Abstract
Human basal-like breast cancer (BLBC) is an enigmatic and aggressive malignancy with a poor prognosis. There is an urgent need to identify therapeutic targets for BLBC because current treatment modalities are limited and not effective. The forkhead box transcription factor FOXC1 has recently been identified as a critical functional biomarker for BLBC. However, how it orchestrates BLBC cells was not clear. Here we show that FOXC1 activates the transcription factor NF-κB in BLBC cells by increasing p65/RelA protein stability. High NF-κB activity has been associated with estrogen receptor-negative breast cancer, particularly BLBC. The effect of FOXC1 on p65/RelA protein stability is mediated by increased expression of Pin1, a peptidyl-prolyl isomerase. FOXC1 requires NF-κB for its regulation of cell proliferation, migration, and invasion. Notably, FOXC1 overexpression renders breast cancer cells more susceptible to pharmacologic inhibition of NF-κB. These results suggest that BLBC cells may rely on FOXC1-driven NF-κB signaling. Interventions of this pathway may provide modalities for the treatment of BLBC.
Keywords: basal-like breast cancer, FOXC1, NF-κB, p65/RelA, Pin1, protein stability
Introduction
Although first reported more than 20 years ago on the basis of immunohistochemical (IHC) detection of high molecular weight basal cytokeratins (CKs) in a small subgroup of breast cancers (1, 2), the basal-like subtype again became notable after global gene expression analysis confirmed it as a distinct molecular entity within breast cancer (3). Basal-like breast cancers (BLBCs) express genes characteristic of basal/myoepithelial cells in the normal mammary gland and comprise up to 25% of all breast cancers (4, 5). They underexpress estrogen receptor (ERα), progesterone receptor (PR), and HER2, and are associated with high histological grade and aggressive clinical behavior (6). However, BLBC is not synonymous with the ER-/PR-/HER2-triple-negative phenotype (TNP). Whereas ERα and HER2 guide targeted treatment of luminal and HER2-positive breast cancers, respectively, chemotherapy is still the only modality of systemic therapy for BLBC. The high mortality of BLBC reflects its rapid growth rate (7, 8) and aggressive migration/invasion (9). Perhaps not surprisingly, BLBC is overrepresented among the interval breast cancers arising between mammograms (10).
Recently, the forkhead box transcription factor FOXC1 has been identified as a potential pivotal biomarker for BLBC, and high expression correlates with poor overall survival in breast cancer (11–13). Forkhead box transcription factors, characterized by a common 100-amino acid winged-helix DNA-binding domain, play important roles in cell growth, survival, differentiation, migration, and longevity (14). FOXC1 has been postulated to control the development of embryonic mesenchymal tissue (15). FOXC1 is mutated in the autosomal dominant disease Axenfeld-Rieger syndrome (AR) (16), which is characterized by ocular defects, cardiac disease, and cranio-facial abnormalities. FOXC1 homozygous knockout mice die at birth with hydrocephalus, skeletal, and eye defects (17). Further examination of FOXC1 heterozygous knockout mice showed that these mice have anterior eye segment malformations similar to those found in human patients with FOXC1 mutations (18). Mechanistically, how FOXC1 exerts these effects is not well understood.
Previously, it was shown that ectopic overexpression of FOXC1 in breast cancer cells induced aggressive phenotypes such as epithelial-mesenchymal transition, and increased cell proliferation, migration, and invasion (12). Knockdown of FOXC1 using shRNA in breast cancer cells with high endogenous levels of FOXC1 demonstrated the opposite effects with loss of aggressive phenotypic features. However, the mechanism underlying the role of FOXC1 in BLBC cells is not clear. To address this question, this study was designed to determine how FOXC1 enlists or interacts with other basal-like tumor-associated signaling pathways to control cancer cell functions. By characterizing the molecular actions of FOXC1, our results demonstrate that FOXC1 orchestrates BLBC-associated phenotypes by regulating Pin1/NF-κB signaling.
