Abstract
Background
Chlamydia trachomatis is globally the predominant infectious cause of blindness and one of the most common bacterial causes of sexually transmitted infection. Infections of the conjunctiva cause the blinding disease trachoma, an immuno-pathological disease that is characterised by chronic conjunctival inflammation and fibrosis. The polymorphic Killer-cell Immunoglobulin-like Receptors (KIR) are found on Natural Killer cells and have co-evolved with the Human Leucocyte Antigen (HLA) class I system. Certain genetic constellations of KIR and HLA class I polymorphisms are associated with a number of diseases in which modulation of the innate responses to viral and intracellular bacterial pathogens is central.
Methodology
A sample of 134 Gambian pedigrees selected to contain at least one individual with conjunctival scarring in the F1 generation was used. Individuals (n = 830) were genotyped for HLA class I and KIR gene families. Family Based Association Tests and Case Pseudo-control tests were used to extend tests for transmission disequilibrium to take full advantage of the family design, genetic model and phenotype.
Principle findings
We found that the odds of trachomatous scarring increased with the number of genome copies of HLA-C2 (C1/C2 OR = 2.29 BHP-value = 0.006; C2/C2 OR = 3.97 BHP-value = 0.0004) and further increased when both KIR2DL2 and KIR2DL3 (C2/C2 OR = 5.95 BHP-value = 0.006) were present.
Conclusions
To explain the observations in the context of chlamydial infection and trachoma we propose a two-stage model of response and disease that balances the cytolytic response of KIR expressing NK cells with the ability to secrete interferon gamma, a combination that may cause pathology. The data presented indicate that HLA-C genotypes are important determinants of conjunctival scarring in trachoma and that KIR2DL2/KIR2DL3 heterozygosity further increases risk of conjunctival scarring in individuals carrying HLA-C2.
Author Summary
Chlamydia trachomatis is a pathogen that causes sexually transmitted infections (STIs) and the blinding disease trachoma. Natural Killer (NK) cells are part of the host immune system's first line of defence against infection. NK cell functions are genetically encoded and differences between individuals mean that some people are better able to respond to infections than others. We found that in certain combinations, specific variants of the gene HLA-C (Human Leucocyte Antigen, C) and of a complex set of genes called the Killer-cell Immunoglobulin-like Receptors (KIR) were associated with a six-fold increase in the relative risk of scarring tissue damage resulting from ocular C. trachomatis infection (trachoma). This combination of genetic variants may reduce the host's ability to effectively resolve infections and result in a harmful immune response that ultimately leads to tissue damage and scarring. KIR+ NK cells are potential cellular mediators of the damaging immune response. Previous studies have identified that the same HLA-KIR genetic constellation that associates with trachoma is actually protective against infectious diseases such as malaria and tuberculosis. The high frequency of the trachoma-associated constellation in African populations may therefore be explained by the evolutionary benefits of protection from the complications of severe disease.
Introduction
Chlamydia trachomatis (Ct) is an obligate intracellular bacterium [1] which causes significant morbidity as the causative factor of around 106 million new sexually transmitted infections per annum [2]. As the cause of trachoma, the same bacterium is the most common infectious cause of blindness [3]. Ct serovars exhibit highly specific tissue tropism, with serovars A–C being limited to the mucosal epithelium of the ocular conjunctiva. The remaining serovars are sexually transmitted, but whilst serovars D–K are limited to the mucosal epithelia of the genitourinary tract and rectum, the strains L1–L3 are able to invade other tissues including the lymph nodes. Ocular infection in trachoma is spread among young persons through exposure to secretions from the infected eye via direct physical contact, on fomites or by eye-seeking flies [4]. Repeated and prolonged cycles of infection and inflammation have been identified as the main factors that lead to the progressive formation of fibrotic scars on the tarsal conjunctiva, which ultimately becomes deformed. This can cause entropion and trachomatous trichiasis (TT), a condition where the eyelashes turn inwards and irreversibly damage the cornea by scratching the globe of the eye. If left unchecked, TT causes corneal opacity, visual impairment and blindness.
Active trachoma is frequently found in the absence of detectable Ct infection and both tissue damage and scarring are thought to be the result of a chronic immuno-pathological reaction [5]. Human conjunctival transcriptome studies in trachoma suggest that in addition to T cell and innate responses of epithelial cells, the activation and cytotoxic responses of natural killer (NK) cells is an important determinant of the severity of active trachoma [6], [7]. NK cells are a rich source of multiple chemokines and cytokines, including interferon gamma (IFNγ), a cytokine that is central to the control of chlamydial intracellular development and growth. IFNγ also has anti-fibrotic properties that can counteract the effects of TGF-β and inhibit fibroblast proliferation and collagen synthesis [8], but when inappropriately expressed may cause immunopathology. NK cells in mucosal-associated lymphoid tissues are known to be important in the maintenance of epithelial cell integrity via production of the cytokine IL-22 [9]. NK cells therefore have the potential to fulfil multiple roles that encompass tissue homeostasis, tissue re-modelling and immunity.
Early studies in murine chlamydial model infections found that NK cell depletion exacerbated disease, delayed clearance and limited the development of specific T cell responses [10], [11]. Subsequent studies have confirmed that in response to chlamydial stimulation, NK cells are promoters of T cell immunity and a major source of IFNγ [10], [12] but their role as lytic effector cells is less clear. Although Ct infected cell lines are lysed in vitro, NK cells purified from the peripheral blood of individuals with current chlamydial infection had diminished lytic activity (and reduced IFNγ) compared with uninfected controls [13]. Population diversity in the highly polymorphic genes that encode the variable NK receptors and their ligands [14] along with functional heterogeneity in the NK cell repertoire may account for these findings [15].
Trachoma is a complex inflammatory fibrotic disease in which host polymorphism in immune response genes plays a significant role [16]–[18]. The conjunctival epithelial surface is compromised in trachoma [5] as a result of the host response to the causative bacterium, which occupies an intracellular niche. Therefore the mechanisms used by NK cells in the control of other intracellular infections such as Hepatitis B [19], Hepatitis C [20] and HIV [21]–[23] might also be effective against intracellular Ct.
NK cells become activated when they are released from inhibition that is normally bound by interaction of specific HLA class I ligands with inhibitory Killer-cell Immunoglobulin-like Receptors (KIRs) [24]. The ligands of several inhibitory KIR have been described including HLA-A3 and HLA-A11 alleles, which are ligands of the KIR3DL2 receptor [25], [26] and the HLA-Bw4 public epitope which is the ligand of KIR3DL1 [27], [28]. HLA-C alleles can be classified (according to a functional dimorphism at amino acid position 80) as carrying one of two KIR binding epitopes, which are known as HLA-C1 and HLA-C2 [29]. The HLA-C2 group of alleles (HLA-C*02/04/05/06…) are ligands of the inhibitory receptor KIR2DL1 [30]–[32] and its activating counterpart KIR2DS1 [33]. The HLA-C1 group alleles (HLA-Cw*01/03/07/08…) are ligands of both KIR2DL2 and KIR2DL3 [30]–[32], however, the latter KIR are both able to cross-react (with differing avidities) with a small number of HLA-C2 and HLA-B allotypes [34]. Although germ-line encoded, the KIR gene system is highly polymorphic and exhibits extensive diversity both between individuals and between populations [35]–[38]. KIRs exhibit haplotype diversity such that different individuals possess variable gene contents. Since KIR and HLA are also found on different chromosomes, individuals can possess a KIR for which they have no cognate ligand, or vice versa. The extensive polymorphism in the KIR system culminates in a repertoire of NK cells within an individual that is more or less sensitive to release from inhibition under appropriate physiological conditions [39]. The strength of the signals mediated by interactions between specific HLA and KIR alleles is also highly variable [29], [40], [41] and this further limits overall NK cell responsiveness [42]. In part the responsiveness might be predicted by the presence of type ‘A’ and type ‘B’ KIR haplotypes. Type A haplotypes carry genes encoding predominantly inhibitory KIRs. B haplotypes contain some or all of the same genes found on A haplotypes, but additionally may carry the inhibitory KIR2DL2 and KIR2DL5 genes and numerous activating KIRs [36]. KIR haplotypes can be separated in to two variable regions, defined by their orientation towards the centromeric (Cen) or telomeric (Tel) regions of the chromosome [43]. The KIR A and B haplotypes are present in all populations studied to date and are thought to be maintained by the balancing selection pressures of infection, immunopathology and healthy reproduction [44]–[47]. In recent human history, a wide range of infectious diseases may have reduced the balancing effects in African populations, leading to more directional selection and a unique pattern of HLA and KIR diversity in this region [38], [47]. We therefore assessed the extent to which host genotypes at the HLA and KIR loci were associated with trachomatous scarring in a trachoma endemic population from The Gambia.
