Abstract
Bartonella bacilliformis is the etiologic agent of Carrion's disease. This disease has two well established phases, the most relevant being the so called Oroya Fever, in which B. bacilliformis infect the erythrocytes resulting in severe anemia and transient immunosuppression, with a high lethality in the absence of adequate antibiotic treatment. The presence of B. bacilliformis was studied in 113 blood samples suspected of Carrion’s disease based on clinical criteria, despite the absence of a positive thin blood smear, by two different PCR techniques (using Bartonella-specific and universal 16S rRNA gene primers), and by bacterial culture. The specific 16S rRNA gene primers revealed the presence of 21 B. bacilliformis and 1 Bartonella elizabethae, while universal primers showed both the presence of 3 coinfections in which a concomitant pathogen was detected plus Bartonella, in addition to the presence of infections by other microorganisms such as Agrobacterium or Bacillus firmus. These data support the need to implement molecular tools to diagnose Carrion’s disease.
Introduction
The genus Bartonella includes at least 24 different species of bacteria. Currently, at least 9 of these microorganisms have been associated with infectious diseases in humans and some, such as Bartonella bacilliformis, Bartonella quintana or Bartonella henselae have been fully identified as causative agents of disease [1], [2].
B. bacilliformis are Gram-negative cocco bacilli which infects red blood cells and endothelial cells, being the etiologic agent of Bartonellosis also known as Carrion's disease, which is a biphasic disorder. Thus, two well established phases have been described in this disease, the first being the acute phase, the so-called Oroya Fever, in which B. bacilliformis microorganisms infect the erythrocytes resulting in severe anemia and transient immunosuppression [3], [4] which may result in death in the absence or delay of adequate treatment. The second or chronic phase, also named ‘Verruga Peruana’ or ‘Peruvian warts’, is characterized by the development of nodular dermal eruptions [5]. Asymptomatic carriers have also been described in endemic areas.
Although B. bacilliformis can be cultured from blood samples, including those stored at 4°C over a long period of time [6] or biopsies of eruptive or subcutaneous nodules [7], the growth of these bacteria has been shown to be difficult and slow, requiring at least 2–6 weeks of culture.
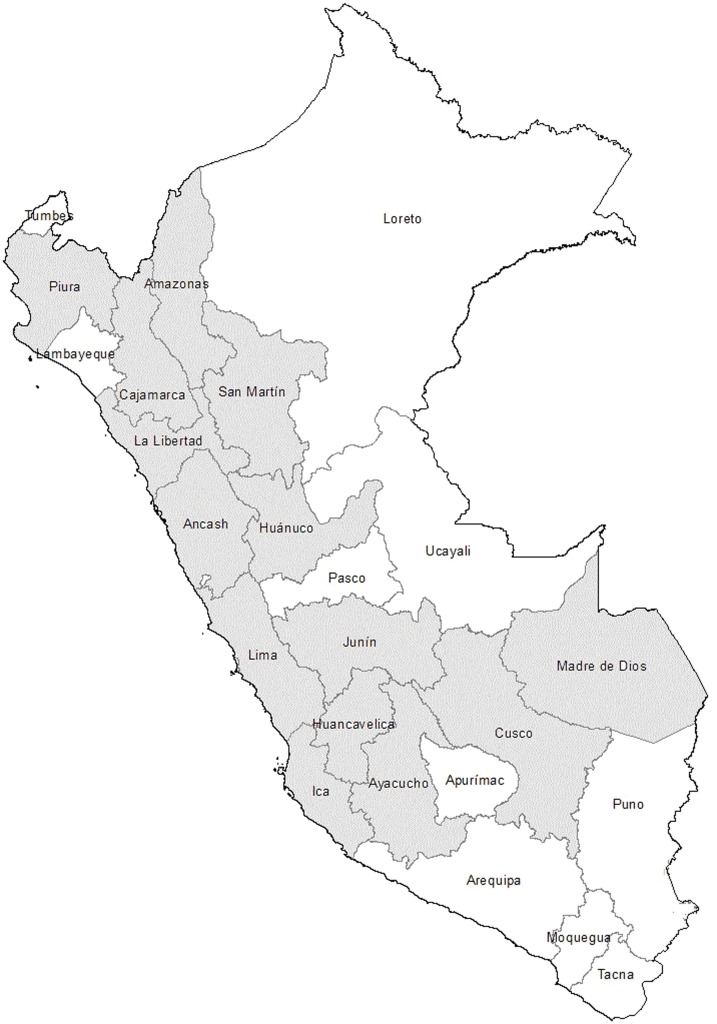
This pathogen is transmitted by the bite of members of the genus Lutzomyia including L. verrucarum, L. peruensis and L. pescei [8], which are distributed on the Western side of the Cordillera of the Andes, affecting Ecuador, Colombia and Peru, and is also sporadically reported in Bolivia and Chile [8] at 800 to 3200 meters above sea level [3], [9], [10]. Carrion’s disease is endemic and widespread in Peru, being present in at least 14 of the 24 departments of the country: Piura, Cajamarca, Amazonas, San Martin, La Libertad, Ancash, Lima, Huancavelica, Huánuco, Ica, Junín, Ayacucho, Madre de Dios and Cuzco; with the department of Ancash, being the most important endemic area (Figure 1) [1], [7], [11]. At present, human Bartonellosis is considered as an emerging/reemerging disease which is influenced by climate changes caused by the “Niño phenomenon” [7], [12].