Results and Discussion
To date, the genetic profile and biologic basis of BLBC are poorly understood. Recent studies have implicated several signaling pathways such as MEK/PI3K (19, 20), integrin (21), Notch pathways (22), and NF-κB (23) in BLBC development and progression. To identify BLBC-specific signaling features and to confirm the relevance of these and other pathways reported to impact breast cancer, we used the Ingenuity IPA platform and publicly available cDNA microarray datasets to conduct unbiased, systematic screening analysis of signaling networks in human breast cancer. As illustrated in Figure 1A, NF-κB was revealed as one of the most distinctive pathways in basal-like tumors. This is consistent with previous reports that sustained NF-κB activation exists mostly in human BLBC cell lines (23, 24) and ER-negative breast cancer (25) with its highest activity found in triple-negative tumors (26). NF-κB activity was also found to be essential for the proliferation and survival of BLBC cells (23).
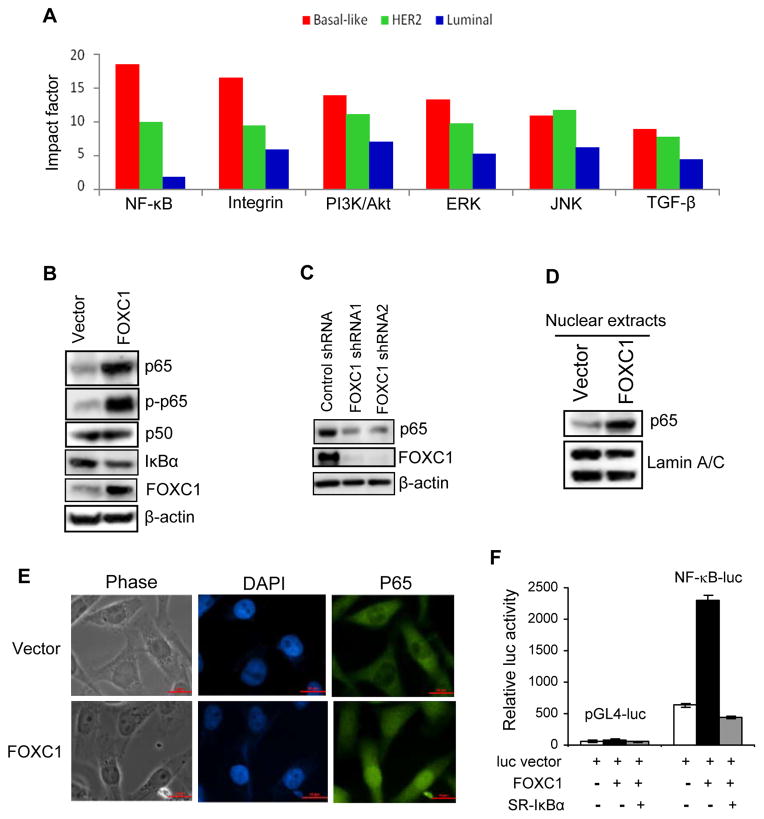
Figure 1. FOXC1 induces NF-κB activity in breast cancer cells.
(A) Significant canonical signaling pathways in basal-like (red), HER2 (green) and luminal (blue) breast cancers from the Richardson et al. dataset were identified using Ingenuity Pathway Analysis and ranked by the impact factor (see Supplementary Information for detailed methods). (B) Immunoblotting of NF-κB components in MDA-MB-231 cells overexpressing FOXC1 or the vector. (C) Immunoblotting of p65 in control or FOXC1 shRNA-expressing BT-549 breast cancer cells. (D) Nuclear proteins were isolated from vector- or FOXC1-overexpressing MDA-MB-231 cells, followed by immunoblotting of p65 and a nuclear marker Lamin A/C. (E) Nuclear localization of p65 protein was visualized by fluorescence microscopy (green, p65; blue, nuclear DNA staining by DAPI). (F) MDA-MB-231 cells were transiently transfected with NF-κB-luc or the vector pGL4-luc, FOXC1, and IκBα-SR. NF-κB activity was assessed by luciferase assays. Datarepresent mean ± SD (n = 3).