Methods
Ethics statement
The study was conducted in accordance with the tenets of the Declaration of Helsinki. The Ethics Committee of the Gambian Government/Medical Research Council Unit, and the ethics committee of the London School of Hygiene and Tropical Medicine approved the study (MRC SCC1177). Individual written informed consent was obtained from all adult participants. Written informed consent was obtained from a parent/guardian on behalf of those subjects aged <18 years who wished to take part in the study. All samples were anonymised.
Study population, sampling and ascertainment
We selected a family study design and identified probands at a relatively early age for clinical signs of conjunctival scarring. This maximised statistical power whilst controlling for population stratification through the use of related control samples. The study population came from multiple regions of The Gambia and included multiplex families of mixed ethnic background. Families were ascertained through the identification of probands in which there were signs of trachomatous scarring at an early age (age ≤30 years). This approach maximised the extent to which genetic rather than environmental factors could be expected to have contributed significantly to the probands' phenotypes. We recruited first-degree relations of the probands. In most cases this meant that we sampled both biological parents of the probands and all their (self-described) full siblings. Samples for DNA analysis were collected from buccal mucosae using sterile cyto-brushes (Part Number F-440151, SLS, Nottingham, UK). After collection, brushes were returned to their original packaging and stored dry at room temperature for up to 6 months [48] before DNA extraction was performed using a salting out procedure.
Sample size and power calculation
An average of 4 offspring per family was assumed with a population prevalence of scarring in those <30 years of age in The Gambia of ∼2% [49]. The Pedigree Based Association Test (PBAT) v3.6 program [50] was used to calculate the power of the study to detect with 95% confidence (α<0.05) a genetic association with odds ratios 1.5, 2 and 3 when the hypothetical disease allele had a frequency between 0.01 and 0.50. Figure S1 shows the estimated power of this study to detect genetic associations with trachomatous scarring at a range of allele frequencies and effect magnitudes, given the sample size. We had >90% power to detect an effect size greater than an odds ratio (OR) = 3 when the allele frequency was ≥0.05 and similar power to detect an effect size of OR = 2 when the allele frequency was ≥0.19.
Trachoma phenotypes
Trachoma was graded in the field using the WHO simplified grading system by field supervisors certified for trachoma grading with regular performance checks as described by the PRET clinical trial manual of operations [51]. Left and right tarsal conjunctivae of all subjects were photographed as described by Derrick et al. [52]. Photographs were subsequently reviewed by two ophthalmologists with experience of grading trachoma and a final grade agreed. Subjects were assigned to the ‘scarred’ group if there were any signs of trachomatous scarring, in either eye. Individuals where phenotypes could not be confirmed for reasons of poor quality photography (n = 5) did not contribute to the statistical tests of association.
Genotyping and pedigree tests for Mendelian inheritance
KIR genotyping for the presence or absence of 17 KIR genes was performed by PCR using the set of sequence specific primers described by Vilches et. al. [53]. The genotyping method was validated by participation in the UCLA Immunogenetics Center KIR exchange programme (http://www.hla.ucla.edu/cellDna.htm). Medium resolution HLA-A, -B and -C genotyping was performed using LABtype sequence specific oligonucleotide probes (OneLambda, Canoga Park, CA. USA) on a Luminex platform (Luminexcorp, Austin, TX. USA). Medium resolution HLA typing data generates strings of possible allele combinations. Information from the HLA genotypes of family members was used to reduce the length of the strings of possible allele pairs and to eliminate alleles that were not compatible with Mendelian inheritance within a given pedigree. Strings were further shortened where possible to include only common and well-defined alleles [54]. In order to maximise statistical power, highly sequence similar HLA alleles were combined in to groups (table S1) before FBAT. KIR ligands of HLA (HLA-A*03/11/Bw4, HLA-B-Bw4, HLA-C1/C2) were inferred from the full HLA genotypes of individual specimens rather than the reduced strings. The HLA-C*16:01 (HLA-C1) and HLA-C*16:02 (HLA-C2) alleles were frequently ambiguous and where this was the case alleles were assigned to HLA-C*16:01 because the HLA-C*16:02 allele has not been observed in other West African populations whilst HLA-C*16:01 is very common (data from allelefrequencies.net). HLA types were used to identify cases of parental mis-assignment and inconsistent parent-offspring genotypes. KIR phenotypes (presence/absence) were tested for Mendelian inconsistencies. KIR2DL5, KIR2DS3 and KIR2DS5 were not included in the association tests as they can segregate to both Cen-B and Tel-B regions and confound haplotype assignments.
Statistical analysis
KIR gene frequencies were compared to those of other world populations using data from allelefrequencies.net and PCA using R. Family based tests of HLA association were carried out using FBAT v.2.0.3 [55] performing a series of bi-allelic tests (i.e. association of an index allele against all other alleles) under an additive genetic model and the null hypothesis of no linkage and no association of any factor of the HLA system with trachomatous scarring. This approach is robust to effects of population structure [55], [56] and is applicable to a data set with samples originating in mixed ethnic backgrounds. We tested for associations between scarring and all HLA alleles with a sample frequency greater than 0.05 with an offset value of 0.02 (population prevalence of scarring in persons ≤30 years of age) to allow the unaffected siblings to contribute to the test statistic. All FBAT p-values were adjusted using a conservative Bonferroni correction. Significant associations were tested again using a case/pseudo-control conditional logistic regression (CLR) [57], which generated estimates of odds ratios and associated p-values. To test for independence between the disease-associated alleles, we included all alleles that had a corrected p<0.05 in a multivariate CLR model. To establish whether significant HLA associations were restricted to F1 subjects with specific KIR genotypes we tested the full data set under a genotype model [58], [59], using CLR, in different subsets of the F1 data where the population was limited by the KIR genotype. Because of the high linkage disequilibrium between factors of the KIR system, these tests were not considered to be independent and test statistics were corrected using the Benjamini-Hochberg method.
Results
Sample population
We sampled 830 individuals from 134 pedigrees and 146 nuclear families in which scarring trachoma had been identified in the first filial (F1) generation. The self-described ethnic background of the parental (P0) population (n = 260) was approximately 40% Mandinka, 23% Fula, 15% Jola, 15% Wolof, 5% Bambara and 2% other minority ethnic groups. There were 570 persons in the F1 generation, where the gender distribution was 52% (n = 296) male and 48% (n = 274) female. The median number of offspring per pedigree was 4 (range 1–11). Eight families had one missing parent. There were 180 (∼32%) cases of trachomatous scarring in the F1 generation and of these, 72 (40%) were female and 108 (60%) were male. Three hundred and eighty six (∼67.8%) F1 individuals were unaffected and phenotypic status could not be confirmed for 4 (<1%). Table 1 gives a detailed description of the phenotype distribution in the families. Detailed examination of photographs revealed that 12 probands did not have sufficient signs of trachomatous scarring. One proband could not be graded. In all the families where there was no photography confirmed scarred proband, at least one sibling was identified who was under 30 years of age and had signs of scarring. HLA genotyping identified paternal misassignment in 63 F1 individuals (11%) who were reassigned to an unknown father but were otherwise retained for analysis.
Table 1. Clinical and Demographic features of the sample.
