Summary
Protein kinase R (PKR) is a component of the innate immunity antiviral pathway. PKR is activated upon binding to dsRNA to undergo dimerization and autophosphorylation. Adenovirus virus-associated RNA I (VAI) is a short, non-coding transcript whose major function is to inhibit the activity of PKR. VAI contains three domains: an apical stem-loop, a highly structured central domain, and a terminal stem. Previous studies have localized PKR binding to the apical stem and central domain. However, the molecular mechanism for inhibition of PKR is not known. We have characterized the stoichiometry and affinity of PKR binding to VAI and several domain constructs using analytical ultracentrifugation and correlated VAI binding and PKR inhibition. Although PKR binding to simple dsRNAs is not regulated by divalent ion, analysis of the interaction of the isolated dsRNA binding domain with VAI reveals that the binding affinity is enhanced by divalent ion. Dissection of VAI into its constituent domains indicates that none of the isolated domains retains the PKR binding affinity or inhibitory potency of the full length RNA. PKR is capable of binding the isolated terminal stem, but deletion of this domain from VAI does not affect PKR binding or inhibition. These results indicate that the apical stem and the central domain are both required to form a high affinity PKR binding site. Our data support a model whereby VAI functions as a PKR inhibitor because it binds a monomer tightly but does not facilitate dimerization.
Keywords: Analytical ultracentrifugation, innate immunity, protein-nucleic acid interactions, protein kinase, DMS probing
Introduction
Protein kinase R (PKR) is a component of the innate immune pathway and plays a critical role in the antiviral response in eukaryotes.1 PKR binds to and is activated by dsRNA, which is a byproduct of transcription in positive-strand RNA and DNA viruses.2 Activated PKR undergoes autophosphorylation and phosphorylates the α subunit of the translational initiation factor, eIF2, leading to an arrest of viral protein synthesis in the host cell. Many viruses have developed sophisticated strategies to inhibit PKR and thereby permit replication.3 In particular, Epstein-Barr virus and adenovirus each produce non-coding RNAs which act as RNA decoys and sequester PKR but do not activate, thereby allowing viral replication to proceed.
PKR contains two domains: an N-terminal dsRNA binding domain (dsRBD)* and a C-terminal protein kinase. These domains are connected by a long (~90 residue) unstructured linker. The dsRBD contains two tandem RNA binding motifs, dsRBM1 and dsRBM2, joined by a short (20 residue) linker. Both of these modules adopt an αβββα fold which is typically found in dsRNA binding motifs.4 The crystal structure of a dsRNA binding motif bound to dsRNA reveals that the protein spans about 16 bp and primarily interacts with the 2’-hydroxyls and the phosphate backbone of an A-form dsRNA helix.5 These observations explain why PKR binds to dsRNA in a sequence-independent manner. However, there is evidence for sequence specific dsRNA recognition by the dsRBMs from ADAR26 and Aquifex aeolicus RNase III7 and shape specific recognition of a tetraloop by yeast RNase III.8 Although the tandem dsRBMs from PKR show strong sequence and structural homology, dsRBM1 binds to dsRNA with higher affinity than dsRBM2.9 The presence of dsRBM2 enhances the binding affinity of dsRBM19,10 and interacts with RNA when PKR binds to longer dsRNA sequences.9–11
The crystal structure of the catalytic domain of PKR in complex with eIF2α reveals a bilobal structure that is typical of many protein kinases.12 Interestingly, the kinase domain crystallizes as dimer. Dimerization plays a key role in the mechanism of PKR activation12,13 and the structure suggests a possible pathway linking the dimer interface to the catalytic site. A minimum dsRNA length of 30–33 bp is required to activate PKR and to form a catalytically competent dimer. 14– 16
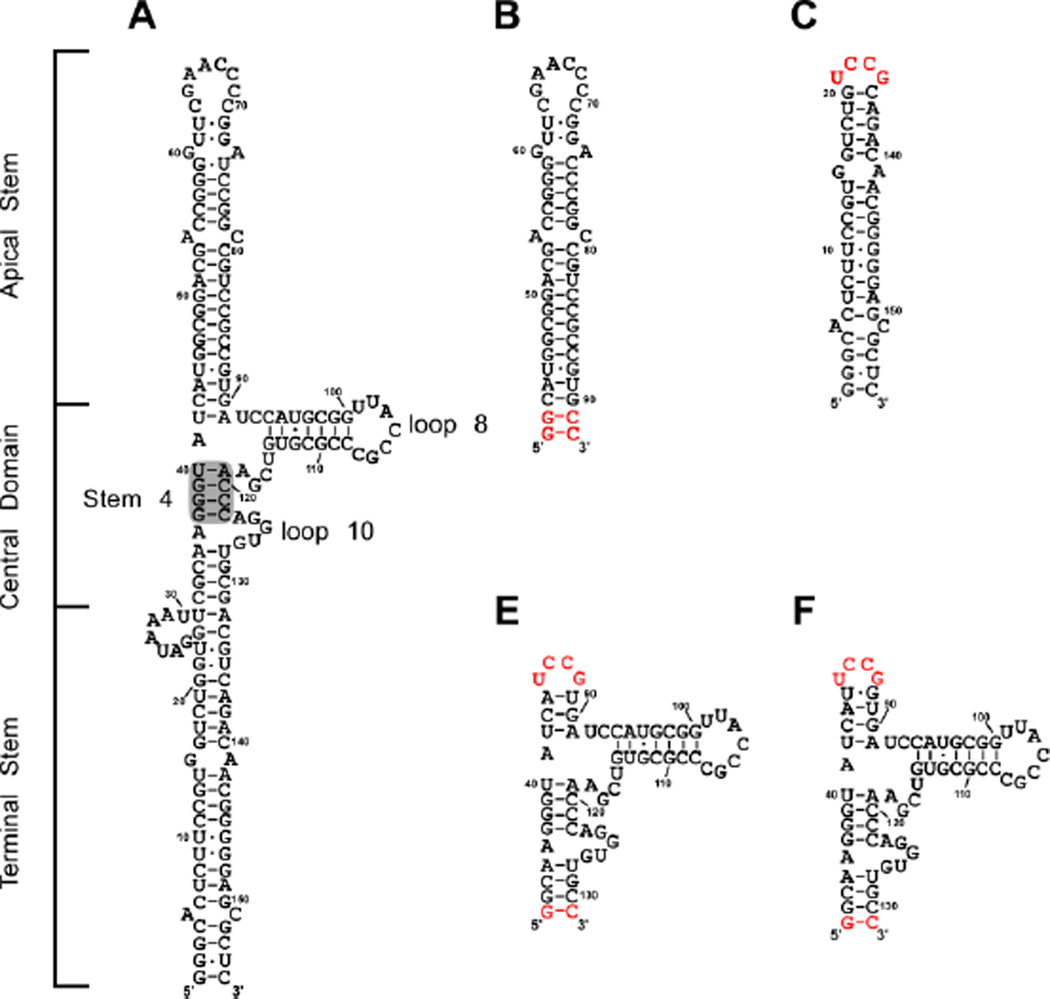
Adenovirus synthesizes two non-coding RNAs, adenovirus-associated RNA (VAI) and adenovirus-associated RNA II (VAII). VAI is produced at high (micromolar) levels in the host cell during late stages of infection and its primary function is inhibition of PKR.17–19 It has also been implicated in the suppression of the RNA interference pathway,20,21 the induction of interferon,22 and the regulation of 2',5'-oligoadenylate synthetase.23,24 VAI RNAs from different serotypes of adenovirus exhibit limited sequence homology and vary in length.25 However, extensive phylogenetic analysis26 and enzymatic and chemical probing experiments25,27,28 reveal a conserved secondary structure consisting of three distinct domains: an apical stem, a highly structured central domain, and a terminal stem (Figure 1A). The apical stem represents the primary PKR binding site28–31 and consists of a 20 basepair stem loop interrupted by two mismatches. Enzymatic structure probing indicates a minor population of VAI with slightly altered base pairing within the apical stem exists in equilibrium with the secondary structure depicted in Figure 1A.32 The terminal stem contains a shorter duplex with two mismatches and a seven base bulge adjacent to the central domain. This stem is thought to stabilize the central domain.25,27 Deletion of the terminal stem does not affect PKR binding or inhibition.27,33 The central domain has a complex secondary structure centered on a three way junction and represents a secondary binding site for PKR.28–31 This region is believed to play a role in PKR inhibition.28,34–38 The central domain contains a conserved tetranucleotide pair within stem 4 that is critical for PKR inhibition and may be involved in tertiary interactions.25,38,39 Other evidence for tertiary structure are protection of bases in loops 8 and 10 in enzymatic and chemical probing experiments39 and involvement of a protonated base in stabilizing the structure of the central domain.27
Figure 1.