An interesting and axiomatic issue arising from these findings is whether FOXC1 coordinates specific BLBC-associated signaling pathways. Because some Fox transcription factors reportedly modulate NF-κB in non-malignant conditions (27), we postulated that FOXC1 might modulate NF-κB function in BLBC cells. Using immunoblotting, we found that the NF-κB p65 subunit and its phosphorylation at Ser-546 (by IκB kinase [IKK]) were markedly induced by FOXC1 overexpression in basal-like MDA-MB-231 breast cancer cells (Figure 1B) and luminal MCF-7 breast cancer cells (Supplemental Figure 1A), which harbor low and undetectable endogenous FOXC1 levels, respectively (12). Notably, levels of the NF-κB inhibitor IκBα were moderately but consistently reduced, which could be attributed to modestly higher IKK levels in these cells (data not shown). Conversely, knockdown of FOXC1 by its shRNA, which reduced FOXC1 levels by > 90%, suppressed p65 expression in basal-like BT-549 (human, Figure 1C) and 4T1 (mouse, Supplemental Figure 1B) breast cancer cells, both of which possess high endogenous FOXC1 levels (12).
Next we examined the nuclear localization of p65, an indicator of activated NF-κB. Immunoblotting with nuclear extracts of vector- and FOXC1-overexpressing MDA-MB-231 and MCF-7 cells indicated that FOXC1 promoted p65 translocation into the nucleus (Figure 1D and Supplemental Figure 1C). This was corroborated by increased immunofluorescence staining of nuclear p65 in FOXC1-overexpressing cells (Figure 1E). In agreement with this result, TransAM ELISA with oligonucleotides comprising consensus NF-κB-binding sequences showed that increase of FOXC1 expression potentiated the DNA-binding activity of p65 without affecting that of p50 in breast cancer cells (Supplemental Figure 1D). Conversely, FOXC1 knockdown by its shRNAs reduced p65 DNA-binding activity (Supplemental Figure 1E). To further substantiate that FOXC1 enhances NF-κB activity, we used a NF-κB-responsive luciferase reporter construct. As expected, FOXC1 overexpression robustly increased NF-κB-driven luciferase activity in MDA-MB-231 in (Figure 1F) and MCF-7 cells (Supplemental Figure 1F), and ectopic overexpression of the IκBα S32A/S36A super-repressor (IκBα-SR) abolished this FOXC1 effect. Supporting the above findings, FOXC1 overexpression upregulated NF-κB-inducible interleukin-6 (IL-6) expression in MDA-MB-231 (Supplemental Figure 1G), whereas FOXC1 knockdown downregulated IL-6 expression in BT-549 and 4T1 cells (Supplemental Figure 1H). Taken together, these results demonstrate that FOXC1 is a potent inducer of NF-κB activity in breast cancer cells and provides clues to why NF-κB is hyperactive in BLBC.
We then explored the potential mechanisms underlying FOXC1-mediated upregulation of p65. Whereas p65 mRNA levels were similar across breast cancer subgroups in microarray analysis (data not shown) and were not altered by FOXC1 overexpression, IκBα transcription was enhanced (Supplemental Figure 2), reflecting a known negative feedback mechanism for regulating increased NF-κB activity. Thus we postulated that p65 expression might be regulated at the protein level. Pin1, a peptidyl-prolyl isomerase that binds to and isomerizes specific phosphorylated Ser or Thr, was of particular interest due to its pivotal role in the control of p65 protein stability and activity (28) and its involvement in cancer development (29). It binds to p65 and thereby blocks its association with IκBα and SOCS-1, a ubiquitin ligase for p65, leading to inhibition of p65 proteolysis. Indeed, immunoblotting and quantitative RT-PCR showed that Pin1 was upregulated by FOXC1 overexpression in MDA-MB-231 cells (Figure 2A and B), while downregulated by FOXC1 shRNAs in BT-549 cells (Supplemental Figure 3). In addition, FOXC1 potentiated the luciferase reporter activity driven by a Pin1 promoter (Figure 2C).