Figure 1. Geographical distribution of Carrion’s disease in Peru.
*Carrion’s disease has also been sporadically described in other departments such as some specific areas of Loreto.
In endemic areas the diagnosis of the acute phase of the disease is currently mainly based on clinical symptoms and by the detection of the bacteria in peripheral blood smears, which is a useful, extensively used technique in health centers, especially in remote areas. Nonetheless, this method is “expertise-dependent” having a low sensitivity ranging from 24–36% [8]. Despite having a high specificity, the microbiological culture has a very low sensitivity because of the aforementioned difficulty to culture this microorganism. Moreover, the clinical utility of this method is limited because of the slow growth of these bacteria [7]. In this context, the development of various new molecular biology techniques for rapid diagnosis of this pathogen is of crucial relevance [8].
The aim of this study was therefore to develop and implement a PCR method as a rapid diagnostic tool for the detection of Bartonella spp. in blood samples, by analyzing a series of clinically suspected B. bacilliformis infections with a negative thin blood smear.
Methods
Patients and sampling
One hundred thirty-six venous blood samples of patients submitted to the Regional Laboratory of Cajamarca – Peru from January 2010 to December 2011 with clinical suspicion of Carrion’s disease but with a negative B. bacilliformis thin blood smear, were sent to the Instituto de Investigaciones Nutricionales (IIN, Lima, Peru) for diagnosis by molecular tools. The results obtained were reported to physicians for clinical management of the patients.
Relevant medical information of the patients was recorded on a form developed by the Instituto Nacional de Salud (INS) - Peru. Venous blood was collected in tubes containing EDTA and citrate and sent to the IIN facilities where they were stored at 4°C until processing.
Samples were collected for diagnosis purposes following an explanation to each of the participants by the physician responsible for the health center, in all cases, an informed consent for testing was obtained from patients, samples were coded to preserve the anonymate of the patients, and results were timely reported to local physicians.
Molecular diagnostic analysis of these samples was carried out as part of febrile syndromic surveillance of the Cajamarca department, conducted by the Regional Directorate of Health (DIRESA-Cajamarca) which is the regulatory center under a collaborative agreement with the IIN to improve the diagnostic of the Carrion Disease.
The Cajamarca Regional Directorate has no ethics committee. thus, despite the encoding and the diagnostic nature of the samples, and them exempt to be approved by an Human Subjects Committe, the studies were submitted, revised and approved by the Ethics and Research Committees of the Hospital Clinic of Barcelona.
Bacterial culture conditions
Microorganisms were cultured as previously described [6], [13] with slight modifications. Briefly, 2 ml of the blood sample were grown in Columbia agar medium supplemented with 10% sheep blood and incubated at 28°C for approximately 45 days under anaerobic conditions. The plates were visually inspected at 24, 48 and 72 hours to detect contamination, and then each 7 days for bacterial growth. Colonies were identified based on the characteristic morphology and Gram staining. Bartonella spp. were confirmed by amplification and sequencing of a 438 bp region of the 16S rRNA gene using 16S rRNA gene-specific primers for the genus Bartonella. When no amplification product was obtained, 16S rRNA gene-universal primers were used, and the product obtained was also sequenced. The conditions of both cases are described below.