| Scarring (C) Grade (n [%]) | ||||||
| Group | Age in years (Median [min - max]) | C0 | C1 | C2 | C3 | No FPC Grading |
| F1 Generation | ||||||
| Total (n = 570) | 08 [0.1–40] | 386 (67.8) | 48 (8.4) | 112 (19.6) | 20 (3.5) | 4 (0.7) |
| Male (n = 296) | 08 [0.1–28] | 188 (63.6) | 25 (8.4) | 71 (24.0) | 12 (4.0) | 0 (0.0) |
| Female (n = 274) | 08 [0.1–40] | 198 (72.2) | 23 (8.4) | 41 (15.0) | 8 (2.9) | 4 (1.5) |
| Probands (n = 134) | 05 [0.3–22] | 12 (9.0) | 12 (9.0) | 93 (69.4) | 16 (11.9) | 1 (0.7) |
| Siblings (n = 436) | 10 [0.1–40] | 374 (85.7) | 36 (8.3) | 19 (4.4) | 4 (0.9) | 3 (0.7) |
| P0 Generation | ||||||
| Total (n = 260) | 39 [18–72] | 121 (46.6) | 65 (25.0) | 69 (26.5) | 4 (1.5) | 1 (0.4) |
| Female (n = 132) | 34 [18–65] | 77 (58.2) | 22 (16.7) | 31 (23.5) | 1 (0.8) | 1 (0.8) |
| Male (n = 128) | 45 [23–72] | 44 (34.5) | 43 (33.6) | 38 (29.7) | 3 (2.3) | 0 (0.0) |
| Families | Minimum | 1st Quantile | Median | 3rd Quantile | Maximum | |
| Number persons F1 | 1 | 3 | 4 | 5 | 11 | |
HLA and KIR genotypes
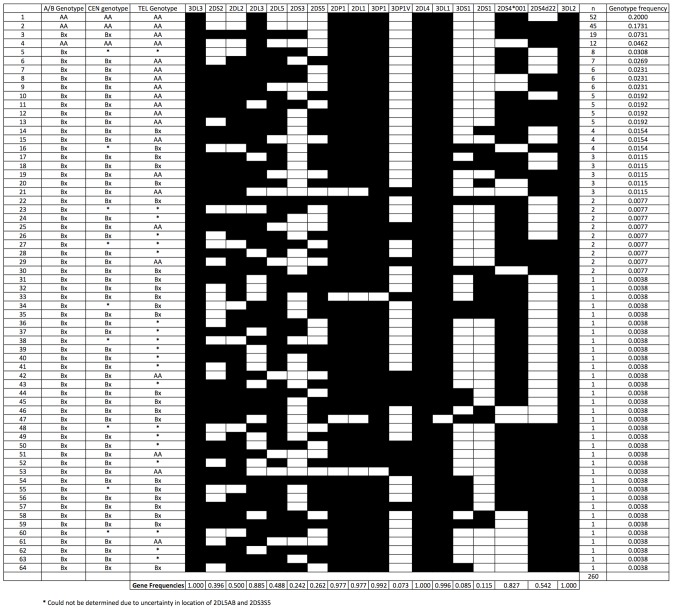
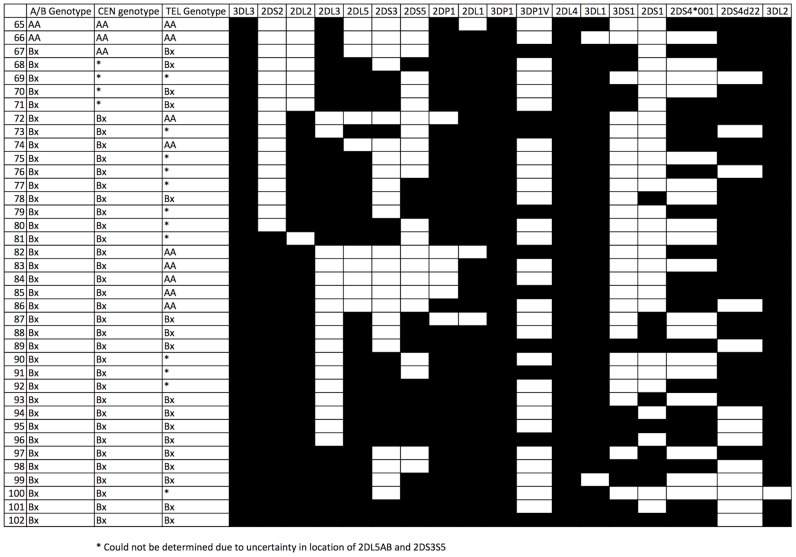
Table 2 shows the Family Based Association Test (FBAT) estimates of the HLA allele and KIR epitope frequencies in the sample population. Figure 1 describes the 64 unique KIR genotypes that were observed in the P0 generation. Thirty-eight additional KIR genotypes were revealed by re-assortment of the parental haplotypes in the F1 generation (Figure 2). All observed genotypes were assigned as either the ‘AA’ or ‘Bx’ genotypes (where Bx includes both AB and BB genotypes) for the full KIR region and where possible, for each of the Cen and Tel regions. A number of unusual genotypes were identified in this population, most notably, 10.4% of P0 individuals (n = 27/260) possessed KIR2DL2 but not KIR2DS2.
Table 2. HLA Class I allele frequencies and FBAT tests of association.
| Locus | Allele Name* | Allele frequency (P0) | # informative families (n) | S-E(S) | Var(S) | Z | P | Corrected P |
| HLA-A | A*02:01 | 0.067 | 37 | −4.66 | 10.767 | −1.42 | 0.155552 | 1 |
| A*23:01 | 0.164 | 67 | −0.56 | 24.675 | −0.113 | 0.910241 | 1 | |
| A*26:01 | 0.063 | 32 | −2.74 | 8.366 | −0.947 | 0.343476 | 1 | |
| A*30:02 | 0.065 | 28 | −0.54 | 8.752 | −0.183 | 0.855165 | 1 | |
| A*33:01 | 0.138 | 62 | 2.557 | 18.996 | 0.587 | 0.557473 | 1 | |
| A*68:01 | 0.05 | 21 | 2.95 | 7.209 | 1.099 | 0.27191 | 1 | |
| A*68:02 | 0.052 | 27 | 3.18 | 8.352 | 1.1 | 0.271184 | 1 | |
| HLA_B | B*07:02 | 0.067 | 33 | 2.66 | 10.335 | 0.827 | 0.407994 | 1 |
| B*08:01 | 0.069 | 41 | −12.18 | 11.786 | −3.548 | 0.000388 | 0.01 | |
| B*35:01 | 0.133 | 51 | 6.77 | 17.467 | 1.62 | 0.105266 | 1 | |
| B*15:03 | 0.071 | 35 | −0.14 | 12.033 | −0.04 | 0.967807 | 1 | |
| B*53:01 | 0.123 | 48 | 9.62 | 16.341 | 2.38 | 0.017324 | 0.49 | |
| B*58:01 | 0.075 | 40 | 2.28 | 12.467 | 0.646 | 0.518451 | 1 | |
| B*78:01 | 0.056 | 29 | −0.53 | 8.268 | −0.184 | 0.853761 | 1 | |
| HLA_C | C*02:02 | 0.114 | 53 | −0.29 | 16.851 | −0.071 | 0.943679 | 1 |
| C*03:04 | 0.083 | 45 | −11.888 | 13.791 | −3.201 | 0.001369 | 0.04 | |
| C*04:01 | 0.186 | 66 | 11.629 | 23.828 | 2.382 | 0.017205 | 0.49 | |
| C*06:02 | 0.089 | 33 | 5.52 | 10.042 | 1.742 | 0.081515 | 1 | |
| C*07:01 | 0.114 | 55 | −2.403 | 17.979 | −0.567 | 0.570849 | 1 | |
| C*16:01/02 | 0.12 | 52 | −7 | 14.981 | −1.809 | 0.070526 | 1 | |
| C*17:01 | 0.054 | 27 | 2.834 | 7.569 | 1.03 | 0.302871 | 1 | |
| HLA-A3/11 | No KIR epitope | 0.772 | 81 | −2.41 | 31.893 | −0.427 | 0.669564 | 1 |
| Bw4_80I | 0.183 | 69 | −0.06 | 26.511 | −0.012 | 0.990703 | 1 | |
| HLA-Bw4/Bw6 | Bw6 | 0.625 | 108 | −3.907 | 42.358 | −0.6 | 0.548333 | 1 |
| Bw4_80I | 0.318 | 101 | 3.867 | 41.641 | 0.599 | 0.549034 | 1 | |
| Bw4_80T | 0.056 | 28 | 0.04 | 10.116 | 0.013 | 0.989966 | 1 | |
| HLA-C1C2 | C2 | 0.499 | 99 | 23.08 | 40.6 | 3.622 | 0.000292 | 0.008 |
*Named alleles may indicate the first allele identifier in a longer string of related alleles, but these have been shortened for ease of reading. Full details can be found in table S1.
Figure 1. KIR genotypes and observed frequencies in the P0 population (n = 260).
Figure 2. KIR genotypes and that were uniquely identified in the F1 population.
Linkage disequilibrium
Pairwise linkage disequilibrium data (LD) for the KIR genes were calculated (figure S2). Contrary to data from other studied human populations [58], [60], [61] and consistent with other findings within Africa [47], we observed reduced LD between KIR genes. We did not identify any pairs of KIR genes that were in perfect LD (r2 = 1 : only two of the four possible haplotypes observed), although a number of KIR genes were found to be in complete LD (D′ = 1 : only three of the four possible haplotypes observed). The extent of LD was insufficient for high confidence imputation of missing KIR genotypes for use in FBAT [58].
Association tests
Any HLA alleles and KIR epitopes with estimated frequencies above 0.05 were included in the FBAT. Three sets of HLA alleles were significantly associated with trachomatous scarring (Table 2). These were HLA-B*08:01 (Z = −3.548, p = 0.0004, corrected p = 0.01), HLA-C*03:04 (Z = −3.201, p = 0.0014, corrected p = 0.04) and the KIR epitope HLA-C1/C2 (Z = 3.622, p = 0.0003, corrected p = 0.008). Only HLA-C1/C2 remained significant (HLA-C2, OR = 1.684 p = 0.0033) in a multivariate case/pseudo-control, additive model that included all three factors (Table 3), indicating that the HLA-C1/C2 epitope was the only significant independent factor of the HLA system that was associated with trachomatous scarring. In line with previous study designs and analyses we divided the data into several subsets [58], [59]. We identified that in the majority of subsets, as with the unselected sample, the relative risk of scarring increased with the number of genomic copies of the HLA-C2 epitope in an additive manner (Table 4). The association of the HLA-C2 homozygote genotype with trachomatous scarring was restricted to the subsets of offspring who were KIR2DL2 + and KIR2DL3 + (Cen-AB) (OR = 5.95, p = 0.0025, BH corrected p = 0.006) and to those who were KIR3DL1 + KIR3DS1 − and KIR2DS1− (Tel-AA) (HLA-C2 homozygote OR = 4.89, p = 0.00006, BH corrected p 0.0004). Elevated odds ratios were observed in sensitivity analyses (Table 4) in F1 samples where the case definition was restricted to those with moderate or severe (WHO FPC grade C2 or C3) rather than evidence of any (C1, C2 or C3) scarring.
