Abstract
β-Catenin has a dual function in cells: fortifying cadherin-based adhesion at the plasma membrane and activating transcription in the nucleus. We found that in melanoma cells, WNT5A stimulated the disruption of N-cadherin and β-catenin complexes by activating the guanosine triphosphatase adenosine diphosphate ribosylation factor 6 (ARF6). Binding of WNT5A to the Frizzled 4–LRP6 (low-density lipoprotein receptor–related protein 6) receptor complex activated ARF6, which liberated β-catenin from N-cadherin, thus increasing the pool of free β-catenin, enhancing β-catenin–mediated transcription, and stimulating invasion. In contrast to WNT5A, the guidance cue SLIT2 and its receptor ROBO1 inhibited ARF6 activation and, accordingly, stabilized the interaction of N-cadherin with β-catenin and reduced transcription and invasion. Thus, ARF6 integrated competing signals in melanoma cells, thereby enabling plasticity in the response to external cues. Moreover, small-molecule inhibition of ARF6 stabilized adherens junctions, blocked β-catenin signaling and invasiveness of melanoma cells in culture, and reduced spontaneous pulmonary metastasis in mice, suggesting that targeting ARF6 may provide a means of inhibiting WNT/β-catenin signaling in cancer.
INTRODUCTION
The canonical function of WNTs has been largely attributed to the stabilization of the cytoplasmic pool of β-catenin, leading to nuclear translocation and activation of transcription (1). In addition to transcription, β-catenin has a distinct structural role at the plasma membrane in adherens junctions in linking cadherins to the actin cytoskeleton and stabilizing cell-cell contacts (2). Although adhesion and transcription can share the same pool of β-catenin, our understanding of the mechanisms by which junctional β-catenin feeds into canonical signaling is limited (2, 3). The release of β-catenin from cadherin potentially has dual roles in promoting tumor cell invasion: (i) weakening cell-cell contacts by destabilizing adherens junctions and (ii) enhancing transcription by augmenting the nuclear pool of β-catenin. Among the WNTs, WNT5A has emerged as a key mediator of tumor cell invasion (4), yet its role has been attributed to β-catenin– independent, noncanonical signaling mechanisms. WNT5A can stimulate β-catenin signaling, depending on receptor context (5–10), but whether this occurs naturally in mammalian cells or in the setting of cancer is unknown.
Adenosine diphosphate (ADP)–ribosylation factor 6 (ARF6) is a small guanosine triphosphatase (GTPase) that is a critical mediator of endocytosis and recycling of cadherin-catenin complexes at the cell surface (11). In the endothelium, we have shown that the ligand SLIT and its receptor ROBO induce GTPase-activating proteins (GAPs) to convert ARF6 to the inactive, guanosine diphosphate (GDP)–bound state (12), enhancing the localization of vascular endothelial–cadherin to the cell surface and promoting stability of cell-cell interactions (13). In epithelial cells, hepatocyte growth factor activates ARF6 to promote internalization of E-cadherin and cell motility (11). Likewise, in breast cancer, the epidermal growth factor receptor induces guanine exchange proteins (GEFs) to induce guanosine 5′-triphosphate (GTP) loading and activation of ARF6 (ARF6-GTP), reducing E-cadherin at the cell surface and promoting an invasive phenotype (14). Thus, ARF6 is at the center of opposing signals that influence cellular motility by regulating adherens junctions. Whether ARF6 is also central to the mechanism controlling the relationship between junctional and nuclear pools of β-catenin has not been explored.
Here, we showed in melanoma cells that ARF6 acts as a molecular switch to control the shuttling of β-catenin between the plasma membrane and the cytoplasm. This switch is controlled by two competing signals, WNT5A and SLIT2. WNT5A activates ARF6, leading to the disruption of N-cadherin–β-catenin complexes, accumulation of cytoplasmic and nuclear β-catenin, increased transcription, and tumor cell invasion. In contrast, SLIT2 inactivates ARF6, thus stabilizing N-cadherin–β-catenin interactions and reducing transcription and invasion. Hence, the activation state of ARF6 controls the intracellular location of β-catenin, which directly stimulates tumor cell invasion. Our work indicates that a WNT can induce the disruption of cadherin-catenin interactions and that endogenous WNT5A signaling augments canonical signaling. Our data support a mechanism in which ARF6 is critical in the WNT5A signaling cascade and explain how junctional and nuclear β-catenin pools are related. Furthermore, we show that inhibition of this ARF6 mechanism impedes spontaneous melanoma metastasis in vivo.
RESULTS
ARF6 controls the release of β-catenin from N-cadherin, affecting β-catenin transactivation
Because activated ARF6 increases endothelial and epithelial cell motility by decreasing the surface localization of cadherins (11–14), we hypothesized that ARF6 might promote tumor cell invasion by a similar mechanism. To this end, we evaluated the role of ARF6 in invasion of melanoma cells. Both N-cadherin (15–20) and ARF6 (21–23) have been implicated in melanoma invasion, but a relationship between the two has not been investigated. N-cadherin abundance in plasma membrane fractions was unchanged after small interfering RNA (siRNA)–mediated knockdown of ARF6 in LOX melanoma cells (fig. S1). In contrast, ARF6 knockdown in LOX cells increased both the association of β-catenin with N-cadherin (Fig. 1A and figs. S2A and S3A) and the membrane fraction of β-catenin (Fig. 1B and figs. S2B and S3B), with a concomitant decrease in cytoplasmic and nuclear β-catenin (Fig. 1B and figs. S2B and S3B). Immunofluorescence staining of LOX cells showed that N-cadherin localization was relatively unchanged in ARF6 knockdown cells (fig. S3C), whereas membrane localization of β-catenin increased (fig. S3D) and active β-catenin in the nucleus decreased (fig. S3E). ARF6 silencing did not alter total β-catenin or the amount of unphosphorylated, stabilized, active β-catenin (fig. S4A), suggesting that the ARF6-dependent increase in cytoplasmic and nuclear β-catenin was unrelated to mechanisms controlling β-catenin protein stability. In support of this, silencing of ARF6 did not affect the phosphorylation of low-density lipoprotein receptor–related protein 6 (LRP6) (fig. S4A), a marker for the sequestration of the β-catenin degradation complex by Dishevelled (24). In addition, ARF6 activation was not altered by chemical manipulation of the β-catenin degradation complex. Specifically, the amount of ARF6-GTP was constant in LOX cells exposed to the AXIN stabilizer endo-IWR 1, the tankyrase inhibitor X AV-939, or the glycogen synthase kinase 3β (GSK3β) inhibitor BIO (fig. S4B). In addition, the amount of β-catenin bound to N-cadherin was not altered by AXIN stabilization or inhibition of GSK3β (fig. S4C), suggesting that the N-cadherin–associated pool of β-catenin was distinct from the canonical, WNT-generated pool of β-catenin. These data suggest that ARF6 controls the disassembly of cadherin and β-catenin, converting junctional β-catenin into cytoplasmic pools available for nuclear translocation.
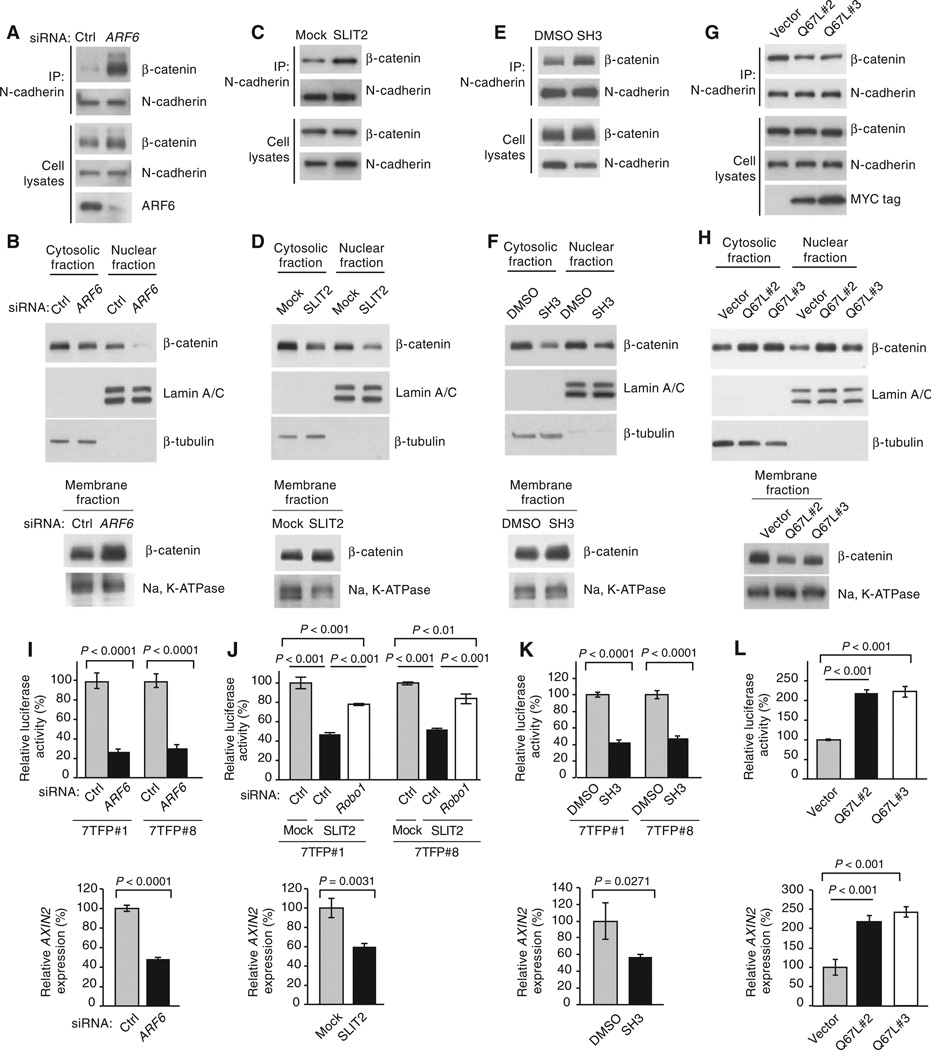
Fig. 1.
ARF6-GTP induces release of β-catenin from N-cadherin and augments β-catenin transactivation. (A) Coimmunoprecipitation of N-cadherin and β-catenin in LOX cells treated with control (Ctrl) or ARF6 siRNAs. (B) Subcellular fractionation of β-catenin in LOX cells treated with Ctrl or ARF6 siRNAs. (C) Coimmuno-precipitation of N-cadherin and β-catenin in LOX cells treated with Mock or SLIT2. (D) Subcellular fractionation of β-catenin in LOX cells treated with control Mock or SLIT2. (E) Coimmunoprecipitation of N-cadherin and β-catenin in LOX cells treated with dimethyl sulfoxide (DMSO) or SecinH3 (SH3). (F) Subcellular fractionation of β-catenin in LOX cells treated with DMSO or SH3. (G) Coimmunoprecipitation of N-cadherin and β-catenin in LOX cells stably transformed with empty vector or Myc-tagged ARF6-GTP (Q67L, stable clones #2 and #3). (H) Subcellular fractionation of β-catenin in LOX cells stably transformed with vector or Q67L #2 or #3. (I) β-Catenin transactivation in LOX cells treated with Ctrl or ARF6 siRNAs (two-tailed t test). (J) β-Catenin transactivation in LOX cells treated with Mock or SLIT2. Upper panel: Rescue of β-catenin–mediated luciferase activity with Ctrl or ROBO1 siRNA–transfected LOX cells treated with Mock or SLIT2 [upper panel, one-way analysis of variance (ANOVA) and Tukey’s post hoc test; lower panel, two-tailed t test]. (K) β-Catenin transactivation in LOX cells treated with DMSO or SH3 (upper panel, one-way ANOVA and Tukey’s post hoc test; lower panel, two-tailed t test). (L) β-Catenin transactivation in LOX cells stably transformed with vector or Q67L #2 or #3. Stable 7TFP-luciferase clones, 7TFP#1 and 7TFP#8 (I to K), were used for all luciferase assays except in (L). For (L), stable Q67L clones were infected with 7TFP lentivirus before the luciferase assay (one-way ANOVA and Dunnett’s post hoc test). For all, error bars represent SD and n ≥ 3 independent experiments. See fig. S2 (A to H) for quantification of blots. See related experiments in figs. S3 to S7.