All the bacteria isolated and included in the study are disposable for scientific non-commercial purposes.
DNA extraction
The DNA was extracted from 200 μl of blood samples using a commercial extraction kit (High Pure Kit Preparation template, Roche Applied Science, Mannheim, Germany). Bacterial DNA obtained after extraction was eluted in 100 μl of nuclease free water and then processed or stored at –20°C until use.
Amplification of a Bartonella spp.-specific 16S rRNA gene fragment
A 438 bp fragment of the 16S rRNA gene was amplified both in blood samples and in the microorganisms isolated adapting Bartonella spp. 16S rRNA gene-specific primers (Table 1) and PCR conditions designed by Garcia-Esteban et al. [14]. The final volume of the PCR mixture was 50 μl, distributed as follows: 25 μl of enzyme mix (Taq polymerase, 2.5 mM MgCl2, 15 mM Tris/HCl pH 8.3, 50 mM KCl, 200 μM of each deoxynucleotide), 20 pmol of each primer (Macrogen, Seoul, Korea), and 5 μl of DNA extraction. Thus, the PCR conditions were: 95°C for 10 min, followed by 45 cycles of 94°C for 1 min, 55°C for 1 min and 72°C for 1 min, with a final elongation of 10 min at 72°C. The amplified DNA products were analyzed by gel electrophoresis on 2% agarose (FMC, Rockland, ME) gel containing ethidium bromide (3 mg/L). Amplified products were gel recovered, purified (SpinPrep™ Gel DNA Kit, San Diego, USA) and sent to be sequenced (Macrogen, Seoul, Korea).
Table 1. The primers and conditions used in this study.
| Primers | Sequence | Size (bp) | Reference |
| P24Emod | CCTTCAGTTMGGCTGGATC | 438 | [14] |
| 16S-R | GCCYCCTTGCGGTTAGCACA† | ||
| 8F | AGAGTTTGATCCTGGCTCAG | 1503 | [15] |
| 1510R | GGTTACCTTTGTTACGACTT |
The primer described in reference 14 has three more bases (GCCYCCTTGCGGTTAGCACAGCA).
Amplification of 16S rRNA gene fragments using universal primers
This protocol was performed using the aforementioned PCR conditions and the universal primers described previously [15] and was applied in all blood samples as well as in microorganisms in which no positive PCR was obtained using Bartonella spp.-specific primers.
Data analysis
DNA sequences were analyzed with the BLAST analysis tool and compared with the GenBank database. Statistical significance was established using the Fisher exact test. Differences were considered as significant with a p-value <0.05.
Results
One hundred thirty-six blood samples from patients clinically diagnosed with Oroya Fever were collected, but those with no positive thin blood smear were included in the study. Of these, 23 were discarded for different reasons, including no collection of clinical data or problems with sample transportation, among others. Of the 113 samples included in the study nearly half (47%) belonged to people under 20 years of age, while no significant differences in sex were observed. PCR confirmed that B. bacilliformis infections were mainly detected in men (13 cases, 62%), while only 7 women (33%) were positive. Additionally, in one case data regarding sex was not specified. In positive cases the age ranged from 3 to 55 years, being mainly recovered from the age range from 26–35 years (35%), followed by age groups of 19–25 and <10 years (23% and 22%, respectively) (Table 2).
Table 2. Characteristics of the population studied.
| Age (years old) | Frequency | Percentage (%) | PCR-positive† |
| ≤ 10 | 18 | 15.93 | 4 (22.0%)†† |
| 11 – 18 | 29 | 25.66 | 4 (13.8%) |
| 19 – 25 | 13 | 11.50 | 3 (23.0%) |
| 26 – 35 | 20 | 17.70 | 7 (35.0%) |
| 36 –45 | 17 | 15.04 | 1 (5.8%) |
| 46 – 55 | 7 | 06.20 | 1 (14.3%) |
| ≥ 56 | 8 | 07.08 | 1 (12.5%) |
| No data | 1 | 00.88 | 1 (100%) |
| Area of Origin | |||
| Cajamarca | 109 | 96.46 | |
| Lima | 2 | 01.77 | |
| Ancash | 2 | 01.77 | |
| TOTAL | 113 | 100% | |
Positive samples using specific primers for Bartonella 16S rRNA.
