with higher specificity.
In summary, CRISPR/Cas mediated genome editing represents an efficient and simple method of generating sophisticated genetic modifications in mice such as conditional alleles and endogenous reporters in one step. The principles described in this study could be directly adapted to other mammalian species, opening the possibility of sophisticated genome engineering in many species where ES cells are not available.
EXPERIMENTAL PROCEDURES
Production of Cas9 mRNA and sgRNA
Bicistronic expression vector px330 expressing Cas9 and sgRNA (Cong et al., 2013) was digested with BbsI and treated with Antarctic Phosphatase, and the linearized vector was gel-purified. A pair of oligos (Table S3) for each targeting site was annealed, phosphorylated, and ligated to the linearized vector.
T7 promoter was added to Cas9 coding region by PCR amplification using primer Cas9 F and R (Table S4). T7-Cas9 PCR product was gel-purified and used as the template for in vitro transcription (IVT) using mMESSAGE mMACHINE T7 ULTRA kit (Life Technologies). T7 promoter was added to sgRNAs template by PCR amplification using primer listed in Table S4. The T7-sgRNA PCR product was gel-purified and used as the template for IVT using MEGAshortscript T7 kit (Life Technologies). Both the Cas9 mRNA and the sgRNAs were purified using MEGAclear kit (Life Technologies) and eluted in RNase-free water.
Single stranded and double stranded DNA donors
All single stranded oligos were ordered as Ultramer DNA oligos from Integrated DNA Technologies. Nanog-2A-mCherry vector was modified from previously published targeting vector Nanog-2A-mCherry-PGK-Neo (Faddah et al., 2013). Nanog-2A-mCherry-PGK-Neo was digested with PacI and AscI to drop out the PGK-Neo cassette, the 9.7kb fragment was gel purified and blunt-ended using T4 DNA polymerase (New England Biolabs), then self-ligated using T4 DNA ligase (New England Biolabs). Oct4-IRES-eGFP-PGK-Neo vector is previously published (Lengner et al., 2007).
Suveryor assay and RFLP analysis for genome modification
Suveryor assay was performed as described (Guschin et al., 2010). Genomic DNA from targeted and control mice or embryos was extracted and PCR was performed using gene specific primers (Table S4) under the following conditions: 95°C for 5 min; 35× (95°C for 30 s, 60°C for 30 s, 68°C for 40 s); 68°C for 2 min; hold at 4°C. PCR products were then denatured, annealed, and treated with Suveryor nuclease (Transgenomic). DNA concentration of each band was measured on an ethidium bromide-stained 10% acrylamide Criterion TBE gel (BioRad) and quantified using Image J software. For RFLP analysis, 10μl of Tet1, Tet2, Mecp2-R1, R2 PCR product was digested with EcoRI, 10μl of Mecp2-L1, L2 PCR product was digested with NheI. Digested DNA was separated on an ethidium bromide-stained agarose gel (2%). For sequencing, PCR products were cloned using the Original TA Cloning Kit (Invitrogen), and mutations were identified by Sanger sequencing.
One cell embryo injection
All animal procedures were performed according to NIH guidelines and approved by the Committee on Animal Care at MIT. B6D2F1 (C57BL/6 X DBA2) female mice and ICR mouse strains were used as embryo donors and foster mothers, respectively. Super-ovulated female B6D2F1 mice (7–8 weeks old) were mated to B6D2F1 stud males, and fertilized embryos were collected from oviducts. Different concentrations of Cas mRNA, sgRNA and oligos or plasmid vectors were mixed and injected into the cytoplasm or pronucleus of fertilized eggs with well-recognized pronuclei in M2 medium (Sigma). The injected zygotes were cultured in KSOM with amino acids at 37 °C under 5% CO2 in air until blastocyst stage by 3.5 days. Thereafter, 15–25 blastocysts were transferred into uterus of pseudopregnant ICR females at 2.5 dpc.
Southern blotting
Genomic DNA was separated on a 0.8% agarose gel after restriction digests with the appropriate enzymes, transferred to a nylon membrane (Amersham) and hybridized with 32P random primer (Stratagene)-labeled probes. Between hybridizations, blots were stripped and checked for complete removal of radioactivity before rehybridization with a different probe.
In vitro Cre recombination
A 20-μl reaction containing 1 μg of genomic DNA and 10 units of recombinant Cre recombinase (New England Biolabs) in 1x buffer was incubated at 37 °C for one hour. For all targets, 1 μl of the Cre reaction mix was used as template for PCR reactions with gene-specific primers. For each target, primers DF and DR were used for detecting the deletion products, and primers CF and CR were used to detect the circle product. All products were sequenced.
Immunostaining and Western blot analysis
For immunostaining, cells in 24-well were fixed in PBS supplemented with 4% paraformaldehyde for 15 min at room temperature (RT). The cells were then permeabilized using 0.2% Triton X-100 in PBS for 15 min at RT. The cells were blocked for 30 min in 1% BSA in PBS. Primary antibody against V5 (ab9137, abcam) was diluted in the same blocking buffer and incubated with the samples overnight at 4 °C. The cells were treated with a fluorescently coupled secondary antibody and then incubated for 1 hr at RT. The nuclei were stained with Hoechst 33342 (Sigma) for 5 min at RT.