Secondary structure of VAI and domain constructs. A) Secondary structure of full length VAI as proposed in reference 35. Stem 4, loop 8 and loop 10 are annotated as indicated in reference 30. Shaded region indicates conserved tetranucleotide pair. Secondary structures of B) apical stem (AS), C) terminal stem (TS), D) central domain-1 (CD1), E) central domain-2 (CD2). All domain constructs are numbered according to full length VAI. Bases in depicted in red denote those added for efficient in vitro transcription.
The detailed molecular mechanism for inhibition of PKR by VAI is not well understood. Early studies proposed that VAI inhibits PKR by binding a single monomer, thereby preventing dimerization on the RNA and subsequent activation.18,40 Although two PKR monomers sequentially bind to VAI in the absence of divalent ion, a single PKR binds to VAI in the presence of Mg2+, supporting the model whereby VAI functions as a inhibitor by binding a PKR monomer.33 Other studies reported that the isolated apical stem functions as a PKR activator and that other regions of VAI mediate inhibition.41 It has also been proposed that VAI inhibits by disrupting PKR dimerization.42 In the present study, we have refined our analysis of the contribution of divalent ion to PKR-VAI interactions and dissected the contributions of each region of VAI to PKR binding and inhibition. We find that divalent ion differentially affects PKR and dsRBD binding to VAI. In the presence of Mg2+, PKR binds weakly to the isolated domains of VAI. The apical stem and central domain cooperatively form a high affinity PKR binding site.
Results
Divalent ion modulates PKR binding to VAI RNA
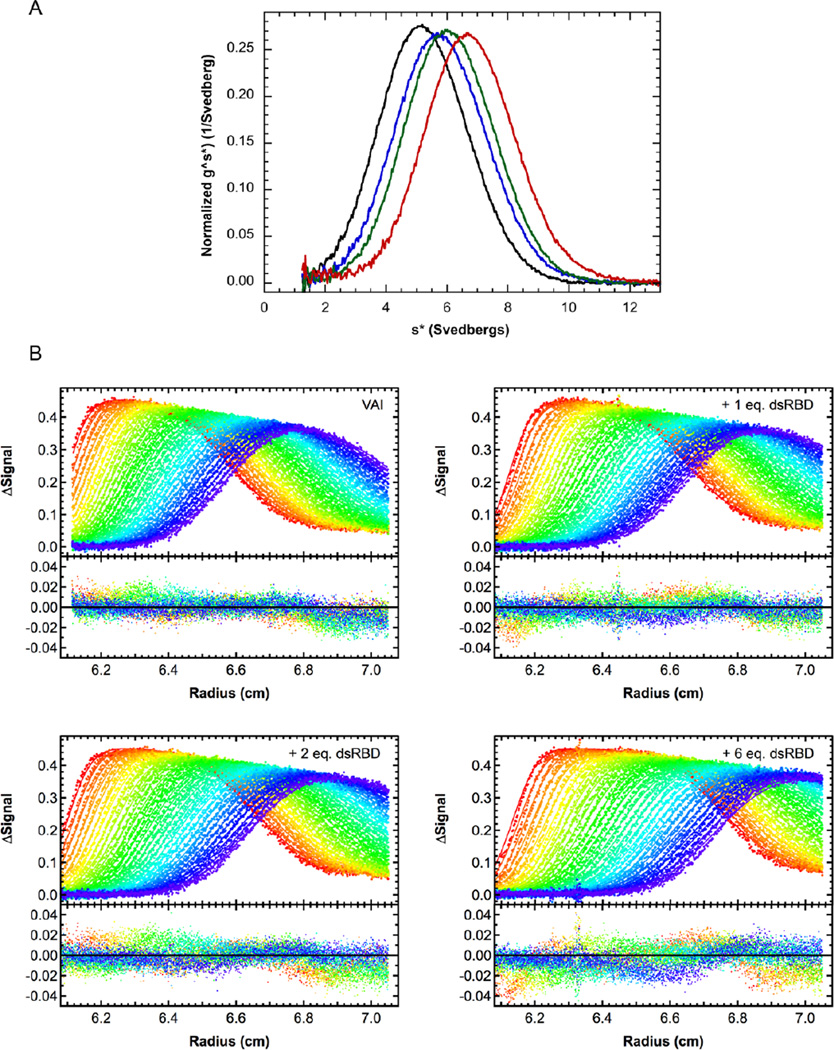
In order to further understand the strong effect of divalent ion on PKR-VAI interactions we have characterized binding of the PKR dsRBD to VAI as a function of Mg2+. Figure 2A shows an overlay of g(s*) distributions obtained by titrating VAI with increasing concentrations of dsRBD in the absence of divalent ion. The peak gradually shifts to higher S with increasing dsRBD concentration, consistent with weak binding of a single dsRBD. Indeed, in global analysis the combined data fit well to a 1:1 binding model with Kd = 950 nM (Figure 2B, Table 1). Thus, under similar conditions, dsRBD binds more weakly than full length PKR33 and with a reduced stoichiometry. The Kd for dsRBD binding to VAI is higher than previously reported41 due to the higher salt concentration in the present work. Sedimentation velocity measurements were repeated using dsRBM1 and dsRBM2 constructs, but the binding affinities were too low to obtain meaningful results (data not shown). To determine whether the presence of the kinase domain in PKR affects binding of a second monomer in the absence of Mg2+, we also investigated the interaction of the isolated kinase domain with VAI. However, this construct did not bind VAI under these conditions (data not shown).