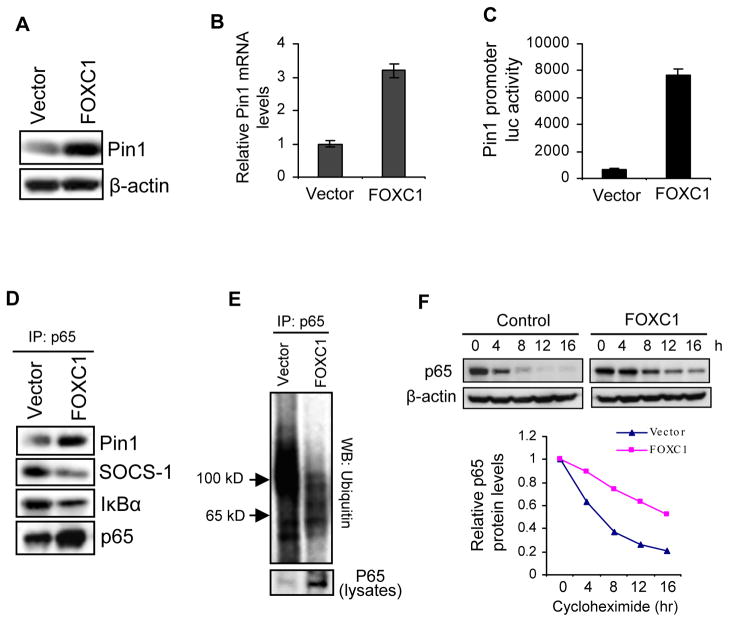
Figure 2. FOXC1 increases p65 protein stability by upregulating Pin1 in breast cancer cells.
(A) Immunoblotting of Pin1 in MDA-MB-231 cells overexpressing FOXC1 or the vector. (B) Real-time RT-PCR analysis of Pin1 mRNA in the same cells. The Pin primers are 5′-TGGGTGCCTTCAGCAGAGGTCAG-3′ and 5′-CCGGAATCCGTGAACACGGGC-3′ (see Supplementary Information for detailed methods). (C) A 2.3 kb Pin1 promoter-luciferase reporter construct was co-transfected into MDA-MB-231 cells with FOXC1 or the vector, followed by luciferase assays. (D) Lysates from FOXC1- or vector-overexpressing MDA-MB-231 cells were immunoprecipitated with an anti-p65 antibody, followed by immunoblotting of IκBα, Pin1, and SOCS-1. (E) Same cells were transfected with a ubiquitin construct, treated with 10 μM MG-132, and subjected to immunoprecipitation with an anti-p65 antibody, followed by immunoblotting of ubiquitin. (F) Same cells were treated with cycloheximide (10 μg/ml) and harvested at the indicated time points, followed by immunoblotting and densitometry of protein bands. Band intensities were normalized to that of actin, then normalized to the t = 0 controls.
To examine whether FOXC1 enhances the association between Pin1 and p65, we performed immunoprecipitation with an anti-p65 antibody followed by immunoblotting. As presented in Figure 2D, FOXC1 increased the binding of p65 to Pin1, but decreased its binding to IκBα and SOCS-1. As SOCS-1 facilitates p65 degradation (28), we reasoned that ubiquitin-mediated proteolysis is involved in the FOXC1 effect on p65 protein levels. To address this question, we immunoprecipitated p65 and performed immunoblotting of ubiquitin after treating MDA-MB-231 cells with the proteasome inhibitor MG-132. As illustrated in Figure 2E, FOXC1 overexpression attenuated the ubiquitination of p65. Similar results were obtained when ubiquitinated proteins were immunoprecipitated, followed by immunoblotting of p65 (Supplemental Figure 4). To corroborate these results, we treated the same cells with the translation inhibitor cycloheximide for different time periods. Immunoblot analysis of cells with inhibited de novo protein synthesis demonstrated that FOXC1 enhanced p65 protein stability (Figure 2F). In line with these findings, knockdown of Pin1 by its siRNA reduced NF-κB-responsive luciferase reporter activity in parental MDA-MB-231 cells and abolished FOXC1-induced increase of NF-κB-responsive luciferase activity and p65 levels (Supplemental Figure 5). Collectively, these data indicate that Pin1 is involved in the activation of NF-κB by FOXC1.