Table 3. Significant results of case/pseudo-control CLR analysis of total family data set.
| Allele | Odds Ratio | P Value | |
| Multivariate CLR TEST | HLA-B*08:01/…* | 0.694 | 0.7000 |
| HLA-C*03:04/…** | 0.500 | 0.1500 | |
| HLA-C EPITOPE C2 | 1.684 | 0.0033 |
Likelihood ratio test = 23.1 on 3 df, p = 0.0000379 n = 580, number of informative events = 152.
Table 4. Subset analysis of HLA-C1C2 genotype associations with scarring.
| Offspring Genotype | Genotype test HLA-C1/C2 | BH Corrected P | Genotype test HLA-C2/C2 | BH Corrected P | n | Number of events |
| Unselected | OR = 2.29 p = 0.0026 | 0.0061 | OR = 3.97 p = 0.000051 | 0.0004 | 636 | 159 |
| KIR2DL2 − KIR2DL3 + (Cen-AA) | OR = 1.94 p = 0.08 | 0.120 | OR = 2.00 p = 0.15 | 0.191 | 296 | 74 |
| KIR2DL2 + KIR2DL3 + (Cen-AB) | OR = 2.33 p = 0.057 | 0.100 | OR = 5.95 p = 0.0025 | 0.006 | 240 | 60 |
| KIR2DL2 + KIR2DL3 − (Cen-BB) | OR = 1.5 p = 0.73 | 0.786 | OR = 6.00 p = 0.13 | 0.182 | 76 | 19 |
| KIR3DL1 + KIR3DS1 − KIR2DS1 − (Tel-AA) | OR = 2.86 p = 0.0013 | 0.006 | OR = 4.89 p = 0.00006 | 0.0004 | 524 | 131 |
| KIR3DS1 + and/or KIR2DS1 + (Tel-Bx) | OR = 0.52 p = 0.29 | 0.338 | OR = 0.90 p = 0.89 | 0.890 | 88 | 22 |
| Affected cases defined by more severe scarring (WHO FPC score C2 or C3) & KIR2DL2 + KIR2DL3 + (Cen-AB) | OR = 2.07 p = 0.026 | 0.052 | OR = 3.57 p = 0.0017 | 0.006 | 444 | 111 |
Comparison of Gambian KIR gene frequencies to other human populations
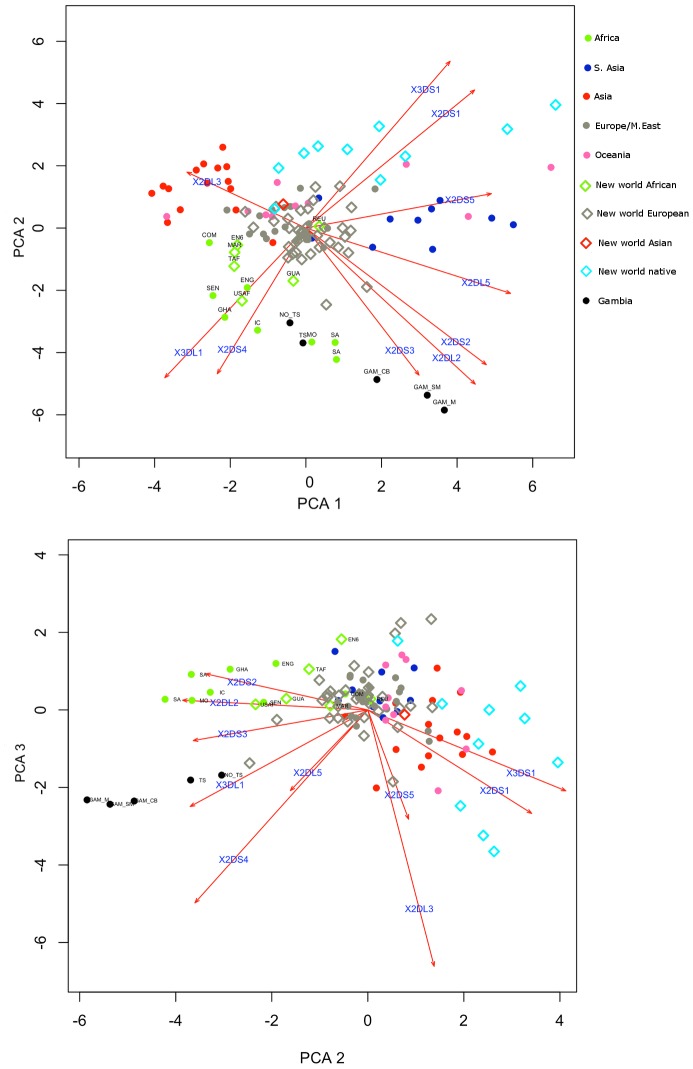
We used Principle Components Analysis (PCA) to compare the KIR gene frequencies observed in the P0 generation of the Gambian trachoma families to those observed in other populations where data was available (allelefrequencies.net database, (Figure 3)). The proportions of the total variance explained by the first three principle components were 0.42 (σ = 2.05), 0.28 (σ = 1.69) and 0.11 (σ = 1.03). The P0 specimens clustered with other populations of African descent, which could be recognised by the observation of high frequencies of the genes defining the Cen-B (KIR2DS2, KIR2DL2) and Tel-A (KIR3DL1 and KIR2DS4) haplotypes.
Figure 3. Principle Components Analysis of Gambian KIR frequencies and other world populations.
African populations are characterised by high frequencies of the Cen-B (KIR2DS2∼KIR2DL2, KIR2DL2) and Tel-A (KIR3DS1∼KIR2DS4) haplotypes. Gambian samples, including the Po specimens and malaria cases cluster together and have some of the highest observed frequencies of Cen-B and Tel-A. The proportions of the total variance explained by the first three principal components were respectively 0.42, 0.28 and 0.11. Footnote to figure 3. NO_TS : Parents from Gambian trachoma families, unaffected, TS : Parents from Gambian trachoma families, affected, GAM_SM : Gambian severe malaria cases, GAM_M : Gambian uncomplicated Malaria cases, GCB : Gambian cord bloods, COM : Comoros, ENG : Equatorial New Guinea, GHA : Ghana, IC : Ivory Coast, MO : Morocco, SEN : Senegal, SA : South Africa, EN6 : England – West Midlands Afro-Caribbean, GUA : Guadaloupe, MAR : Martinique, REU : Reunion, TAF (Trinidad Africans), USAF : USA Californian African Americans.
Discussion
We identified three factors of the HLA system (HLA-C1/C2, HLA-B*08:01 and HLA-C*03:04) that associated with trachomatous scarring. However, the protective associations of HLA-B*08:01 and HLA-C*03:04 observed by univariate analysis were not independent of the HLA-C1/C2 association under a multivariate model. This dependence can be explained by the presence of a common HLA-B*08∼C*03 haplotype, which we estimate to have a frequency of around 5.7% in the Gambian trachoma families. In Senegalese Mandinka, this haplotype has an estimated population frequency of 5.5% [62] whilst in African-Americans the frequency is estimated to be as high as 22% [63]. The associations that were observed between HLA-B*08 and HLA-C*03 (an HLA-C1 group allele) appear to be proxies for the association of the HLA-C1/C2 epitope, which reached statistical significance in univariate analysis because of their high population frequencies and the relatively large contribution they therefore made to the HLA-C epitope data.
We observed patterns of transmission disequilibrium in our sample of families that suggest HLA and KIR genotypes associate with high magnitude increases in the relative risk of scarring in trachomatous disease. Through sensitivity analysis (Table 4) we demonstrated the robustness of the analysis and consistency of the results when the cases were defined by more stringently defined phenotypes relating to severity of scarring.
HLA-C2 epitopes potentially impair NK cell responses and facilitate chronic Ct infection
The chlamydial protease, CPAF has been reported to interfere with the surface presentation of HLA class I molecules [14], [64]–[66], but recently this has been called in to question by Chen et al. [67]. Kägebein et al. [68] then demonstrated that Ct infection does not lead to alteration in normal MHC Class-I expression, maturation or surface presentation. This implies that Ct infected cells are unlikely to be targets for missing-self reactions mediated by NK cells which selectively monitor down-regulation or loss of self-type MHC class I on target cells. Instead it is more likely that cytotoxic NK responses in chlamydial infections are controlled by dynamic changes in the expression levels of activating NK receptors. These changes may occur as a result of infection and other environmental triggers [69], [70] and might overwhelm the inhibitory effects of the more strictly expression-regulated [69] NK inhibitory pathways.