Figure 2.
Sedimentation velocity analysis of dsRBD binding to VAI in the absence of Mg2+. A) Normalized g^(s*) distributions of 0.4 µM VAI (black), VAI + 1 eq. dsRBD (blue), VAI + 2 eq. dsRBD (green) and VAI + 6 eq. dsRBD (red). B) Global analysis of sedimentation velocity difference curves for dsRBD binding to VAI. The top panels show the data (points) and fit (solid lines) and the bottom panels show the residuals (points). The best fit parameters are shown in Table 1.
Table 1.
Protein - VAI RNA interaction parameters derived from sedimentation velocity analysis.
| Protein | [Mg2+] (mM) |
Binding Model |
Kd1 (nM) | Kd2 (nM) | s(RP)a | s(RP2)a | RMSb |
|---|---|---|---|---|---|---|---|
| PKRc | 0 | R + P ↔ RP RP + P ↔ RP2 |
14 (3,41) | 601 (359, 1200) | 6.78 (6.63, 7.04) | 8.54 (8.30, 9.00) | 0.00841 |
| PKRc | 5 | R + P ↔ RP | 334 (278, 401) | -- | 7.18 (7.07, 7.29) | -- | 0.00691 |
| dsRBD | 0 | R + P ↔ RP | 950 (811, 1137) | -- | 7.56 (7.42, 7.75) | -- | 0.00697 |
| dsRBD | 5 | R + P ↔ RP | 505 (412, 622) | -- | 6.64 (6.56, 6.74) | -- | 0.00656 |
Parameters were obtained by global nonlinear least square analysis of sedimentation velocity measurements. The values in parentheses represent the 95% joint confidence intervals obtained using the F-statistic.
Uncorrected sedimementation coefficient (Svedbergs).
Root mean square deviation of the fit in absorbance units.
Data were previously reported in reference 29.
In the presence of 5 mM Mg2+ the Kd for binding of dsRBD to VAI decreases about two-fold to 505 nM (Table 1), which is close the Kd we previously reported for full length PKR.33 This enhancement in binding affinity upon addition of divalent ion is unusual as the affinity of PKR for regular duplex RNA decreases about three fold in the presence of 5 mM Mg2+.33 The hydrodynamic parameters for the dsRBD-VAI complex are also affected by divalent ion. The corrected sedimentation coefficient (s20,w) of the complex decreases from 7.93 S to 6.98 S upon addition of 5 mM Mg2+, corresponding to an increase the frictional ratio (f/fo) from 1.21 to 1.38. Divalent ion is present under physiological conditions. Given the substantial effects of divalent ion on PKR-VAI interactions, subsequent analysis of PKR binding to VAI domain constructs was performed in the presence of 5 mM Mg2+.
Contribution of VAI domains to PKR binding
We synthesized VAI domain constructs to define the contribution of each region to PKR binding and inhibition (Figure 1). The AS construct contains the entire apical stem (nucleotides 43–90) with two additional G-C base pairs to promote T7 RNA polymerase transcription. The TS construct contains the duplex region of the terminal stem (nucleotides 1–20 and 136–155) with a highly stable UCCG tetraloop52 appended. Analysis of these constructs by denaturing and native gel electrophoresis and sedimentation velocity analysis indicates that they are homogeneous and monomeric (data not shown). The central domain has a complex secondary structure centered on a three-way junction. In order to stabilize the fold of the central domain in isolation, constructs were engineered to contain either three (CD1) or four (CD2) base pairs of the apical stem capped by a UCCG tetraloop. An additional G-C base pair was appended to the terminal region to enhance transcription by T7 RNA polymerase.
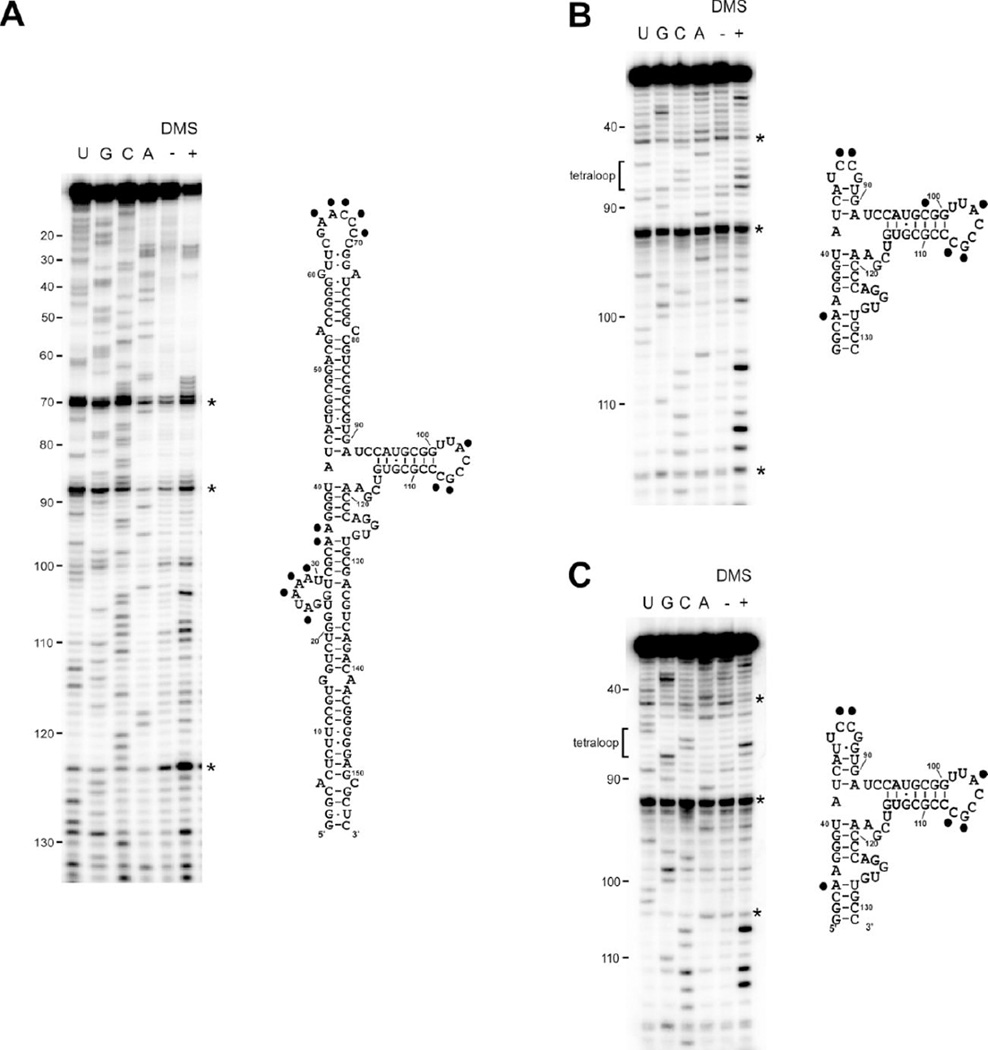
DMS structure probing was used to confirm that the central domain constructs adopt the native fold found within full length VAI. DMS methylates the base-pairing faces of unpaired adenosines and cytosines.53 As expected, the two cytosines within the designed UCCG tetraloop in CD1 and CD2 are modified by DMS (Figures 3B and 3C). However, loop 8 shows significant protection from DMS modification. Both cytosine 104 and 105 are protected whereas adenine 103 and cytosine 107 are modified. The same modification pattern within loop 8 is observed in full length VAI (Figure 3A). Consistent with the present results, several bases in loop 8 are protected from single-strand specific nuclease digestion and it has been proposed that loop 8 engages in tertiary interactions with loop 10.39 In addition to modification of adenosines and cytosines, DMS is also capable of methylating N7 of guanosines.53 This modification is usually silent because it does not block reverse transcriptase, but we observe a stop at guanosine 106 for all of the VAI constructs. There are some minor differences in the pattern of DMS modification of VAI and the central domain constructs. In full length VAI, both adenosine 35 and 36 are methylated by DMS but only A35 is modified in CD1 and CD2. Conversely, C98 is modified in CD1 even though it lies within a duplex region. Taken together, the DMS modification data indicate that both of the central constructs adopt the native VAI fold.