Consistent with a previous study showing a trend towards a correlation of Pin1 protein levels with ER-negative breast cancer (30), analysis of two cDNA microarray datasets of 51 human breast cancer cell lines revealed that Pin1 mRNA levels were significantly higher in BLBC cells than in luminal cells (31, 32) (Supplemental Figure 6A), which was confirmed by immunoblotting (Supplemental Figure 6B). Surprisingly, we did not find a statistically significant association between Pin1 mRNA levels and tumors of the basal-like subgroup in cDNA microarray analysis (data not shown), although high Pin1 expression correlated with worse recurrence-free survival (Supplemental Figure 6C) and higher tumor grade, which are commonly associated with basal-like tumors. We also observed a trend towards decreased overall survival in breast cancer patients with high Pin1 levels (Supplemental Figure 7). Because Pin1 is essential for breast cancer development, its overexpression across all subgroups of breast cancer might mask detection of higher Pin1 levels in BLBC. Nevertheless, our findings may implicate Pin1 in the regulation of BLBC. How Pin1 expression is controlled by FOXC1 remains to be determined.
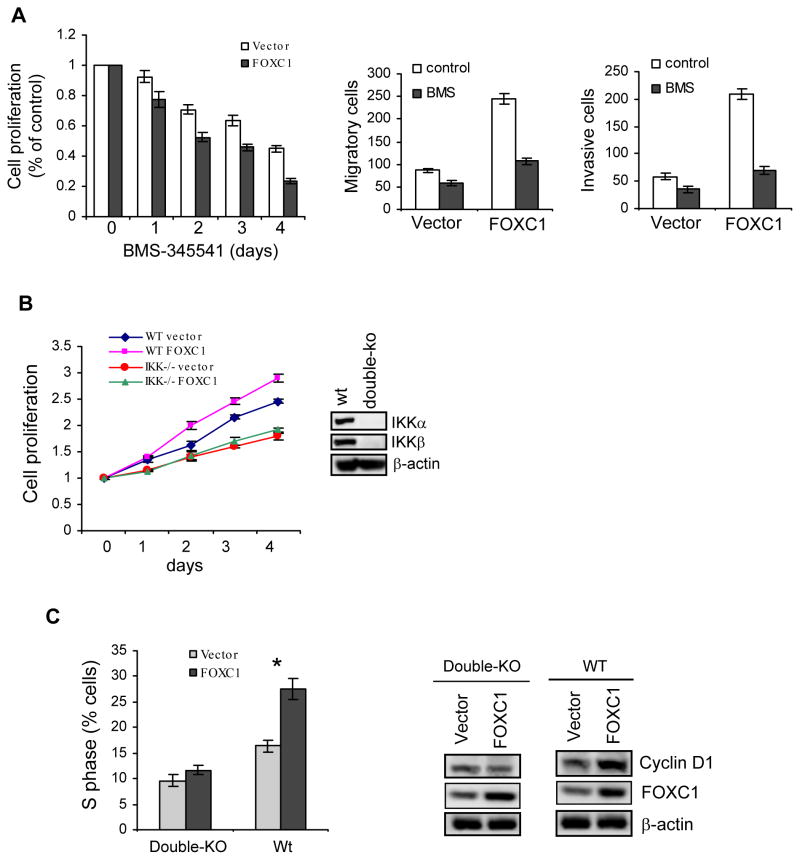
Because of a critical role of NF-κB in cancer cell functions, there has been great interest in targeting NF-κB for development of anticancer therapy. To determine whether NF-κB blockade impairs FOXC1-induced cell phenotypes, we treated vector- and FOXC1-overexpressing MDA-MB-231 cells with small-molecule NF-κB inhibitors. As illustrated in Figure 3A, increase of FOXC1 sensitized MDA-MB-231 cells to pharmacologic inhibition of NF-κB by the IKK inhibitor BMS-345541 in cell proliferation, migration, and invasion assays. Similar results were found with other NF-κB inhibitors such as BAY-117082 (data not shown). Coupled with previous findings that FOXC1 is critically involved in BLBC cell functions (12), these data suggest that FOXC1 exploits NF-κB to promote aggressive cellular traits commonly associated with BLBC (7, 9). To further corroborate that NF-κB is involved in the effects of FOXC1, we overexpressed FOXC1 in IKKα/IKKβ double-knockout and p65 knockout mouse embryonic fibroblasts (MEFs). MTT assays showed that increased FOXC1 did lead to enhanced cell proliferation