Includes a B. elizabethae isolate.
The main clinical presentations reported by clinicians were general discomfort (57.66%), fever (52.25%) and headache (51.30%), in addition to other symptoms including myalgia and joint pain, among others (Table 3). Regarding the PCR-confirmed cases the main symptoms reported were headache (76.19%) followed by fever, chills and joint pain (66.70%). The use of the Fisher test showed the presence of significant differences among clinical symptoms between PCR confirmed and non confirmed Bartonella cases. Thus, loss of appetite, pollakiuria (p<0.0001), chills, joint pain, pallor, cough and ictericia (p<0.05) were significantly more frequent among PCR-confirmed Bartonella cases. Additionally, a trend towards significance (p = 0.066) was observed with respect to abdominal pain in patients with confirmed B. bacilliformis. No data regarding erythrocyte counts were available.
Table 3. Signs and symptoms.* .
| Total Patients (N: 113) | PCR-positive B. bacilliformis cases (N: 21) | PCR-negative cases (N: 92) | ||||
| Signs and symptoms | No | Percentage (%) | No | Percentage (%) | No | Percentage (%) |
| Discomfort | 64 | 57.66 | 12 | 57.14 | 52 | 56.52 |
| Fever | 58 | 52.25 | 14 | 66.70 | 44 | 47.83 |
| Headache | 57 | 51.30 | 16 | 76.19 | 41 | 44.57 |
| Chills | 55 | 49.54 | 14†† | 66.70 | 41†† | 44.57 |
| Myalgia | 50 | 44.70 | 12 | 57.14 | 38 | 41.30 |
| Joint pain | 48 | 43.24 | 14†† | 66.70 | 34†† | 36.96 |
| Pallor | 36 | 32.43 | 12 | 57.14 | 24 | 26.08 |
| Abdominal pain | 34 | 30.63 | 10 | 47.62 | 24 | 26.08 |
| Nausea | 30 | 27.02 | 6 | 28.57 | 24 | 26.08 |
| Dizziness | 24 | 21.61 | 2 | 09.52 | 12 | 13.04 |
| Cough | 24 | 21.61 | 9†† | 42.86 | 15†† | 16.30 |
| Vomiting | 21 | 18.90 | 3 | 14.29 | 18 | 19.56 |
| Diarrhea | 21 | 18.90 | 6 | 28.57 | 15 | 16.30 |
| Loss of appetite | 13 | 11.70 | 13† | 61.90 | 0† | 0 |
| Sore throat | 13 | 11.70 | 0 | 00.00 | 13 | 14.13 |
| Dyspnea | 13 | 11.70 | 3 | 14.29 | 10 | 10.87 |
| Pollakiuria | 10 | 09.00 | 10† | 47.62 | 0† | 0 |
| Ictericia | 10 | 09.00 | 5†† | 23.81 | 5†† | 5.43 |
| Warts | 7 | 06.30 | 1 | 04.76 | 6 | 6.52 |
| Dysuria | 7 | 06.30 | 2 | 09.52 | 5 | 5.43 |
| Others¥ | Less than1% | |||||
p<0.001††p<0.05 (Statistical significance established using the Fisher exact test).
* Symptoms considered on the basis of standard solicited data for the surveillance of Carrion’s disease.
¥Others: cyanosis, drowsiness, itching, back pain, chest pain, petechiae, congestion.
The samples were analyzed using two different techniques: bacterial culture and two PCR schemes. Thirteen samples (11.71%) were culture positive. Of these, 11 were identified as B. bacilliformis, one as Bartonella elizabethae and the remaining sample was identified to be Bacillus firmus. Twenty-two (19.46%) blood samples were positive on amplification of a specific fragment of the Bartonella 16S rRNA gene; of these 21 were B. bacilliformis (18.58%) and 1 B. elizabethae as confirmed when sought in GenBank as showing 100% of identity with sequences NR_074203.1 (B. bacilliformis) and L01260.1 (B. elizabethae). Regarding bacterial culture, amplification of a specific fragment of the Bartonella 16S rRNA gene showed a sensitivity of 100%, and a specificity of 100%. Finally, the use of universal 16S rRNA gene primers revealed the presence of 33 (29.20%) samples presenting bacterial microorganisms: B. bacilliformis (18 cases; 15.93%), Rhodoccus spp. (3 cases; 2.70%), B. firmus (3 cases; 2.70%) and others such as B.elizabethae, Achromobacter spp, Agrobacterium spp., Artrobacter spp., Bacilus cereus or Blastococcus spp., among others, accounting for one each (0.9%), respectively (Tables 4 and 5).