Figure 3.
DMS structure probing of VAI (A), CD1 (B) and CD2 (C). Gels are shown on the left and secondary structures diagrams are shown on the right with modified bases indicated by a black dot. Each gel includes a sequencing ladder, a control lane without DMS (−) and a lane with DMS (+). Artifacts arising from reverse transcriptase pause sites are marked with an asterisk.
Sedimentation velocity titrations were used to analyze PKR binding to the VAI domain constructs. For both AS and TS, the g^(s*) distributions gradually shift to the right with increasing concentrations of PKR, consistent with weak binding (Figure 4). The small magnitude of the shifts is consistent with binding of a single PKR monomer. Global analysis of these data indicate that PKR binds somewhat weakly to the TS stem-loop with Kd = 5.53 µM (Table 1). PKR binds with somewhat higher affinity to AS with Kd = 1.71 µM.
Figure 4.
Sedimentation velocity analysis of PKR binding to VAI domain constructs. Normalized g^(s*) distributions for AS (A), TS (B), CD1 (C) and CD2 (D). For each construct the curves are labeled as follows: RNA alone (black), RNA + 1 eq. PKR (blue), RNA + 2 eq. PKR (green) and RNA + 6 eq. PKR (red).
In contrast to AS and TS, there is no distinct shift in the g^(s*) distributions for either CD1 or CD2, indicating the absence of substantial population of a complex. The slight shift to the left upon addition of 6 eq. PKR is due to the contribution of free PKR with s ~ 3.3 S. Note that these plots do not exclude the possibility of very weak interaction of PKR with CD1 and CD2 and the data were globally analyzed to estimate Kd. Because of the low population of the complex, it was necessary to fix the sedimentation coefficients assuming binding of a single PKR. This parameter was estimated by assuming a frictional ratio of f/f0 = 1.4, which is typical for complexes of PKR with RNAs of this size.11,16,33 Although the global fits converge to very weak Kd values of 134 µM and 63 µM for CD1 and CD2, respectively, the narrow confidence intervals indicate that these parameters are statistically reliable (Table 1).
Inhibition of PKR by VAI domains
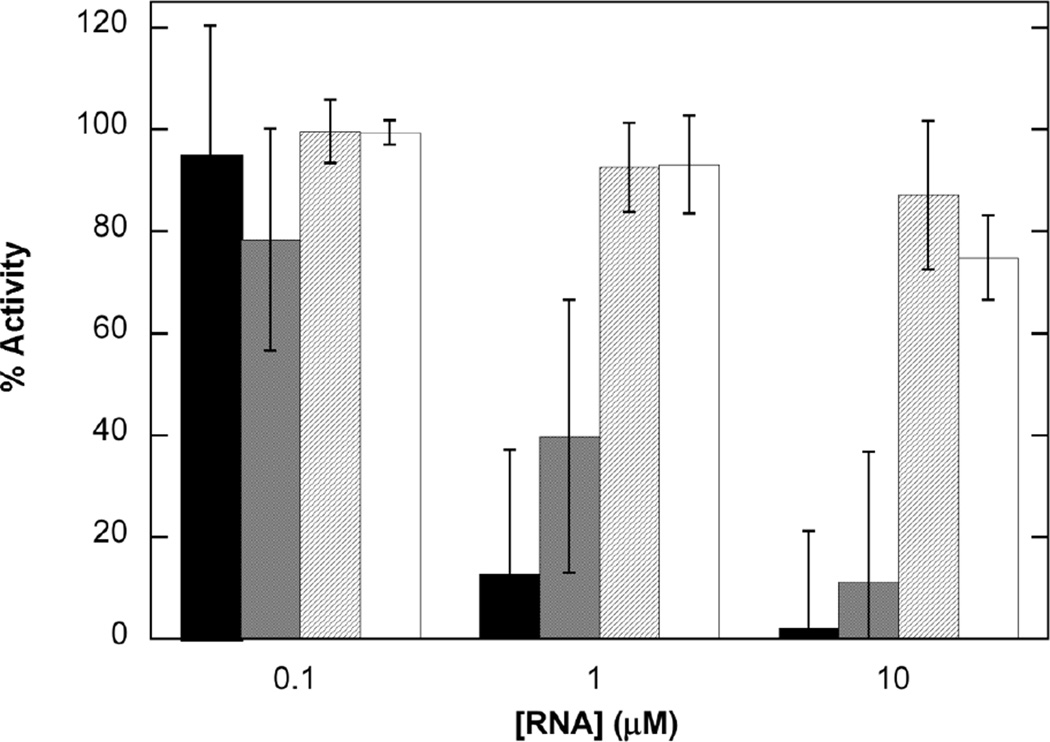
In order to correlate RNA binding with function, we have measured inhibition of PKR activation by the VAI domain constructs using autophosphorylation assays. The assays were performed in the same buffer as the analytical ultracentrifugation experiments. As we previously reported,33 VAI itself is a potent inhibitor of PKR autophosphorylation induced by dsRNA and reduces activity below 20% at 1 µM (Figure 5). Although AS inhibits PKR, it is considerably weaker and only reaches a comparable level of inhibition at 10 µM. TS does not significantly inhibit PKR activation over the concentration range examined and CD2 shows only very slight inhibition at 10 µM. Thus, the ability of the VAI domain constructs to inhibit PKR correlates with their binding affinity. In control experiments performed in the absence of poly (rI:rC), CD2 showed some activation of PKR but the other VAI constructs did not exhibit significant background activation (data not shown). Because PKR binds CD2 with Kd >> 10 µM, the activation of PKR by CD2 is most likely due to the presence of a monomeric or dimeric impurity at concentrations below the limit of detection for our methods.