To determine whether the activated form of ARF6 is necessary for the release of β-catenin from N-cadherin, we inhibited ARF6 activation in LOX cells by either activating ARF6 GAPs or inhibiting ARF6 GEFs. SLIT2, which is a ligand for ROBO receptors, inhibits ARF6 activation in endothelial cells by stimulating ARF6 GAP activity in a ROBO4-dependent manner (12). Similarly, ROBO1 was necessary for the inhibitory effects of SLIT2 on ARF6 activation in LOX cells (fig. S5, A and B). ARF6 GEFs, such as cytohesins and IQSECs (for example, GEP100, which is also known as IQSEC1), and the PSD families convert inactive ARF6-GDP to active ARF6-GTP, and some ARF6 GEFs can be inhibited with the small-molecule SecinH3 (25). SecinH3 inhibited ARF6 activation in LOX cells (fig. S5C). Treatment of LOX cells with either SLIT2 (Fig. 1, C and D, and fig. S2, C and D) or SecinH3 (Fig. 1, E and F, and fig. S2, E and F) mimicked the effects of ARF6 knockdown on β-catenin localization, causing an increase in N-cadherin–bound β-catenin at the plasma membrane and a decrease in cytoplasmic and nuclear β-catenin. This shift in β-catenin from the nucleus to the membrane in response to SLIT2 or SecinH3 was observed after 3 hours and persisted up to 24 hours (fig. S6, A and B). These data suggested that ARF6-GTP switched β-catenin localization from the cell surface to the cytoplasm where it could then translocate to the nucleus. Consistent with this interpretation, ectopic expression of the constitutively active mutant of ARF6 (Q67L) reduced N-cadherin– bound β-catenin and the membrane fraction of β-catenin and increased both cytoplasmic and nuclear β-catenin (Fig. 1, G and H, and fig. S2, G and H). Together, these data indicate that ARF6-GTP induces the release of β-catenin from N-cadherin, thus increasing the pool of free β-catenin that is available for nuclear translocation.
Given that changes in the activation state of ARF6 altered the amount of nuclear β-catenin, we reasoned that ARF6 may also affect β-catenin– mediated transcription. Indeed, ARF6 knockdown (Fig. 1I and fig. S7, A and B) or inhibition of ARF6 activation by SLIT2-ROBO1 (Fig. 1J) or SecinH3 (Fig. 1K) significantly blunted both the activity of a reporter gene for β-catenin–mediated transcriptional activity (7TFP-luciferase) and the expression of AXIN2, a β-catenin target. In contrast, ectopic expression of ARF6 Q67L increased luciferase activity and AXIN2 expression (Fig. 1L). Together, these data demonstrated that activated ARF6 could activate β-catenin–mediated transcription. This study demonstrates that ARF6 controls the cadherin-bound pool of β-catenin and that, when released, this source of β-catenin can activate transcription.
WNT5A activates ARF6 through Frizzled 4 and LRP6
Because SLIT2-ROBO1 inactivates ARF6 and opposes β-catenin signaling in melanoma cells, we reasoned that there must be a source of ARF6 activation in these cells. The ability of ARF6 to affect β-catenin transactivation suggests that WNTs may activate ARF6. LOX melanoma cells produce generous amounts of WNT5A (fig. S7C), consistent with their invasive and metastatic behavior (21, 26). WNT5A is a key mediator of tumor cell invasion (4), and thus far, its role in invasion has been attributed only to β-catenin–independent (noncanonical) signaling mechanisms (27–30). In LOX cells, silencing WNT5A reduced the amount of ARF6-GTP (Fig. 2A and fig. S2I). In contrast, after serum starvation and cell washing to remove endogenous WNT5A, treatment with recombinant WNT5A increased ARF6-GTP (Fig. 2B and fig. S2J). Among the nine human melanoma cell lines we evaluated, four produced endogenous WNT5A (fig. S7C). In A2058 and Yusac-2 melanoma cells, WNT5A was present in relatively low amounts. Treatment of these cells with recombinant WNT5A resulted in an increase in ARF6-GTP (Fig. 2B and fig. S2I). A375 melanoma cells produced the highest amounts of WNT5A among the cell lines we tested (fig. S7C). Regardless of the relative amounts of endogenous WNT5A, knockdown of WNT5A reduced ARF6-GTP abundance (Fig. 2C and fig. S2K). LOX cells also produced low amounts of WNT2, but WNT3A and WNT7A were not detected (fig. S7D). WNT5A knockdown did not reduce WNT2 expression (fig. S7E). ARF6 activation was reduced by WNT2 knockdown (fig. S8A), but knockdown of WNT2 did not affect WNT5A expression (fig. S8B). Together, these data demonstrate that ARF6 is activated by endogenous WNT5A in multiple melanoma cell lines but that other WNTs may be upstream of ARF6.
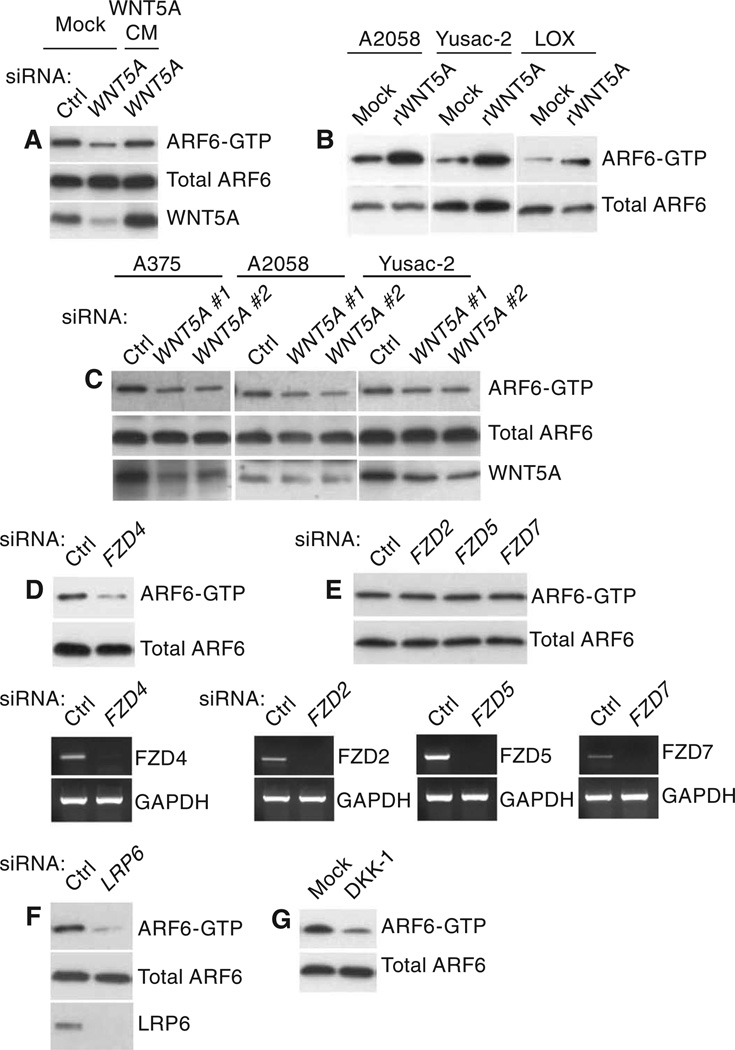
Fig. 2.
WNT5A activates ARF6 through FZD4 and LRP6. (A) ARF6-GTP amounts in LOX melanoma cells treated with control (Ctrl) or WNT5A siRNAs and rescue of ARF6-GTP amounts with WNT5A conditioned medium (CM). (B) ARF6-GTP in A2058, Yusac-2, or LOX melanoma cells treated with Mock or recombinant WNT5A (rWNT5A). (C) ARF6-GTP amounts in A375, A2058, or Yusac-2 cells treated with Ctrl or WNT5A #1 or #2 siRNAs. (D) ARF6-GTP amounts in LOX cells treated with Ctrl or FZD4 siRNAs. Lower panel, reverse transcription polymerase chain reaction (RT-PCR) for FZD4 mRNA. (E) ARF6-GTP amounts in LOX cells treated with Ctrl, FZD2, FZD5, or FZD7 siRNAs. Lower panels, RT-PCR for FZD mRNAs. (F) ARF6-GTP amounts in LOX cells treated with Ctrl or LRP6 siRNAs. (G) ARF6-GTP amounts in LOX cells treated with Mock or DKK-1. For all, n = 3 independent experiments. See fig. S2 (I to O) for quantification of blots. See related experiments in figs. S9 and S10.
Because the repertoire of Frizzled receptors present at the cell surface can influence whether WNT5A activates β-catenin signaling (5), we sought to determine which WNT5A receptors were responsible for ARF6 activation. Among the Frizzled (FZD) proteins, FZD2 (31), FZD4 (32), FZD5 (33), and FZD7 (9) have been reported to be WNT5A receptors, but only FZD4 and the co-receptor LRP5 allow WNT5A to activate β-catenin–mediated transcription when ectopically expressed in human embryonic kidney (HEK) 293 cells (5). Although LOX cells express FZD1 to FZD10 (fig. S8C), silencing of FZD4 (Fig. 2D and figs. S2L and S9), but not FZD2, FZD5, or FZD7 (Fig. 2E and fig. S2M), reduced the accumulation of ARF6-GTP in LOX cells. Silencing of the WNT co-receptor LRP6 (Fig. 2F and fig. S2N) or inhibition of LRP6 with DKK-1 (Figs. 2G and fig. S2O) also prevented ARF6 activation. These data indicate that ARF6 activation by WNT5A requires FZD4 and LRP6.