HLA-C1:KIR2DL3 inhibited NK cells have weaker inhibitory signals than other HLA-C inhibited cells [29] and may have a lower threshold for activation. Khakoo et al. [40] reported that the HLA-C1/C1 KIR2DL3/2DL3 genotype constellation increased probability of clearance of early stage Hepatitis C Virus (HCV) infections. Ahlenstiel et al. [42] provided evidence that HLA-C1 homozygotes might be better able to challenge early infections by showing that the proportion of the total NK cell repertoire that is educated and inhibited by HLA-C is ∼50% greater in this group than that in HLA-C2 homozygotes [42]. The same study showed that HLA-C1 inhibited NK cells are better able to mount rapid, intense responses to infection through degranulation and IFNγ secretion [42]. In Ct infections, HLA-C1/C1 individuals may be able to limit chronicity by controlling the early stages of Ct infections with an NK response that is easily activated, and involves a more substantial component of the NK repertoire than in HLA-C2/C2 individuals. This may also be true of HLA-C2+ individuals who possess only weakly responsive KIR2DL1 alleles, such as those alleles that are found on the commonest B haplotypes in Caucasian populations [41]. However, in the Ga-Adangbe population of Ghana, there was a great diversity B haplotypes, none of which were found at high frequency and many of which carried non-attentuated KIR2DL1 alleles [38]. Any assumption about how the presence of Cen-B might indicate reduced cellular inhibition in Gambians should therefore be made with some caution.
A role for within-person inhibitory KIR diversity in influencing immunopathology
The role of KIR in mediating NK cytotoxic responses is well studied, but it is now clear that KIR expressing NK cells are also a major source of IFNγ [71]. The ability of NK cells to produce IFNγ in response to microbial stimuli is related to the density of NCAM-1 (CD56) expressed on their surface, their KIR genotype and the degree of stimulus by accessory cells. An indication of the strength of regulation imposed by the KIR genotype can be estimated as a ratio, known as the ‘DIM factor’, between the response of the CD56dim (KIR-HLA dependent) and CD56bright (KIR-HLA independent) IFNγ responding populations [71]. The majority of human NK cells in the periphery are CD56dim, express KIR and are susceptible to inhibition through KIR-HLA interaction. KIR genotype directly influences the DIM factor, but the exact genotypic conformation that defines this has yet to be elucidated. It has been proposed that the NK cell IFNγ response will be higher in individuals with more KIR educated NK cells, a situation found when there is a greater diversity of within-person inhibitory KIR genes. Experimentally, IFNγ production in CD56dim NK cells showed least inhibition (and the highest DIM factor) in KIR AB heterozygotes [71]. In HLA-C2 homozygotes, we observed a significant KIR2DL2/L3 heterozygote (Cen-AB) disadvantage (Table 4) and an increased relative risk in those with the Tel-AA genotype. The number of persons with Tel-B genotypes was very low in this study, which reflects the low diversity in the Tel region that was reported in another West African population [38]. The high phenotypic frequency of KIR3DL1 (Tel-A) in this Gambian population (∼99.6%) indicates that most individuals with the Cen-AB genotype possess at least one Tel-A haplotype. The Cen-AB, Tel-A+ genotype represents a full complement of the known MHC specific inhibitory KIRs (KIR2DL1, KIR2DL2, KIR2DL3, KIR3DL1 and KIR3DL2) and this genotype might define a high DIM factor [71]. NK cell clones with a Cen-AB genotype would therefore be relatively resistant to inhibition (DIM factor >1) and would retain the potential for high IFNγ production.
Common tropical infectious diseases drive selection of high frequencies of trachoma risk genotype constellations
The KIR system exhibits extensive diversity in African populations [47], [72], [73] possibly driven by a high burden of life threatening infectious diseases, that have exerted strong (diversifying) selective pressures on each population [46], [47], [74]. The high prevalence of Ct STIs in some African populations has been implicated as a contributory factor to the high incidences of infection related infertility that are observed in Africa [75]. It is therefore surprising that Ct disease associated KIR and HLA genotypes are enriched in Africa. One explanation is that opposing selection pressures from other infectious diseases negate selection by Ct. Our sample was selected based on disease phenotype and we found KIR gene frequencies similar to other African populations (Figure 3). The Gambian samples are clearly separated from those in other geographical regions by high frequencies of the genes that define the Cen-B and Tel-A haplotypes (Figure 3) [72]. The frequency of HLA-C2 epitopes is reported to be higher in African populations than in other populations [46], [76], [77] and the HLA-C epitope frequencies that we observed are similar to those previously described [77]. Yindom et al. [78] reported that the proportion of persons with the constellation HLA-C1 and KIR2DL2/KIR2DS2 (Cen-B) is higher in cases of malaria than in population matched, cord-blood controls [78]. In a study of a South-East Asian population, Hirayasu et al. [79] reported that natural selection may have reduced the frequency of the HLA-C1 and KIR2DL3 (Cen-A) because this genotype associates with cases of cerebral malaria. Both studies identify HLA-C1 in association with malarial disease, but they implicate different KIR Cen haplotypes. In the Gambian trachoma families, we observed that many Cen-B haplotypes lacked KIR2DS2, whilst maintaining KIR2DL2. This genotype has previously been identified in an African population [73] and its presence could be explained if KIR2DS2, rather than KIR2DL2, were mediating the Cen-B risk effect. The combined evidence of several TB studies shows that KIR Cen-A [80]–[82] and Tel-B [82], [83] haplotypes associate with TB cases. We therefore suggest that the HLA-C2 homozygous, Cen-AB, Tel-A+ population are more resistant to the complications of both malaria and TB, but more susceptible to trachomatous scarring and that trachomatous scarring (and possibly reduced fertility) is the penalty of increased survival.
A dual role for KIR-HLA restricted NK cells in trachoma and its implications for chlamydial vaccine development
We identified KIR-HLA interactions as an important contributory factor to risk of scarring. The HLA-C2 homozygous, KIR2DL2 +, KIR2DL3 + genotype associates with high relative risk of scarring. We suggest a model that may explain the data in which HLA-C2 may favour chronic infection, whilst KIR2DL2/L3 heterozygosity favours chronic inflammation (Figure 4). In some aspects this is similar to the model put forward by Hollenbach et al. to explain the observation of HLA-C1/C1, KIR2DL2/L3 heterozygote risk in Crohn's disease [84], which like trachoma is characterised by chronic inflammation and fibrotic immunopathology. It is possible that the high burden of trachomatous scarring, TT and infection related infertility in observed in Africa can be explained in part by unusually high frequencies of HLA-C2 and KIR2DL2/L3 heterozygosity and the effects of NK cell responsiveness. The therapeutic consequences of such a theory would impact on vaccine immune-therapies and we would expect that current efforts in the development of chlamydial vaccines, adjuvants and immunisation schedules would additionally monitor the boosting or modulating effects on the NK cell compartment. As early as the 1990 it was shown that vaccination with influenza virus was able to elicit NK cell responses [85]. More recent work has demonstrated that many vaccination regimes against viruses boost not only adaptive T and B cell response but also lead to repetitive expansions of NK cells [86]–[88]. Some immunologists have termed these “memory-like NK cell responses” and have now begun to consider the role of these responses in vaccine induced immunity [89]. The effectiveness of NK cells as targets of vaccine immuno-therapy has been described in oncology [90]. Efforts are now required to investigate the role of NK cells in immunity following vaccination with a wider spectrum of bacterial vectors and in natural immunity to infectious diseases such as trachoma.
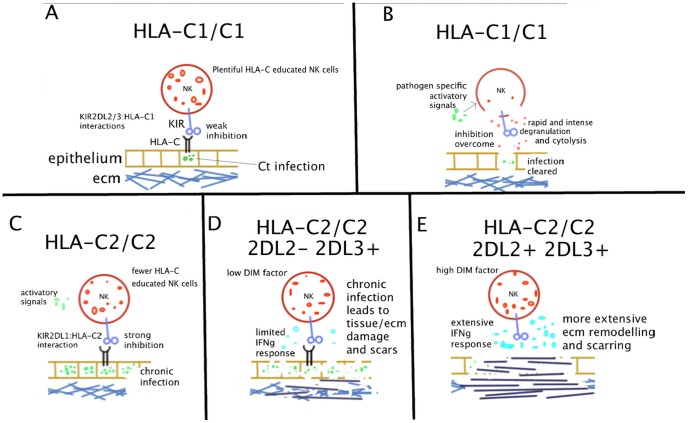
Figure 4. Model of NK cell mediated scarring in trachoma.