Figure 5.
Inhibition of PKR activation by VAI domain constructs. VAI (black), AS (gray), TS (diagonal lines), and CD2 (white). PKR was incubated with VAI domain constructs at the indicated concentration prior to addition of 10 µg/ml poly(rI:rC) and 0.4 mM ATP containing 4 µCi of γ-32P[ATP]. The percent inhibition was calculated relative to a sample containing no inhibitory RNA. The data represent the average of three experiments with error bars indicated.
Discussion
Although PKR binding to simple duplex RNAs is not regulated by divalent ion, apart from nonspecific salt effects,33,54 the interaction with VAI is modulated by Mg2+. ITC and sedimentation velocity data indicate that two PKR monomers sequentially bind to VAI in the absence of divalent ion, with Kd1 = 14 nM and Kd2 = 601 nM, whereas only a single PKR binds with Kd = 334 nM in the presence of 5 mM Mg2+.33 Interestingly, the interaction of the dsRBD with VAI shows a different dependence: a single dsRBD binds in the absence and presence of divalent ion and the binding affinity of dsRBD is enhanced about two-fold upon addition of Mg2+. It is noteworthy that Kd2 for PKR binding to VAI in the absence of Mg2+ is about two-fold greater than Kd in the presence of Mg2+ and the Kd values for PKR and dsRBD converge to about the same value in the presence of divalent ion (Table 1). Thus, the divalent ion sensitivity and affinity for binding of the second PKR to VAI closely correlates with the binding behavior for dsRBD, suggesting that they correspond to equivalent interactions. In this model, Mg2+ prevents binding of the first PKR and slightly enhances binding of the second, lower affinity PKR. Previous studies have also reported that two PKRs bind to VAI in the absence of Mg2+.27,55 However, they associated the lower affinity binding event with nonspecific interactions. In contrast, our results indicate that the lower-affinity PKR corresponds to the PKR that binds in the presence of Mg2+ and is thus physiologically relevant. The dsRBD does not exhibit the high affinity interaction with VAI in the absence of Mg2+, suggesting that other regions of PKR contribute to this interaction. Our data indicate that the kinase domain alone does not bind to VAI; however, the linker region lying between the dsRBD and kinase domain may be involved.
The hydrodynamic parameters for the dsRBD-VAI complex are also affected by divalent ion. In the presence of Mg2+ the sedimentation coefficient of the complex decreases with a corresponding increase in the frictional ratio. These data indicate that the dsRBD-VAI complex becomes more asymmetric or undergoes a change in hydration in the presence of divalent ion. Note that the frictional ratio and Rg of VAI itself are not significantly affected by Mg2+,33 suggesting that the effects of divalent ion on dsRBD or PKR binding to VAI, and the hydrodynamic properties of the resulting complexes, are not due to large scale changes in VAI conformation. However, addition of Mg2+ does induce some changes in the pattern of nuclease and chemical cleavages in the central domain that may be associated with local structural rearrangements.28 In contrast, using DMS or 1-methyl-7-nitroisatoic anhydride modification, we do not detect changes in VAI structure upon addition of Mg2+ (K. Launer-Felty and J.L. Cole, unpublished results). Experiments are in progress to characterize the site of Mg2+ binding. Given that PKR kinase activity is dependent on divalent ion and Mg2+ is present in the cytoplasm, the PKR-VAI interactions parameters determined in the presence of divalent ion are more physiologically-relevant than those obtained in the absence of Mg2+.
Dissection of VAI into AS, TS and CD1/2 constructs reveals that none of the isolated domains retains the PKR binding affinity or inhibitory potency of the full length RNA. Even though the primary site of PKR binding has previously been mapped to the apical stem,28–31 PKR exhibits a ~6-fold lower binding affinity for AS relative to full length VAI. Secondary structure defects, such as internal loops and bulges, typically reduce PKR activation and may also reduce binding affinity.56–58 However, the lower affinity binding of PKR to AS is unlikely to be caused by the two mismatches in this domain. PKR binds to a 20 bp perfect RNA duplex with similar affinity (Kd = 1.63 µM) under the same conditions.).33 In addition, replacement of these mismatches with Watson-Crick pairs does not alter PKR binding in the context of a full length VAI construct (K. Launer-Felty, E. Stassen and J. Cole, unpublished results). Thus, PKR binds to AS nonspecifically. Our results do not support the previous report that the isolated apical stem functions as a PKR activator.41
Deletion of the terminal stem does not affect PKR binding affinity or inhibition27,33 and footprinting studies have not detected PKR binding to the terminal stem in the context of full length VAI. Our measurements show that PKR has the potential to bind to TS, albeit rather weakly. PKR binds to a construct that contains both the central domain and terminal stem with approximately the same affinity as TS (K. Launer-Felty and J.L. Cole, unpublished results). PKR binding affinity increases with length of dsRNA,10,11,14 and the reduced affinity of PKR for TS relative to AS may be due to the shorter duplex in the TS construct. As in the case of AS, we see no evidence for any specificity for PKR binding despite the two mismatches present in TS.
Although the isolated central domain adopts the same fold as found in full length VAI, PKR binds extremely weakly to the CD1 and CD2 constructs. In support of this observation, a previous study demonstrated that PKR binds weakly to the central domain; however, this construct lacked loop 10.41 The weak binding affinity for the central domain may be due to the absence of extended duplex RNA. In addition, our DMS probing results and previous studies39 suggest that the central domain is stabilized by tertiary interactions that may impede PKR binding. Footprinting28,31 and affinity cleavage29,30 measurements suggest that PKR interacts with a region of the central domain proximal to the apical stem. Indeed, the PKR binding affinity of full length VAI or a truncation lacking the terminal stem33 is significantly greater than either the apical stem or central domain alone. Thus, the central domain and apical stem interact to mediate high affinity PKR binding. In addition to perfect RNA duplexes, PKR recognizes RNAs containing symmetrical defects, A-form dsRNA mimics, and coaxial stacks of helices.59 Possibly, the central domain and apical stem adopt a structure that mimics an extended duplex.