WNT5A-FZD4-LRP6 stimulate the release of β-catenin from N-cadherin, augmenting β-catenin signaling
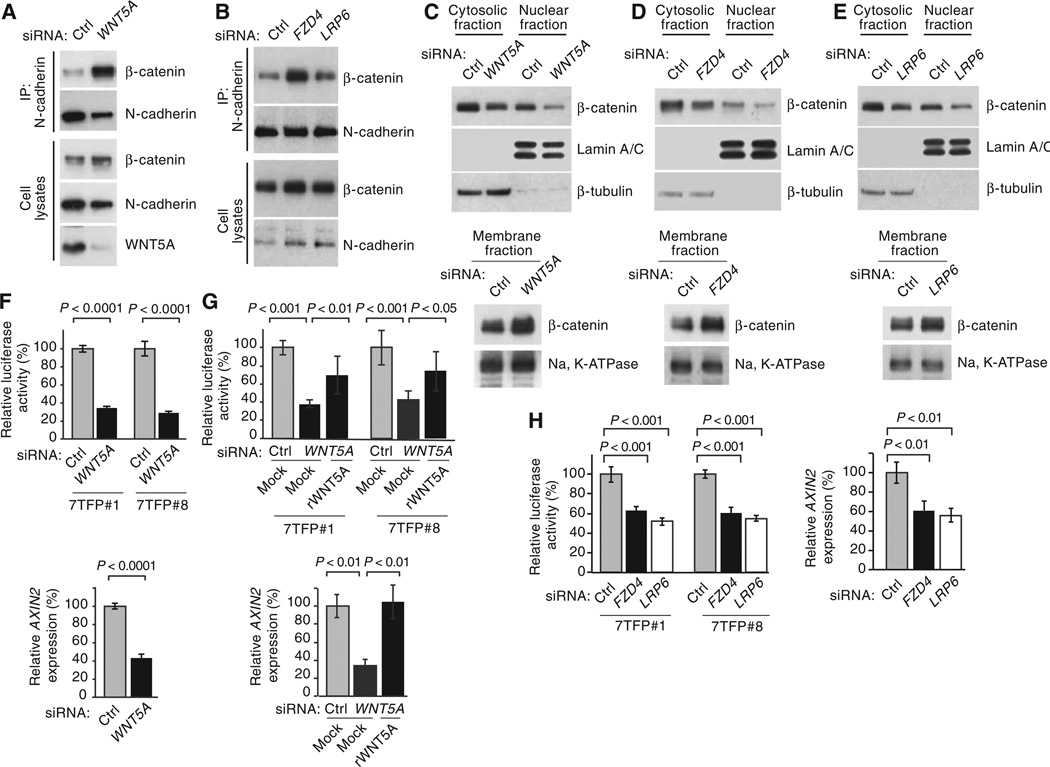
We next tested whether knockdown of WNT5A, FZD4, and LRP6 increased the pool of junctional β-catenin. Silencing of WNT5A, FZD4, or LRP6 in LOX cells caused a redistribution of β-catenin, with an increase in β-catenin associated with N-cadherin and a simultaneous decrease in cytosolic and nuclear β-catenin (Fig. 3, A to E, and fig. S2, P to S), a result that mirrors knockdown or inhibition of ARF6 (Fig. 1 and fig. S3). Silencing of WNT5A caused a significant decrease in both the activity of 7TFP-luciferase and AXIN2 expression (Fig. 3F). Luciferase activity was partially rescued and AXIN2 expression was completely rescued by treatment with recombinant WNT5A in WNT5A knockdown cells (Fig. 3G), demonstrating that β-catenin–mediated transcription was specifically activated by WNT5A. Endogenous WNT5A also activated β-catenin in other melanoma cells (fig. S10). Specifically, knockdown of ARF6 or WNT5A in A375, A2058, or Yusac-2 cells significantly reduced 7TFP-luciferase activity. WNT5A siRNA treatment had no effect on 7TFP-luciferase activity in MeWo melanoma cells (fig. S10), which do not produce WNT5A (fig. S7C). These data indicate that endogenous WNT5A– ARF6–β-catenin signaling is present in multiple human melanoma cell lines. In LOX cells, silencing of FZD4 or LRP6 reduced luciferase activity and AXIN2 expression (Fig. 3H). These data confirm that WNT5A, FZD4, and LRP6 stimulate β-catenin signaling in melanoma cells, similar to activated ARF6. Thus, these results implicate WNT5A in disrupting adherens junctions and driving junctional β-catenin into the canonical pathway.
Fig. 3.
WNT5A/FZD4/LRP6 promotes disassembly of N-cadherin–β-catenin complexes and activation of β-catenin–mediated transcription. (A to H) LOX cells treated with control (Ctrl), WNT5A, FZD4, or LRP6 siRNAs. (A and B) Coimmunoprecipitation of N-cadherin and β-catenin. (C to E) Subcellular fractionation of β-catenin. (F to H) β-Catenin transactivation. Rescue of β-catenin transactivation with recombinant WNT5A (rWNT5A) in LOX cells treated with Ctrl or WNT5A siRNAs (G). 7TFP#1 and 7TFP#8: stable 7TFP luciferase clones. For all, error bars represent SD and n ≥ 3 independent experiments. For (F), two-tailed t test. For (G) and (H), one-way ANOVA with Tukey’s (G) or Dunnett’s (H) post hoc test. See fig. S2 (P to S) for quantification of blots.
Activated ARF6 restores β-catenin signaling and invasion when WNT5A is silenced
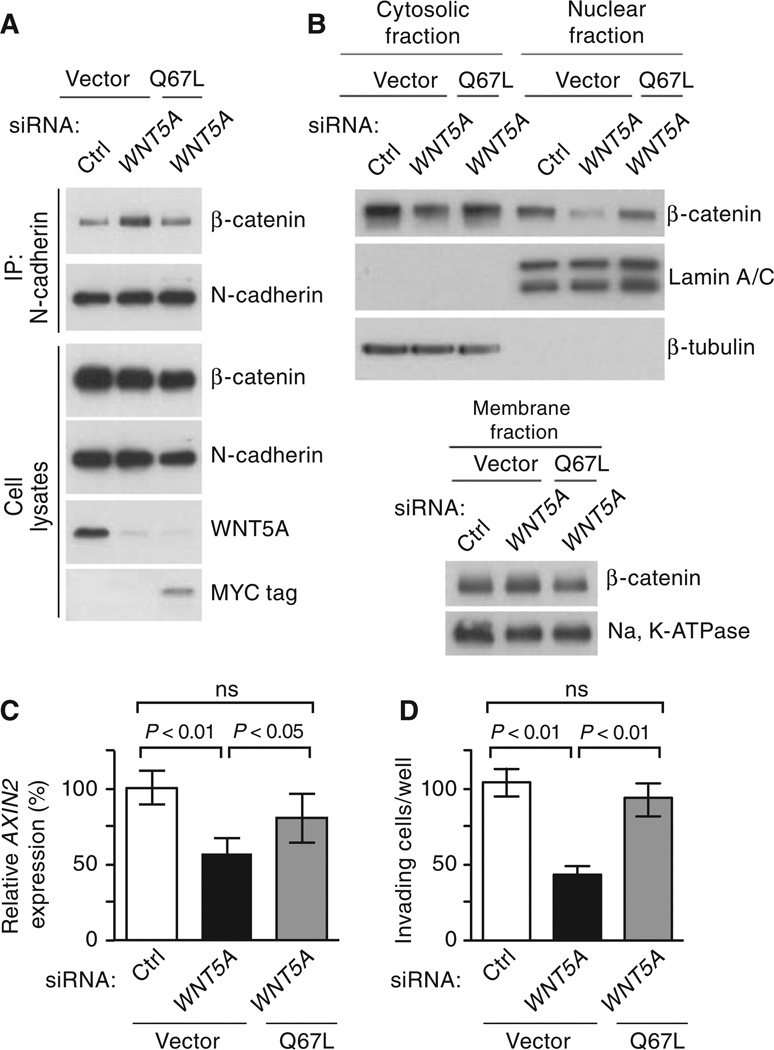
Because ARF6 was necessary for WNT5A–meditated β-catenin signaling, we asked whether activated ARF6 was sufficient to rescue β-catenin signaling when WNT5A is silenced. In WNT5A–depleted LOX cells, expression of ARF6 Q67L disrupted the N-cadherin–β-catenin interaction (Fig. 4A and fig. S2T) and rescued cytoplasmic and nuclear accumulation of β-catenin (Fig. 4B and fig. S2U), as well as AXIN2 expression (Fig. 4C). WNT5A stimulates motility (27) and invasion in several other melanoma cell lines (28, 29). Silencing of WNT5A inhibited invasion of LOX cells, a result that was reversed by ectopic expression of ARF6 Q67L (Fig. 4D). Although β-catenin–independent WNT5A pathways have been described in melanoma (27–30), our data indicate that WNT5A stimulates ARF6 to release cadherin-bound β-catenin, increasing the pool of β-catenin available for nuclear translocation, transactivation, and tumor cell invasion.
Fig. 4.
Activated ARF6 restores β-catenin signaling and melanoma invasion when WNT5A is silenced. (A to D) LOX melanoma cells transfected with control (Ctrl) or WNT5A siRNAs in the presence or absence of stable expression of Myc-tagged ARF6 Q67L. (A) Coimmunoprecipitation of N-cadherin and β-catenin. (B) Subcellular fractionation of β-catenin. (C) Relative AXIN2 expression. (D) Matrigel invasion (average number of invaded cells per well, counted at ×200 magnification). Error bars represent SD (C) or SEM (D). For all, n ≥ 3 independent experiments. For (C) and (D), one-way ANOVA with Tukey’s post hoc test. See fig. S2 (T and U) for quantification of blots.
ARF6 and stabilized β-catenin promote invasion and facilitate invadopodia activity
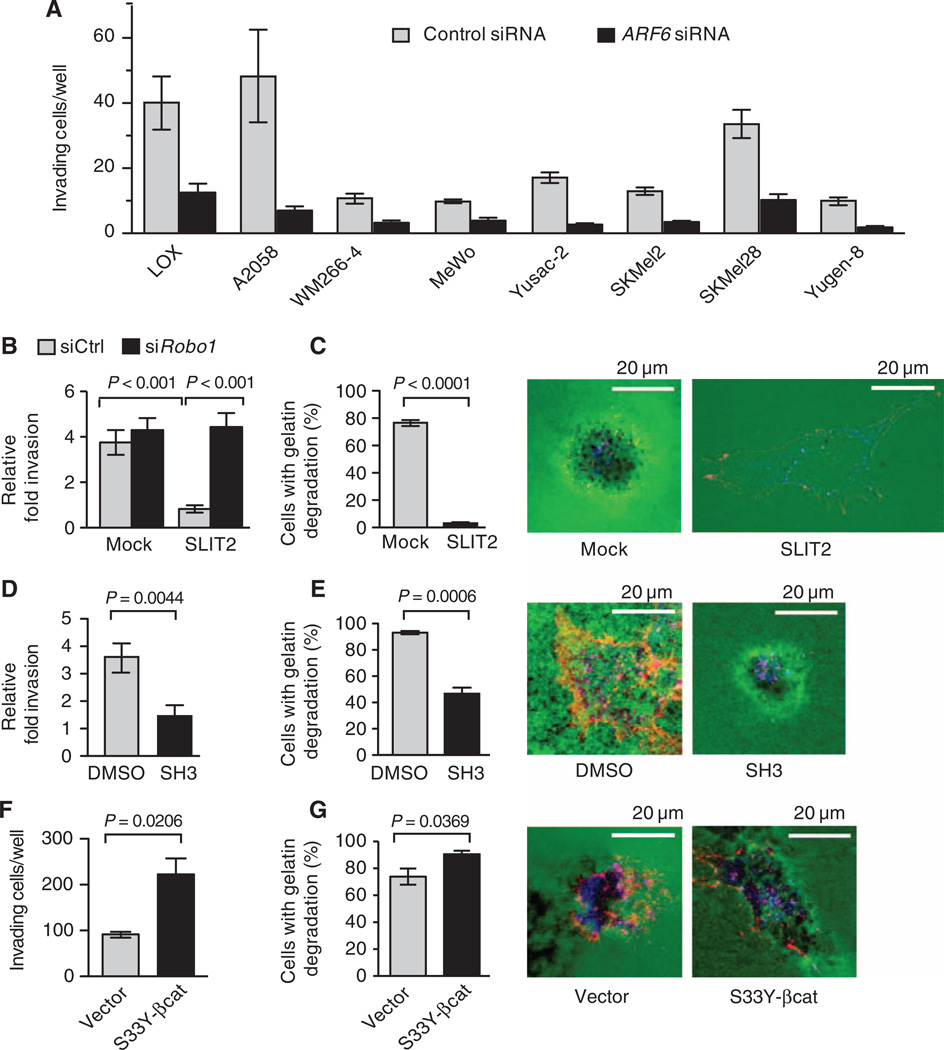
To determine the requirement of ARF6 for melanoma invasion, we silenced ARF6 in eight melanoma cell lines. ARF6 knockdown consistently inhibited invasion (Fig. 5A), a result that is consistent with studies showing that overexpression of an inactive mutant ARF6 inhibited invasion, whereas a constitutively active mutant ARF6 enhanced invasion of LOX cells (21–23).
Fig. 5.