For western blot, Cell pellets were lysed on ice in Laemmli buffer (62.5 mM Tris-HCl, pH 6.8, 2% sodium dodecyl sulfate, 5% beta-mercaptoethanol, 10% glycerol and 0.01% bromophenol blue) for 30 min in presence of protease inhibitors (Roche Diagnostics), boiled for 5–7 min at 100 °C, and subjected to western blot analysis. Primary antibodies: V5 (1:1,000, ab9137, abcam), beta-actin (1:2,000). Blots were probed with anti-goat, or anti-rabbit IgG-HRP secondary antibody (1:10,000) and visualized using ECL detection kit (GE Healthcare).
ESC derivation and differentiation
Morulas or blastocysts were selected to generate ES cell lines. The zona pellucida was removed using acid Tyrode solution. Each embryo was transferred into one well of a 96-well plate seeded with ICR embryonic fibroblast feeders in ESC medium supplemented with 20% knockout serum replacement, 1,500 U/ml leukemia inhibitory factor (LIF), 3 μM CHIR99021, and 1 μM PD0325901. After 4–5 days in culture, the colonies were trypsinized and transferred to a 96-well plate with a fresh feeder layer in fresh medium. Clonal expansion of the ESCs proceeded from 48-well plates to 6-well plates with feeder cells and then to 6-well plates for routine culture.
For ESC differentiation, cells were harvested by trypsinization and transferred to bacterial culture dishes in the ES medium without or LIF. After 3 days, aggregated cells were plated onto gelatin-coated tissue culture dishes and incubated for another 3 days.
Prediction of potential off-targets
Potential off-targets were predicted by searching the mouse genome (mm9) for matches to the 20-nt sgRNA sequence allowing for up to 4 mismatches (Nanog) or 3 mismatches (Sox2, Oct4, Mecp2-L2 and Mecp2-R1) followed by NGG PAM sequence. Matches were ranked first by ascending number of mismatches, then by ascending distance from the PAM sequence.
Supplementary Material
Research Highlights.
One step generation of mice with reporters in endogenous genes
One step generation of conditional mutant mice
Off-target analysis suggests high specificity of the CRISPR/Cas9 system
Acknowledgments
We thank Ruth Flannery and Kibibi Ganz for support with animal care and experiments. We thank Jaenisch lab members for helpful discussions on the manuscript. We are also grateful to Dina A. Faddah for the help of editing the manuscript. AWC is supported by a Croucher scholarship. R.J. is an adviser to Stemgent and a cofounder of Fate Therapeutics. This work was supported by NIH grants HD 045022 and R37CA084198 to RJ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brown AJ, Fisher DA, Kouranova E, McCoy A, Forbes K, Wu Y, Henry R, Ji D, Chambers A, Warren J, et al. Whole-rat conditional gene knockout via genome editing. Nat Methods. 2013;10(7):638–40. doi: 10.1038/nmeth.2516. [DOI] [PubMed] [Google Scholar]
- Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6:507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- Carbery ID, Ji D, Harrington A, Brown V, Weinstein EJ, Liaw L, Cui X. Targeted genome modification in mice using zinc-finger nucleases. Genetics. 2010;186:451–459. doi: 10.1534/genetics.110.117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Ji D, Fisher DA, Wu Y, Briner DM, Weinstein EJ. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat Biotechnol. 2011;29:64–67. doi: 10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- Faddah DA, Wang H, Cheng AW, Katz Y, Buganim Y, Jaenisch R. Single-Cell Analysis Reveals that Expression of Nanog Is Biallelic and Equally Variable as that of Other Pluripotency Factors in Mouse ESCs. Cell Stem Cell. 2013;13:23–29. doi: 10.1016/j.stem.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013 doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013 doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner CJ, Camargo FD, Hochedlinger K, Welstead GG, Zaidi S, Gokhale S, Scholer HR, Tomilin A, Jaenisch R. Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell. 2007;1:403–415. doi: 10.1016/j.stem.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013a doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013b;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, de Angelis MH, Wurst W, Kuhn R. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc Natl Acad Sci U S A. 2010;107:15022–15026. doi: 10.1073/pnas.1009424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, Zhang X, Zhang P, Huang X. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;23(5):720–3. doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, Jeong D, Kim JS, Lee HW. Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol. 2013;31:23–24. doi: 10.1038/nbt.2477. [DOI] [PubMed] [Google Scholar]
- Tesson L, Usal C, Menoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L, et al. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29:695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wefers B, Meyer M, Ortiz O, Hrabe de Angelis M, Hansen J, Wurst W, Kuhn R. Direct production of mouse disease models by embryo microinjection of TALENs and oligodeoxynucleotides. Proc Natl Acad Sci U S A. 2013;110:3782–3787. doi: 10.1073/pnas.1218721110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.