The ability of VAI domains to inhibit PKR activation by dsRNA parallels their PKR binding affinity, supporting the model whereby VAI inhibits PKR simply by sequestering the enzyme as a monomer and thus preventing activation via dimerization and autophosphorylation.18,40 Our data do not support earlier proposals that the central domain specifically functions as an inhibitory region. However, the fact that the stoichiometry for PKR binding to each of the constituent domains of VAI is not additive in the full length RNA suggests steric hindrance where binding of a PKR to the preferred site comprising the apical stem and central domain prevents binding of a second PKR at the terminal stem. The sequestration of PKR by adenovirus VAI represents one of the many mechanisms employed by viruses to inhibit this enzyme3 and underscores the significance of PKR in the innate immune response to viral infection.
Materials and Methods
All reagents purchased were of reagent grade. Unphosphorylated PKR43 and the kinase domain of PKR44 were purified as previously described. The dsRBD (containing dsRBM1 and dsRBM2)45, and dsRBM1 alone10 were purified as described with some modifications. Following Sepharose-SP and heparin Sepharose FF chromatography, dsRBD and dsRBM1 were further purified on Superdex 200 and Sephacryl S-100 columns, respectively. dsRBM2 was expressed and purified using the same protocol used for dsRBM1, except that the Sephacryl S-100 column was replaced with Superdex 75. All size exclusion chromatography columns were equilibrated in either AU200 (20 mM HEPES, 200 mM NaCl, 0.1mM EDTA, 0.1 mM TCEP) or AU200 + 5 mM MgCl2. The extinction coefficients for all proteins were calculated using SEDNTERP.46
RNAs were synthesized by in vitro transcription using plasmid templates and T7 RNA polymerase. DNA sequences encoding VAI and VAI domains (Figure 1) were ligated into a plasmid containing a 3’ modified HDV ribozyme sequence.47 Plasmids were linearized with either DraI or XbaI and transcribed as previously described.33 Transcripts were purified by denaturing gel electrophoresis and extracted from the gel by either electroelution or crush-and-soak. Purified RNAs were annealed by diluting to 1 µM in 1X TE buffer (10 mM Tris-HCl, 0.1 mM EDTA, pH 7.5) and heating to 95°C for 3 minutes followed by snap cooling on ice. Extinction coefficients were measured by digesting each RNA with ribonuclease (Riboshredder, Epicentre, Chicago, IL) for 20 minutes and comparing the absorbance at 260 nm for the digested and undigested samples. The extinction coefficients are: VAI, 1.19×106 M−1cm−1; AS, 3.66×105 M−1cm−1; TS, 3.32×105 M−1cm−1; CD1, 5.28×105 M−1cm−1; and CD2, 5.22×105 M−1cm−1.
Prior to analytical ultracentrifugation, proteins and RNAs were buffer exchanged into AU200 or AU200 + 5 mM MgCl2 using Biogel P6 spin columns. RNAs were diluted to concentrations corresponding to an absorbance of ~ 0.5 OD at 260 nm for a 1.2 cm pathlength and 1, 2 and 6 equivalents of protein were added. To better elucidate the low binding affinity for TS 7 equivalents of protein was also added to the analyses. Sedimentation velocity measurements were performed using a Beckman-Coulter XL-I analytical ultracentrifuge with an An50-Ti rotor at 20°C. Samples were loaded into cells containing double sector aluminum-epon or charcoal-epon centerpieces with quartz windows. Rotor speeds were: 35,000 RPM (VAI + dsRBD), 40,000 RPM (CD1 + PKR, CD2 + PKR), 42,000 RPM (TS + PKR, AS + PKR) and 44,000 RPM (VAI + dsRBM1, VA1 + dsBM2). In each experiment, at least 80 scans were collected using absorbance optics at 260 nm. Sedimentation coefficients of the free RNAs were obtained from single discrete species analysis using Sedfit.48 Sedimentation velocity data for protein-RNA mixtures were initially analyzed using a model-independent approach by generating g(s*) distributions with the program DCDT+.49 Subsequently, global analysis was performed using SEDANAL50 to obtain protein-RNA dissociation constants and sedimentation coefficients for the complexes. Frictional ratios were calculated using SEDNTERP.46
RNA structure analysis was performed using DMS probing. Prior to analysis, RNAs were annealed at 95°C for 3 min in 20 mM phosphate, 0.1 mM EDTA, pH 7.5 and snap-cooled on ice. 200 mM NaCl and 5 mM MgCl2 were added and the RNAs were allowed to fold for 30 min at 37°C. RNAs were treated with 38.4 mM of DMS dissolved in ethanol for 15 min at room temperature and the reactions were quenched by adding 51.8 mM β-mercaptoethanol. Control samples were prepared by substituting 1 µL 100% ethanol for the DMS solution. Samples were ethanol precipitated, dried, and resuspended in 0.5 × TE. DNA primers were 5’-end labeled using polynucleotide kinase (New England BioLabs) and γ-32P-ATP and were annealed to the RNAs by heating to 65°C for 5 min, 35°C for 5 min, and 4°C for at least 1 min. Reverse transcription was performed according to a previous protocol.51 Samples were loaded on an 8% (VAI) or 12% (CD1 and CD2) TBE-urea sequencing gels and the dried gels were exposed to a phosphor screen overnight and imaged using a Typhoon Trio (GE Life Sciences).
PKR inhibition assays were performed as previously described33 except that the inhibiting RNAs were preincubated with 100 nM PKR for 15 minutes. The solution was then incubated with poly(rI:rC) for 15 minutes before addition of ATP. Fixed and dried gels were exposed to a phosphor storage screen overnight. Images were scanned using a Typhoon Trio and band intensities were measured using ImageQuant TL software. Experiments were performed in triplicate.
Table 2.
PKR - RNA interaction parameters derived from sedimentation velocity analysis in the presence of 5 mM Mg2+.
| RNA | Kd (µM) | s(RP)a | RMSb |
|---|---|---|---|
| VAIc | 0.334 (0.278, 0.401) | 7.18 (7.07, 7.29) | 0.00691 |
| AS | 1.71 (1.54, 1.90) | 4.82 (4.76, 4.89) | 0.00703 |
| TS | 5.04 (4.18, 6.10) | 4.03 (3.94, 4.12) | 0.00887 |
| CD1 | 134 (97.0, 212) | 5.37 (fixed)d | 0.00672 |
| CD2 | 62.9 (53.7, 75.4) | 5.44 (fixed)d | 0.00561 |
Parameters were obtained by global nonlinear least square analysis of sedimentation velocity measurements. The values in parentheses represent the 95% joint confidence intervals obtained using the F-statistic.
Uncorrected sedimementation coefficient (Svedbergs).
Root mean square deviation of the fit in absorbance units
Data were previously reported in reference 29.
The sedimentation coefficients of the complexes could not be determined by global analysis and were fixed assuming a frictional ratio of f/f0 = 1.4.
Highlights.
Divalent ion regulates PKR binding to VAI RNA
PKR binds weakly to the isolated VAI domains
The apical stem and central domain together form a high affinity PKR binding site.