Inhibition of ARF6 prevents invasion, whereas stabilized β-catenin promotes invasion of melanoma cells. (A) LOX, A2058, WM266-4, MeWo, Yusac-2, SKMel2, SKMel28, and Yugen-8 melanoma cells were transiently transfected with control (Ctrl) or ARF6 siRNAs and assayed for Matrigel invasion. For LOX cells, P = 0.0028. For A2058 cells, P = 0.0092. For all other cells, P < 0.0001. Statistical significance was determined by a two-tailed t test. (B) Matrigel invasion of LOX cells trans-fected with Ctrl or ROBO1 siRNAs and treated with Mock or SLIT2 (one-way ANOVA, Tukey’s post hoc test). (C) Invadopodia assay in LOX cells treated with Mock or SLIT2. (D and E) Matrigel invasion (D) and invadopodia assay (E) of LOX cells treated with DMSO or SecinH3 (SH3). (F and G) Matrigel invasion (F) and invadopodia assay (G) of LOX cells stably expressing empty vector or S33Y-β-catenin (S33Y-β-cat). Invasion data are presented as the average number of invading cells per well (A and F) or the fold invasion relative to unstimulated cells (B and D), counted at 320× (A, B, and D) or 200× (F) magnification. (C, E, and G) Invadopodia activity is presented as the percent of cells demonstrating degradation (black) of gelatin (green). Individual cells were visualized with cortactin (blue) and actin (red) staining. For all, n ≥ 3 experiments, and error bars represent SEM. For (C) to (G), two-tailed t test.
Expression profiling of uveal melanomas indicates that reduced ROBO1 expression is among the 12 expression markers that predict progression to metastatic disease (34). As indicated above, in LOX cells, SLIT2 inhibited ARF6 activation in a ROBO1-dependent manner (fig. S5A). SLIT2 also reduced invasion of LOX cells in a ROBO1-dependent manner (Fig. 5B). Furthermore, invadopodia activity was abrogated by SLIT2 (Fig. 5C). A similar phenotype was observed upon inhibition of ARF6 activation with SecinH3 (Fig. 5, D and E), confirming that this small molecule phenocopied the effects of either ARF6 protein loss or inactivation.
Although the disruption of the cadherin–β-catenin complex is one mechanism by which WNT5A and ARF6-GTP can stimulate invasion, we sought to determine whether stabilized, active β-catenin also contributes to invasion. Ectopic expression of stabilized β-catenin (S33Y-β-catenin) increased the invasive properties of LOX cells (Fig. 5, F and G). The effect of S33Y-β-catenin had a greater effect on invasion than invadopodia activity, suggesting that β-catenin might control tumor motility to a greater extent than matrix degradation. Regardless, these data support a role for nuclear β-catenin in LOX melanoma cell invasion and are consistent with findings in vitro (19, 20, 35) and in vivo (36) whereby transcriptionally active β-catenin increases melanoma invasion and metastasis, respectively.
GEP100 is necessary for WNT5A-induced ARF6 activation, β-catenin transactivation, and invasion
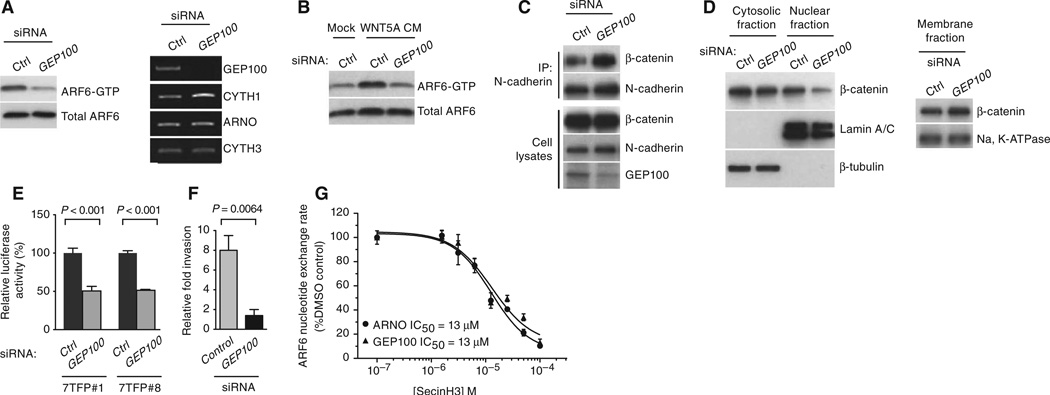
To further understand the mechanism by which WNT5A activates ARF6 to release junctional β-catenin and promote invasion, we sought to identify the GEF responsible for activation of ARF6 downstream of WNT5A. Cytohesins are ARF GEFs that are targets of SecinH3 (25). Because SecinH3 inhibits ARF6 activation (fig. S5C) and the subsequent downstream effects on β-catenin signaling and invasion (Figs. 1, E, F, and K, and 5, D and E, and fig. S2, E and F), we reasoned that cytohesins were responsible for ARF6 activation in LOX cells. Silencing of cytohesin-1, cytohesin-2 (also called ARNO), or cytohesin-3 failed to reduce ARF6-GTP abundance in LOX cells (fig. S11). In contrast, GEP100 knockdown significantly reduced ARF6 activation (Fig. 6A and fig. S2V) and prevented recombinant WNT5A from activating ARF6 (Fig. 6B and fig. S2W) in LOX cells. Silencing of GEP100 mimicked WNT5A and ARF6 knockdown, fortifying N-cadherin–β-catenin interactions, reducing nuclear β-catenin and transcription, and melanoma invasion (Fig. 6, C to F, and fig. S2, X and Y). GEP100 catalyzed the activation of ARF6 in nucleotide exchange assays carried out in vitro (37), and SecinH3 exhibited comparable inhibitory potencies for ARF6 nucleotide exchange in the presence of GEP100 or ARNO (Fig. 6G). Together, these data identify GEP100 as a critical member of a signaling pathway consisting of WNT5A, ARF6, and β-catenin that leads to melanoma invasion.
Fig. 6.
GEP100 is necessary for the activation of ARF6 by WNT5A. (A to F) LOX cells treated with control (Ctrl) or GEP100 siRNAs. (A) ARF6-GTP amounts (left panels). Expression of mRNAs encoding ARF-GEFs as detected by RT-PCR (right panels). (B) ARF6-GTP in LOX cells treated with Mock or WNT5A conditioned medium (CM) transfected with Ctrl or GEP100 siRNAs. (C) Coimmunoprecipitation of N-cadherin and β-catenin. (D) Subcellular fractionation of β-catenin. (E) β-Catenin–mediated transcription (7TFP-luciferase reporter). (F) Matrigel invasion. (G) Nucleotide exchange assay using ARF6, ARNO, or GEP100 recombinant proteins. (E to G) Error bars represent SD. For all, n = 3 independent experiments. (E and F) Two-tailed t test. See fig. S2 (V to Y) for quantification of blots.
Pharmacologic inhibition of ARF6 activation reduces spontaneous pulmonary metastasis
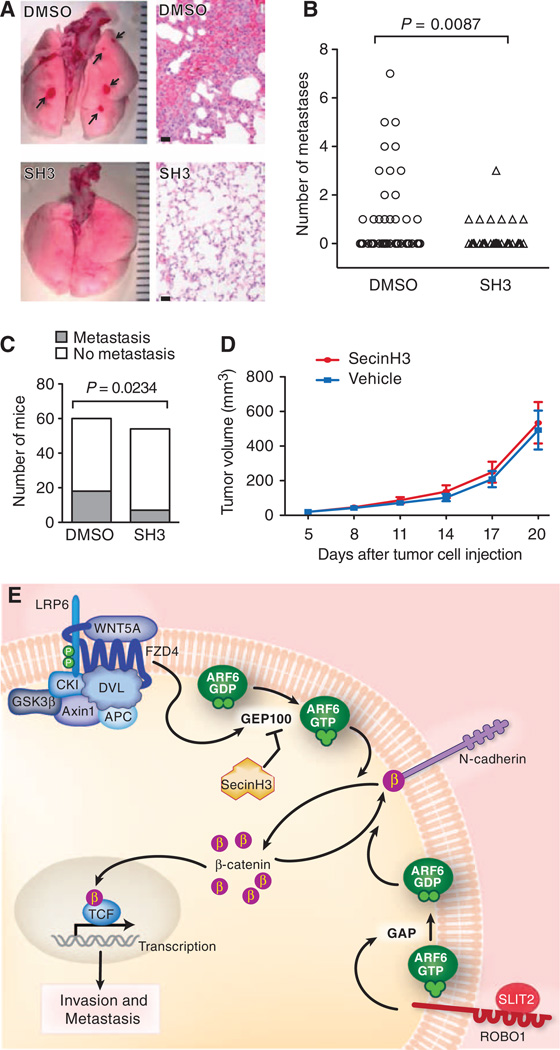
On the basis of the data presented thus far, we hypothesized that metastasis would be inhibited by blocking the signaling pathway consisting of WNT5A, FZD4, LRP6, GEP100, ARF6, and β-catenin. Because pharmacologic inhibition of ARF6 activation with SecinH3 (fig. S5C) strengthened the N-cadherin–β-catenin interaction (Fig. 1E and fig. S2E), reduced cytoplasmic and nuclear pools of β-catenin (Fig. 1F and fig. S2F), reduced β-catenin signaling (Fig. 1K), and suppressed invasion (Fig. 5, D and E), we sought to test whether SecinH3 had antimetastatic activity in vivo. Subcutaneous LOX xenograft tumors produce spontaneous pulmonary micrometastases that are identified grossly as discrete, round, hemorrhagic foci (26) (Fig. 7A). Systemic treatment with SecinH3 significantly reduced both the number of metastatic foci (Fig. 7B) and the number of mice with metastasis (Fig. 7C). There was no difference in overall primary tumor growth between the two groups (Fig. 7D). We frequently observed reduced nuclear β-catenin staining in primary tumors from SecinH3-treated mice but not in those from control mice (fig. S12). These data support a role for ARF6 in melanoma metastasis in vivo and suggest that ARF6 or ARF6 GEFs (or both) may be viable targets for systemic intervention in patients, particularly those with WNT/β-catenin–driven cancers.
Fig. 7.
Pharmacologic inhibition of ARF6-GTP with SecinH3 reduces spontaneous pulmonary metastasis of melanoma in vivo. (A) Gross and hematoxylin and eosin (H&E) images (400×) of lungs from mice with LOX xenograft tumors, treated with either vehicle or SecinH3 (SH3) daily. Ruler hatch marks (gross): 1 mm. Scale bars (microscopic): 20 µm. Arrows: hemorrhagic foci of microme-tastases (upper left panel) with clusters of melanoma cells (upper right panel). (B) Number of lung metastases per mouse (Mann-Whitney test, one-tailed). (C) Total number of mice with lung metastasis (Fisher’s exact test, one-tailed). (>D>) Primary tumor growth. Error bars = SEM. (E) Model of ARF6-dependent WNT5A/β-catenin signaling in melanoma invasion and metastasis. In the presence of WNTs, the degradation complex consisting of APC, Axin1, CK1, and GSK3β is sequestered by Dishevelled (DSH), allowing β-catenin stabilization. WNT5A activates ARF6 through FZD4 and LRP6 by stimulating GEP100, a GEF for ARF6. ARF6-GTP facilitates release of junctional β-catenin to augment transactivation of β-catenin, invasion, and metastasis. This process can be antagonized by SLIT2-ROBO1, which stimulates GAPs to convert ARF6-GTP to ARF6-GDP. Pharmacologic inhibition of ARF6 activation with SecinH3, an ARF6 GEF inhibitor, also prevents WNT5A/β-catenin signaling, invasion, and metastasis.
DISCUSSION
Here, we define a signaling cascade consisting of WNT5A, FZD4, LRP6, GEP100, ARF6, and β-catenin that promotes melanoma invasion and metastasis (Fig. 7E). GEP100 and ARF6 link WNT5A stimulation to the release of β-catenin from N-cadherin, ultimately increasing β-catenin transactivation. Thus, WNT5A and ARF6-GTP facilitate the switch in β-catenin function from adhesion to transcription. BCL9 (38) and FoxM1 (39) mediate β-catenin nuclear transport, but unlike these proteins, ARF6 localizes to the plasma membrane and endosomes (40) and may not be directly involved in nuclear translocation of β-catenin. Activated ARF6 mediates invasion of diverse malignant lineages, including melanoma, breast carcinoma, and glioma (21–23). In breast cancer cells, GEP100 facilitates ARF6 activation to promote invasion and metastasis (14). The proinvasive activities of ARF6 have been attributed to its role in vesicular transport and remodeling of the actin cytoskeleton (40–42) but have not been linked to β-catenin function. ARF6 may shuttle β-catenin between the plasma membrane and cytosol through endocytic mechanisms.