Table 4. Comparison of the techniques used to identify Bartonella spp.
| Microbiological culture N(%) | Direct blood PCR of 16S rRNA gene Bartonella spp. N(%) | Direct blood PCR of 16S rRNA gene Universal | |
| Positive | 13 (11.5) | 22 (19.5) * | 33 (29.2) † |
| Negative | 100 (88.5) | 91 (80.5) | 80 (70.8) |
| TOTAL | 113 (100%) | 113 (100%) | 113 (100%) |
* Twenty-one B. bacilliformis, 1 B. elizabethae.
Also include other microorganisms: Rhodoccus spp. (3 isolates) Bacillus firmus (3 isolates), Achromobacter xylosoxidans, Agrobacterium spp., Artrhobacter spp, Bacillus cereus, Blastococcus spp., Pseudomonas putida and Staphylococcus pettenkoferi.
Table 5. Information about culture and direct blood PCR in positive Samples.
| Direct Blood PCR | |||
| Sample | Culture* | 16S rRNA-Bartonella | 16S rRNA Universal |
| 1 | + | B.bacilliformis | B. bacilliformis |
| 2 | + | B.bacilliformis | B. bacilliformis |
| 3 | + | B.bacilliformis | B. bacilliformis |
| 8 | + | B.bacilliformis | B. bacilliformis |
| 11 | – | B.bacilliformis | Arthrobacter sp |
| 14 | + | B.bacilliformis | B. bacilliformis |
| 22 | + | B.bacilliformis | B. bacilliformis |
| 28 | + | B. elizabethae | A.xylosoxidans |
| 32 | + | B.bacilliformis | B. bacilliformis |
| 35 | + | B.bacilliformis | B. bacilliformis |
| 36 | + | B.bacilliformis | B. bacilliformis |
| 38 | – | B.bacilliformis | B. bacilliformis |
| 39 | – | B.bacilliformis | B. bacilliformis |
| 43 | – | B.bacilliformis | B. bacilliformis |
| 44 | + | B.bacilliformis | B. bacilliformis |
| 48 | – | B.bacilliformis | B. bacilliformis |
| 50 | – | – | Achromobacter spp. |
| 54 | + | B.bacilliformis | B. firmus |
| 56 | – | – | Rhodococcus spp |
| 59 | – | – | Rhodococcus spp |
| 61 | – | – | Bacillus cereus |
| 67 | – | – | Agrobacterium spp |
| 68 | – | – | Bacillus firmus |
| 76 | + | – | Bacillus firmus |
| 77 | – | – | Blastococcus spp. |
| 78 | – | – | S. pettenkoferi |
| 89 | – | – | Rhodococcus spp. |
| 100 | – | B.bacilliformis | B. bacilliformis |
| 103 | – | B.bacilliformis | B. bacilliformis |
| 124 | – | – | P. putida |
| 125 | – | B. bacilliformis | B. bacilliformis |
| 129 | – | B. bacilliformis | B. bacilliformis |
| 136 | – | B. bacilliformis | B. bacilliformis |
* All positive cultures but No. 76 were identified as B. bacilliformis or B. elizabethae (culture No. 28). Culture 76 was identified as Bacillus firmus, in accordance with direct blood PCR assays.
The analysis of concordance between the techniques used showed that in all cases in which Bartonella positive culture was obtained, both PCR schemes also detected its presence. Besides, in 9 cases the PCR techniques detected the presence of a Bartonella spp. in the absence of a positive culture. In all cases but three both PCR techniques detected B. bacilliformis in the same samples. In these three non-concordant cases B. bacilliformis was detected amplifying the specific 16S rRNA fragment, while another microorganism was identified using the 16S rRNA universal primers being interpreted as coinfections. Thus, analysis of the results showed the presence of B. bacilliformis plus B.firmus, B.bacilliformis plus Artrhobacter spp. and B. elizabethae plus Achromobacter xylososidans.