Acknowledgements
The authors would like to thank O. Federova and A.M. Pyle for guidance in developing DMS probing studies and E. Stassen for assistance in purification of apical stem mutants of VAI. The plasmid containing a 3’ HDV ribozyme sequence was a generous gift from G. Conn. This work was supported by grant number AI-53615 from the National Institutes of Health to J.L.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: DMS, dimethyl sulfate; dsRBD, double-stranded RNA binding domain; dsRBM, double-stranded RNA binding motif; VAI, adenovirus-associated RNA I; VAII, adenovirus-associated RNA II; AS, apical stem; TS, terminal stem; CD1, central domain construct 1; CD2, central domain construct 2.
References
- 1.Unterholzner L, Bowie AG. The interplay between viruses and innate immune signaling: recent insights and therapeutic opportunities. Biochem. Pharmacol. 2008;75:589–602. doi: 10.1016/j.bcp.2007.07.043. [DOI] [PubMed] [Google Scholar]
- 2.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. Journal of Virology. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langland JO, Cameron JM, Heck MC, Jancovich JK, Jacobs BL. Inhibition of PKR by RNA and DNA viruses. Virus Research. 2006;119:100–110. doi: 10.1016/j.virusres.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 4.Nanduri S, Carpick BW, Yang Y, Williams BR, Qin J. Structure of the double-stranded RNA binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stefl R, Oberstrass FC, Hood JL, Jourdan M, Zimmermann M, Skrisovska L, Maris C, Peng L, Hofr C, Emeson RB, Allain FH-T. The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove. Cell. 2010;143:225–237. doi: 10.1016/j.cell.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gan J, Tropea JE, Austin BP, Court DL, Waugh DS, Ji X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. doi: 10.1016/j.cell.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Hartman E, Roy K, Chanfreau G, Feigon J. Structure of a Yeast RNase III dsRBD Complex with a Noncanonical RNA Substrate Provides New Insights into Binding Specificity of dsRBDs. Structure. 2011;19:999–1010. doi: 10.1016/j.str.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian B, Mathews MB. Functional characterization of and cooperation between the double-stranded RNA-binding motifs of the protein kinase PKR. J Biol Chem. 2001;276:9936–9944. doi: 10.1074/jbc.M007328200. [DOI] [PubMed] [Google Scholar]
- 10.Ucci JW, Kobayashi Y, Choi G, Alexandrescu AT, Cole JL. Mechanism of interaction of the double-stranded RNA (dsRNA) binding domain of protein kinase R with short dsRNA sequences. Biochemistry. 2007;46:55–65. doi: 10.1021/bi061531o. [DOI] [PubMed] [Google Scholar]
- 11.Husain B, Mukerji I, Cole JL. Analysis of high-affinity binding of protein kinase R to double-stranded RNA. Biochemistry. 2012;51:8764–8770. doi: 10.1021/bi301226h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dar A, Dever T, Sicheri F. Higher-Order Substrate Recognition of eIF2α by the RNA-Dependent Protein Kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 13.Cole JL. Activation of PKR: an open and shut case? Trends Biochem Sci. 2007;32:57–62. doi: 10.1016/j.tibs.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manche L, Green SR, Schmedt C, Mathews MB. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng X, Bevilacqua PC. Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA. 2004;10:1934–1945. doi: 10.1261/rna.7150804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemaire PA, Anderson E, Lary J, Cole JL. Mechanism of PKR Activation by dsRNA. J Mol Biol. 2008;381:351–360. doi: 10.1016/j.jmb.2008.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Malley RP, Mariano TM, Siekierka J, Mathews MB. A mechanism for the control of protein synthesis by adenovirus VA RNAI. Cell. 1986;44:391–400. doi: 10.1016/0092-8674(86)90460-5. [DOI] [PubMed] [Google Scholar]
- 18.Kitajewski J, Schneider RJ, Safer B, Munemitsu SM, Samuel CE, Thimmappaya B, Shenk T. Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2 alpha kinase. Cell. 1986;45:195–200. doi: 10.1016/0092-8674(86)90383-1. [DOI] [PubMed] [Google Scholar]
- 19.Akusjärvi G, Svensson C, Nygård O. A mechanism by which adenovirus virus-associated RNAI controls translation in a transient expression assay. Mol Cell Biol. 1987;7:549–551. doi: 10.1128/mcb.7.1.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sano M, Kato Y, Taira K. Sequence-specific interference by small RNAs derived from adenovirus VAI RNA. FEBS Lett. 2006;580:1553–1564. doi: 10.1016/j.febslet.2006.01.085. [DOI] [PubMed] [Google Scholar]
- 21.Andersson MG, Haasnoot PCJ, Xu N, Berenjian S, Berkhout B, Akusjärvi G. Suppression of RNA interference by adenovirus virus-associated RNA. Journal of Virology. 2005;79:9556–9565. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber F, Wagner V, Kessler N, Haller O. Induction of interferon synthesis by the PKR-inhibitory VA RNAs of adenoviruses. J Interferon Cytokine Res. 2006;26:1–7. doi: 10.1089/jir.2006.26.1. [DOI] [PubMed] [Google Scholar]
- 23.Meng H, Deo S, Xiong S, Dzananovic E, Donald LJ, van Dijk CW, McKenna SA. Regulation of the interferon-inducible 2“-5-”oligoadenylate synthetases by adenovirus VA(I) RNA. J Mol Biol. 2012;422:635–649. doi: 10.1016/j.jmb.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 24.Desai SY, Patel RC, Sen GC, Malhotra P, Ghadge GD, Thimmapaya B. Activation of interferon-inducible 2“-5” oligoadenylate synthetase by adenoviral VAI RNA. J Biol Chem. 1995;270:3454–3461. doi: 10.1074/jbc.270.7.3454. [DOI] [PubMed] [Google Scholar]
- 25.Ma Y, Mathews MB. Structure, function, and evolution of adenovirus-associated RNA: a phylogenetic approach. Journal of Virology. 1996;70:5083–5099. doi: 10.1128/jvi.70.8.5083-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma Y, Mathews MB. Comparative analysis of the structure and function of adenovirus virus-associated RNAs. Journal of Virology. 1993;67:6605–6617. doi: 10.1128/jvi.67.11.6605-6617.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wahid AM, Coventry VK, Conn GL. Systematic deletion of the adenovirus-associated RNAI terminal stem reveals a surprisingly active RNA inhibitor of double-stranded RNA-activated protein kinase. J Biol Chem. 2008;283:17485–17493. doi: 10.1074/jbc.M802300200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clarke PA, Mathews MB. Interactions between the double-stranded RNA binding motif and RNA: definition of the binding site for the interferon-induced protein kinase DAI (PKR) on adenovirus VA RNA. RNA. 1995;1:7–20. [PMC free article] [PubMed] [Google Scholar]