ARF6 as an integrative switch
Our data reveal the importance of ARF6 in the integration of competing signals that help drive plasticity in β-catenin function and the melanoma cell response. WNT5A activates ARF6 to shift the cadherin pool of β-catenin into the nucleus, whereas SLIT2 inactivates ARF6, fortifying the association of β-catenin and N-cadherin at the plasma membrane. SLIT2-ROBO1 inhibits canonical WNT signaling in mammary myoepithelial cells, affecting duct branching during maturation (43). Together with our data, it appears that SLIT-ROBO may be a natural antagonist of WNT signaling in multiple cell types. In melanoma, both WNT5A and SLIT2-ROBO1 appear to be clinically relevant pathways. Detection of WNT5A in patient tumors correlates with disease progression and reduced survival (29, 44), and loss of ROBO1 expression in uveal melanoma helps identify patients at high risk for metastasis (34). Somatic loss or mutations of the SLIT2, ROBO1, and ROBO2 genes have been observed in pancreatic ductal adenocarcinoma (45), providing further evidence that these genes have a tumor suppressor role.
Our data show that both WNT5A and SLIT2-ROBO1 control invasion by regulating the activation state of ARF6 and, as a consequence, the integrity of adherens junctions and β-catenin signaling. This is analogous to the opposing effects of SLIT2-ROBO4 on inflammatory signals whereby SLIT2 promotes endothelial barrier function by inhibiting ARF6 activation and stabilizing cadherin-catenin interactions at the cell surface (46). Hence, SLIT-ROBO opposes the effects of distinct promigratory signals in inflammation and cancer by inactivating ARF6. In addition, we have demonstrated that the proinflammatory cytokines interleukin-1β and tumor necrosis factor–α activate ARF6 and that inhibiting ARF6 activation in mouse models of rheumatoid arthritis and inflammation reduces vascular leak and inflammation (47). The present study shows that the activation state of ARF6 can control the intracellular location of β-catenin in melanoma cells, thereby influencing cancer cell invasion. Combined, these studies suggest that ARF6 forms a molecular convergent point between inflammatory cytokine signaling cascades and cancer-promoting pathways such as WNT/β-catenin signaling. Future studies will need to explore whether inflammation disrupts surface cadherin-catenin complexes and increases intra-cellular pools of β-catenin in neoplastic cells, potentially rendering them more sensitive to perturbations of WNT/β-catenin signaling.
WNT5A and β-catenin signaling
Our study describes an endogenous WNT5A pathway in human melanoma cells that facilitates β-catenin signaling. Although previous investigations into the mechanism of WNT5A-mediated invasion have uncovered only noncanonical pathways (27–30), Wnt5a can stimulate canonical signaling in Xenopus (6–10) and in mammalian cells (5). Thus, the dichotomy between canonical and noncanonical pathways might be less distinct than had been appreciated. These data have been received with some caution, however, because of the use of overexpressed proteins (1). Wnt5a ectopic expression induces β-catenin transactivation in the developing mouse embryo (48). Our data provide evidence for a native WNT5A/β-catenin pathway in human cancer cells. We showed that WNT5A used the cadherin-bound pool of β-catenin for canonical signaling through ARF6. Whether ARF6 is also important for noncanonical signaling remains to be investigated. In the cell lines we studied, noncanonical mechanisms might be important for invasion, and our data do not rule out this possibility. Our data both support the model advocated by van Amerongen and Nusse (1), in which WNTs can initiate various interrelated signaling events, and provide information that explains how β-catenin signaling can be propagated by a prototypical noncanonical WNT.
The ability of ARF6 to facilitate WNT5A/ β-catenin signaling raises the question whether ARF6 meditates other WNT/β-catenin pathways. On the basis of our observation that WNT2 also activates ARF6 (fig. S8), we suspect that ARF6 may be a general effector in WNT signaling. Previous reports indicate that QS11, an ARF GAP inhibitor that increases ARF6-GTP abundance, can boost WNT3A-dependent TOPFlash activity in HEK293 cells (49), and ARF1 and ARF6 are activated by WNT3A conditioned medium in HEK293T cells (50). Our data show the mechanism by which ARF6 mediates an endogenous WNT5A/β-catenin pathway that is biologically active in cancer invasion. These findings imply that ARF6 may impinge on various WNT-dependent processes in development and cancer, which will need to be addressed using in vivo genetic approaches.
β-Catenin in melanoma
In our system, stabilized, active β-catenin induced invasion of melanoma cells (Fig. 5, F and G), which agrees with a study demonstrating that stabilized β-catenin enhances melanoma metastasis in mice (36). In contrast, β-catenin may also suppress invasion of melanoma cells (51). In this latter study, silencing of β-catenin increased invasion in vitro. There are several possible explanations for these discordant results. First, melanoma is a heterogeneous disease. β-Catenin function may be context-dependent and, therefore, may vary between cell lines and model systems. Second, knockdown of β-catenin will decrease the abundance of β-catenin not only in the nucleus but also at the membrane. The loss of β-catenin could destabilize adherens junctions and increase cell motility. This is supported by reports showing that in melanoma cells, β-catenin interacts with N-cadherin during the initial contacts with en-dothelial cells but must be released from N-cadherin for transendothelial migration to occur (19, 20). Once released from N-cadherin, β-catenin accumulates in the nucleus of melanoma cells and activates transcription, facilitating transendothelial migration (19). This study highlights the fact that invasion is a dynamic process and that β-catenin function alternates between the membrane and the nucleus as cells make and break contacts during this process. Thus, drawing conclusions about β-catenin cellular function based on subcellular localization requires an evaluation in migrating cells.
Given the dynamic nature of β-catenin localization during invasion, care must be exercised when interpreting β-catenin staining patterns at a single point in time, such as in patient samples. Correlative clinical studies suggest that nuclear β-catenin staining is a useful prognostic marker, predicting a more favorable outcome in melanoma (52–54). Although these studies do not establish causality, they may hint at a biphasic population of malignant cells within the tumors. Chien et al. (52) showed that proliferation is inversely related to nuclear β-catenin staining in both primary and metastatic lesions. Hence, nuclear β-catenin staining may be a surrogate for nonproliferating cells in these samples and may, in fact, be highlighting the nonproliferative, invasive population within these tumors.
During cancer progression, tumor cells may switch between invasive and proliferative states, and nuclear β-catenin may favor one phenotype over the other. Phenotype switching is a poorly understood process, and the role of β-catenin in melanoma is unclear. The presence of LEF1 or TCF4 has been shown by one study (55) to determine the proliferative or invasive phenotypes of melanoma cells, respectively. These data reveal that β-catenin function can be context-dependent. Future studies of phenotype switching may shed more light on β-catenin function. For now, we and others have observed that nuclear β-catenin promotes invasion. We recognize that activation of β-catenin signaling is not the only means of stimulating invasion downstream of WNT5A. Noncanonical WNT5A signaling is important, but our data indicate that β-catenin signaling is also active downstream of WNT5A and is dependent on activated ARF6.
Clinical implications
We have shown that pharmacologic inhibition of ARF6 activation fortifies adherens junctions, effectively inhibiting β-catenin signaling (Fig. 1, E, F, and K, and fig. S2, E and F), melanoma invasion (Fig. 5, D and E), and spontaneous pulmonary metastasis of melanoma (Fig. 7). In cancer, ARF6 is necessary for invasion of tumor cells originating from diverse cell lineages (14, 21–23, 56, 57), indicating that migrating malignant cells readily recruit ARF6. As a molecular switch, ARF6 is ideally suited for controlling the dynamic changes in cell function that are required for invasion and metastasis. Clinically, inhibitors of the metastatic process are needed to improve therapeutic outcomes (58). This is particularly important for melanoma because, despite substantial progress in the development of immunotherapy and therapies that target activating mutations in the kinase BRAF, few patients achieve a sustainable response that affords long-term remission (59–61). We have exploited the aggressive nature of melanoma to elucidate a role for WNT5A, FZD4, LRP6, GEP100, and ARF6 in β-catenin function and show that this signaling pathway controls invasion and metastasis. It will be important to evaluate whether targeting ARF6 is an effective strategy in other WNT/β-catenin–driven cancers.
MATERIALS AND METHODS
Cell lines
LOX, SKMel28, Yugen-8, and Yusac-2 cells were provided by D. Grossman (Huntsman Cancer Institute, University of Utah), and A375 cells were a gift from S. Holmen (Huntsman Cancer Institute, University of Utah). HEK293T, A2058, MeWo, SKMel2, and WM266-4 cells were purchased from the American Type Culture Collection (ATCC). All cells except SKMel28 and A375 were maintained in Dulbecco’s modified Eagle’s medium + GlutaMAX supplemented with 1 mM sodium pyruvate. SKMel28 cells were maintained in modified Eagle’s medium + GlutaMAX and 1 mM sodium pyruvate. A375 cells were maintained in RPMI 1640 medium including GlutaMAX and 1 mM sodium pyruvate. LOX, Yugen-8, and Yusac-2 were maintained with 5% fetal bovine serum (FBS), whereas the other cells received 10% FBS.
Plasmids and transfections
FLAG-tagged S33Y-β-catenin in pcDNA3.1 was provided by C. Murtaugh (Department of Human Genetics, University of Utah). Murine Wnt2, Wnt5a, and Wnt7a, and human WNT3A coding sequences were individually cloned into the pCI vector under the control of the cytomegalovirus promoter. HEK293T cells were transiently transfected with 10 µg of plasmid using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. For establishing stable cell lines, LOX cells were transfected with 2 µg of pcDNA3.1 (Invitrogen) vector, ARF6 Q67L, or S33Y β-catenin using Metafectene (Biontex Laboratories) according to the manufacturer’s instructions and selected and maintained with G418 (800 µg/ml; Gibco) for 2 weeks. Myc-tagged ARF6 Q67L in pcDNA3.1 was generated by PCR-based mutagenesis from the full-length coding sequence of human ARF6. Briefly, two fragments of overlapping ARF6 sequence were amplified. The first fragment was amplified with primers hARF6-F-Kpn I (5′-cggGGTACCgccaccatggggaaggtgctatcca-3′) and hARF6-Q67L–R (5′-gagcggccggatcttgtccaggccgcccacatcccatac-3′) and contained base pairs 1 to 219. The second fragment was amplified with primers hARF6-Q67L–F (5′-gtatgggatgtgggcggcctggacaagatccggccgctc-3′) and hARF6-R-Not I (5′-ataagaatGCGGCCGCcaagatttgtagttagagg-3′) and contained base pairs 181 to 526. The two DNA fragments were mixed in equal amounts for fusion PCR. The full-length human ARF6 Q67L coding sequence was amplified from the fusion template with primers hARF6-F-Kpn I and hARF6-R-Not I and sequenced for verification.
Lentiviral transduction
For lentiviral transduction, LOX, MeWo, A375, A2058, and Yusac-2 cells were plated at 10,000 cells per well on a 96-well plate in growth medium and incubated at 37°C/5% CO2 overnight. Cells were then treated with lentiviral particles at a multiplicity of infection of 3 to 5 in growth medium containing hexadimethrine bromide (8 µg/ml) and incubated for 20 hours at 37°C/CO2. Medium was replaced with fresh growth medium, and the cells were incubated overnight and then split into 24-well plates. Forty-eight hours after removal of the lentivirus-containing medium, selection for cells containing stable integration of the lentiviral construct was begun using growth medium containing puromycin (0.5 µg/ml; Invitrogen) for LOX cells and puromycin (1 µg/ml) for MeWo, A375, A2058, and Yusac-2. Selection was complete by 3 days of treatment. However, cells were expanded for 2 weeks on selection medium before being used for cellular assays.