Discussion
The reporting of B. bacilliformis in Peru is based on the analysis of microbiological culture or thin blood smear. This last technique is the most frequently used in endemic areas, but it has a series of limitations including a low sensitivity. Laboratory confirmation by culture and isolation of the causative organism is generally considered to be the gold standard for the diagnosis of most bacterial infections. However, Bartonella spp. are fastidious and fragile organisms which are difficult to culture and require a period of growth of 15 to 45 days, depending on the species.
Thus, the use of other techniques for adequate disease identification is necessary [8], [14]. Some techniques have been proposed, including ELISA, immunoblotting, indirect fluorescence antibody tests and PCR [8], [14]. However, some of these techniques present a series of limitations including false positive results. It has been proposed the use of Real Time PCR in the diagnosis of infections due to different Bartonella species [16]–[18], however, as other aforementioned tests, this technique may be expensive and cumbersome for routine implementation in the main endemic areas, most of which are located in remote low-income remote regions of Peru.
On the other hand, PCR for the diagnosis of Bartonella spp. infections is not-cumbersome and is a sensitive, specific and rapid molecular technique, with a relatively low cost [8. 14]. Additionally, if combined with restriction analysis, may result in the exact specie identification [19]. Nonetheless the characteristics of PCR do not allow its implementation in low-income rural areas, but it does have potential for use in regional healthcare centers which may act as reference centers providing early results to local physicians. Therefore, PCR may be an alternative to improve the diagnosis of infection in these areas [20]. Indeed, the use of a specific PCR may result in better diagnosis of Bartonella infections, while the use of universal 16S rRNA gene primers may facilitate the detection of other bacterial pathogens causing febrile syndromes as well as coinfections between Bartonella and other microorganisms such as what occurred in the present report. Thus, the combined use of the present two PCR techniques may also have the advantage of detecting both B. Bacilliformis as well as other microorganisms present either as the main infective cause or as a concomitant opportunistic infection. In fact, the use of universal 16S rRNA gene primers has been explored in order to facilitate diagnosis of central nervous system infections [21].
A series of samples clinically suspected of having B. bacilliformis, but with no positive blood smear, were collected in the area of Cajamarca. In these samples the amplification of a specific fragment of the B. bacilliformis 16S rRNA gene showed the presence of this microorganism in 21 out of 113 samples, again showing both the strong limitations of the thin blood smear to detect B. bacilliformis and the need to implement more efficient techniques for achieving correct diagnoses in endemic areas. This result is in accordance with previous studies which have demonstrated that the PCR technique detects B. bacilliformis with a higher sensitivity than blood smear [22].
It is of note that the presence of B. elizabethae in a case of suspected Oroya Fever, suggesting the potential role of other members of the Bartonella genus to cause Oroya Fever-like illness, as has been previously described in the case of Bartonella rochalimae [23], and supporting the descriptions of patients with compatible symptoms in other distant areas, such as Thailand or Guatemala [8].
A series of other pathogens was also detected in blood samples using universal 16S rRNA primers, including one Agrobacterium spp., an environmental microorganism with a close phylogenetic relation to the Bartonella genus. This microorganism has been described as causing illness in immunocompromised patients [24]. Most of the remaining microorganisms detected are also considered to be opportunistic pathogens or have only been detected as causing infections in immunocompromised patients [25]–[28]. The presence of this kind of pathogen, together with the ability of B. bacilliformis to produce transient immunosuppression [4] which facilitates the presence of concomitant infections, may suggest the presence of undetected B. bacilliformis. Although no direct data on erythrocyte counts were reported, the clinical symptoms of the suspicious PCR-positive Bartonella cases were suggestive of a status of anemia. Patients with confirmed Bartonella present a significantly higher prevalence of paleness and ictericia. Regarding other symptoms a series of significant differences were also observed between PCR-positive Bartonella and PCR-negative Bartonella. These differences together with those in ictericia and pallor support the contention that these two populations represent two different infectious processes and therefore support the utility of the amplification of a specific fragment of the Bartonella 16S rRNA gene to achieve a more precise etiologic diagnosis. Further studies in a larger number of Carrion disease-positive cases are needed to confirm these clinical findings.