(A) An HLA-C1/C1 NK cell interacts with HLA class I molecules on a Ct infected epithelial cell via interactions between KIR2DL2/L3 and HLA-C1. HLA-C educated NK cells represent a greater proportion of the total NK repertoire in HLA-C1 homozygotes, compared to those who carry HLA-C2 (B) The inhibitory signals are overcome by loss of the inhibitory HLA-C molecule and/or by activatory signals, which may be from intracellular or extracellular pathogen derived moieties. The NK cytotoxic response is triggered and the target cell is lysed, which culminates in effective resolution of the infection. (C) An HLA-C2/C2 NK cell interacts with MHC class I molecules on Ct infected epithelial cell via interactions between KIR2DL1 and HLA-C2. A smaller proportion of the NK cell repertoire is HLA-C educated in this setting. Cytotoxic and IFNγ responses are less likely to be triggered and responses are less intense than in HLA-C1/C1 individuals. Chronic infection is established (D) KIR2DL2 −, KIR2DL3 + individuals have a low DIM factor and NK cell release of IFNγ is more limited. Chronic infection leads to some damage to epithelium and extracellular matrix (ECM) but anti-fibrotic properties of IFNγ are maintained. (E) KIR2DL2/KIR2DL3 heterozygous individuals have a high DIM factor and NK cells release larger quantities of IFNγ. Chronic infection is coupled with pathologically high levels of IFNγ. HLA-C1/C2 genotypes confer intermediate risks that do not significantly differ from the homozygous individuals. Relative risk of scarring was increased in KIR2DL2 homozygotes, but this was not significant.
Supporting Information
PBAT estimates of power in this study to detect genetic associations with trachomatous scarring at a range of allele frequencies and effect magnitudes.
(TIF)
Evidence for (a) complete [D′ = 1] but (b) not perfect [R2 = 1] linkage disequilibrium between pairs of KIR genes. LD was insufficiently strong to be used to reconstruct missing genotype data in the family study. In (a) dark grey indicates strong evidence of linkage, light grey is uninformative and white indicates strong evidence of recombination. D′ values below 1 are shown. In (b) white indicates R2 = 0, shades of grey indicate 0<R2<1 and black indicates R2 = 1. R squared values below 1 are shown. 2DS4d22 indicates alleles of KIR2DS4 carrying a 22 bp deletion. 3DP1V indicates alleles of KIR3DP1 carrying exon 2.
(TIF)
HLA Allele strings used in FBAT and associated frequencies. For the purposes of grouping for FBAT and in order to maximise statistical power, individual HLA genotypes were assigned to the ‘identifier’ groups if they possessed any or all of the alleles shown in the full genotype string.
(DOCX)
Acknowledgments
The authors wish to thank Omar Camera and Omar Ceesay for their work in the field. Dr. Elizabeth Trachtenberg and Dr. Kathleen Houtchens for confirmatory genotyping. Prof. Mary Carrington, Prof. Eleanor Riley, Dr. Paul Norman, Dr. Jill Hollenbach, Dr. Taane Clark, Dr. James Traherne, Dr. Becca Asquith and Marina Collins for technical advice, discussions and critical reading. Thanks also to the members of the Partnership for the Rapid Elimination of Trachoma (PRET) team, MRC laboratories The Gambia and to the families and individuals who consented to participate in this study.
Funding Statement
This study was funded by grants from the Wellcome Trust (079246/Z/06/Z and GR079246MA). The funders had no part in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication.
References
- 1. Abdelrahman YM, Belland RJ (2005) The chlamydial developmental cycle. FEMS Microbiol Rev 29: 949–959 doi:10.1016/j.femsre.2005.03.002 [DOI] [PubMed] [Google Scholar]
- 2. Rowley J, Toskin I, Ndowa F (2008) Global Incidence and Prevalence of Selected Curable Sexually Transmitted Infections, 2008. World Health Organisation [Google Scholar]
- 3.Resnikoff S, Pascolini D, Etya D, Kocur I, Pararajasegaram R, et al. (2004) Policy and Practice Global data on visual impairment in the year 2002. 012831. [PMC free article] [PubMed]
- 4. Harding-Esch EM, Edwards T, Mkocha H, Munoz B, Holland MJ, et al. (2010) Trachoma prevalence and associated risk factors in the gambia and Tanzania: baseline results of a cluster randomised controlled trial. PLoS Negl Trop Dis 4: e861 doi:10.1371/journal.pntd.0000861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hu VH, Holland MJ, Burton MJ (2013) Trachoma: protective and pathogenic ocular immune responses to Chlamydia trachomatis. PLoS Negl Trop Dis 7: e2020 doi:10.1371/journal.pntd.0002020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hu VH, Weiss HA, Ramadhani AM, Tolbert SB, Massae P, et al. (2012) Innate immune responses and modified extracellular matrix regulation characterize bacterial infection and cellular/connective tissue changes in scarring trachoma. Infect Immun 80: 121–130 doi:10.1128/IAI.05965-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Natividad A, Freeman TC, Jeffries D, Burton MJ, Mabey DCW, et al. (2010) Human conjunctival transcriptome analysis reveals the prominence of innate defense in Chlamydia trachomatis infection. Infect Immun 78: 4895–4911 doi:10.1128/IAI.00844-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gurujeyalakshmi G, Giri SN (1995) Molecular mechanisms of antifibrotic effect of interferon gamma in bleomycin-mouse model of lung fibrosis: downregulation of TGF-beta and procollagen I and III gene expression. Exp Lung Res 21: 791–808 doi:10.3109/01902149509050842 [DOI] [PubMed] [Google Scholar]
- 9. Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, et al. (2009) A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature 457: 722–725 doi:10.1038/nature07537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nagarajan UM, Sikes J, Prantner D, Andrews CW, Frazer L, et al. (2011) MyD88 deficiency leads to decreased NK cell gamma interferon production and T cell recruitment during Chlamydia muridarum genital tract infection, but a predominant Th1 response and enhanced monocytic inflammation are associated with infection resolution. Infect Immun 79: 486–498 doi:10.1128/IAI.00843-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tseng CT, Rank RG (1998) Role of NK cells in early host response to chlamydial genital infection. Infect Immun 66: 5867–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gall A, Horowitz A, Joof H, Natividad A, Tetteh K, et al. (2011) Systemic effector and regulatory immune responses to chlamydial antigens in trachomatous trichiasis. Front Microbiol 2: 10 doi:10.3389/fmicb.2011.00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Holland MJ, Bailey RL, Conway DJ, Culley F, Miranpuri G, et al. (1996) T helper type-1 (Th1)/Th2 profiles of peripheral blood mononuclear cells (PBMC); responses to antigens of Chlamydia trachomatis in subjects with severe trachomatous scarring. Clin Exp Immunol 105: 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ibana JA, Aiyar A, Quayle AJ, Schust DJ (2012) Modulation of MICA on the surface of Chlamydia trachomatis-infected endocervical epithelial cells promotes NK cell-mediated killing. FEMS Immunol Med Microbiol 65: 32–42 doi:10.1111/j.1574-695X.2012.00930.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, et al. (1997) Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity 7: 739–751. [DOI] [PubMed] [Google Scholar]
- 16. Natividad A, Wilson J, Koch O, Holland MJ, Rockett K, et al. (2005) Risk of trachomatous scarring and trichiasis in Gambians varies with SNP haplotypes at the interferon-gamma and interleukin-10 loci. Genes Immun 6: 332–340 doi:10.1038/sj.gene.6364182 [DOI] [PubMed] [Google Scholar]
- 17. Natividad A, Holland MJ, Rockett KA, Forton J, Faal N, et al. (2008) Susceptibility to sequelae of human ocular chlamydial infection associated with allelic variation in IL10 cis-regulation. October 323–329 doi:10.1093/hmg/ddm310 [DOI] [PubMed] [Google Scholar]
- 18. Natividad a, Hanchard N, Holland MJ, Mahdi OSM, Diakite M, et al. (2007) Genetic variation at the TNF locus and the risk of severe sequelae of ocular Chlamydia trachomatis infection in Gambians. Genes Immun 8: 288–295 doi:10.1038/sj.gene.6364384 [DOI] [PubMed] [Google Scholar]
- 19. Lu Z, Zhang B, Chen S, Gai Z, Feng Z, et al. (2008) Association of KIR genotypes and haplotypes with susceptibility to chronic hepatitis B virus infection in Chinese Han population. Cell Mol Immunol 5: 457–463 doi:10.1038/cmi.2008.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, et al. (2004) HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science 305: 872–874 doi:10.1126/science.1097670 [DOI] [PubMed] [Google Scholar]