in wildtype MEFs, but not in IKKα/IKKβ-null (Figure 3B) and p65-null MEFs (Supplemental Figure 8). Consistent with these results, FOXC1 overexpression increased levels of the NF-κB and Pin1 target cyclin D1 and the percentage of cells in S phase of the cell cycle in wildtype MEFs but not in IKKα/IKKβ-null MEFs (Figure 3C). Taken together, these data suggest that the effect of FOXC1 requires intact NF-κB activity.
Figure 3. NF-κB mediates the effects of FOXC1 on cell proliferation, migration, and invasion.
(A) FOXC1- or vector-overexpressing MDA-MB-231 cells were treated with the NF-κB inhibitor BMS-345541 (2 μM), followed by MTT assays (left), transwell migration assays (middle), and transwell invasion assays (right). Data represent mean ± SD (n = 3). (B) Wild-type (wt) and IKKα/IKKβ-null MEFs were transfected with FOXC1 or the vector, followed by cell proliferation MTT assays at the indicated time points (left). Deficiency of IKK expression in knockout MEFs is shown by immunoblotting (right). (C) Wild-type (wt) and IKKα/IKKβ-null MEFs were transfected with FOXC1 or the vector, followed by cell cycle analysis using flow cytometry (left). *, P < 0.05. CyclinD1 expression was assessed by immunoblotting (right).
In summary, these findings uncover a functional link between FOXC1 expression and NF-κB signaling in BLBC cells. The FOXC1-NF-κB pathway, which involves increased expression of Pin1 and possibly IKK, might be key for acquisition of aggressive cellular traits of BLBC, and targets on this pathway might serve as the basis for therapeutic interventions in BLBC. Further studies will determine whether FOXC1 also contributes to activation or mediation of other BLBC-associated pathways.
Supplementary Material
Acknowledgments
We thank Yixian Zheng for the HA-ubiquitin plasmid, Fred Miller for 4T1 breast cancer cells, Inder M. Verma for IKK double-knockout MEFs, Amer A. Beg for p65-null MEFs, and Dave Hoon and Myles Cabot for technical support. This work was supported by National Institutes of Health (CA151610), QVC and the Fashion Footwear Association of New York Charitable Foundation, and the Avon Foundation (02-2010-068) to XC.
Footnotes
Conflict of Interest
Dr. Wang, Dr. Ray, Dr. Bagaria, and Dr. Cui are named inventors on patent applications regarding the role of FOXC1 in cancer.