B. bacilliformis was detected in three samples by amplifying the specific fragment of the Bartonella 16S rRNA gene, while another microorganism was detected using the universal 16S rRNA primers. Although the possibility of contamination can not be ruled out, the lack of a negative control demonstrating a contamination band leads to a possible scenario of a coinfection. This differential amplification might be related to the specific amount of each pathogen in blood as well as to the intracellular nature of B. bacilliformis. Both of these factors may result in better amplification of one pathogen with respect another. Although DNA sequencing is not a feasible technique to be implemented in most endemic rural areas, the present data suggest that the use of both techniques may provide a better diagnosis.
In the present report, most of the cases of suspected Oroya Fever belonged to young patients. This fact may be related to the age-structure of the populations of the areas as well as the partial immunity developed after B. bacilliformis exposure which may provide a degree of protection against further infections by this microorganism as has been previously proposed [1].
In summary, the present data show the urgent need to introduce better diagnostic tools in the areas in which Carrion’s disease is endemic in order to improve the diagnosis and management of this illness and to avoid possible misdiagnoses.
Funding Statement
This work has been partially supported by Optimus Foundation and by personal funds of JdVM. JR is supported by the program I3, of the ISCIII (grant number: CES11/012) and by ISCIII grant PI11/00983. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Breña Chávez JP, Maguiña Vargas CP, Hernández Diaz HR, Castillo Díaz ME, Pozo Tovar WE (2006) Bartonelosis aguda en niños: estudio de 32 casos en el Instituto Especializado de Salud del Niño y el Hospital Nacional Cayetano Heredia (Período 1993–2003). Rev Med Hered 17: 122–131. [Google Scholar]
- 2. Eicher S, Dehio C (2012) Bartonella entry mechanisms into mammalian host cells. Cell Microbiol 14: 1166–1173. [DOI] [PubMed] [Google Scholar]
- 3. Ihler GM (1996) Bartonella bacilliformis: dangerous pathogen slowly emerging from deep background. FEMS Microbiol Lett 144: 1–11. [DOI] [PubMed] [Google Scholar]
- 4. Ticona E, Huaroto L, Garcia Y, Vargas L, Madariaga MG (2010) The pathophysiology of the acute phase of human bartonellosis resembles AIDS. Med Hypotheses 74: 45–49. [DOI] [PubMed] [Google Scholar]
- 5. Maguiña C, Gotuzzo E (2000) Bartonellosis - new and old. Infect Dis Clin North Am 14: 1–22. [DOI] [PubMed] [Google Scholar]
- 6. Ruiz J, Silva W, Pons MJ, del Valle LJ, Tinco CR, et al. (2012) Long time survival of Bartonella bacilliformis in blood stored at 4°C. Blood Transfus 10: 563–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pachas P (2000) Epidemiología de la Bartonelosis en el Perú. Serie de módulos técnicos. Lima (Peru): Oficina General de Epidemiología – Instituto Nacional de Salud
- 8. Sanchez Clemente N, Ugarte-Gil CA, Solórzano N, Maguiña C, Pachas P, et al. (2012) Bartonella bacilliformis: a systematic review of the literature to guide the research agenda for elimination PLoS Negl Trop Dis. 6: e1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Benson LA, Kar S, McLaughlin G, Ihler GM (1986) Entry of Bartonella bacilliformis into erythrocytes. . Infect Immun. 54: 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rolain JM, Fournier PE, Raoult D, Bonerandi JJ (2003) First isolation and detection by immunofluorescence assay of Bartonella koehlerae in erythrocytes from a French cat. J Clin Microbiol 41: 4001–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chamberlin J, Laughlin LW, Romero S, Solórzano N, Gordon S, et al. (2001) Epidemiology of endemic Bartonella bacilliformis: a prospective cohort study in a Peruvian mountain valley community. J Infect Dis 186: 983–990.. [DOI] [PubMed] [Google Scholar]