RNA interference, recombinant proteins, and SecinH3
All siRNA and short hairpin RNA (shRNA) sequences are listed in table S1. siRNA duplexes (20 nM) were transfected into LOX cells using HiPerFect (Qiagen) according to the manufacturer’s instructions. Forty-eight hours after transfection, cells were split to 50% confluency, retrans-fected, and grown for another 24 hours before cellular assays. Recombinant WNT5A was obtained from R&D Systems and PeproTech. Control (Mock) and WNT5A conditioned media were collected from L cells (mouse fibroblasts) (ATCC) grown according to ATCC instructions. For ARF6-GTP pull-downs, cells were serum-starved overnight, washed in phosphate-buffered saline (PBS) to remove endogenous WNT5A, and then treated with recombinant WNT5A (100 ng/ml) or WNT5A conditioned medium for 3 hours. For luciferase assays, recombinant WNT5A (10 ng/ml) was given in complete medium for 6 hours in WNT5A knockdown LOX cells. Human DKK-1 was purchased from R&D Systems, and cells were treated with 300 ng/ml in complete medium for 3 hours. For recombinant SLIT2 generation, LOX cells were infected with either empty adenovirus (Mock) or adenovirus containing a SLIT2 expression construct, and either SLIT2 was salt-extracted from the conditioned medium as previously described (62) or the conditioned medium was directly used without salt extraction. For ARF6-GTP pull-downs, cells were treated with 20 nM SLIT2 for 3 hours. For fractionation, 7TFP-luciferase, and AXIN2 experiments, cells were treated with 20 nM SLIT2 for 24 hours. For Matrigel invasion and invadopodia activity, cells were treated with 10 nM SLIT2 conditioned medium, or a single dose (20 nM) of salt-extracted SLIT2 was added to the medium. SecinH3 was purchased from Calbiochem and Albany Molecular Research Inc. For ARF6-GTP pull-downs, cells were treated with 30 µM SecinH3 for 3 hours in complete medium. For fractionation, luciferase, and AXIN2 experiments, cells were treated with 30 µM SecinH3 for 24 hours. For Matrigel invasion and invadopodia activity, cells were treated with one dose of 30 µM SecinH3 over the entire course of the experiment. endo-IWR 1 and XAV-939 were purchased from Calbiochem. BIO was obtained from Tocris Bioscience. For ARF6-GTP pull-downs, LOX cells were treated with endo-IWR 1 (25 µM), XAV-939 (2 µM), or BIO (5 µM) for 3 hours in complete medium. For immunoprecipitation with β-catenin and N-cadherin, cells were treated with 25 µM endo-IWR 1, 2 µM XAV-939, or 5 µM BIO for 18 hours.
Western blots, immunoprecipitation, cell fractionation, and ARF6-GTP pull-downs
With the exception of the GGA3 ARF6-GTP pull-downs and cell fractionations, all cell lysates were prepared with 25 mM Hepes (pH 7.4), 150 mM NaCl, 1% Triton X-100, and 1% sodium deoxycholate, plus phosphatase and protease inhibitors. For immunoprecipitation, lysates were centrifuged at 12,000g for 10 min, precleared with protein A/G (GE Healthcare) coupled to agarose beads for 2 hours, and then incubated for 1 hour at 4°C with N-cadherin antibody and protein A/G–agarose beads. For immunoblotting, primary antibodies were diluted in 5% nonfat dry milk in PBS + 0.1% Tween 20 and incubated overnight at 4°C. For cell fractionation, whole-cell lysates were prepared in radioimmunoprecipitation assay buffer [50 mM tris-Cl (pH 7.4), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS] plus protease and phosphatase inhibitors. Quantification was by scanning densitometry whereby changes were normalized to loading controls or input and represents an amalgamation of all independent experiment replicates (n > 3 each).
Antibodies against ARF6, active β-catenin (Millipore), WNT5A, WNT3A, LRP6, phospho-LRP6, lamin A/C, β-actin, Na,K-ATPase (Cell Signaling), WNT2, WNT7A, β-tubulin, Myc (Santa Cruz Biotechnology), N-cadherin, and β-catenin (BD Biosciences) were used for immunoblotting and/or immunoprecipitation. GGA3 ARF6-GTP pull-downs were performed with Arf6 Activation Assay Kit (Cell Biolabs) according to the manufacturer’s instructions. Cytoplasmic and nuclear fractions were prepared with NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific). Membrane and plasma membrane fractions were isolated with Plasma Membrane Protein Extraction Kit (Abcam) according to the manufacturer’s instructions.
Quantification of immunoblots and statistical analysis
Quantitative values were obtained by scanning densitometry. Each band on the immunoblots was normalized to its paired internal control protein. After normalization, the ratio of each experimental treatment to its paired control treatment was obtained, and the geometric means and 95% confidence intervals were calculated.
RNA extraction and quantitative RT-PCR
RNA was prepared with NucleoSpin RNA II (Macherey-Nagel). Complementary DNA was synthesized from 5 µg of total RNA with Oligo(dT) primers and Superscript III reverse transcriptase (Invitrogen). Quantitative RT-PCR was performed with the Applied Biosystems 7900HT and QuantiTect SYBR Green PCR Kits (Qiagen) with the primers listed in table S2. All samples were run in triplicate and normalized to GAPDH. AXIN2 and GAPDH primers were validated for equal amplification efficiencies, and the relative expression of these genes was determined with the ΔΔCT method. All primer sequences are listed in table S2.
Luciferase assay
Luciferase activity was assayed with lentivirally transduced cells that stably express the TOPFlash-based 7TFP reporter (Addgene) (63). Twenty micrograms of lysate was assayed for firefly luciferase with Promega’s Luciferase Assay System according to the manufacturer’s instructions. Twelve replicates (wells) were assayed for each experimental condition. Stable Q67L clones were infected with 7TFP lentivirus 48 hours before the luciferase assay.
Matrigel invasion
BD Biocoat Matrigel Invasion Chambers (24-well plates) were purchased from BD Biosciences, and cells were assayed according to the manufacturer’s instructions, with minor modifications. Briefly, cells were pretreated with mitomycin C (10 µg/ml) for 2 hours to prevent cell division, washed, and resuspended in medium with 0.2% serum. For most assays, cells were plated at a density of 5 × 104 cells per well. Cells stably expressing ARF6 Q67L were plated at 2.5 × 104 cells per well. Invasion was induced with complete medium in the lower chamber. Cells were fixed with 5% glutar-aldehyde for 20 min and stained with 0.5% toluidine blue in 3% NaHCO3 for 40 min. Five separate fields were counted per well (four wells per experimental variable).
Invadopodia assay
LOX cells were evaluated for invadopodia activity as previously described (21). Invadopodia activity was scored by counting the percentage of total cells that show pericellular gelatin degradation.
Immunofluorescence staining
LOX cells were plated for 24 hours at a density of 8 × 104 cells per well in complete medium on collagen-coated eight-well cover glasses, washed twice in PBS, and fixed. For intracellular staining, monolayers were fixed in 1:1 methanol/acetone at −20°C and permeabilized in 0.5% Triton X-100 for 10 min. For cell surface staining of N-cadherin, monolayers were fixed in 4% paraformaldehyde for 10 min at room temperature, followed by 10 min in 100% methanol at −20°C. Both fixation conditions were blocked in 5% bovine serum albumin for 1 hour at room temperature. Primary antibodies against active β-catenin (Millipore), β-catenin (Invitrogen), and N-cadherin (BD Transduction Laboratories) were diluted according to the manufacturer’s specifications and applied overnight at 4°C. Signals were detected by Alexa Fluor–conjugated anti–immunoglobulin G (IgG) diluted to 10 µg/ml. Fields for imaging were randomly selected in the 4’,6-diamidino-2-phenylindole channel and imaged at 600× on an Olympus FV1000 confocal microscope.
Four-micrometer sections of formalin-fixed, paraffin-embedded LOX xenograft tumors were subjected to heat-induced epitope retrieval in citrate buffer (pH 6.0) before immunostaining. The β-catenin primary antibody (clone 14, BD Transduction Laboratories; 1:50) was preincubated with secondary antibody (rabbit anti-mouse Fab2, Dako; 1:100) for 30 min. To reduce background staining of mouse tissue, 5 µl of mouse serum was added and incubated for 30 min, creating a primary/secondary cocktail. Sections were incubated with this cocktail for 2 hours at 37°C. A biotinylated tertiary antibody (goat anti-rabbit IgG, Sigma, 1:100) was applied for 32 min. Development of the alkaline phosphate red signal was performed with the Ventana XT detection kit. Levamisole was applied for 8 min as a blocker, and slides were counterstained with hematoxylin for 4 min. Using a BioView Duet imaging system, we chose three random 400× fields per tumor on bright field and then imaged them on a fluorescence microscope. Investigators were blinded to the treatment groups when evaluating β-catenin staining.
Xenograft melanoma model
Athymic nude and nonobese diabetic/severe combined immunodeficient mice were purchased from Jackson Laboratories. Mice were injected sub-cutaneously with 2 × 106 to 3 × 106 LOX cells in 0.1 ml of Dulbecco’s PBS. Tumors were allowed to establish for 5 days before initiating daily intraperitoneal injections of 0.1 ml of 5 mM SecinH3 or vehicle (25% DMSO + 3.75% dextrose). Investigators were blinded to treatment groups for evaluation of primary tumor size and scoring of metastasis. Primary subcutaneous tumors were measured every 3 days. Tumor volume was calculated using the ellipsoid formula: V = 4/3 × π × length/2 × width/ 2 × depth/2. On day 21 after tumor cell injection, mice were sacrificed by isoflurane inhalation and evaluated for spontaneous pulmonary metastasis. Lungs were removed and rinsed thoroughly in PBS. Discrete, round, and red metastatic foci were identified and counted. Primary tumors were evaluated histologically by H&E staining.
Biochemical nucleotide exchange assay
Nucleotide exchange was assayed in vitro using ARF6 (amino acids 14 to 175, which lacks the N-terminal autoinhibitory region), as well as ARNO (also known as cytohesin2) (amino acids 50 to 255), or GEP100 (amino acids 391 to 602) protein fragments that include Sec7 domains. All recombinant proteins were expressed as N-terminally His-tagged fusions in Escherichia coli system using pET28a vector and purified to apparent homogeneity by immobilized metal affinity chromatography. The nucleotide exchange reaction was carried out with 50 mM tris-HCl (pH 7.5), 1 mM MgCl2, 0.001% Triton X-100, 2 mM β-mercaptoethanol, 1% DMSO, and 50 nM GTP-BODIPY FL supplemented with 50 nM GEF protein and 200 nM GDP-bound form of ARF6 in the absence or presence of SecinH3. Replacement of ARF6-bound GDP with GTP-BODIPY FL was monitored by measuring increases in fluorescence intensity that result from the relief of intramolecular fluorescence quenching of the latter fluorogenic nucleotide upon binding to the GTPase (64). Specifically, fluorescence intensities were determined in real-time mode with a Synergy 4 plate reader (BioTek) at excitation and emission wavelengths of 490 and 520 nm, respectively.
Statistical analysis
Excel or Prism was used to assess statistical significance. When two groups were compared, a Student’s t test was performed. When more than two groups were compared, one-way ANOVA followed by either Tukey’s or Dunnett’s post hoc test was performed. When one-way ANOVA was performed, P values shown in figures represent results from the post hoc test. When data were not normally distributed, nonparametric Mann-Whitney test was used. Fisher’s exact test was performed to analyze data from contingency tables.
Supplementary Material
Acknowledgments
We thank J. Christian, A. Welm, C. Murtaugh, R. Dorsky, D. Jones, S. Holmen, and R. Stewart for critical reading of the manuscript. We thank T. Greene (Study Design and Biostatistics Center, University of Utah) for support with statistical analyses. We thank C. Murtaugh and D. Grossman for reagents and technical advice, H. S. Kim for technical guidance, R. Zhang for technical support, and D. Lim for graphical assistance.