- 21. Flores-Villanueva PO, Yunis EJ, Delgado JC, Vittinghoff E, Buchbinder S, et al. (2001) Control of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proc Natl Acad Sci U S A 98: 5140–5145 doi:10.1073/pnas.071548198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martin MP, Qi Y, Gao X, Yamada E, Martin JN, et al. (2007) Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet 39: 733–740 doi:10.1038/ng2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martin MP, Gao X, Lee J-H, Nelson GW, Detels R, et al. (2002) Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet 31: 429–434 doi:10.1038/ng934 [DOI] [PubMed] [Google Scholar]
- 24. Moretta A, Vitale M, Bottino C, Orengo AM, Morelli L, et al. (1993) P58 molecules as putative receptors for major histocompatibility complex (MHC) class I molecules in human natural killer (NK) cells. Anti-p58 antibodies reconstitute lysis of MHC class I-protected cells in NK clones displaying different specificities. J Exp Med 178: 597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Döhring C, Colonna M, Dohring C (1996) Human natural killer cell inhibitory receptors bind to HLA class I molecules. Eur J Immunol 26: 365–369 doi:10.1002/eji.1830260215 [DOI] [PubMed] [Google Scholar]
- 26. Pende D, Biassoni R, Cantoni C, Verdiani S, Falco M, et al. (1996) The natural killer cell receptor specific for HLA-A allotypes: a novel member of the p58/p70 family of inhibitory receptors that is characterized by three immunoglobulin-like domains and is expressed as a 140-kD disulphide-linked dimer. J Exp Med 184: 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Colonna M, Samaridis J (1995) Cloning of Immunoglobulin-Superfamily Members Associated with HLA-C and HLA-B Recognition by Human Natural Killer Cells. Science (80-) 268: 405–408. [DOI] [PubMed] [Google Scholar]
- 28. Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P (1995) The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med 181: 1133–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N (1998) Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol 161: 571–577. [PubMed] [Google Scholar]
- 30. Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL (1993) HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc Natl Acad Sci U S A 90: 12000–12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Colonna M, Brooks EG, Falco M, Ferrara GB, Strominger JL (1993) Generation of allospecific natural killer cells by stimulation across a polymorphism of HLA-C. Science (80-) 260: 1121–1124. [DOI] [PubMed] [Google Scholar]
- 32. Ciccone E, Pende D, Vitale M, Nanni L, Di Donato C, et al. (1994) Self class I molecules protect normal cells from lysis mediated by autologous natural killer cells. Eur J Immunol 24: 1003–1006. [DOI] [PubMed] [Google Scholar]
- 33. Stewart CA, Laugier-Anfossi F, Vely F, Saulquin X, Riedmuller J, et al. (2005) Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci U S A 102: 13224–13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, et al. (2008) Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol 180: 3969–3979. [DOI] [PubMed] [Google Scholar]
- 35. Uhrberg M, Parham P, Wernet P (2002) Definition of gene content for nine common group B haplotypes of the Caucasoid population: KIR haplotypes contain between seven and eleven KIR genes. Immunogenetics 54: 221–229 doi:10.1007/s00251-002-0463-7 [DOI] [PubMed] [Google Scholar]
- 36. Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, et al. (1997) Human diversity in killer cell inhibitory receptor genes. Immunity 7: 753–763. [DOI] [PubMed] [Google Scholar]
- 37. Wilson MJ, Torkar M, Haude A, Milne S, Jones T, et al. (2000) Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A 97: 4778–4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Norman PJ, Hollenbach JA, Nemat-Gorgani N, Guethlein LA, Hilton HG, et al. (2013) Co-evolution of human leukocyte antigen (HLA) class I ligands with killer-cell immunoglobulin-like receptors (KIR) in a genetically diverse population of sub-Saharan Africans. PLoS Genet 9: e1003938 doi:10.1371/journal.pgen.1003938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moretta A, Sivori S, Vitale M, Pende D, Morelli L, et al. (1995) Existence of both inhibitory (p58) and activatory (p50) receptors for HLA-C molecules in human natural killer cells. J Exp Med 182: 875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Khakoo SI, Thio CL, Martin MP, Collin R, Gao X, et al. (2008) HLA and NK Cell Inhibitory Receptor Genes in Resolving Hepatitis C Virus Infection. Science (80-) 872 doi:10.1126/science.1097670 [DOI] [PubMed] [Google Scholar]
- 41. Bari R, Bell T, Leung W-H, Vong QP, Chan WK, et al. (2009) Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine 245. Blood 114: 5182–5190 doi:10.1182/blood-2009-07-231977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ahlenstiel G, Martin MP, Gao X, Carrington M, Rehermann B (2008) Distinct KIR/HLA compound genotypes affect the kinetics of human antiviral natural killer cell responses. J Clin Invest 118: 1017–1026 doi:10.1172/JCI32400 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hsu KC, Chida S, Geraghty DE, Dupont B (2002) The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol Rev 190: 40–52. [DOI] [PubMed] [Google Scholar]
- 44. Gendzekhadze K, Norman PJ, Abi-Rached L, Layrisse Z, Parham P (2006) High KIR diversity in Amerindians is maintained using few gene-content haplotypes. Immunogenetics 58: 474–480 doi:10.1007/s00251-006-0108-3 [DOI] [PubMed] [Google Scholar]
- 45. Gendzekhadze K, Norman PJ, Abi-Rached L, Graef T, Moesta AK, et al. (2009) Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc Natl Acad Sci U S A 106: 18692–18697 doi:10.1073/pnas.0906051106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Parham P, Moffett A (2013) Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat Rev Immunol 13: 133–144 doi:10.1038/nri3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, et al. (2007) Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet 39: 1092–1099 doi:10.1038/ng2111 [DOI] [PubMed] [Google Scholar]
- 48. Saftlas AF, Waldschmidt M, Logsden-sackett N, Triche E, Field E (2004) Optimizing buccal cell DNA yields in mothers and infants for human leukocyte antigen genotyping. Am J Epidemiol 160: 77–84 doi:10.1093/aje/kwh171 [DOI] [PubMed] [Google Scholar]
- 49. Dolin PJ, Faal H, Johnson GJ, Ajewole J, Mohamed a a, et al. (1998) Trachoma in the Gambia. Br J Ophthalmol 82: 930–933 doi:10.1136/bjo.82.8.930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Van Steen K, Lange C (2005) PBAT: a comprehensive software package for genome-wide association analysis of complex family-based studies. Hum Genomics 2: 67–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stare D, Harding-Esch E, Munoz B, Bailey R, Mabey D, et al. (2011) Design and baseline data of a randomized trial to evaluate coverage and frequency of mass treatment with azithromycin: the Partnership for Rapid Elimination of Trachoma (PRET) in Tanzania and The Gambia. Ophthalmic Epidemiol 18: 20–29 doi:10.3109/09286586.2010.545500 [DOI] [PubMed] [Google Scholar]
- 52. Derrick T, Roberts C h, Rajasekhar M, Burr SE, Joof H, et al. (2013) Conjunctival MicroRNA Expression in Inflammatory Trachomatous Scarring. PLoS Negl Trop Dis 7: e2117 doi:10.1371/journal.pntd.0002117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vilches C, Castano J, Gomez-Lozano N, Estefania E, Castan J, et al. (2007) Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens 70: 415–422 doi:10.1111/j.1399-0039.2007.00923.x [DOI] [PubMed] [Google Scholar]
- 54. Cano P, Klitz W, Mack SJ, Maiers M, Marsh SGE, et al. (2007) Common and well-documented HLA alleles: report of the Ad-Hoc committee of the american society for histocompatiblity and immunogenetics. Hum Immunol 68: 392–417. [DOI] [PubMed] [Google Scholar]
- 55. Horvath S, Xu X, Laird NM (2001) The family based association test method: strategies for studying general genotype–phenotype associations. Eur J Hum Genet 9: 301–306 doi:10.1038/sj.ejhg.5200625 [DOI] [PubMed] [Google Scholar]
- 56. Laird NM, Horvath S, Xu X (2000) Implementing a unified approach to family-based tests of association. Genet Epidemiol 19 Suppl 1: S36–42 doi:+;10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M [DOI] [PubMed] [Google Scholar]
- 57. Cordell HJ, Barratt BJ, Clayton DG (2004) Case/pseudocontrol analysis in genetic association studies: A unified framework for detection of genotype and haplotype associations, gene-gene and gene-environment interactions, and parent-of-origin effects. Genet Epidemiol 26: 167–185 doi:10.1002/gepi.10307 [DOI] [PubMed] [Google Scholar]
- 58. Besson C, Roetynck S, Williams F, Orsi L, Amiel C, et al. (2007) Association of killer cell immunoglobulin-like receptor genes with Hodgkin's lymphoma in a familial study. PLoS One 2: e406 doi:10.1371/journal.pone.0000406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Malik S, Abel L, Tooker H, Poon A, Simkin L, et al. (2005) Alleles of the NRAMP1 gene are risk factors for pediatric tuberculosis disease. Proc Natl Acad Sci U S A 102: 12183–12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Middleton D, Meenagh A, Gourraud PA (2007) KIR haplotype content at the allele level in 77 Northern Irish families. Immunogenetics 59: 145–158 doi:10.1007/s00251-006-0181-7 [DOI] [PubMed] [Google Scholar]