References
- 1.Dairkee SH, Puett L, Hackett AJ. Expression of basal and luminal epithelium-specific keratins in normal, benign, and malignant breast tissue. J Natl Cancer Inst. 1988;80(9):691–5. doi: 10.1093/jnci/80.9.691. Epub 1988/07/06. [DOI] [PubMed] [Google Scholar]
- 2.Wetzels RHW, Holland R, Haelst UJGMv, Lane EB, Leigh IM, Ramaekers FCS. Detection of Basement Membrane Components and Basal Cell Keratin 14 in Noninvasive and Invasive Carcinomas of the Breast. Am J Pathol. 1989;134(3):571–9. [PMC free article] [PubMed] [Google Scholar]
- 3.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 4.Kreike B, van Kouwenhove M, Horlings H, Weigelt B, Peterse H, Bartelink H, et al. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res. 2007;9(5):R65. doi: 10.1186/bcr1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol. 2008;26(15):2568–81. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- 6.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. Jama. 2006;295(21):2492–502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 7.Seewaldt VL, Scott V. Images in clinical medicine. Rapid progression of basal-type breast cancer. N Engl J Med. 2007;356(13):e12. doi: 10.1056/NEJMicm063760. Epub 2007/03/30. [DOI] [PubMed] [Google Scholar]
- 8.Livasy CA, Karaca G, Nanda R, Tretiakova MS, Olopade OI, Moore DT, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19(2):264–71. doi: 10.1038/modpathol.3800528. Epub 2005/12/13. [DOI] [PubMed] [Google Scholar]
- 9.Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68(4):989–97. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 10.Collett K, Stefansson IM, Eide J, Braaten A, Wang H, Eide GE, et al. A basal epithelial phenotype is more frequent in interval breast cancers compared with screen detected tumors. Cancer Epidemiol Biomarkers Prev. 2005;14(5):1108–12. doi: 10.1158/1055-9965.EPI-04-0394. Epub 2005/05/17. [DOI] [PubMed] [Google Scholar]
- 11.Dejeux E, Ronneberg JA, Solvang H, Bukholm I, Geisler S, Aas T, et al. DNA methylation profiling in doxorubicin treated primary locally advanced breast tumours identifies novel genes associated with survival and treatment response. Mol Cancer. 2010;9:68. doi: 10.1186/1476-4598-9-68. Epub 2010/03/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ray PS, Wang J, Qu Y, Sim MS, Shamonki J, Bagaria SP, et al. FOXC1 is a potential prognostic biomarker with functional significance in basal-like breast cancer. Cancer Res. 2010;70(10):3870–6. doi: 10.1158/0008-5472.CAN-09-4120. Epub 2010/04/22. [DOI] [PubMed] [Google Scholar]
- 13.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A. 2010;107(35):15449–54. doi: 10.1073/pnas.1004900107. Epub 2010/08/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishimura DY, Swiderski RE, Alward WL, Searby CC, Patil SR, Bennet SR, et al. The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat Genet. 1998;19(2):140–7. doi: 10.1038/493. Epub 1998/06/10. [DOI] [PubMed] [Google Scholar]
- 15.Kume T, Deng KY, Winfrey V, Gould DB, Walter MA, Hogan BL. The forkhead/winged helix gene Mf1 is disrupted in the pleiotropic mouse mutation congenital hydrocephalus. Cell. 1998;93(6):985–96. doi: 10.1016/s0092-8674(00)81204-0. Epub 1998/07/11. [DOI] [PubMed] [Google Scholar]
- 16.Mears AJ, Jordan T, Mirzayans F, Dubois S, Kume T, Parlee M, et al. Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld-Rieger anomaly. Am J Hum Genet. 1998;63(5):1316–28. doi: 10.1086/302109. Epub 1998/10/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kidson SH, Kume T, Deng K, Winfrey V, Hogan BL. The forkhead/winged-helix gene, Mf1, is necessary for the normal development of the cornea and formation of the anterior chamber in the mouse eye. Dev Biol. 1999;211(2):306–22. doi: 10.1006/dbio.1999.9314. Epub 1999/07/09. [DOI] [PubMed] [Google Scholar]