Funding: D.Y.L. is supported by grants from the Huntsman Cancer Institute, University of Utah CRR Program, National Heart, Lung, and Blood Institute (NHLBI), Juvenile Diabetes Research Foundation, Rocky Mountain Regional Center for Excellence in Biodefense and Emerging Infectious Disease, American Asthma Foundation, and Department of Defense. A.H.G. was supported by the R. L. Kirschstein NRSA (2T32HL007576-26) from the NHLBI. Additional grants (C.D.S.-S.) are from the National Cancer Institute and the University of Notre Dame SAPC Program. J.C. is an Indiana-CTSI predoctoral fellow and A.S. is supported by a University of Notre Dame Graduate Assistantship.
Footnotes
Author contributions: A.H.G., J.H.Y., S.J.O., and D.Y.L. were responsible for project conceptualization, experimental design, data analysis, and manuscript preparation. A.H.G. generated in vitro and in vivo data, provided pathology expertise, and was primarily responsible for writing the manuscript. J.H.Y. generated biochemical and in vitro data. J.C., A.S., and C.D.S.-S. provided in vitro data and helped in the critical reading of the manuscript. L.K.S. generated in vitro data and immunocytofluorescence images. Z.T. and K.R.T. provided several plasmid constructs and adenoviruses. K.R.T. provided critical review of the manuscript. K.O. provided data from GTP exchange assays. A.R. provided in vitro data. K.F.G. provided expert opinion in melanoma medical oncology and helped prepare the manuscript. S.R.T. performed immunofluorescence staining and image acquisition of tumors. S.J.O. generated in vivo data and performed statistical analyses.
Competing interests: The authors declare competing financial interests. The University of Utah has filed intellectual property concerning targeting signaling pathways to treat cancer and licensed technology to Navigen, a biotechnology company owned in part by the University of Utah Research Foundation. D.Y.L. and K.R.T. have equity interest, management or advisory relationships, and paid consulting relationship with Navigen. Those relationships are related to patent applications surrounding small-molecule inhibition of Arf6 that predate this article and are licensed from the University of Utah by Navigen. Navigen is owned in part by the University of Utah Research Foundation.
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/6/265/ra14/DC1
Fig. S1. ARF6 knockdown does not alter total N-cadherin protein at the plasma membrane.
Fig. S2. Quantification of immunoblots.
Fig. S3. ARF6 knockdown drives β-catenin from the nucleus to N-cadherin.
Fig. S4. ARF6, junctional β-catenin, and the canonical destruction complex.
Fig. S5. SLIT2-ROBO1 and SecinH3 inhibit ARF6 activation.
Fig. S6. Time course of β-catenin relocalization after ARF6 inhibition.
Fig. S7. ARF6-dependent β-catenin transactivation and relative WNT production in melanoma cell lines.
Fig. S8. WNT2 knockdown reduces ARF6 activation.
Fig. S9. FZD4 knockdown reduces ARF6 activation.
Fig. S10. WNT5A, ARF6, and β-catenin signaling in multiple human melanoma cell lines.
Fig. S11. Cytohesin knockdown does not reduce ARF6 activation.
Fig. S12. β-Catenin immunostaining of LOX melanoma xenograft tumors.
Table S1. siRNA and shRNA sequences.
Table S2. Primer sequences for RT-PCR.
REFERENCES AND NOTES
- 1.van Amerongen R, Nusse R. Towards an integrated view of Wnt signaling in development. Development. 2009;136:3205–3214. doi: 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
- 2.Heuberger J, Birchmeier W. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb. Perspect. Biol. 2010;2:a002915. doi: 10.1101/cshperspect.a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson WJ, Nusse R. Convergence of Wnt, β-catenin and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kikuchi A, Yamamoto H, Sato A, Matsumoto S. Wnt5a: Its signalling, functions and implication in diseases. Acta Physiol. 2012;204:17–33. doi: 10.1111/j.1748-1716.2011.02294.x. [DOI] [PubMed] [Google Scholar]
- 5.Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits β-catenin–TCF signaling depending on receptor context. PLoS Biol. 2006;4:e115. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cha SW, Tadjuidje E, Tao Q, Wylie C, Heasman J. Wnt5a and Wnt11 interact in a maternal Dkk1-regulated fashion to activate both canonical and non-canonical signaling in Xenopus axis formation. Development. 2008;135:3719–3729. doi: 10.1242/dev.029025. [DOI] [PubMed] [Google Scholar]
- 7.Cha SW, Tadjuidje E, White J, Wells J, Mayhew C, Wylie C, Heasman J. Wnt11/5a complex formation caused by tyrosine sulfation increases canonical signaling activity. Curr. Biol. 2009;19:1573–1580. doi: 10.1016/j.cub.2009.07.062. [DOI] [PubMed] [Google Scholar]
- 8.Tao Q, Yokota C, Puck H, Kofron M, Birsoy B, Yan D, Asashima M, Wylie CC, Lin X, Heasman J. Maternal wnt11 activates the canonical wnt signaling pathway required for axis formation in Xenopus embryos. Cell. 2005;120:857–871. doi: 10.1016/j.cell.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 9.Umbhauer M, Djiane A, Goisset C, Penzo-Méndez A, Riou JF, Boucaut JC, Shi DL. The C-terminal cytoplasmic Lys-thr-X-X-X-Trp motif in frizzled receptors mediates Wnt/β-catenin signalling. EMBO J. 2000;19:4944–4954. doi: 10.1093/emboj/19.18.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He X, Saint-Jeannet JP, Wang Y, Nathans J, Dawid I, Varmus H. A member of the Frizzled protein familymediating axis induction by Wnt-5A. Science. 1997;275:1652–1654. doi: 10.1126/science.275.5306.1652. [DOI] [PubMed] [Google Scholar]
- 11.Palacios F, Price L, Schweitzer J, Collard JG, D’Souza-Schorey C. An essential role for ARF6-regulated membrane traffic in adherens junction turnover and epithelial cell migration. EMBO J. 2001;20:4973–4986. doi: 10.1093/emboj/20.17.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones CA, Nishiya N, London NR, Zhu W, Sorensen LK, Chan AC, Lim CJ, Chen H, Zhang Q, Schultz PG, Hayallah AM, Thomas KR, Famulok M, Zhang K, Ginsberg MH, Li DY. Slit2-Robo4 signalling promotes vascular stability by blocking Arf6 activity. Nat. Cell Biol. 2009;11:1325–1331. doi: 10.1038/ncb1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.London NR, Zhu W, Bozza FA, Smith MC, Greif DM, Sorensen LK, Chen L, Kaminoh Y, Chan AC, Passi SF, Day CW, Barnard DL, Zimmerman GA, Krasnow MA, Li DY. Targeting Robo4-dependent Slit signaling to survive the cytokine storm in sepsis and influenza. Sci. Transl. Med. 2010;2 doi: 10.1126/scitranslmed.3000678. 23ra19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morishige M, Hashimoto S, Ogawa E, Toda Y, Kotani H, Hirose M, Wei S, Hashimoto A, Yamada A, Yano H, Mazaki Y, Kodama H, Nio Y, Manabe T, Wada H, Kobayashi H, Sabe H. GEP100 links epidermal growth factor receptor signalling to Arf6 activation to induce breast cancer invasion. Nat. Cell Biol. 2008;10:85–92. doi: 10.1038/ncb1672. [DOI] [PubMed] [Google Scholar]
- 15.Hsu MY, Meier FE, Nesbit M, Hsu JY, Van Belle P, Elder DE, Herlyn M. E-cadherin expression in melanoma cells restores keratinocyte-mediated growth control and down-regulates expression of invasion-related adhesion receptors. Am. J. Pathol. 2000;156:1515–1525. doi: 10.1016/S0002-9440(10)65023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu MY, Wheelock MJ, Johnson KR, Herlyn M. Shifts in cadherin profiles between human normal melanocytes and melanomas. J. Investig. Dermatol. Symp. Proc. 1996;1:188–194. [PubMed] [Google Scholar]
- 17.Li G, Satyamoorthy K, Herlyn M. N-cadherin-mediated intercellular interactions promote survival and migration of melanoma cells. Cancer Res. 2001;61:3819–3825. [PubMed] [Google Scholar]
- 18.Augustine CK, Yoshimoto Y, Gupta M, Zipfel PA, Selim MA, Febbo P, Pendergast AM, Peters WP, Tyler DS. Targeting N-cadherin enhances antitumor activity of cytotoxic therapies in melanoma treatment. Cancer Res. 2008;68:3777–3784. doi: 10.1158/0008-5472.CAN-07-5949. [DOI] [PubMed] [Google Scholar]
- 19.Qi J, Chen N, Wang J, Siu CH. Transendothelial migration of melanoma cells involves N-cadherin-mediated adhesion and activation of the β-catenin signaling pathway. Mol. Biol. Cell. 2005;16:4386–4397. doi: 10.1091/mbc.E05-03-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi J, Wang J, Romanyuk O, Siu CH. Involvement of Src family kinases in N-cadherin phosphorylation and β-catenin dissociation during transendothelial migration of melanoma cells. Mol. Biol. Cell. 2006;17:1261–1272. doi: 10.1091/mbc.E05-10-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tague SE, Muralidharan V, D’Souza-Schorey C. ADP-ribosylation factor 6 regulates tumor cell invasion through the activation of the MEK/ERK signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9671–9676. doi: 10.1073/pnas.0403531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muralidharan-Chari V, Clancy J, Plou C, Romao M, Chavrier P, Raposo G, D’Souza-Schorey C. ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr. Biol. 2009;19:1875–1885. doi: 10.1016/j.cub.2009.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muralidharan-Chari V, Hoover H, Clancy J, Schweitzer J, Suckow MA, Schroeder V, Castellino FJ, Schorey JS, D’Souza-Schorey C. ADP-ribosylation factor 6 regulates tumorigenic and invasive properties in vivo. Cancer Res. 2009;69:2201–2209. doi: 10.1158/0008-5472.CAN-08-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bilic J, Huang YL, Davidson G, Zimmermann T, Cruciat CM, Bienz M, Niehrs C. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science. 2007;316:1619–1622. doi: 10.1126/science.1137065. [DOI] [PubMed] [Google Scholar]
- 25.Hafner M, Schmitz A, Grüne I, Srivatsan SG, Paul B, Kolanus W, Quast T, Kremmer E, Bauer I, Famulok M. Inhibition of cytohesins by SecinH3 leads to hepatic insulin resistance. Nature. 2006;444:941–944. doi: 10.1038/nature05415. [DOI] [PubMed] [Google Scholar]
- 26.Shoemaker RH, Dykes DJ, Plowman J, Harrison SD, Jr, Griswold DP, Jr, Abbott BJ, Mayo JG, Fodstad O, Boyd MR. Practical spontaneous metastasis model for in vivo therapeutic studies using a human melanoma. Cancer Res. 1991;51:2837–2841. [PubMed] [Google Scholar]
- 27.Witze ES, Litman ES, Argast GM, Moon RT, Ahn NG. Wnt5a control of cell polarity and directional movement by polarized redistribution of adhesion receptors. Science. 2008;320:365–369. doi: 10.1126/science.1151250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Connell MP, Fiori JL, Xu M, Carter AD, Frank BP, Camilli TC, French AD, Dissanayake SK, Indig FE, Bernier M, Taub DD, Hewitt SM, Weeraratna AT. The orphan tyrosine kinase receptor, ROR2, mediates Wnt5A signaling in metastatic melanoma. Oncogene. 2010;29:34–44. doi: 10.1038/onc.2009.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weeraratna AT, Jiang Y, Hostetter G, Rosenblatt K, Duray P, Bittner M, Trent JM. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell. 2002;1:279–288. doi: 10.1016/s1535-6108(02)00045-4. [DOI] [PubMed] [Google Scholar]