- 61. Martin MP, Single RM, Wilson MJ, Trowsdale J, Carrington M (2008) KIR haplotypes defined by segregation analysis in 59 Centre d'Etude Polymorphisme Humain (CEPH) families. Immunogenetics 60: 767–774 doi:10.1007/s00251-008-0334-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sanchez-Mazas A, Steiner Q-G, Grundschober C, Tiercy J-M (2000) The molecular determination of HLA-Cw alleles in the Mandenka (West Africa) reveals a close genetic relationship between Africans and Europeans. Tissue Antigens 56: 303–312 doi:10.1034/j.1399-0039.2000.560402.x [DOI] [PubMed] [Google Scholar]
- 63. Maiers M, Gragert L, Klitz W (2007) High-resolution HLA alleles and haplotypes in the United States population. Hum Immunol 68: 779–788 doi:10.1016/j.humimm.2007.04.005 [DOI] [PubMed] [Google Scholar]
- 64. Ibana JA, Schust DJ, Sugimoto J, Nagamatsu T, Greene SJ, et al. (2011) Chlamydia trachomatis immune evasion via downregulation of MHC class I surface expression involves direct and indirect mechanisms. Infect Dis Obstet Gynecol 2011: 420905 doi:10.1155/2011/420905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhong G, Liu L, Fan T, Fan P, Ji H (2000) Degradation of transcription factor RFX5 during the inhibition of both constitutive and interferon gamma-inducible major histocompatibility complex class I expression in chlamydia-infected cells. J Exp Med 191: 1525–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhong G, Fan P, Ji H, Dong F, Huang Y (2001) Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J Exp Med 193: 935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chen AL, Johnson KA, Lee JK, Sütterlin C, Tan M (2012) CPAF: a Chlamydial protease in search of an authentic substrate. PLoS Pathog 8: e1002842 doi:10.1371/journal.ppat.1002842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kägebein D, Gutjahr M, Große C, Vogel AB, Rödel J, et al. (2013) Chlamydia-infected epithelial cells and fibroblasts retain the ability to express surface-presented MHC class I molecules. Infect Immun 1–31 doi:10.1128/IAI.01473-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, et al. (2013) Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med 5: 208ra145 doi:10.1126/scitranslmed.3006702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Leavy O (2013) Natural killer cells: a virtual pick and mix. Nat Rev Immunol 13: 844–845 doi:10.1038/nri3566 [DOI] [PubMed] [Google Scholar]
- 71. Korbel DS, Norman PJ, Newman KC, Horowitz A, Gendzekhadze K, et al. (2009) Killer Ig-like receptor (KIR) genotype predicts the capacity of human KIR-positive CD56dim NK cells to respond to pathogen-associated signals. J Immunol 182: 6426–6434 doi:10.4049/jimmunol.0804224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hollenbach JA, Nocedal I, Ladner MB, Single RM, Trachtenberg EA (2012) Killer cell immunoglobulin-like receptor (KIR) gene content variation in the HGDP-CEPH populations. Immunogenetics 64: 719–737 doi:10.1007/s00251-012-0629-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Norman PJ, Carrington C V, Byng M, Maxwell LD, Curran MD, et al. (2002) Natural killer cell immunoglobulin-like receptor (KIR) locus profiles in African and South Asian populations. Genes Immun 3: 86–95. [DOI] [PubMed] [Google Scholar]
- 74. Qutob N, Balloux F, Raj T, Liu H, Marion de Procé S, et al. (2012) Signatures of historical demography and pathogen richness on MHC class I genes. Immunogenetics 64: 165–175 doi:10.1007/s00251-011-0576-y [DOI] [PubMed] [Google Scholar]
- 75. Cates W, Farley T, Rowe P (1985) Worldwide Patterns Of Infertility: Is Africa Different? Lancet 5800: 596–598. [DOI] [PubMed] [Google Scholar]
- 76. Parham P, Norman PJ, Abi-Rached L, Guethlein LA (2012) Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules. Philos Trans R Soc Lond B Biol Sci 367: 800–811 doi:10.1098/rstb.2011.0266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Single RM, Martin MP, Gao X, Meyer D, Yeager M, et al. (2007) Global diversity and evidence for coevolution of KIR and HLA. Nat Genet 39: 1114–1119 doi:10.1038/ng2077 [DOI] [PubMed] [Google Scholar]
- 78. Yindom L-M, Forbes R, Aka P, Janha O, Jeffries D, et al. (2012) Killer-cell immunoglobulin-like receptors and malaria caused by Plasmodium falciparum in The Gambia. Tissue Antigens 79: 104–113 doi:10.1111/j.1399-0039.2011.01818.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hirayasu K, Ohashi J, Kashiwase K, Hananantachai H, Naka I, et al. (2012) Significant association of KIR2DL3-HLA-C1 combination with cerebral malaria and implications for co-evolution of KIR and HLA. PLoS Pathog 8: e1002565 doi:10.1371/journal.ppat.1002565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mendez A, Granda H, Meenagh A, Contreras S, Zavaleta R, et al. (2006) Study of KIR genes in tuberculosis patients. Tissue Antigens 68: 386–389 doi:TAN685 [pii] 10.1111/j.1399-0039.2006.00685.x [doi] [DOI] [PubMed] [Google Scholar]
- 81. Mahfouz R, Halas H, Hoteit R, Saadeh M, Shamseddeen W, et al. (2011) Study of KIR genes in Lebanese patients with tuberculosis. Int J Tuberc Lung Dis 15: 1688–1691 doi:10.5588/ijtld.11.0138 [DOI] [PubMed] [Google Scholar]
- 82. Pydi SS, Sunder SR, Venkatasubramanian S, Kovvali S, Jonnalagada S, et al. (2013) Killer cell immunoglobulin like receptor gene association with tuberculosis. Hum Immunol 74: 85–92 doi:10.1016/j.humimm.2012.10.006 [DOI] [PubMed] [Google Scholar]
- 83. Lu C, Shen Y-J, Deng Y-F, Wang C-Y, Fan G, et al. (2012) Association of killer cell immunoglobulin-like receptors with pulmonary tuberculosis in Chinese Han. Genet Mol Res 11: 1370–1378 doi:10.4238/2012.May.15.7 [DOI] [PubMed] [Google Scholar]
- 84. Hollenbach Ja, Ladner MB, Saeteurn K, Taylor KD, Mei L, et al. (2009) Susceptibility to Crohn's disease is mediated by KIR2DL2/KIR2DL3 heterozygosity and the HLA-C ligand. Immunogenetics 61: 663–671 doi:10.1007/s00251-009-0396-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Schapiro JM, Segev Y, Rannon L, Alkan M, Rager-Zisman B (1990) Natural killer (NK) cell response after vaccination of volunteers with killed influenza vaccine. J Med Virol 30: 196–200. [DOI] [PubMed] [Google Scholar]
- 86. Horowitz A, Hafalla JCR, King E, Lusingu J, Dekker D, et al. (2012) Antigen-specific IL-2 secretion correlates with NK cell responses after immunization of Tanzanian children with the RTS,S/AS01 malaria vaccine. J Immunol 188: 5054–5062 doi:10.4049/jimmunol.1102710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Horowitz A, Behrens RH, Okell L, Fooks AR, Riley EM (2010) NK cells as effectors of acquired immune responses: effector CD4+ T cell-dependent activation of NK cells following vaccination. J Immunol 185: 2808–2818 doi:10.4049/jimmunol.1000844 [DOI] [PubMed] [Google Scholar]
- 88. Hall LJ, Clare S, Dougan G (2010) NK cells influence both innate and adaptive immune responses after mucosal immunization with antigen and mucosal adjuvant. J Immunol 184: 4327–4337 doi:10.4049/jimmunol.0903357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Rölle A, Pollmann J, Cerwenka A (2013) Memory of infections: an emerging role for natural killer cells. PLoS Pathog 9: e1003548 doi:10.1371/journal.ppat.1003548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Woo CY, Clay TM, Lyerly HK, Morse MA, Osada T (2006) Role of natural killer cell function in dendritic cell-based vaccines. Expert Rev Vaccines 5: 55–65 doi:10.1586/14760584.5.1.55 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PBAT estimates of power in this study to detect genetic associations with trachomatous scarring at a range of allele frequencies and effect magnitudes.
(TIF)
Evidence for (a) complete [D′ = 1] but (b) not perfect [R2 = 1] linkage disequilibrium between pairs of KIR genes. LD was insufficiently strong to be used to reconstruct missing genotype data in the family study. In (a) dark grey indicates strong evidence of linkage, light grey is uninformative and white indicates strong evidence of recombination. D′ values below 1 are shown. In (b) white indicates R2 = 0, shades of grey indicate 0<R2<1 and black indicates R2 = 1. R squared values below 1 are shown. 2DS4d22 indicates alleles of KIR2DS4 carrying a 22 bp deletion. 3DP1V indicates alleles of KIR3DP1 carrying exon 2.
(TIF)
HLA Allele strings used in FBAT and associated frequencies. For the purposes of grouping for FBAT and in order to maximise statistical power, individual HLA genotypes were assigned to the ‘identifier’ groups if they possessed any or all of the alleles shown in the full genotype string.
(DOCX)