- 29.Spanggord RJ, Vuyisich M, Beal PA. Identification of binding sites for both dsRBMs of PKR on kinase-activating and kinase-inhibiting RNA ligands. Biochemistry. 2002;41:4511–4520. doi: 10.1021/bi0120594. [DOI] [PubMed] [Google Scholar]
- 30.Spanggord RJ, Beal PA. Selective binding by the RNA binding domain of PKR revealed by affinity cleavage. Biochemistry. 2001;40:4272–4280. doi: 10.1021/bi002512w. [DOI] [PubMed] [Google Scholar]
- 31.Circle DA, Neel OD, Robertson HD, Clarke PA, Mathews MB. Surprising specificity of PKR binding to delta agent genomic RNA. RNA. 1997;3:438–448. [PMC free article] [PubMed] [Google Scholar]
- 32.Wahid AM, Coventry VK, Conn GL. The PKR-binding domain of adenovirus VA RNAI exists as a mixture of two functionally non-equivalent structures. Nucleic Acids Research. 2009;37:5830–5837. doi: 10.1093/nar/gkp595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Launer-Felty K, Wong CJ, Wahid AM, Conn GL, Cole JL. Magnesium-dependent interaction of PKR with adenovirus VAI. J Mol Biol. 2010;402:638–644. doi: 10.1016/j.jmb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clarke PA, Pe'ery T, Ma Y, Mathews MB. Structural features of adenovirus 2 virus-associated RNA required for binding to the protein kinase DAI. Nucleic Acids Research. 1994;22:4364–4374. doi: 10.1093/nar/22.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furtado MR, Subramanian S, Bhat RA, Fowlkes DM, Safer B, Thimmappaya B. Functional dissection of adenovirus VAI RNA. Journal of Virology. 1989;63:3423–3434. doi: 10.1128/jvi.63.8.3423-3434.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mellits KH, Kostura M, Mathews MB. Interaction of adenovirus VA RNAl with the protein kinase DAI: nonequivalence of binding and function. Cell. 1990;61:843–852. doi: 10.1016/0092-8674(90)90194-j. [DOI] [PubMed] [Google Scholar]
- 37.Ghadge GD, Malhotra P, Furtado MR, Dhar R, Thimmapaya B. In vitro analysis of virus-associated RNA I (VAI RNA): inhibition of the double-stranded RNA-activated protein kinase PKR by VAI RNA mutants correlates with the in vivo phenotype and the structural integrity of the central domain. Journal of Virology. 1994;68:4137–4151. doi: 10.1128/jvi.68.7.4137-4151.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rahman A, Malhotra P, Dhar R, Kewalramani T, Thimmapaya B. Effect of single-base substitutions in the central domain of virus-associated RNA I on its function. Journal of Virology. 1995;69:4299–4307. doi: 10.1128/jvi.69.7.4299-4307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma Y, Mathews MB. Secondary and tertiary structure in the central domain of adenovirus type 2 VA RNA I. RNA. 1996;2:937–951. [PMC free article] [PubMed] [Google Scholar]
- 40.Robertson HD, Mathews MB. The regulation of the protein kinase PKR by RNA. Biochimie. 1996;78:909–914. doi: 10.1016/s0300-9084(97)86712-0. [DOI] [PubMed] [Google Scholar]
- 41.McKenna SA, Kim I, Liu CW, Puglisi JD. Uncoupling of RNA binding and PKR kinase activation by viral inhibitor RNAs. J Mol Biol. 2006;358:1270–1285. doi: 10.1016/j.jmb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 42.McKenna SA, Lindhout DA, Kim I, Liu CW, Gelev VM, Wagner G, Puglisi JD. Molecular framework for the activation of RNA-dependent protein kinase. J Biol Chem. 2007;282:11474–11486. doi: 10.1074/jbc.M700301200. [DOI] [PubMed] [Google Scholar]
- 43.Anderson E, Pierre-Louis WS, Wong CJ, Lary JW, Cole JL. Heparin Activates PKR by Inducing Dimerization. J Mol Biol. 2011;413:973–984. doi: 10.1016/j.jmb.2011.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson E, Cole JL. Domain stabilities in protein kinase R (PKR): evidence for weak interdomain interactions. Biochemistry. 2008;47:4887–4897. doi: 10.1021/bi702211j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ucci JW, Cole JL. Global analysis of non-specific protein-nucleic interactions by sedimentation equilibrium. Biophys Chem. 2004;108:127–140. doi: 10.1016/j.bpc.2003.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laue TM, Shah B, Ridgeway TM, Pelletier SL. Computer-aided interpretation of sedimentation data for proteins. Analytical Ultracentrifugation in Biochemistry and Polymer Science. 1992:90–125. [Google Scholar]
- 47.Walker SC, Avis JM, Conn GL. General plasmids for producing RNA in vitro transcripts with homogeneous ends. Nucleic Acids Research. 2003;31:e82. doi: 10.1093/nar/gng082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 50.Stafford WF, Sherwood PJ. Analysis of heterologous interacting systems by sedimentation velocity: curve fitting algorithms for estimation of sedimentation coefficients, equilibrium and kinetic constants. Biophys Chem. 2004;108:231–243. doi: 10.1016/j.bpc.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 51.Wilkinson KA, Merino EJ, Weeks KM. Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nature Protocols. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 52.Dale T, Smith R, Serra MJ. A test of the model to predict unusually stable RNA hairpin loop stability. RNA. 2000;6:608–615. doi: 10.1017/s1355838200992495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tijerina P, Mohr S, Russell R. DMS footprinting of structured RNAs and RNA–protein complexes. Nature Protocols. 2007;2:2608–2623. doi: 10.1038/nprot.2007.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bevilacqua PC, Cech TR. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry. 1996;35:9983–9994. doi: 10.1021/bi9607259. [DOI] [PubMed] [Google Scholar]
- 55.McKenna SA, Lindhout DA, Shimoike T, Aitken CE, Puglisi JD. Viral dsRNA inhibitors prevent self-association and autophosphorylation of PKR. J Mol Biol. 2007;372:103–113. doi: 10.1016/j.jmb.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng X, Bevilacqua PC. Straightening of bulged RNA by the double-stranded RNA-binding domain from the protein kinase PKR. Proc Natl Acad Sci USA. 2000;97:14162–14167. doi: 10.1073/pnas.011355798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heinicke LA, Nallagatla SR, Hull CM, Bevilacqua PC. RNA helical imperfections regulate activation of the protein kinase PKR: effects of bulge position, size, and geometry. RNA. 2011;17:957–966. doi: 10.1261/rna.2636911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patel S, Blose JM, Sokoloski JE, Pollack L, Bevilacqua PC. Specificity of the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR for double-stranded RNA: insights from thermodynamics and small-angle X-ray scattering. Biochemistry. 2012;51:9312–9322. doi: 10.1021/bi300935p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nallagatla SR, Toroney R, Bevilacqua PC. Regulation of innate immunity through RNA structure and the protein kinase PKR. Curr Opin Struct Biol. 2011;21:119–127. doi: 10.1016/j.sbi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]