- 30.Dissanayake SK, Wade M, Johnson CE, O’Connell MP, Leotlela PD, French AD, Shah KV, Hewitt KJ, Rosenthal DT, Indig FE, Jiang Y, Nickoloff BJ, Taub DD, Trent JM, Moon RT, Bittner M, Weeraratna AT. The Wnt5A/protein kinase C pathway mediates motility in melanoma cells via the inhibition of metastasis suppressors and initiation of an epithelial to mesenchymal transition. J. Biol. Chem. 2007;282:17259–17271. doi: 10.1074/jbc.M700075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slusarski DC, Corces VG, Moon RT. Interaction of Wnt and a Frizzled homologue triggers G-protein-linked phosphatidylinositol signalling. Nature. 1997;390:410–413. doi: 10.1038/37138. [DOI] [PubMed] [Google Scholar]
- 32.Chen W, ten Berge D, Brown J, Ahn S, Hu LA, Miller WE, Caron MG, Barak LS, Nusse R, Lefkowitz RJ. Dishevelled 2 recruits β-arrestin 2 to mediate Wnt5A–stimulated endocytosis of Frizzled 4. Science. 2003;301:1391–1394. doi: 10.1126/science.1082808. [DOI] [PubMed] [Google Scholar]
- 33.Sen M, Chamorro M, Reifert J, Corr M, Carson DA. Blockade of Wnt-5A/frizzled 5 signaling inhibits rheumatoid synoviocyte activation. Arthritis Rheum. 2001;44:772–781. doi: 10.1002/1529-0131(200104)44:4<772::AID-ANR133>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 34.Onken MD, Worley LA, Tuscan MD, Harbour JW. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J. Mol. Diagn. 2010;12:461–468. doi: 10.2353/jmoldx.2010.090220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinnberg T, Menzel M, Ewerth D, Sauer B, Schwarz M, Schaller M, Garbe C, Schittek B. β-Catenin signaling increases during melanoma progression and promotes tumor cell survival and chemoresistance. PLoS One. 2011;6:e23429. doi: 10.1371/journal.pone.0023429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Damsky WE, Curley DP, Santhanakrishnan M, Rosenbaum LE, Platt JT, Gould Rothberg BE, Taketo MM, Dankort D, Rimm DL, McMahon M, Bosenberg M. β-Catenin signaling controls metastasis in Braf-activated Pten-deficient melanomas. Cancer Cell. 2011;20:741–754. doi: 10.1016/j.ccr.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Someya A, Sata M, Takeda K, Pacheco-Rodriguez G, Ferrans VJ, Moss J, Vaughan M. ARF-GEP100, a guanine nucleotide-exchange protein for ADP-ribosylation factor 6. Proc. Natl. Acad. Sci. U.S.A. 2001;98:2413–2418. doi: 10.1073/pnas.051634798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brembeck FH, Schwarz-Romond T, Bakkers J, Wilhelm S, Hammerschmidt M, Birchmeier W. Essential role of BCL9-2 in the switch between β-catenin’s adhesive and transcriptional functions. Genes Dev. 2004;18:2225–2230. doi: 10.1101/gad.317604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang N, Wei P, Gong A, Chiu WT, Lee HT, Colman H, Huang H, Xue J, Liu M, Wang Y, Sawaya R, Xie K, Yung WK, Medema RH, He X, Huang S. FoxM1 promotes β-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer Cell. 2011;20:427–442. doi: 10.1016/j.ccr.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D’Souza-Schorey C, Chavrier P. ARF proteins: Roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 41.Schweitzer JK, Sedgwick AE, D’Souza-Schorey C. ARF6-mediated endocytic recycling impacts cell movement, cell division and lipid homeostasis. Semin. Cell Dev. Biol. 2011;22:39–47. doi: 10.1016/j.semcdb.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muralidharan-Chari V, Clancy JW, Sedgwick A, D’Souza-Schorey C. Microvesicles: Mediators of extracellular communication during cancer progression. J. Cell Sci. 2010;123:1603–1611. doi: 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macias H, Moran A, Samara Y, Moreno M, Compton JE, Harburg G, Strickland P, Hinck L. SLIT/ROBO1 signaling suppresses mammary branching morphogenesis by limiting basal cell number. Dev. Cell. 2011;20:827–840. doi: 10.1016/j.devcel.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Da Forno PD, Pringle JH, Hutchinson P, Osborn J, Huang Q, Potter L, Hancox RA, Fletcher A, Saldanha GS. WNT5A expression increases during melanoma progression and correlates with outcome. Clin. Cancer Res. 2008;14:5825–5832. doi: 10.1158/1078-0432.CCR-07-5104. [DOI] [PubMed] [Google Scholar]
- 45.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, Chang DK, Cowley MJ, Gardiner BB, Song S, Harliwong I, Idrisoglu S, Nourse C, Nourbakhsh E, Manning S, Wani S, Gongora M, Pajic M, Scarlett CJ, Gill AJ, Pinho AV, Rooman I, Anderson M, Holmes O, Leonard C, Taylor D, Wood S, Xu Q, Nones K, Fink JL, Christ A, Bruxner T, Cloonan N, Kolle G, Newell F, Pinese M, Mead RS, Humphris JL, Kaplan W, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chou A, Chin VT, Chantrill LA, Mawson A, Samra JS, Kench JG, Lovell JA, Daly RJ, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N, Kakkar N, Zhao F, Wu YQ, Wang M, Muzny DM, Fisher WE, Brunicardi FC, Hodges SE, Reid JG, Drummond J, Chang K, Han Y, Lewis LR, Dinh H, Buhay CJ, Beck T, Timms L, Sam M, Begley K, Brown A, Pai D, Panchal A, Buchner N, De Borja R, Denroche RE, Yung CK, Serra S, Onetto N, Mukhopadhyay D, Tsao MS, Shaw PA, Petersen GM, Gallinger S, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Schulick RD, Wolfgang CL, Morgan RA, Lawlor RT, Capelli P, Corbo V, Scardoni M, Tortora G, Tempero MA, Mann KM, Jenkins NA, Perez-Mancera PA, Adams DJ, Largaespada DA, Wessels LF, Rust AG, Stein LD, Tuveson DA, Copeland NG, Musgrove EA, Scarpa A, Eshleman JR, Hudson TJ, Sutherland RL, Wheeler DA, Pearson JV, McPherson JD, Gibbs RA, Grimmond SM. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.London NR, Li DY. Robo4-dependent Slit signaling stabilizes the vasculature during pathologic angiogenesis and cytokine storm. Curr. Opin. Hematol. 2011;18:186–190. doi: 10.1097/MOH.0b013e328345a4b9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu W, London NR, Gibson CC, Davis CT, Tong Z, Sorensen LK, Shi DS, Guo J, Smith MCP, Grossmann AH, Thomas KR, Li DY. Interleukin receptor activates a MyD88-ARNO-ARF6 cascade to disrupt vascular stability. Nature. 2012;492:252–255. doi: 10.1038/nature11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Amerongen R, Fuerer C, Mizutani M, Nusse R. Wnt5a can both activate and repress Wnt/β-catenin signaling during mouse embryonic development. Dev. Biol. 2012;369:101–114. doi: 10.1016/j.ydbio.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Q, Major MB, Takanashi S, Camp ND, Nishiya N, Peters EC, Ginsberg MH, Jian X, Randazzo PA, Schultz PG, Moon RT, Ding S. Small-molecule synergist of the Wnt/β-catenin signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7444–7448. doi: 10.1073/pnas.0702136104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim W, Kim SY, Kim T, Kim M, Bae DJ, Choi HI, Kim IS, Jho E. ADP-ribosylation factors 1 and 6 regulate Wnt/β-catenin signaling via control of LRP6 phos-phorylation. Oncogene. 2012 doi: 10.1038/onc.2012.373. [DOI] [PubMed] [Google Scholar]
- 51.Arozarena I, Bischof H, Gilby D, Belloni B, Dummer R, Wellbrock C. In melanoma, beta-catenin is a suppressor of invasion. Oncogene. 2011;30:4531–4543. doi: 10.1038/onc.2011.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chien AJ, Moore EC, Lonsdorf AS, Kulikauskas RM, Rothberg BG, Berger AJ, Major MB, Hwang ST, Rimm DL, Moon RT. Activated Wnt/β-catenin signaling in melanoma is associated with decreased proliferation in patient tumors and a murine melanoma model. Proc. Natl. Acad. Sci. U.S.A. 2009;106:1193–1198. doi: 10.1073/pnas.0811902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kageshita T, Hamby CV, Ishihara T, Matsumoto K, Saida T, Ono T. Loss of β-catenin expression associated with disease progression in malignant melanoma. Br. J. Dermatol. 2001;145:210–216. doi: 10.1046/j.1365-2133.2001.04336.x. [DOI] [PubMed] [Google Scholar]
- 54.Bachmann IM, Straume O, Puntervoll HE, Kalvenes MB, Akslen LA. Importance of P-cadherin, β-catenin, and Wnt5a/frizzled for progression of melanocytic tumors and prognosis in cutaneous melanoma. Clin. Cancer Res. 2005;11:8606–8614. doi: 10.1158/1078-0432.CCR-05-0011. [DOI] [PubMed] [Google Scholar]
- 55.Eichhoff OM, Weeraratna A, Zipser MC, Denat L, Widmer DS, Xu M, Kriegl L, Kirchner T, Larue L, Dummer R, Hoek KS. Differential LEF1 and TCF4 expression is involved in melanoma cell phenotype switching. Pigment Cell Melanoma Res. 2011;24:631–642. doi: 10.1111/j.1755-148X.2011.00871.x. [DOI] [PubMed] [Google Scholar]
- 56.Hu B, Shi B, Jarzynka MJ, Yiin JJ, D’Souza-Schorey C, Cheng SY. ADP-ribosylation factor 6 regulates glioma cell invasion through the IQ-domain GTPase-activating protein 1-Rac1–mediated pathway. Cancer Res. 2009;69:794–801. doi: 10.1158/0008-5472.CAN-08-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hashimoto S, Onodera Y, Hashimoto A, Tanaka M, Hamaguchi M, Yamada A, Sabe H. Requirement for Arf6 in breast cancer invasive activities. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6647–6652. doi: 10.1073/pnas.0401753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mina LA, Sledge GW., Jr. Rethinking the metastatic cascade as a therapeutic target. Nat. Rev. Clin. Oncol. 2011;8:325–332. doi: 10.1038/nrclinonc.2011.59. [DOI] [PubMed] [Google Scholar]
- 59.Atkins MB, Lotze MT, Dutcher JP, Fisher RI, Weiss G, Margolin K, Abrams J, Sznol M, Parkinson D, Hawkins M, Paradise C, Kunkel L, Rosenberg SA. High-dose recombinant interleukin 2 therapy for patients with metastatic melanoma: Analysis of 270 patients treated between 1985 and 1993. J. Clin. Oncol. 1999;17:2105–2116. doi: 10.1200/JCO.1999.17.7.2105. [DOI] [PubMed] [Google Scholar]
- 60.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, Akerley W, van den Eertwegh AJ, Lutzky J, Lorigan P, Vaubel JM, Linette GP, Hogg D, Ottensmeier CH, Lebbé C, Peschel C, Quirt I, Clark JI, Wolchok JD, Weber JS, Tian J, Yellin MJ, Nichol GM, Hoos A, Urba WJ. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, Lorigan P, Lebbe C, Jouary T, Schadendorf D, Ribas A, O’Day SJ, Sosman JA, Kirkwood JM, Eggermont AM, Dreno B, Nolop K, Li J, Nelson B, Hou J, Lee RJ, Flaherty KT, McArthur GA. BRIM-3 Study Group, Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones CA, London NR, Chen H, Park KW, Sauvaget D, Stockton RA, Wythe JD, Suh W, Larrieu-Lahargue F, Mukouyama YS, Lindblom P, Seth P, Frias A, Nishiya N, Ginsberg MH, Gerhardt H, Zhang K, Li DY. Robo4 stabilizes the vascular network by inhibiting pathologic angiogenesis and endothelial hyperpermeability. Nat. Med. 2008;14:448–453. doi: 10.1038/nm1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fuerer C, Nusse R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS One. 2010;5:e9370. doi: 10.1371/journal.pone.0009370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McEwen DP, Gee KR, Kang RR, Neubig RR. Fluorescent BODIPY-GTP analogs: Real-time measurement of nucleotide binding to G proteins. Anal. Biochem. 2001;291:109–117. doi: 10.1006/abio.2001.5011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.