- 18.Smith RS, Zabaleta A, Kume T, Savinova OV, Kidson SH, Martin JE, et al. Haploinsufficiency of the transcription factors FOXC1 and FOXC2 results in aberrant ocular development. Hum Mol Genet. 2000;9(7):1021–32. doi: 10.1093/hmg/9.7.1021. Epub 2000/04/18. [DOI] [PubMed] [Google Scholar]
- 19.Moulder SL. Does the PI3K Pathway Play a Role in Basal Breast Cancer? Clin Breast Cancer. 2010;10:S66–71. doi: 10.3816/CBC.2010.s.014. Epub 2010/12/01. [DOI] [PubMed] [Google Scholar]
- 20.Rexer BN, Ghosh R, Arteaga CL. Inhibition of PI3K and MEK: it is all about combinations and biomarkers. Clin Cancer Res. 2009;15(14):4518–20. doi: 10.1158/1078-0432.CCR-09-0872. Epub 2009/07/09. [DOI] [PubMed] [Google Scholar]
- 21.Lu S, Simin K, Khan A, Mercurio AM. Analysis of integrin beta4 expression in human breast cancer: association with basal-like tumors and prognostic significance. Clin Cancer Res. 2008;14(4):1050–8. doi: 10.1158/1078-0432.CCR-07-4116. Epub 2008/02/19. [DOI] [PubMed] [Google Scholar]
- 22.Lee CW, Simin K, Liu Q, Plescia J, Guha M, Khan A, et al. A functional Notch-survivin gene signature in basal breast cancer. Breast Cancer Res. 2008;10(6):R97. doi: 10.1186/bcr2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamaguchi N, Ito T, Azuma S, Ito E, Honma R, Yanagisawa Y, et al. Constitutive activation of nuclear factor-kappaB is preferentially involved in the proliferation of basal-like subtype breast cancer cell lines. Cancer Sci. 2009;100(9):1668–74. doi: 10.1111/j.1349-7006.2009.01228.x. Epub 2009/06/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakshatri H, Bhat-Nakshatri P, Martin DA, Goulet RJ, Jr, Sledge GW., Jr Constitutive activation of NF-kappaB during progression of breast cancer to hormone-independent growth. Mol Cell Biol. 1997;17(7):3629–39. doi: 10.1128/mcb.17.7.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biswas DK, Iglehart JD. Linkage between EGFR family receptors and nuclear factor kappaB (NF-kappaB) signaling in breast cancer. J Cell Physiol. 2006;209(3):645–52. doi: 10.1002/jcp.20785. Epub 2006/09/27. [DOI] [PubMed] [Google Scholar]
- 26.Gershtein ES, Scherbakov AM, Platova AM, Tchemeris GY, Letyagin VP, Kushlinskii NE. The expression and DNA-binding activity of NF-kappaB nuclear transcription factor in the tumors of patients with breast cancer. Bull Exp Biol Med. 2010;150(1):71–4. doi: 10.1007/s10517-010-1072-3. Epub 2010/12/17. [DOI] [PubMed] [Google Scholar]
- 27.Peng SL. Interplay between the NF-kappaB and forkhead transcription factors. Cell Death Differ. 2005;12(7):699–701. doi: 10.1038/sj.cdd.4401640. Epub 2005/04/30. [DOI] [PubMed] [Google Scholar]
- 28.Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, et al. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol Cell. 2003;12(6):1413–26. doi: 10.1016/s1097-2765(03)00490-8. Epub 2003/12/24. [DOI] [PubMed] [Google Scholar]
- 29.Lu KP. Prolyl isomerase Pin1 as a molecular target for cancer diagnostics and therapeutics. Cancer Cell. 2003;4(3):175–80. doi: 10.1016/s1535-6108(03)00218-6. Epub 2003/10/03. [DOI] [PubMed] [Google Scholar]
- 30.Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Petkova V, et al. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. Embo J. 2001;20(13):3459–72. doi: 10.1093/emboj/20.13.3459. Epub 2001/07/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoeflich KP, O’Brien C, Boyd Z, Cavet G, Guerrero S, Jung K, et al. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin Cancer Res. 2009;15(14):4649–64. doi: 10.1158/1078-0432.CCR-09-0317. Epub 2009/07/02. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.