- 12. Chinga-Alayo E, Huarcaya E, Nasarre C, del Aguila R, Llanos-Cuentas A (2004) The influence of climate on the epidemiology of bartonellosis in Ancash, Peru. Trans R Soc Trop Med Hyg 98: 116–124. [DOI] [PubMed] [Google Scholar]
- 13. del Valle LJ, Flores L, Vargas M, García-de-la-Guarda R, Quispe RL, et al. (2010) Bartonella bacilliformis, endemic pathogen of the Andean region, is intrinsically resistant to quinolones. Int J Infect Dis 14: 506–510. [DOI] [PubMed] [Google Scholar]
- 14. García-Esteban C, Gil H, Rodríguez-Vargas M, Gerrikagoitia X, Barandika J, et al. (2008) Molecular method for Bartonella species identification in clinical and environmental samples. J Clin Microbiol 46: 776–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Salazar de Vegas EZ, Nieves B, Araque M, Velasco E, Ruiz J, et al. (2006) Outbreak caused by Acinetobacter strain RUH 1139 in an intensive care unit. Infect Control Hosp Epidemiol 27: 397–403. [DOI] [PubMed] [Google Scholar]
- 16. Liberto MC, Lamberti AG, Marascio N, Matera G, Quirino A, et al. (2011) Molecular identification of Bartonella quintana infection using species-specific real-time PCR targeting transcriptional regulatory protein (bqtR) gene. Mol Cell Probes 25: 238–242. [DOI] [PubMed] [Google Scholar]
- 17. Liberto MC, Matera G, Lamberti AG, Quirino A, Barreca GS, et al. (2013) Diagnosis and follow-up of Bartonella henselae infection in the spleen of an immunocompetent patient by real-time quantitative PCR. J Med Microbiol 62: 1081–1085. [DOI] [PubMed] [Google Scholar]
- 18. Smit PW, Peeling RW, Garcia PJ, Torres LL, Pérez-Lu JE, et al. (2013) Dried blood spots for qPCR diagnosis of acute Bartonella bacilliformis infection. . Am J Trop Med Hyg. 89: 988–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. del Valle LJ, Jaramillo ML, Talledo M, Pons MJ, Flores L, et al. (2014) Development of a 16S rRNA PCR-RFLP assay for Bartonella identification: . applicability in the identification of species involved in human infections Univers J Microbiol Res 2: 15–22. [Google Scholar]
- 20. Henriquez C, Infante B, Merello J, Gal’lino M, Santivañez L, et al. (2002) Identificación de Bartonella bacilliformis por métodos moleculares. Rev Med Hered 13: 58–63. [Google Scholar]
- 21. Lu J-J, Perng C-L, Lee S-Y, Wan C-C (2000) Use of PCR with universal primers and restriction endonuclease digestions for detection and identification of common bacterial pathogens in cerebrospinal fluid. J Clin Microbiol 38: 2076–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ellis BA, Rotz LD, Leake JA, Samalvides F, Bernable J, et al. (1999) An outbreak of acute bartonellosis (Oroya fever) in the Urubamba region of Peru, 1998. Am J Trop Med Hyg 61: 344–349. [DOI] [PubMed] [Google Scholar]
- 23. Eremeeva ME, Gerns HL, Lydy SL, Goo JS, Ryan ET, et al. (2007) Bacteremia, fever, and splenomegaly caused by a newly recognized Bartonella species. N Engl J Med 356: 2381–2387. [DOI] [PubMed] [Google Scholar]
- 24. Papithou NI, Rolston KV (2003) Catheter-related bacteremia caused by Agrobacterium radiobacter in a cancer patient: case report and literature review. Infection 31: 421–424. [DOI] [PubMed] [Google Scholar]
- 25. Drobniewski FA (1993) Bacillus cereus and related species. Clin Microbiol Rev 6: 324–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gray KJ, French N, Lugada E, Watera C, Gilks CF (2000) Rhodococcus equi and HIV-1 infection in Uganda. J Infect 41: 227–231. [DOI] [PubMed] [Google Scholar]
- 27. Song SH, Park JS, Kwon HR, Kim SH, Kim HB, et al. (2009) Human bloodstream infection caused by Staphylococcus pettenkoferi. J Med. Microbiol 58: 270–272. [DOI] [PubMed] [Google Scholar]
- 28. Tokuyasu H, Fukushima T, Nakazaki H, Shimuzu E (2012) Infective endocarditis caused by Achromobacter xylosoxidans: a case report and review of the literature. Intern Med 51: 1133–1138. [DOI] [PubMed] [Google Scholar]